Open Access Article
Open Access ArticleApplication of nanomaterials in organoid culture and cryopreservation
Wanjuan
He
,
Qinlin
Sun
,
Dan
Ge
,
Yang
Liu
 * and
Bingbing
Sun
* and
Bingbing
Sun
 *
*
State Key Laboratory of Fine Chemicals, School of Chemical Engineeringand Frontiers Science Center for Smart Materials Oriented Chemical Engineering, Dalian University of Technology, 116024 Dalian, China
First published on 6th November 2025
Abstract
Organoids are three-dimensional, self-organizing microtissues obtained from stem cells, primary tissues, or patient-derived tumors through in vitro culture, which are invaluable models for studying organ development, disease mechanism modelling, drug screening, and regenerative medicine. Nanomaterials, with unique surface properties and excellent biocompatibility, have emerged as powerful tools in biomedical research, particularly in the fields of organoid construction (e.g., microenvironment modulation and advanced engineering cultivation) and cryopreservation (e.g., intracellular delivery, nanowarming, and ice-inhibition). This review comprehensively discusses the recent advancements in nanomaterial-based strategies for organoid fabrication and cryopreservation while addressing their challenges such as biocompatibility, scalability and long-term safety. Finally, we envision the prospects in the development of advanced nanomaterial-based platforms for organoid construction and cryopreservation with higher biocompatibility and standardization.
1. Introduction
Organoids are micro-organs formed by stem cells (including embryonic stem cells, pluripotent stem cells and adult stem cells), tumor cells or primary patient-derived cells through three-dimensional (3D) in vitro culture. These structures closely emulate the complex architecture of native organs and recapitulate their physiological functions.1 In recent years, organoids have demonstrated significant potential across various applications including disease modeling, drug screening, personalized therapy, and regenerative medicine.2–4 For example, intestinal organoids have been successfully employed to study the pathogenesis of Crohn's disease.5 Furthermore, animal studies have shown that transplantation of intestinal organoids facilitates colon repair after injury.6 Additionally, the FDA has explored the use of hepatic organoid chips to model human food-borne diseases and evaluate their potential as a replacement for animal models in compound toxicity testing.7Compared with traditional two-dimensional (2D) cell models, organoids exhibit high similarity to the in vivo microenvironment.8 However, their generation and maintenance remain challenging due to immature protocols, which often lead to poor microenvironmental regulation and central necrosis.9 Therefore, the development of new technologies to precisely control organoid development and function is now a key research focus. Nanomaterials offer a novel strategy to address these challenges due to their unique chemical and physical properties, including programmable surface functionalities and biocompatibility. Nanomaterials can optimize the developmental microenvironment in multiple ways. For instance, Abdel et al. found that variations in the concentration of magnetic nanomaterials could precisely control the elastic modulus of Matrigel.10 This property enables Matrigel to adapt to the microenvironmental requirements of organoids at different growth stages, thereby regulating cellular behaviors such as differentiation. Functional nanomaterials such as mesoporous silica nanoparticles can be used to deliver growth factors and other bioactive molecules, enabling dynamic regulation of stem cell differentiation and organoid morphogenesis.11 By mimicking the natural extracellular matrix (ECM), nanofiber scaffolds significantly enhance cell adhesion and provide essential mechanical support.12
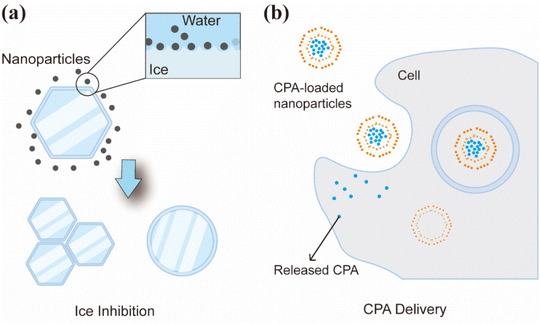
Simultaneously, the establishment of a stable and efficient cryopreservation system is critical to ensure a continuous supply of organoids and facilitate their practical application. Nevertheless, the cryopreservation of organoids remains challenging. A primary difficulty is their size, which limits the rapid transfer of heat and mass. Moreover, most traditional cryoprotectants (CPAs) exhibit cytotoxicity. Nanomaterials offer a promising strategy to overcome these challenges. Nanomaterials such as mesoporous silica and graphene oxide (GO) can serve as heterogeneous nucleation sites, reducing the degree of supercooling and inhibiting the formation of ice crystals. Moreover, magnetic nanoparticles (MNPs) enable uniform thawing under an external physical field, which significantly enhances the post-thaw survival rate of organoids, as demonstrated in heart models.13
Given these unique attributes, nanomaterials have emerged as a key medium for bridging micro-scale cellular behavior and macro-scale tissue function, demonstrating potentials for promoting organoid technology. Herein, we review the current advances and outline the future perspectives of nanomaterials in organoid construction and cryopreservation.
2. Nanomaterial-assisted organoid culture construction
Organoids can be generated from various sources including adult organ-derived cells, stem cells and tumor tissues. Their development is critically dependent on the staged culture conditions that mimic a specific development path. The developmental cycle varies significantly among organoids from different sources. Stem cell-derived organoids undergo a multi-stage process involving cell proliferation, early differentiation, globular structure formation and maturation. In contrast, organoids derived from primary cells or tumors are more dependent on microenvironmental and signaling cues, resulting in a shorter development cycle.14 Although current technology enables the construction of 3D organoids, several significant challenges remain. For stem cell-derived organoids, an in vitro system can hardly simulate the synergistic effect of multiple growth factors present in vivo. This often results in deviations in differentiation stage and direction, incomplete cellular maturation, and consequent functional defects.8 Organoids derived from primary cells and tumor cells often face gradual loss of function and reduced heterogeneity, respectively.15,16 In addition, different organoid types face several common challenges. These include poor control over size, shape and cell composition, leading to low experimental repeatability.17 Furthermore, during long-term culture or upon exceeding a critical size, the lack of a vascular network results in internal hypoxia and subsequent necrosis.18,19 Organoid culture is critically dependent on the ECM. However, commonly used hydrogel scaffolds such as Matrigel have significant limitations including an unclear composition and limited tunability of mechanical properties. These shortcomings prevent them from fully recapitulating the complex physical microenvironment in vivo.Nanomaterials provide a breakthrough strategy for addressing the aforementioned challenges. Specifically, a cell aggregation pattern can be controlled via the dynamic magnetic field based on the MNP-assisted 3D suspension culture technology, reducing the structural heterogeneity.20 By mimicking the native electrophysiological microenvironment, conductive nanofiber scaffolds promote the synapse formation in brain organoids, and thereby address the issue of insufficient maturity in stem cell-derived organoids.21 The nanofiber scaffolds can simulate the spatial conditions for organoid growth, prevent uncontrolled cellular proliferation, and help maintain the heterogeneity of tumor organoids.22 Nanomaterials also function as versatile carriers of growth factors to regulate cell behavior and induce vascularization, thereby alleviating common problems such as internal necrosis.19Table 1 outlines various nanomaterials used for organoid culture construction.
| Nanomaterials | Organoids | References | |
|---|---|---|---|
| a ECM: Extracellular matrix; AuNRs: Gold nanoribbons; e-SiNWs: Electrically conductive silicon nanowires; SiNPs: Silicon nanoparticles; NPs: Nanoparticles; BFP-1: Bone formation petitude-1; MSNs: Mesoporous silica nanoparticles; MNPs: Magnetic nanoparticles; MSC: Mesenchymal stem cell; PLGA: Poly(lactic-co-glycolide); PLLA: Poly(L-lactic acid); PEG: Poly(ethylene glycol); PLL: Poly-L-lysine; hFOB: Human fetal osteoblast; ECs: Endothelial cells; HSCs: Hematopoietic stem cells; PTFE: Polytetrafluoroethylene. | |||
| ECM properties modification | Nanocellulose hydrogel | Mouse-derived intestinal organoids | 23 |
| Calcium silicate nanowire-containing hydrogel | Mouse-derived intestinal organoids | 24 | |
| AuNRs | Human cardiac organoids | 25 | |
| Collagen-carbon nanodots | Mouse-derived neural progenitor spheroid | 26 | |
| e-SiNWs | Human cardiac organoids | 27 | |
| SiNP | Mouse-derived B cell follicle organoid | 11 | |
| BFP-1-laden MSNs | Human bone organoid | 28 | |
| DNA microbeads | Medaka-derived retinal organoids | 29 | |
| 3D culture | Magnetic iron oxide (Fe3O4), AuNPs | Human glioblastoma tumor spheroid | 20 |
| Carbon-encapsulated cobalt MNPs | Mouse-derived breast cancer spheroid and colorectal cancer spheroid | 30 | |
| Magnetic iron oxide (FeO3) | Human MSC spheroid | 31 | |
| Fe3O4 | Human MSC spheroid | 32 | |
| Magnetic iron oxide (Fe3O4)-encapsulated PLGA microparticles/PLLA-b-PEG-folate NPs | Human epidermoid tumor spheroid | 33 | |
| Nanoparticle assembly of Fe2O3 and Au NPs cross-linked with PLL | Mouse-derived adipose tissue organoids | 34 | |
| Nanoparticle assembly of Fe2O3 and Au NPs cross-linked with PLL | Pig-derived salivary adenoid organoid | 35 | |
| hFOB spheroid | 36 | ||
| Human pseudo-islets | 37 | ||
| 3D multicellular spheroid composed of human MSCs, human ECs and human HSCs | 38 | ||
| Human lung spheroid granuloma model | 39 | ||
| Human prostate cancer cell spheroid | 40 | ||
| Liquid marble | PTFE powder particles | Human pancreatic mini-organoids | 41 |
2.1 Construction and static modulation of the extracellular microenvironment
The ECM is an acellular 3D network of macromolecules, composed of collagens, proteoglycans and other glycoproteins.42 As a core component of the cellular microenvironment, it not only provides essential physical support for cells and tissues, but also regulates critical processes such as proliferation, differentiation, and apoptosis through cell membrane-mediated signalling pathways.43 Therefore, the modulation of ECM properties can direct cell orientation and differentiation, influencing their fate and tissue morphology. A major drawback of conventional ECMs such as Matrigel is their poorly defined composition and simplistic mechanical properties. These materials cannot fully simulate the complex in vivo physical microenvironment, which ultimately constrains organoid development.44 Through methods such as directional arrangement, nanomaterials can effectively simulate the natural ECM and modulate the mechanical properties of matrix adhesives, providing new solutions to the above-mentioned problems.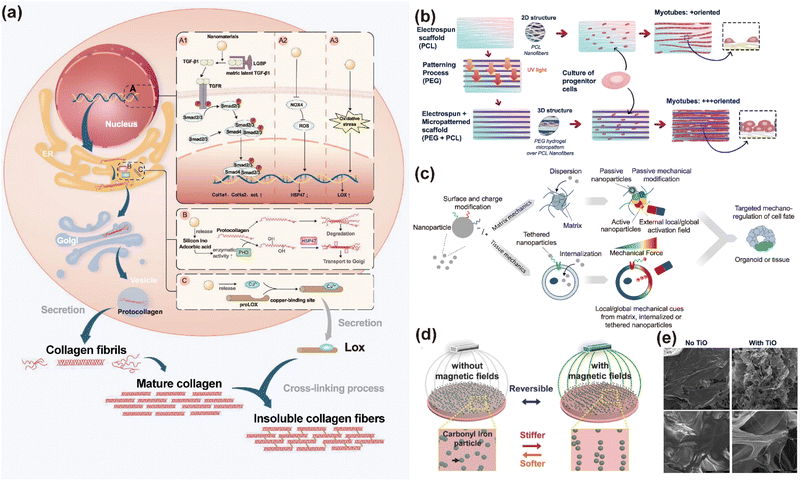 | ||
| Fig. 1 Role of nanomaterials in regulating the in vitro microenvironment. (a) Mechanism by which nanomaterials influence the synthesis and degradation of ECM-related proteins to remodel the cellular microenvironment. (A) Nanomaterials modulate collagen gene expression through multiple pathways. (A1) Activation of the TGF-β/smad pathway. (A2, A3) Transcriptional regulation of HSP47 and lysyl oxidase (LOX) by inhibiting oxidative stress. (B) Nanomaterials facilitate collagen post-translational modification within the endoplasmic reticulum (ER). (C) Nanomaterials modulate extracellular collagen by regulating LOX. This figure has been adapted/reproduced from ref. 48 with permission from the American Chemical Society, copyright 2024. (b) Magnetic nanoparticles (MNPs) can modulate the physical properties of Matrigel through active or passive mechanisms. Under an external magnetic field, activated NPs generate localized mechanical forces, whereas inactive NPs contribute to altering the local matrix stiffness passively. Additionally, magnetized cells experience forces when subjected to an external field and transmit these mechanical stresses to the surrounding extracellular matrix (ECM). This figure has been adapted/reproduced from ref. 47 with permission from Frontiers, copyright 2020. (c) Synthesis and functional schematic of a multiscale scaffold composed of polyethylene glycol and poly(ε-caprolactone) nanofibers for muscle tissue engineering. The aligned nanofibers mimic the nanoscale architecture of the native muscle ECM. This figure has been adapted/reproduced from ref. 10 with permission from MDPI, copyright 2021. (d) In the absence of a magnetic field, the particles remain randomly dispersed, and the hydrogel maintains its soft mechanical properties (left). Under a magnetic field exposure, carbonyl iron particles within the hydrogel align into chain-like structures, resulting in increased stiffness of the material (right). This figure has been adapted/reproduced from ref. 49 with permission from WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. (e) SEM images of collagen scaffolds before (left) and after (right) the incorporation of titanium oxide nanoparticles. After incorporation, the nanocomposite scaffold changes from a dense, sheet layer-like morphology to a three-dimensional structure with a finer, fluffier fibrous network. This figure has been adapted/reproduced from ref. 50 with permission from Elsevier B.V., copyright 2015. | ||
Similarly, the physical properties of nanomaterials, such as hardness and surface topography, are perceived by mechanical stimulation ion channels in cells, leading to the release of signal factors and the expression of related genes.51,52 In the osteogenic differentiation of mesenchymal stem cells (MSCs) into bone organoids, nano-hydroxyphosphine with slightly lower hardness and greater surface roughness has been shown to induce the expression of genes in a better way, which promotes osteogenesis.53 Meanwhile, these nanomaterials can reshape the cellular microenvironment by modulating the synthesis and degradation of ECM-related proteins, thereby indirectly influencing the behavior of surrounding cells.48 Collagen, elastin, and glycosaminoglycans are key components of the natural ECM. Through their synergistic effects, they endow the ECM with diverse biochemical and mechanical properties. Collagen provides the fundamental structural support and mechanical strength for the ECM. Elastin confers elasticity and toughness to tissues, and glycosaminoglycans enhance the buffering capacity of the ECM and regulate permeability through hydration effects. The proportion of these components of the natural ECM varies significantly across different tissues and organs. Carbon nanotubes have been shown to up-regulate type I collagen gene expression, with longer nanotubes demonstrating better effects than the shorter ones.54 Poly(lactic-co-glycolide) (PLGA) nanoparticles can effectively promote the synthesis of elastin.55 Therefore, during organoid cultivation, the physicochemical properties of nanomaterials can be harnessed to precisely regulate the synthesis and assembly of core ECM proteins. This strategy allows for the simulation of the specific ECM component characteristics of target tissue.
In addition, recent studies have demonstrated that incorporating conductive nanomaterials into the ECM can effectively enhance the electrical conductivity, thereby promoting the maturation of certain organs. In a study by Tan et al., the integration of conductive silicon nanowires (e-SiNWs) into cardiac organoids improved the electrical conduction velocity and synchronous contraction capabilities of cardiomyocytes.27 Experiments indicated that e-SiNWs enhance electrical coupling between cells by promoting the expression of gap junction proteins. This enhancement consequently improves the electrophysiological integration efficiency of transplanted cardiac organoids into infarcted myocardium. Similarly, carbon nanodot composites have been shown to significantly promote neurite outgrowth in neural progenitor cell spheres and accelerate the electrical physiological maturation of neurons. These findings offer new strategies and open up new possibilities for functional neural network construction.26
Afting et al. developed nanoscale DNA microbeads for drug delivery. When injected into retinal organoids, these microbeads degrade under ultraviolet physical fields, enabling the gradient release of encapsulated Wnt-surrogate. By adjusting the injection site and photoinitiation parameters, precise spatiotemporal control over organoid development can be achieved, thereby meeting specific mechanical requirements within the organoids.29
2.2 Dynamic regulation via external physical fields
Nanoparticles can respond to external physical fields and undergo directional movement, aggregation and dispersion behavior changes.60,61 Thus, active modulation of the physical field enables the precise control of mechanical forces exerted on organoids. This approach enables stage-specific environmental manipulation that closely mimics physiological growth conditions, while offering a simple and versatile platform to meet the distinct mechanical requirements of each organoid developmental phase.10,62 Simultaneously, the developmental status of organoids can also be monitored in real time by the spatial distribution patterns of MNPs.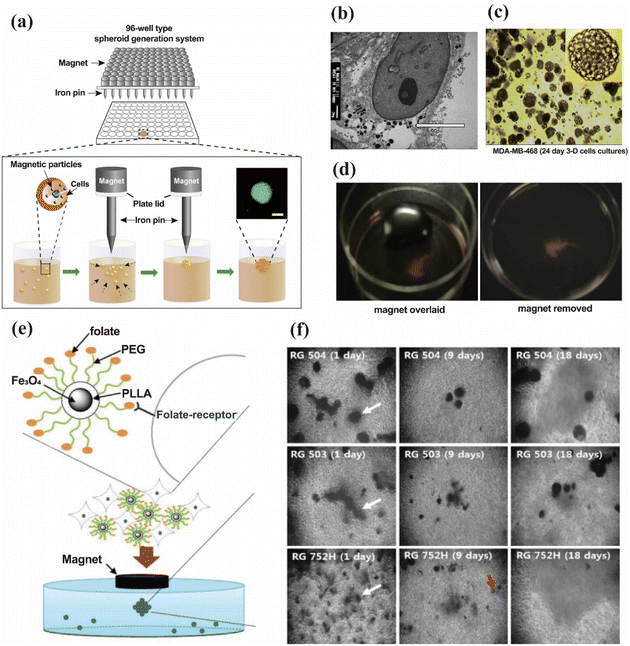 | ||
| Fig. 2 Magnetic levitation culture for organoid generation. (a) Schematic of the formation of spheroids using magnetic levitation technology. Cells internalize magnetic nanoparticles (MNPs) via endocytosis, thereby exhibiting acquired magnetic properties. Under the focused magnetic force from iron pins and magnets, the cells accumulate at specific characteristic positions within the culture medium and gradually assemble into spheroids. This figure has been adapted/reproduced from ref. 70 with permission from Elsevier Ltd, copyright 2013. (b) TEM images of MDA-MB-231 cells containing internalized MNPs. (c) Tumor sphere morphology after 24 days of magnetic suspension culture. (d) Tissue images before and after the removal of the external magnetic field. This figure has been adapted/reproduced from ref. 30 with permission from Elsevier Inc., copyright 2015. (e) Schematic of the magnetic levitation technology mediated by surface adsorption of MNPs. MNPs are bound by electrostatically interacting cell membranes and cultured in a magnetic field for 3D tumor spheroids. (f) Tumor spheroids on different culture days. This figure has been adapted/reproduced from ref. 33 with permission from Elsevier B.V., copyright 2011. | ||
The internalized NP may accumulate to cytotoxic concentrations, degrade into harmful substances, disrupt cellular morphology and signaling pathways, and ultimately induce necrosis or apoptosis.72 To mitigate such cytotoxicity, researchers are actively pursuing surface functionalization strategies to improve nanoparticle biocompatibility. Coating nanoparticles with biocompatible materials such as poly-L-lysine or PVP can significantly mitigate cytotoxicity, ensuring normal growth and functionality.73
Lee et al. successfully engineered human epidermoid tumor spheroids using PLGA particles encapsulated with magnetic iron oxide (Fe3O4).33 The distinction lies in the diameter of the particles: Lee's group utilized larger PLGA particles (20 μm diameter) that remain extracellular throughout the culture process. Instead, cells adhere to the particle surfaces via electrostatic interactions, subsequently proliferating to form 3D clusters (Fig. 2e). This design eliminates the possible toxicity risks of nanomaterials. The magnetic field upregulates N-cadherin expression at the cell membranes and junctions, enhancing intracellular interaction and simulating the in vivo environment. As the PLGA degraded, the magnetic particles were progressively cleansed and cleared, leaving behind the densely packed tumor spheroids (Fig. 2f). Inspired by Lee's work, Rodriguez-Arco synthesized a magnetic nanocomposite with a positively charged core–shell structure, which is designed to bind with negatively charged cell membranes to achieve highly efficient cell magnetization.74 The core consists of an acrylic monomer copolymer coated with a 10–50 nm layer of iron oxide that provides both magnetic and positive surface charges. Furthermore, the outer surface of the iron oxide layer was modified with a poly(ethylene glycol) (PEG) coating to improve the biocompatibility. Gaitán-Salvatella et al. conducted a study on the biocompatibility of a class of nanomaterials that bind to cell membranes via electrostatic interactions.36 The findings demonstrated that these nanomaterials do not interfere with the physiological activities and gene expression of cells; additionally, they are easy to remove, making them suitable for the long-term cultivation of spheroids.
The magnetic levitation culture approach eliminates matrix-induced variability in cell behavior, and the structural diffusion barriers ensure uniform oxygen and nutrient distribution throughout the organoids. Salivary gland organoids cultured via magnetic levitation exhibited autonomous ECM deposition, upregulated expression of N-cadherin and epidermal growth factor receptor (EGFR), and stable 3D structures without central necrosis observed in conventional cultures (Fig. 2b–d).30,35,75
Several research groups have developed hydrogel-assisted magnetic levitation platforms for improved 3D cell culture formation.76,77 These platforms have demonstrated successful generation of diverse spheroids and organoids, with more refined spatial organization and natural structure and function.
2.3 Advanced engineering approaches for organoid cultivation
The standardized quality control and high-throughput culture remain critical challenges limiting the large-scale application of organoids, while nanomaterials present promising solutions to these technological barriers. In 3D bioprinting, the rheological properties of nanocomposite bioinks can be precisely tuned by adjusting the concentration and surface modification of nanoparticles, enabling enhanced digital control of printing process. Microbioreactors modified with nanomaterials not only simulate key aspects of the in vivo physiological microenvironment but also ensure the uniformity of conditions during organoid growth, thereby facilitating high-throughput organoid production.NPs can also play an active role in both labeling and spatial organization. MNPs bound to the cell membrane or internalized within cells enable precise spatial organization of cellular assemblies under external magnetic field guidance, facilitating the formation of geometrically controlled organoids with high structural consistency. Through MNP adhesion to dissociated spinal cord cells, Bowser et al. successfully endowed the cells with magnetic properties and enabled highly reproducible assembly of spinal cord spheres.80 Similar methods have also been successfully adapted for the cultivation of both salivary adenoid epithelial organoids and adipose tissue organoids.81 The use of MNPs establishes a standardized platform for spheroid and organoid formation, ensuring the consistency in size and shape across batches. This technology reduces operator dependency, while significantly improving the reproducibility of 3D biostructure cultivation process.
2.3.2.1 Liquid marbles. Liquid marbles represent a novel class of biological microreactors characterized by a liquid culture core enveloped within a hydrophobic nanoparticle shell, exhibiting typical diameters in the millimeter scale (Fig. 3a). In these bioreactors, nanomaterials serve dual functions: (1) as semi-permeable encapsulation barriers that maintain system isolation, and (2) as gas–exchange interfaces between the liquid core and the external environment. Aalders et al. engineered cardiac organoids by encapsulating a cardiomyocyte-fibroblast suspension within hydrophobic fumed silica nanoparticles.82 Brevini et al. demonstrated that hydrophobic polytetrafluoroethylene (PTFE) nanoparticles could encapsulate cellular suspensions, facilitating the formation of 3D aggregated cell spheres.41 While liquid marble technology promotes cell aggregation and scalability needs, critical gaps remain in manual operation requirements and limited control over the microenvironment.
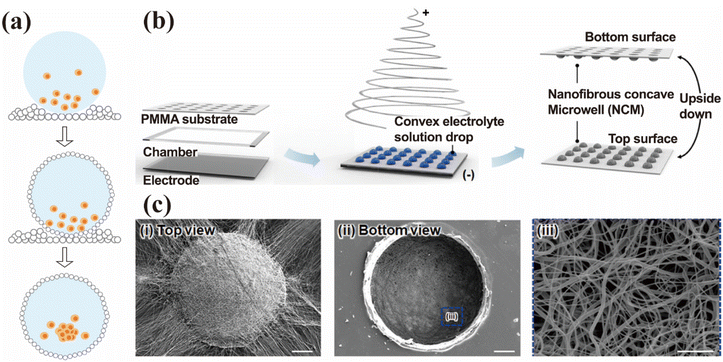 | ||
| Fig. 3 Microreactor for organoid cultivation. (a) Schematic of liquid marble preparation and organoid cultivation. Cell suspensions were placed on the surface of hydrophobic nanoparticles and encapsulated via droplet rolling. The hydrophobic shell enables gas exchange between the droplets and the external environment while promoting cell aggregation and spheroid formation. (b) Schematic of the hydrogel-based U-shaped microwell array fabrication. U-shaped microcavities were fabricated on silicon substrates using a combination of Bosch processes and soft lithography. A PDMS mound was fabricated by replica molding the silicon substrate. The microcavities were then transferred to the hydrogel surface during cross-linking. (c) SEM images of nanofibrous microwells fabricated via electrospinning using both an electrolyte solution and a metal collector. This figure has been adapted/reproduced from ref. 83 with permission from the American Chemical Society, copyright 2018. | ||
2.3.2.2 Nano-microwell culture system. Compared to liquid marble techniques, the microwell chip culture system provides superior control cell distribution in organoid culture and enable high-throughput production. Typically composed of polymer membranes with micropores, the microwell chip culture system incorporates surface-patterned hydrogels (e.g., agarose and meth acryloyl gelatin (GelMA)) fabricated via 3D printing. The uniform-sized pores not only provide a supportive matrix required for cell growth but also constrain pore geometry to promote cell self-assembly and functional tissue formation.84,85 It has been demonstrated to enhance the spheroid formation and enable precise gradient size control.86 The platform facilitates high-throughput organoid cultivation under standardized conditions, thereby significantly improving experimental efficiency and reproducibility. This platform holds substantial promise for preclinical applications and scalable manufacturing, emerging as the predominant methodology for spheroid culture.87 However, reliance on non-physiological polymer substrates may compromise organoid development. Impermeable microporous materials such as PDMS exhibit constrained mass transport properties, significantly limiting their application in organoid culture systems.
In 2016, Shin et al. developed a serum-free, xeno-free microporous 3D culture system utilizing a nanofiber-hydrogel composite for human salivary gland globule organoid cultivation.88,89 The microporous culture system comprises a PCL nanofiber substrate integrated with PEG hydrogel micropore walls. The nanofibers fabricated by electrospinning form a highly porous network, effectively overcoming the mass transport limitations inherent to conventional polymer microporous systems. They can also influence the behavioral response of 3D cells and play a key role in regulating the spheroid size.90 Surface modification of the fiber can enhance the cell adhesion behavior. Subsequent studies expanded on these findings to optimize the microporous reactor design. By employing convex hemispherical electrolyte droplets as a grounded collector for electrospinning, Park et al. developed a method to fabricate nanofiber-based concave micropores (NCM) with tunable shape and size (Fig. 3b and c).83 Uniform and size-controlled human hepatocellular carcinoma cell line (HepG2) spheroids were successfully formed within the NCMs, exhibiting significantly enhanced metabolic activity compared to those cultured in 2D micropores. Kim et al. introduced a nanofiber-based elliptical microporous array, termed the NOVA microporous array.91 The array exhibits an oval geometry with a wide opening, narrow bottom, high aspect ratio (AR) and high pore density. This design not only enhances cell capture efficiency within the micropores but also enables uniform and stable production of numerous viable and functional spheroids.
3. Nanomaterials for the cryopreservation of organoids
Organoids demonstrate significant application potential in fields such as disease modeling and drug screening. However, their clinical translation and large-scale adoption are constrained by challenges in long-term cultivation and storage, specifically during long-term culture and preservation. In particular, during extended expansion, organoids are prone to phenotypic drift and central necrosis.92 Their complex 3D architecture and heterogeneous cellular composition render them highly sensitive to cryopreservation conditions, resulting in issues such as localized supercooling and insufficient cryoprotection. In conventional cryopreservation protocols, the widely used CPA dimethyl sulfoxide (DMSO) exhibits cytotoxicity.93 Moreover, due to their size, organoids require extended periods for CPA permeation, thereby increasing their exposure to toxic damage.94 Second, conventional water bath rewarming exhibits a lower heat transfer efficiency than the required critical warming rate (CWR).95 Ice recrystallization induced by the temperature gradients during warming inflicts severe mechanical damage on cells. Consequently, the development of advanced cryopreservation and rewarming technologies has become a critical priority in the field of organoid research and application.Nanomaterials present a novel strategy to overcome the challenges described above. In contrast to traditional CPAs, nanomaterials can act as auxiliary nucleation sites to regulate ice crystal formation and reduce the risk of supercooling.96 Nanomaterials can also modulate ice crystal growth kinetics and enable the targeted delivery of CPAs such as trehalose, thereby enhancing cryopreservation efficiency. These advantages have allowed nanomaterials to demonstrate promising performance in the cryopreservation of various cell types.97 Furthermore, certain functionalized nanomaterials can enhance cryopreservation outcomes through antioxidant effects and cell stabilization. Additionally, nanomaterials can serve as heating mediators during rewarming, promoting uniform and rapid temperature increase.13 Therefore, elucidating the mechanisms of nanomaterials in organoid cryopreservation and exploring effective application strategies will provide crucial technical support for their long-term stable preservation and promote broader clinical and research application.
3.1 Current advances and challenges in organoid cryopreservation
Cryopreservation represents a core technology driving the large-scale application of organoids. To date, researchers have developed a variety of CPA formulations and corresponding cryopreservation protocols tailored for different functional types of organoids.98 However, the inherent heterogeneity and complex cellular composition of organoids significantly impede the effectiveness of conventional CPAs and uniform heat transfer, presenting major challenges during both the freezing and thawing process.From the application perspective, organoids advance drug development and personalized medicine, offering valuable alternative 2D cell cultures and animal models in high-throughput drug screening.104–106 Studies employing endometrial and breast cancer organoids have enabled rapid screening of drug combinations and identification of effective tumor inhibitors, accelerating novel drug development.107 Moreover, patient-derived organoids preserve genetic traits and reduce disease heterogeneity, providing a platform for developing personalized treatment plans.108,109 For instance, studies utilizing organoid models have successfully identified effective drugs for aggressive diseases such as triple-negative breast cancer.2 Moreover, organoids could function as a bridge therapy to stabilize patients awaiting organ transplantation, exhibiting significant potential in the field of transplant medicine. Compared with allogeneic organ transplantation, organoids derived from autologous stem cells present significantly lower risks of immune rejection. Current research has successfully demonstrated the transplantation of the liver, pancreas, intestine, and other organoid types in mouse models, showing evidence of therapeutic efficacy in disease treatment.110 Nevertheless, the functional mechanism and immunogenic potential of organoids remain incompletely characterized.
Despite the demonstrated potential of organoid technology in both fundamental research and clinical applications, challenges in long-term stable preservation remain a critical limitation for its widespread application.111 Organoids may undergo phenotypic drift and genetic instability due to prolonged in vitro culture conditions and serial passaging, potentially compromising their clinical application. Since many diseases demonstrate progressive pathophysiology, organoid phenotypic instability distorts disease progression studies and causes time-dependent model variations, severely restricting their utility for mechanism exploration. Similarly, phenotypic discrepancies between organoids and native tissues may compromise drug screening accuracy and personalized medicine, leading to unreliable therapeutic predictions.92 The cryopreservation of organoids during early developmental stages minimizes the passage-induced phenotypic drift, improving model consistency.98 Batch-cryopreserved organoids enable reproducible experimentation by allowing repeated utilization of genetically matched models across multiple timepoints, ensuring experimental consistency.
Cryopreservation technology holds a critical value for both organ transplantation and biobanking, enabling long-term storage of viable biological specimens.112 Current culture conditions enable short-term maintenance of organoids, and room temperature storage/transport preserves their active function.113,114 However, the absence of long-term preservation methods leads to loss of cell viability, impaired structural integrity and functional decline of organoids. These limitations significantly compromise organoids' reliability as an experimental model while hindering their translational potential, particularly for multi-institutional studies and large-scale clinical applications. Cryopreservation significantly reduces the cellular metabolic rate and enables on-demand availability of viable biological products for therapeutic use. Cryopreservation is essential for maintaining the viability and functionality of organoids during storage and transport, ensuring their availability for experimental, therapeutic, and clinical applications.
During CPA incubation, organoids exhibited volume changes similar to suspended cells: initial dehydration and shrinkage due to the water efflux, followed by gradual volume recovery as the CPA permeated the cells.116 The key difference is that, due to their large size, outer cells experience a prolonged osmotic equilibration period, making them more susceptible to osmotic damage and cytotoxic effects.94 The cell osmotic pressure in the central region is maintained at a low level, which may impair CPA diffusion. In addition, the mass transfer process involves both intracellular transport and ECM diffusion. The overall process is challenging to control, resulting in inconsistent cryoprotection efficacy that ultimately compromises the 3D structural integrity.117,118 While increasing the CPA concentration and incubation temperature may effectively address these challenges, careful optimization will be required to balance efficacy with potential cytotoxicity. CPA diffusion is driven by the concentration gradient; therefore, increasing the external CPA enhances the driving force, leading to faster CPA penetration rates and shorter incubation times.119 However, this approach must be carefully balanced, as high CPA concentration exhibits significantly greater cytotoxic effects. In addition, the temperature critically influences CPA penetration efficiency, while a lower temperature reduces the intracellular enzyme activity. At room temperature, elevated cellular metabolic activity promotes more efficient material exchange, enabling significant reduction of CPA incubation time.120 However, the CPA cytotoxicity accelerates at room temperature compared to lower temperatures.121,122 Therefore, appropriate CPA concentration, incubation temperature and exposure duration are all crucial for effective cryopreservation.
During the initial stage of cooling, CPA loading is completed, and mass transfer processes, including cellular metabolism, can be neglected. The rapid heat exchange between the outer surface of organoids and the surrounding environment makes the initial cooling rate a critical factor in minimizing cellular damage. Ultra-slow cooling rates may induce mechanical damage due to the excessive ice crystal growth.123 During the ultra-rapid cooling, the osmotic imbalance across the plasma membrane drives excessive water efflux, resulting in cell shrinkage and damage. This may also induce solution supercooling, followed by explosive crystallization at a critical temperature. The subsequent rapid release of latent heat generates substantial internal mechanical stress, potentially causing structural damage.116 During ice formation, the propagating mechanical stress is transmitted through intercellular connections, resulting in the disruption of the 3D architecture of organoids.124,125 Similarly, during rewarming, precise control of thermal gradient is essential for ensuring uniform heating. Notably, the 3D heterogeneous architecture of organoids significantly influences heat conduction rates and temperature distribution. The low inconsistent heat conduction rate within organoids leads to a substantial internal temperature gradient, prolonging the thermal equilibration time and exacerbating the risk of thermal stress-induced damage.126 Therefore, optimizing heat transfer uniformity and temperature distribution homogeneity represents the critical breakthrough needed to resolve dynamic temperature control challenges in organoid cryopreservation.
Furthermore, cellular and dimensional heterogeneity presents a fundamental challenge in organoid cryopreservation. In large-sized organoids, restricted internal mass transfer limits uniform CPA diffusion, significantly increasing the risk of lethal intracellular ice formation during cryopreservation. Small or immature organoids are more sensitive to osmotic stress owing to their elevated metabolic rates, increasing susceptibility to membrane rupture or apoptosis. The cryopreservation requirements of organoids vary significantly across developmental stages, tissue types and physical dimensions, while inherent cellular heterogeneity further complicates protocol optimization.127 Organoids typically comprise heterogeneous cell populations at varying developmental stages with distinct physiological functions. This cellular diversity manifests in differential cell membrane composition, variable CPA permeability coefficients, and dehydration kinetics during cryopreservation.128 For example, the membrane permeability to ethylene glycol (EG) in granulocytes is much higher than that of DMSO and glycerol. Similarly, the permeability of stem cells to glycerol and DMSO is also different.129 The standard intestinal organoid model comprises multiple cellular components including stem cells, epithelial cells, goblet cells, and endocrine cells, each exhibiting distinct cryosentivity.130 During cryopreservation, heterogeneous cell populations within organoids exhibit differential responses to identical freezing conditions. Developing precision cryopreservation protocols that account for this cellular diversity represents a critical challenge requiring innovative solutions.
Although DMSO is an effective cryoprotectant, its inherent cytotoxicity can damage the cell structure and function. Furthermore, DMSO can induce the denaturation of proteins and other biological molecules, adversely affecting the functionality of cells upon thawing.137 Consequently, researchers worldwide have conducted extensive studies aimed at reducing the required concentration of DMSO and identifying less cytotoxic alternative CPAs.138 For example, studies on various cell types have demonstrated that a combined CPA solution containing a lowered concentration of 5% DMSO significantly improves post-thaw cell survival and function compared to higher concentrations.139 However, a DMSO concentration exceeding 15% drastically reduces survival due to extreme osmotic damage and cytotoxicity. Consequently, the development of novel, less toxic CPAs and advanced cryopreservation techniques has become a critical research frontier, essential for enabling the broader applications of organoid technology.
3.2 Cryopreservation mechanism and application of nanomaterials
Nanomaterials have demonstrated a remarkable capability to regulate ice crystal formation and growth, thereby preventing ice crystallization-related damage during cryopreservation. Furthermore, they facilitate the intracellular delivery of non-permeable CPAs such as trehalose (Fig. 4). Recently, research into and application of nanomaterials for cryopreservation have expanded significantly, establishing them as promising emerging strategies to overcome the challenges of conventional cryopreservation protocol.13The presence or absence of impurities distinguishes the two primary mechanisms of ice nucleation: homogeneous nucleation and heterogeneous. During cooling, the CPA solution lowers the freezing point but often lacks sufficient nucleation sites, leading to significant supercooling. While some studies have attempted to induce ice nucleation manually, this approach suffers from poor reproducibility and risk of localized rapid crystallization, which cause damage to cells.140,141 To address this, nanomaterials can serve as heterogeneous nucleating agents by providing a high density of nucleation sites, which promotes uniform ice nucleation and reduces supercooling. The ice nucleating ability of carbon-based nanomaterials was first demonstrated by Whale et al.142 Subsequent investigation studies have revealed that nanomaterials such as nanocrystalline cellulose, MoS2, and silica can act as effective nucleation sites to control ice nucleation, a process influenced by their morphology and size.143,144 The ability to promote ice nucleation is generally stronger in smaller nanoparticles, while larger nanoparticles often exhibit an inhibitory effect. However, the influence of nanoparticle morphology on ice nucleation remains poorly understood and lacks consensus. In addition to promoting ice nucleation, nanomaterials offer additional benefits of limiting ice growth, regulating crystal morphology, and inhibiting recrystallization. Research has shown that nanomaterials with special functional groups and molecular structures such as GO, quasi-carbon nitride quantum dots, and metal–organic framework (MOF) nanoparticles can bind to the basal or prismatic planes of ice crystals. This binding inhibits ice growth and regulates the morphology of crystals (Table 2).145,146
| Nanomaterials | Cells | Function | References | |
|---|---|---|---|---|
| a RBCs: Red blood cells; ASCs: Adipose tissue-derived stem cells; TPP: Tripolyphosphate; GO: Graphene oxide; MSCs: Mesenchymal stem cells; OCNQDs: Oxidized carbon nitride quantum dots; S-OCNQDs: Sulfur-doped carbon nitride quantum dots; G-CDs: Glucose-derived carbon dots; MOF: Metal organic framework; MOLs: Metal–organic layers; Hf: Hafnium; PEG: Poly(ethylene glycol); PGMA: Poly(glycidyl methacrylate); PVA: Poly(vinyl alcohol); PHPMA: Poly(hydroxypropyl methacrylate). | ||||
| Apatite nanoparticles | Sheep RBCs | Trehalose delivery | 147 | |
| pH-responsive genipin-cross-linked pluronic F127-chitosan nanoparticles | Primary human adipose-derived stem cells | 148 | ||
| Cold-responsive trehalose-laden nanoparticles | MDA-MB-231 cancer cells and human ASCs | 149 | ||
| Chitosan-TPP nanoparticles | Natural killer-92 cells | 150 | ||
| Nanohydrogel (acrylamide-type polymer network) | Human umbilical vein endothelial cells | 151 | ||
| Carbon derivatives | GO | Inhibition of ice crystal growth | 96 | |
| Fe3O4 deposited GO | Human MSCs | 152 | ||
| OCNQDs and S-OCNQDs | Sheep RBCs | 153 | ||
| Graphene nanoparticles | — | 154 | ||
| G-CDs | Sheep RBCs | 155 | ||
| Metal nanomaterials | Iron oxide | Mice oocytes | Inhibition of ice crystal growth, uniform rewarming | 156 |
| Ti3C2 | Human MSCs | 157 | ||
| MoS2 | Human lung cancer cells (A549 cells) | 144 | ||
| Organic-inorganic metal materials | Nano-zirconium-based MOF | Human RBCs | Inhibition of ice crystal growth | 158 |
| Two-dimensional MOLs of Hf | Human RBCs | 145 | ||
| PEG-gold hybrid nanoparticles | — | 159 | ||
| Gold nanoparticles grafted with peptide nanofibrils | — | 160 | ||
| Fe3O4@SiO2 nanorods | Porcine artery models | Inhibition of ice crystal growth, uniform rewarming | 161 | |
| Combination of carbon dots and gold nanorods by SiO2 | Hela cells | 146 | ||
| Polymer nanomaterials | PGMA56-PHPMA155/PVA system | Ovine RBCs | Inhibition of ice crystal growth | 162 |
| PVA181-g7-PHPMAn | — | 163 | ||
| Nanomotor with magnesium/palladium covering one side of a silica platform (Mg@Pd@SiO2) | Normal colon epithelial (NCM460) cell | Inhibition of ice crystal growth, reducing oxidative stress, uniform temperature profile | 164 | |
| Ex vivo tissues (small intestine, liver, and kidney) | ||||
The high thermal conductivity of common nanoparticle constituents including metals, metal oxides, and carbon-based materials enhances thermal management during cryopreservation. The incorporation of high-thermal-conductivity nanoparticles into CPAs facilitates the formation of effective heat conduction pathways. These pathways accelerate the thermal transport, which consequently enhance the overall thermal conductivity of the solution.165 In addition, the Brownian motion of nanoparticles makes them more evenly distributed in the solution. This homogeneity helps to eliminate local hot spots, thereby promoting uniform temperature changes throughout the freezing phase.161 The uniform cooling process mitigates the irregular formation of ice crystals and alleviates the possibility of cell damage due to ice crystals.
The generation of reactive oxygen species (ROS) during freezing and thawing processes presents a major threat to cell survival. Therefore, antioxidants such as glutathione are commonly incorporated into commercial organoid cryopreservation reagents to mitigate ROS-induced damage. However, these protective agents themselves can adversely affect essential cellular functions.166,167 Due to their large specific surface area and abundant surface-active sites, nanomaterials can effectively capture free radicals during the freeze-thaw process, thereby preventing oxidative harm.168–170 In addition, these sites can bind to biological macromolecules, such as proteins on the cell membrane, to form a protective layer on the cell surface. This layer provides an additional physical barrier that improves membrane stability, thereby mitigating rapid water loss and exposure to high concentrations of protective agents. This effect reduces osmotic pressure damage;33,171 however, the dosage must be carefully controlled to avoid potential damage to the antioxidant system by excessive nanoparticles.172
Beyond their intrinsic benefits, nanomaterials can act as carriers to deliver non-permeable protective agents such as trehalose into cells, thereby maximizing protection. Trehalose is a natural and widely used non-permeable CPA found in many organisms. However, as mammalian cells cannot synthesize it, trehalose can only provide optimal cryoprotection when present on both sides of the membrane.173 To facilitate intracellular delivery of trehalose, various techniques have been employed, including electroporation, thermal shock, and microinjection.174–176 However, these strategies cause damage to the cell membrane to varying degrees. Furthermore, their strict technical requirements hinder their use in mass cell treatment. Endocytosis-driven nanovector delivery offers a more biocompatible platform for trehalose transport, bypassing the need for additional clearance mechanisms.177
Another major application focuses on controlling ice crystallization and stabilizing outer cell membranes. For instance, Xu et al. studied how ice-suppressing micro–nano materials of different sizes affect the cryopreservation of various cell types, such as human umbilical vein endothelial cells, mouse myoblasts, and bovine mammary epithelial cells. The study revealed that a smaller particle size enhanced the cryoprotective efficacy of the carbon complex particles.179 Compared to spherical particles, asymmetric and flower-like nanomaterials significantly improved cell cryopreservation outcomes. Owing to their good biocompatibility and modifiable surface properties, these nanomaterials can adapt to various conditions and provide new solutions for spheroid and organoid cryopreservation. Gao et al. reported the use of a magnesium/palladium-covered silica nanomotor (Mg@Pd @SiO2) for highly efficient cryopreservation.97 The nanomotor alleviated freezing damage in cells and tissues by inhibiting ice crystallization. The mechanism involves the release of Mg2+ ions and hydrogen gas (H2) upon cooling in solutions. The subsequently promoted adsorption of H2 at the palladium binding site on the cell surface effectively suppresses ice crystal formation. During the thawing process, the nanomotor switched from a dormant state to an active one upon laser activation, enabling the continued release of H2. This approach further restrained ice recrystallization and minimized cell and tissue damage. The team demonstrated that these nanomotor significantly improved both the survival rate and the functional recovery of NCM460 cells. The technology also proved effective in protecting isolated tissues from the small intestine, liver and kidney tissues, demonstrating its potential for organoid cryopreservation.
Despite the multiple advantages that nanomaterials offer for enhancing cell cryopreservation, particular attention must be paid to their permeability when utilized with 3D models such as spheres and organoids. The complex 3D architecture and high cell density of organoids hinder the uniform penetration of CPAs and nanomaterials, currently limiting their application in cryopreservation. In a study, supplementing traditional CPAs with non-permeable chemical nucleating agents effectively improved the survival rate of spheroids after thawing, indicating the significant potential of nucleating agents in cryopreservation.180 Further research has found that anionic nanoparticles can open the tight junctions between intestinal epithelial cells, thereby acting as penetration enhancers for physical and chemical drugs.181 Since this effect is reversible and non-damaging, it presents a new promising approach for enhancing CPA diffusion in intestinal organoids.
3.3 Nanomaterials for organoids thawing
The rewarming of suspended cells is typically performed in a 37 °C water bath. Agitating the cell suspension during thawing promotes efficient heat conduction, shortens the time window for ice crystal formation, and prevents recrystallization. However, for large cell spheroids and organoids, the heat transfer efficiency of water bath rewarming decreases significantly, resulting in a heating rate that falls below the CWR necessary for cells and organ survival. The substantial temperature gradient between the surface and the center promotes ice recrystallization, which causes fatal mechanical damage to the cells within the organoid.182,183 Nevertheless, the conventional approach of thawing in a 37 °C water bath for 2–3 min remains the most commonly used thawing method for globules and organoids. This protocol is a standard practice following both slow freezing and vitrification.132,141,184,185 Although dissociation or sectioning organoids prior to thawing reduces cell damage, it increases subsequent culture time and cost. Therefore, there is an urgent need to develop new rewarming protocols that can achieve a higher CWR for intact organoids.Microwave radio frequency heating rapidly thaws cells, tissues, and organs by exciting water molecules to absorb energy and convert it into heat.182,186 However, the uniformity of heat transfer is often compromised by CPA composition, which may result in local overheating and increased thermal stress damage.183 The limited penetration depth of microwave radiation also prevents uniform rewarming, particularly in large-sized samples, where it creates significant thermal gradients. Advances in nanotechnology have enabled the use of nanoparticles as highly efficient heating media for biomaterials, including laser nano-heating and radio frequency (RF) nano-heating.
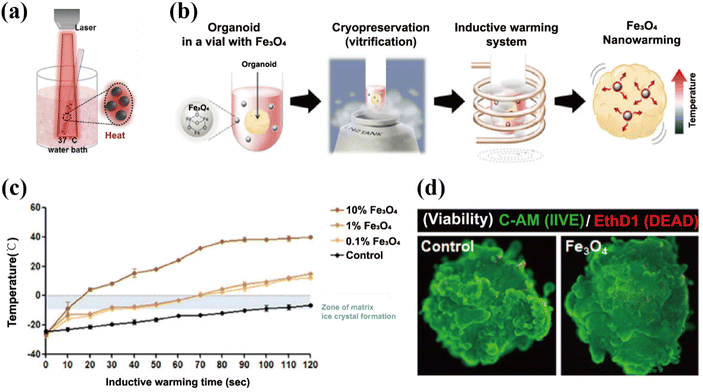 | ||
| Fig. 5 Nano-thawing for organoid cryopreservation. (a) Schematic of laser-nano heating. Cryopreserved samples were placed in a 37 °C warming solution and subjected to a near-infrared laser field at an intensity of 1000 mW cm−2 for 10 s to induce photothermal effects, followed by additional 5 s of rewarming. This figure has been adapted/reproduced from ref. 188 with permission from Elsevier Ltd, copyright 2019. (b) Schematic of radio frequency (RF)-based nanoheating. Organoids were loaded onto vials containing cryoprotectants (CPAs) and Fe3O4 nanoparticles, vitrified in liquid nitrogen, subsequently rewarmed via RF excitation of the Fe3O4 particles surrounding the organoids. (c) Temperature profiles of samples with different concentrations of Fe3O4 (0.1%, 1%, and 10%) measured over the warming period. Data are presented as mean ± SD. n = 3. (d) Viability comparison between 0% and 10% Fe3O4 groups assessed using the LIVE/DEAD assay. n = 5. This figure has been adapted/reproduced from ref. 13 with permission from John Wiley & Sons, Inc., copyright 2023. | ||
As early as 13 years ago, Etheridge et al. demonstrated the use of magnetic nanoparticles to achieve faster and more uniform rewarming in kidney models.193 Further studies revealed that magnetic heating not only reduced internal stress but also reversed its distribution, transferring stress capable of causing cracks toward the outer wall. The residual stress accumulated during the freezing process acts as a buffer, allowing for a higher heating rate by resisting the additional stress generated during rapid rewarming. This mechanism further demonstrates the application potential of magnetic heating in biological tissue thawing. Wakabayashi et al. systematically studied the effects of magnetic field frequency, magnetic nanoparticle concentration, and sample size on nanorewarming.194 The study revealed that the heating rate was promoted by increases in magnetite concentration, applied power, and field frequency, independent of the sample volume. A concentration of 5 mg mL−1 iron oxide nanoparticles achieved the CWR. However, considering the potential complications associated with higher concentrations, they identified the optimal rewarming condition as 5 mg mL−1 nanoparticles, 10 KW power, and 208 kHz frequency. Their research provides a theoretical basis for the application of MNP-mediated rewarming in organoid cryopreservation. Subsequently, Lee et al. successfully applied the previously reported iron oxide nanoparticles to rewarm heart-like organs exceeding 2 mm in diameter. This approach achieved a faster and more uniform heating profile than traditional water baths. This led to significantly improved morphological and functional recovery in the thawed organs (Fig. 5d).13
Building upon iron oxide nanoparticles, Manuchehrabadi et al. developed a coated mesoporous silica structure that effectively prevented the precipitation of magnetic nanoparticles and greatly improved their biocompatibility.198 The experimental results across various cell types and tissue models have further confirmed the superiority of magnetic nanoparticle-mediated heating during rewarming over traditional water bath convection methods. Similarly, PEG-coated superparamagnetic iron oxide nanoparticles synthesized by Chiu-Lam et al.,191 PEG-modified silica-coated iron oxide nanoparticles reported by Karimi,199 and nanoparticles described in other studies all exhibit comparable stability and satisfactory rewarming effects.200 Joshi et al. investigated the mechanical stress induced by silica-coated iron oxide nanoparticles (SiONPs) during the rewarming of rat hearts and human heart models and found variations in the heating rates among different regions of the samples.197 Areas that rewarm more rapidly experience compressive stress, whereas regions heating more slowly are subjected to tensile stress. Such non-uniform thermal stresses pose a greater threat to the structural integrity of vitrified samples. Furthermore, the study indicated that the location and magnitude of the maximum stress during the rewarming vary with the concentration of SiONPs, though no consistent pattern has yet been established. In addition, Liu et al. suggested that encapsulating stem cells within hydrogels embedded with magnetic nanoparticles can prevent direct physical contact between the cells and the nanoparticles. This strategy not only ensures the heating efficiency but also eliminates potential cytotoxicity, showing promising applicability for the rewarming of organoids.201
Hypothermic perfusion also serves as a method to promote the uniform distribution of nanoparticles throughout different regions of biological tissues.202 In contrast to direct incubation, this approach enables homogeneous nanoparticle distribution throughout the organ, enabling heat generation to initiate from within the organoids and diffuse outward. This method facilitates accelerated heating rates and enhanced thermal uniformity, making it particularly suitable for the cryopreservation of large tissues and organs. However, its application depends on the presence of a fully developed vascular system, which limits its use in most organoid models.
4. Summary and future prospects
This review explores the current applications of nanomaterials in organoid culture and cryopreservation and discusses the major challenges associated with the freezing and recovery of organoids. As 3D in vitro models, organoids show great potential for applications in drug screening, disease modelling and other biomedical fields, with the additional advantage of avoiding ethical controversies. However, their broader adoption remains hindered by challenges such as poor reproducibility, difficulties in synchronizing heterogeneous cellular developmental stages, and the limitations of current cryopreservation techniques. Nanomaterials have demonstrated revolutionary potential in both organoid construction and cryopreservation owing to their distinctive physicochemical properties. In organoid fabrication, functionalized nanomaterials can modulate the properties of hydrogel matrices to allow dynamic control over structural stiffness. Furthermore, nanomaterials can guide cells to assemble into specific geometric configuration, thereby facilitating the reproducibility of organoid cultures. In the context of cryopreservation, nanomaterials exhibit considerable application potential by effectively suppressing ice crystal formation, achieving targeted delivery of CPAs and enabling uniform rewarming.Nonetheless, current research still confronts several challenges. First, our understanding of cellular behavior remains inadequate to precisely define the nutritional demands and matrix microenvironment required at various developmental stages. Moreover, the specific impacts of nanomaterial-modified matrix gels on cellular functions and phenotypes have not yet been fully elucidated. Second, the long-term safety of nanomaterials raises another critical concern. Finally, the inherent heterogeneity of organoids means that cryopreservation outcomes are highly dependent on the type and concentration of the nanomaterials used, with results varying not only between organoid types but also within replicates of the same type. The development of precise, tailored cryopreservation methods to address the heterogeneity of organoids constitutes a pivotal challenge that must be urgently addressed.
The application of artificial intelligence (AI) in organoid construction and cryopreservation represents a cutting-edge frontier in biomedical research. Leveraging machine learning (ML) to predict nanomaterial–organoid interactions enables the rational design of materials aimed at reproducible culture and personalized cryopreservation protocols. In addition, the ML models can be trained on data from previous experiments to predict the cytotoxicity and efficiency of novel nanomaterial-based CPA combinations, optimizing for multiple objectives simultaneously, such as maximizing post-thaw viability, minimizing phenotypic drift, and maintaining functionality. AI can also model the complex heat and mass transfer processes during cryopreservation. Enhancing the integration of nanomaterials and biosensors to enable dynamic monitoring of organoid development and cryopreservation would provide deeper insights into the underlying cellular mechanisms. With multidisciplinary collaboration and continuous advancements in materials science and methodology, organoid research is poised to enable broad medical applications and drive significant innovation in healthcare.
Author contributions
Wanjuan He: Reviewing the literature and Writing – major part of the original draft. Qinlin Sun: Writing – part of the original draft. Dan Ge: Writing – review & editing. Bingbing Sun: Writing – review & editing and funding acquisition. Yang Liu: Conceptualization, supervision, writing – review & editing, and funding acquisition.Conflicts of interest
There are no conflicts to declare.Data availability
No primary research results, software or code has been included and no new data were generated or analysed as part of this review.Acknowledgements
This work was supported by the National Key Research and Development Program of China (2022YFC2304305), the Fundamental Research Funds for the Central Universities (DUT20RC(3)035), and the National Natural Science Foundation of China (U22A20455).References
- Z. Zhao, X. Chen, A. M. Dowbaj, A. Sljukic, K. Bratlie, L. Lin, E. L. S. Fong, G. M. Balachander, Z. Chen, A. Soragni, M. Huch, Y. A. Zeng, Q. Wang and H. Yu, Nat. Rev. Methods Primers, 2022, 2, 94 CrossRef CAS PubMed.
- K. P. Guillen, M. Fujita, A. J. Butterfield, S. D. Scherer, M. H. Bailey, Z. Chu, Y. S. DeRose, L. Zhao, E. Cortes-Sanchez, C. Yang, J. Toner, G. Wang, Y. Qiao, X. Huang, J. A. Greenland, J. M. Vahrenkamp, D. H. Lum, R. E. Factor, E. W. Nelson, C. B. Matsen, J. M. Poretta, R. Rosenthal, A. C. Beck, S. S. Buys, C. Vaklavas, J. H. Ward, R. L. Jensen, K. B. Jones, Z. Li, S. Oesterreich, L. E. Dobrolecki, S. S. Pathi, X. Y. Woo, K. C. Berrett, M. E. Wadsworth, J. H. Chuang, M. T. Lewis, G. T. Marth, J. Gertz, K. E. Varley, B. E. Welm and A. L. Welm, Nat. Cancer, 2022, 3, 232–250 CrossRef PubMed.
- X. Guan and S. Huang, Front. Bioeng. Biotechnol., 2022, 10, 1021966 CrossRef PubMed.
- L. Meran, I. Massie, S. Campinoti, A. E. Weston, R. Gaifulina, L. Tullie, P. Faull, M. Orford, A. Kucharska, A. Baulies, L. Novellasdemunt, N. Angelis, E. Hirst, J. König, A. M. Tedeschi, A. F. Pellegata, S. Eli, A. P. Snijders, L. Collinson, N. Thapar, G. M. H. Thomas, S. Eaton, P. Bonfanti, P. De Coppi and V. S. W. Li, Nat. Med., 2020, 26, 1593–1601 CrossRef CAS PubMed.
- T. W. Dennison, R. D. Edgar, F. Payne, K. M. Nayak, A. D. B. Ross, A. Cenier, C. Glemas, F. Giachero, A. R. Foster, R. Harris, J. Kraiczy, C. Salvestrini, G. Stavrou, F. Torrente, K. Brook, C. Trayers, R. Elmentaite, G. Youssef, B. Tél, D. J. Winton, N. Skoufou-Papoutsaki, S. Adler, P. Bufler, A. Azabdaftari, A. Jenke, N. G, N. Thomas, E. Miele, A. Al-Mohammad, G. Guarda, S. Kugathasan, S. Venkateswaran, M. R. Clatworthy, T. Castro-Dopico, O. Suchanek, C. Strisciuglio, M. Gasparetto, S. Lee, X. Xu, E. Bello, N. Han, D. R. Zerbino, S. A. Teichmann, J. Nys, R. Heuschkel, F. Perrone and M. Zilbauer, Gut, 2024, 73, 1464 CrossRef CAS PubMed.
- C. J. Childs, H. M. Poling, K. Chen, Y. H. Tsai, A. Wu, A. Vallie, M. K. Eiken, S. Huang, C. W. Sweet, R. Schreiner, Z. Xiao, R. C. Spencer, S. A. Paris, A. S. Conchola, J. W. Villanueva, M. F. Anderman, E. M. Holloway, A. Singh, R. J. Giger, M. M. Mahe, C. Loebel, M. A. Helmrath, K. D. Walton, S. Rafii and J. R. Spence, Cell Stem Cell, 2025, 32, 640–651 CrossRef CAS PubMed.
- S. Reardon, Nature, 2017 DOI:10.1038/nature.2017.21818.
- S. Chen, X. Chen, Z. Geng and J. Su, Bioact. Mater., 2022, 18, 15–25 CAS.
- X. Qian, Y. Su, C. D. Adam, A. U. Deutschmann, S. R. Pather, E. M. Goldberg, K. Su, S. Li, L. Lu, F. Jacob, T. T. N. Phuong, S. Huh, A. Hoke, S. E. Swinford-Jackson, Z. Wen, X. Gu, R. C. Pierce, H. Wu, L. A. Briand, H. I. Chen, J. A. Wolf, H. Song and G. Ming, Cell Stem Cell, 2020, 26, 766 CrossRef CAS PubMed.
- A. R. Abdel Fattah, S. Ghosh and I. K. Puri, ACS Appl. Mater. Interfaces, 2016, 8, 11018–11023 CrossRef CAS PubMed.
- A. Purwada, M. K. Jaiswal, H. Ahn, T. Nojima, D. Kitamura, A. K. Gaharwar, L. Cerchietti and A. Singh, Biomaterials, 2015, 63, 24–34 CrossRef CAS PubMed.
- D. Xie, B. Chen, W. Wang, W. Guo, Z. Sun, L. Wang, B. Shi, Y. Song and M. Su, ACS Nano, 2025, 19, 12458–12466 CrossRef CAS PubMed.
- S. Lee, J. Kim, J. Seok, M. W. Kim, J. Rhee, G. Song, S. Park, S. Lee, Y. Jeong, H. M. Chung and C. Kim, Biotechnol. J., 2024, 19, 2300311 CrossRef CAS PubMed.
- Z. Peng, X. Lv, P. Zhang, Q. Chen, H. Zhang, J. Chen, X. Ma, B. Ouyang, M. Hao, H. Tong, D. Guo, Y. Luo and S. Huang, Curr. Protein Pept. Sci., 2024, 25, 71–82 CrossRef CAS PubMed.
- T. Thalheim, S. Siebert, M. Quaas, M. Herberg, M. R. Schweiger, G. Aust and J. Galle, Cells, 2021, 10, 1718 CrossRef CAS PubMed.
- M. G. Andrews and A. R. Kriegstein, Annu. Rev. Neurosci., 2022, 45, 23–39 CrossRef CAS PubMed.
- N. Prior, P. Inacio and M. Huch, Gut, 2019, 68, 2228–2237 CrossRef CAS PubMed.
- A. Bhaduri, M. G. Andrews, W. Mancia Leon, D. Jung, D. Shin, D. Allen, D. Jung, G. Schmunk, M. Haeussler, J. Salma, A. A. Pollen, T. J. Nowakowski and A. R. Kriegstein, NATURE, 2020, 578, 142 CrossRef CAS PubMed.
- Y. Shi, X. Han, S. Zou and G. Liu, ACS Nano, 2024, 18, 33276–33292 CrossRef CAS PubMed.
- G. R. Souza, J. R. Molina, R. M. Raphael, M. G. Ozawa, D. J. Stark, C. S. Levin, L. F. Bronk, J. S. Ananta, J. Mandelin, M. Georgescu, J. A. Bankson, J. G. Gelovani, T. C. Killian, W. Arap and R. Pasqualini, Nat. Nanotechnol, 2010, 5, 291–296 CrossRef CAS PubMed.
- T. L. Li, Y. Liu, C. Forro, X. Yang, L. Beker, Z. Bao, B. Cui and S. P. Paşca, Biomaterials, 2022, 290, 121825 CrossRef CAS PubMed.
- Y. Qin, B. Chen, Y. Hu, X. Zhang, Z. Wang, C. Ma, R. Yang, B. Wang, F. Li, S. Niu, Y. Han and D. Lu, Adv. Healthcare Mater., 2025, 14, e2404178 CrossRef PubMed.
- R. Curvello and G. Garnier, Biomacromolecules, 2021, 22, 701–709 CrossRef CAS PubMed.
- W. Ma, Y. Zheng, G. Yang, H. Zhang, M. Lu, H. Ma, C. Wu and H. Lu, Mater. Horiz., 2024, 11, 2957–2973 RSC.
- A. Patino-Guerrero, H. Esmaeili, R. Q. Migrino and M. Nikkhah, RSC Adv., 2023, 13, 16985–17000 RSC.
- D. J. Lomboni, A. Ozgun, T. V. de Medeiros, W. Staines, R. Naccache, J. Woulfe and F. Variola, Adv. Healthcare Mater., 2024, 13, e2301894 CrossRef PubMed.
- Y. Tan, R. C. Coyle, R. W. Barrs, S. E. Silver, M. Li, D. J. Richards, Y. Lin, Y. Jiang, H. Wang, D. R. Menick, K. Deleon-Pennell, B. Tian and Y. Mei, Sci. Adv., 2023, 9, eadf2898 CrossRef CAS PubMed.
- Z. Luo, S. Zhang, J. Pan, R. Shi, H. Liu, Y. Lyu, X. Han, Y. Li, Y. Yang, Z. Xu, Y. Sui, E. Luo, Y. Zhang and S. Wei, Biomaterials, 2018, 163, 25–42 CrossRef CAS PubMed.
- C. Afting, T. Walther, O. M. Drozdowski, C. Schlagheck, U. S. Schwarz, J. Wittbrodt and K. Göpfrich, Nat. Nanotechnol, 2024, 19, 1849–1857 CrossRef CAS PubMed.
- H. L. Bumpers, D. G. Janagama, U. Manne, M. D. Basson and V. Katkoori, J. Surg. Res., 2015, 194, 319–326 CrossRef PubMed.
- N. S. Lewis, E. E. Lewis, M. Mullin, H. Wheadon, M. J. Dalby and C. C. Berry, J. Tissue Eng., 2017, 8, 1545533460 Search PubMed.
- L. Labusca, D. D. Herea, A. E. Minuti, C. Stavila, C. Danceanu, M. Grigoras, G. Ababei, H. Chiriac and N. Lupu, J. Biomed. Mater. Res., Part B, 2021, 109, 630–642 CrossRef CAS PubMed.
- W. R. Lee, K. T. Oh, S. Y. Park, N. Y. Yoo, Y. S. Ahn, D. H. Lee, Y. S. Youn, D. Lee, K. Cha and E. S. Lee, Colloids Surf., B, 2011, 85, 379–384 CrossRef CAS PubMed.
- H. Tseng, A. C. Daquinag, G. R. Souza and M. G. Kolonin, Methods Mol. Biol., 2018, 1773, 147–154 CrossRef CAS PubMed.
- J. N. Ferreira, R. Hasan, G. Urkasemsin, K. K. Ng, C. Adine, S. Muthumariappan and G. R. Souza, J. Tissue Eng. Regener. Med., 2019, 13, 495–508 CrossRef CAS PubMed.
- I. Gaitán-Salvatella, E. O. López-Villegas, P. González-Alva, F. Susate-Olmos and M. A. Álvarez-Pérez, Front. Mol. Biosci., 2021, 8, 672518 CrossRef PubMed.
- M. Urbanczyk, A. Zbinden, S. L. Layland, G. Duffy and K. Schenke-Layland, Tissue Eng. Part A, 2019, 26, 387–399 CrossRef PubMed.
- E. Barreto-Duran, C. C. Mejia-Cruz, L. F. Jaramillo-Garcia, E. Leal-Garcia, A. Barreto-Prieto and V. M. Rodriguez-Pardo, J. Blood Med., 2021, 12, 517–528 CrossRef PubMed.
- L. A. Kotze, C. Beltran, D. Lang, A. G. Loxton, S. Cooper, M. Meiring, C. Koegelenberg, B. W. Allwood, S. T. Malherbe, A. M. Hiemstra, B. Glanzmann, C. Kinnear, G. Walzl and N. du Plessis, mSphere, 2021, 6, e0055221 CrossRef PubMed.
- L. Bonfim, P. de Queiroz Souza Passos, K. de Oliveira Gonçalves, L. C. Courrol, F. R. de Oliveira Silva and D. P. Vieira, Appl. Nanosci., 2019, 9, 1707–1717 CrossRef CAS.
- T. A. L. Brevini, E. F. M. Manzoni, S. Ledda and F. Gandolfi, in Organoids: Stem Cells, Structure, and Function, ed. K. Turksen, Springer New York, New York, NY, 2019, vol. 1576, pp. 291–299 Search PubMed.
- C. Frantz, K. M. Stewart and V. M. Weaver, J. Cell Sci., 2010, 123, 4195–4200 CrossRef CAS PubMed.
- X. Li, S. Sheng, G. Li, Y. Hu, F. Zhou, Z. Geng and J. Su, Adv. Healthcare Mater., 2024, 13, 2400431 CrossRef CAS PubMed.
- Z. Chen, H. Zhang, J. Huang, W. Weng, Z. Geng, M. Li and J. Su, Mater. Today Bio, 2025, 31, 101509 CrossRef CAS PubMed.
- O. Y. Antonova, O. Y. Kochetkova, I. L. Kanev, E. A. Shlyapnikova and Y. M. Shlyapnikov, ACS Chem. Neurosci., 2021, 12, 2838–2850 CrossRef CAS PubMed.
- Y. You, F. Xu, L. Liu, S. Chen, Z. Ding and D. Sun, Polymers, 2024, 16, 2664 CrossRef CAS PubMed.
- M. Beldjilali-Labro, R. Jellali, A. D. Brown, A. Garcia Garcia, A. Lerebours, E. Guenin, F. Bedoui, M. Dufresne, C. Stewart, J. Grosset and C. Legallais, in International Journal of Molecular Sciences, 2022, vol. 23, p. 260 Search PubMed.
- Z. Zhou, Y. Zhang, Y. Zeng, D. Yang, J. Mo, Z. Zheng, Y. Zhang, P. Xiao, X. Zhong and W. Yan, ACS Nano, 2024, 18, 7688–7710 CrossRef CAS PubMed.
- A. A. Abdeen, J. Lee, N. A. Bharadwaj, R. H. Ewoldt and K. A. Kilian, Adv. Healthcare Mater., 2016, 5, 2536–2544 CrossRef CAS PubMed.
- Z. Zhang, S. Gao, Y. Hu, X. Chen, C. Cheng, X. Fu, S. Zhang, X. Wang, Y. Che, C. Zhang and R. Chai, Adv. Sci., 2022, 9, 2203557 CrossRef CAS PubMed.
- M. Bubna-Litic and R. Mayor, Curr. Opin. Cell Biol., 2025, 94, 102514 CrossRef CAS PubMed.
- C. Y. Shen, Q. R. Zhou, X. Wu, X. Y. Han, Q. Zhang, X. Chen, Y. X. Lai, L. Bai, Y. Y. Jing, J. H. Wang, C. L. Wang, Z. Geng and J. C. Su, Mil. Med. Res., 2025, 12, 39 CAS.
- S. Fu, H. Li, Y. Wu and J. Wang, J. Biomed. Mater. Res., Part A, 2024, 112, 193–209 CrossRef CAS PubMed.
- J. Dong and Q. Ma, Nanotoxicology, 2016, 10, 699–709 CrossRef CAS PubMed.
- B. Sivaraman and A. Ramamurthi, Acta Biomater., 2013, 9, 6511–6525 CrossRef CAS PubMed.
- Q. Li, H. Lin, O. Wang, X. Qiu, S. Kidambi, L. P. Deleyrolle, B. A. Reynolds and Y. Lei, Sci. Rep., 2016, 6, 31915 CrossRef CAS PubMed.
- N. Gjorevski, N. Sachs, A. Manfrin, S. Giger, M. E. Bragina, P. Ordóñez-Morán, H. Clevers and M. P. Lutolf, Nature, 2016, 539, 560–564 CrossRef CAS PubMed.
- F. A. Abdel and A. Ranga, Front. Bioeng. Biotechnol., 2020, 8, 240 CrossRef PubMed.
- L. Bao, X. Cui, X. Wang, J. Wu, M. Guo, N. Yan and C. Chen, ACS Nano, 2021, 15, 15858–15873 CrossRef CAS PubMed.
- A. M. Abdalla, T. Majdi, S. Ghosh and I. K. Puri, Mater. Res. Express, 2016, 3, 125014 CrossRef.
- I. C. Hou, L. Li, H. Zhang and P. Naumov, Smart Mol., 2024, 16(1) DOI:10.1002/smo.20230031.
- D. Kokkinis, M. Schaffner and A. R. Studart, Nat. Commun., 2015, 6, 8643 CrossRef PubMed.
- Y. Sapir-Lekhovitser, M. Y. Rotenberg, J. Jopp, G. Friedman, B. Polyak and S. Cohen, Nanoscale, 2016, 8, 3386–3399 RSC.
- M. Filippi, B. Dasen, J. Guerrero, F. Garello, G. Isu, G. Born, M. Ehrbar, I. Martin and A. Scherberich, Biomaterials, 2019, 223, 119468 CrossRef CAS PubMed.
- H. Li, Y. Yin, Y. Xiang, H. Liu and R. Guo, Biomed. Mater., 2020, 15, 45004 CrossRef CAS PubMed.
- Q. Zhang, G. Yang, L. Xue, G. Dong, W. Su, M. J. Cui, Z. G. Wang, M. Liu, Z. Zhou and X. Zhang, ACS Nano, 2022, 16, 21555–21564 CrossRef CAS PubMed.
- L. Ou, T. Wu, B. Qiu, H. Jin, F. Xu, H. Wu, W. Zhang, M. Xue, Z. Zhou, B. Lin, D. Sun and S. Chen, ACS Appl. Mater. Interfaces, 2024, 16, 45861–45870 CrossRef CAS PubMed.
- Z. Lin, W. Wang, R. Liu, Q. Li, J. Lee, C. Hirschler and J. Liu, Nat. Protoc., 2025, 20, 2528–2559 CrossRef CAS PubMed.
- I. A. Marques, C. Fernandes, N. T. Tavares, A. S. Pires, A. M. Abrantes and M. F. Botelho, Int. J. Mol. Sci., 2022, 23, 12681 CrossRef CAS PubMed.
- J. A. Kim, J. Choi, M. Kim, W. J. Rhee, B. Son, H. Jung and T. H. Park, Biomaterials, 2013, 34, 8555–8563 CrossRef CAS PubMed.
- H. Tseng, J. A. Gage, R. M. Raphael, R. H. Moore, T. C. Killian, K. J. Grande-Allen and G. R. Souza, Tissue Eng., Part C, 2013, 19, 665–675 CrossRef CAS PubMed.
- P. Xiong, X. Huang, N. Ye, Q. Lu, G. Zhang, S. Peng, H. Wang and Y. Liu, Adv. Sci., 2022, 9, e2106049 CrossRef PubMed.
- N. Li, X. Fan, K. Tang, X. Zheng, J. Liu and B. Wang, Colloids Surf., B, 2016, 140, 287–296 CrossRef CAS PubMed.
- L. Rodriguez-Arco, I. A. Rodriguez, V. Carriel, A. B. Bonhome-Espinosa, F. Campos, P. Kuzhir, J. D. G. Duran and M. T. Lopez-Lopez, Nanoscale, 2016, 8, 8138–8150 RSC.
- C. Shen, Z. Zhang, X. Li, Y. Huang, Y. Wang, H. Zhou, L. Xiong, Y. Wen, H. Zou and Z. Liu, Front. Immunol., 2023, 14, 1172262 CrossRef CAS PubMed.
- E. Turker, N. Demircak and A. Arslan-Yildiz, Biomater. Sci., 2018, 6, 1996 RSC.
- R. Onbas and A. Arslan Yildiz, ACS Appl. Bio Mater., 2021, 4, 1794–1802 CrossRef CAS PubMed.
- P. Krontiras, P. Gatenholm and D. A. Hägg, J. Biomed. Mater. Res., Part B, 2015, 103, 195–203 CrossRef PubMed.
- K. Markstedt, A. Mantas, I. Tournier, H. M. Avila, D. Hagg and P. Gatenholm, Biomacromolecules, 2015, 16, 1489–1496 CrossRef CAS PubMed.
- D. A. Bowser and M. J. Moore, Biofabrication, 2020, 12, 015002 CrossRef CAS PubMed.
- G. Urkasemsin, S. Rungarunlert and J. N. Ferreira, Methods Mol. Biol., 2020, 2140, 243–249 CrossRef CAS PubMed.
- J. Aalders, L. Leger, D. Piras, J. van Hengel and S. Ledda, Methods Mol. Biol., 2021, 2273, 85–102 CrossRef CAS PubMed.
- S. M. Park, S. J. Lee, J. Lim, B. C. Kim, S. J. Han and D. S. Kim, ACS Appl. Mater. Interfaces, 2018, 10, 37878–37885 CrossRef CAS PubMed.
- G. Fang, Y. Chen, H. Lu and D. Jin, Adv. Funct. Mater., 2023, 33, 2215043 CrossRef CAS.
- N. Brandenberg, S. Hoehnel, F. Kuttler, K. Homicsko, C. Ceroni, T. Ringel, N. Gjorevski, G. Schwank, G. Coukos, G. Turcatti and M. P. Lutolf, Nat. Biomed. Eng., 2020, 4, 863 CrossRef CAS PubMed.
- G. Fang, H. Lu, A. Law, D. Gallego-Ortega, D. Jin and G. Lin, Lab Chip, 2019, 19, 4093–4103 RSC.
- D. Kim, K. Kim and J. Y. Park, Lab Chip, 2021, 21, 1974–1986 RSC.
- H. Shin, Y. Kook, H. J. Hong, Y. Kim, W. Koh and J. Lim, Acta Biomater., 2016, 45, 121–132 CrossRef CAS PubMed.
- H. Shin, H. J. Hong, W. Koh and J. Lim, ACS Biomater. Sci. Eng., 2018, 4, 4311–4320 CrossRef CAS PubMed.
- Y. Y. Choi, B. G. Chung, D. H. Lee, A. Khademhosseini, J. Kim and S. Lee, Biomaterials, 2010, 31, 4296–4303 CrossRef CAS PubMed.
- D. Kim, S. J. Lee, J. Youn, H. Hong, S. Eom and D. S. Kim, Biofabrication, 2021, 13 DOI:10.1088/1758-5090/ac044c.
- F. Papes, A. P. Camargo, J. S. de Souza, V. Carvalho, R. A. Szeto, E. LaMontagne, J. R. Teixeira, S. H. Avansini, S. M. Sánchez-Sánchez, T. S. Nakahara, C. N. Santo, W. Wu, H. Yao, B. Araújo, P. Velho, G. G. Haddad and A. R. Muotri, Nat. Commun., 2022, 13, 2387 CrossRef CAS PubMed.
- Y. Liu, X. Xu, X. Ma, E. Martin-Rendon, S. Watt and Z. Cui, Biotechnol. Prog., 2010, 26(6), 1635–1643 CrossRef CAS PubMed.
- J. D. Finan and F. Guilak, J. Cell. Biochem., 2010, 109, 460–467 CrossRef CAS PubMed.
- Z. Han and J. C. Bishop, CryoLetters, 2020, 41, 185–193 Search PubMed.
- H. Geng, X. Liu, G. Shi, G. Bai, J. Ma, J. Chen, Z. Wu, Y. Song, H. Fang and J. Wang, Angew. Chem., Int. Ed., 2017, 56, 997–1001 CrossRef CAS PubMed.
- R. Gao, W. Wang, Z. Wang, Y. Fan, L. Zhang, J. Sun, M. Hong, M. Pan, J. Wu, Q. Mei, Y. Wang, L. Qiao, J. Liu and F. Tong, Adv. Healthcare Mater., 2024, 13, 2401833 CrossRef CAS PubMed.
- W. Xue, H. Li, J. Xu, X. Yu, L. Liu, H. Liu, R. Zhao and Z. Shao, Cells Rep. Methods, 2024, 4, 100777 CrossRef CAS PubMed.
- R. Dong, Y. Fei, Y. He, P. Gao, B. Zhang, M. Zhu, Z. Wang, L. Wu, S. Wu, X. Wang, J. Cai, Z. Chen and X. Zuo, Adv. Sci., 2025, 12, 2412559 CrossRef CAS PubMed.
- K. Saxena, S. E. Blutt, K. Ettayebi, X. Zeng, J. R. Broughman, S. E. Crawford, U. C. Karandikar, N. P. Sastri, M. E. Conner, A. R. Opekun, D. Y. Graham, W. Qureshi, V. Sherman, J. Foulke-Abel, J. In, O. Kovbasnjuk, N. C. Zachos, M. Donowitz and M. K. Estes, J. Virol., 2015, 90, 43–56 CrossRef PubMed.
- V. Mangena, R. Chanoch-Myers, R. Sartore, B. Paulsen, S. Gritsch, H. Weisman, T. Hara, X. O. Breakefield, K. Breyne, A. Regev, K. Chung, P. Arlotta, I. Tirosh and M. L. Suvà, Cancer Discovery, 2025, 15, 299–315 CrossRef CAS PubMed.
- W. Shao, H. Xu, K. Zeng, M. Ye, R. Pei and K. Wang, Stem Cell Res. Ther., 2025, 16, 27 CrossRef PubMed.
- M. M. A. Verstegen, F. J. M. Roos, K. Burka, H. Gehart, M. Jager, M. de Wolf, M. J. C. Bijvelds, H. R. de Jonge, A. I. Ardisasmita, N. A. van Huizen, H. P. Roest, J. de Jonge, M. Koch, F. Pampaloni, S. A. Fuchs, I. F. Schene, T. M. Luider, H. P. J. van der Doef, F. A. J. A. Bodewes, R. H. J. de Kleine, B. Spee, G. Kremers, H. Clevers, J. N. M. IJzermans, E. Cuppen and L. J. W. van der Laan, Sci. Rep., 2020, 10, 21900 CrossRef CAS PubMed.
- Z. Peng, X. Lv, H. Sun, L. Zhao and S. Huang, Mol. Cancer, 2025, 24, 93 CrossRef CAS PubMed.
- D. Kharaghani, G. M. DeLoid, P. He, B. Swenor, T. H. Bui, N. Zuverza-Mena, C. Tamez, C. Musante, M. Verzi, J. C. White and P. Demokritou, J. Hazard. Mater., 2025, 490, 137714 CrossRef CAS PubMed.
- S. Yang, Y. Ge, T. Zhang, L. Yin, Y. Pu, Z. Chen and G. Liang, Environ. Int., 2025, 195, 109266 CrossRef CAS PubMed.
- P. Boix-Montesinos, P. Carrascosa-Marco, A. Armiñán and M. J. Vicent, J. Controlled Release, 2025, 381, 113584 CrossRef CAS PubMed.
- D. Li and C. Liu, Nat. Chem. Biol., 2021, 17, 237–245 CrossRef CAS PubMed.
- J. Liang, D. Zhao, H. Yin, T. Tian, J. Kang, S. Shen and J. Wang, Nano Today, 2025, 61, 102665 CrossRef CAS.
- S. Yui, T. Nakamura, T. Sato, Y. Nemoto, T. Mizutani, X. Zheng, S. Ichinose, T. Nagaishi, R. Okamoto, K. Tsuchiya, H. Clevers and M. Watanabe, Nat. Med., 2012, 18, 618–623 CrossRef CAS PubMed.
- A. Villaronga-Luque, R. G. Savill, N. López-Anguita, A. Bolondi, S. Garai, S. I. Gassaloglu, R. Rouatbi, K. Schmeisser, A. Poddar, L. Bauer, T. Alves, S. Traikov, J. Rodenfels, T. Chavakis, A. Bulut-Karslioglu and J. V. Veenvliet, Cell Stem Cell, 2025, 32, 759–777 CrossRef CAS PubMed.
- X. Xu, Y. Liu and Z. Cui, J. Tissue Eng. Regener. Med., 2014, 8, 664–672 CrossRef CAS PubMed.
- M. J. Powell-Palm, V. Charwat, B. Charrez, B. Siemons, K. E. Healy and B. Rubinsky, Commun. Biol., 2021, 4, 1118 CrossRef PubMed.
- D. Skubleny, S. Garg, J. Wickware, K. Purich, S. Ghosh, J. Spratlin, D. E. Schiller and G. R. Rayat, Biomedicines, 2023, 11, 151 CrossRef CAS PubMed.
- G. D. Elliott, S. Wang and B. J. Fuller, Cryobiology, 2017, 76, 74–91 CrossRef CAS PubMed.
- P. Kilbride, S. Lamb, S. Gibbons, J. Bundy, E. Erro, C. Selden, B. Fuller and J. Morris, PLoS One, 2017, 12, e0183385 CrossRef PubMed.
- N. Dolezalova, A. Gruszczyk, K. Barkan, J. A. Gamble, S. Galvin, T. Moreth, K. O'Holleran, K. T. Mahbubani, J. A. Higgins, F. M. Gribble, F. Reimann, J. Surmacki, S. Andrews, J. J. Casey, F. Pampaloni, M. P. Murphy, G. Ladds, N. Slater and K. Saeb-Parsy, Sci. Rep., 2021, 11, 10418 CrossRef CAS PubMed.
- K. Peter, M. Julie, M. C. Mira, R. Susan and C. Tessa, in Cryopreservation, ed. Q. Marian, IntechOpen, Rijeka, 2022, p. 5 Search PubMed.
- R. M. Warner, R. Shuttleworth, J. D. Benson, A. Eroglu and A. Z. Higgins, Biophys. J., 2021, 120, 4980–4991 CrossRef CAS PubMed.
- M. Wusteman, M. Robinson and D. Pegg, Cryobiology, 2004, 48, 179–189 CrossRef CAS PubMed.
- G. J. Morris, M. Goodrich, E. Acton and F. Fonseca, Cryobiology, 2006, 52, 323–334 CrossRef CAS PubMed.
- P. Kilbride and G. J. Morris, Cryobiology, 2017, 76, 92–97 CrossRef CAS PubMed.
- S. Bojic, A. Murray, B. L. Bentley, R. Spindler, P. Pawlik, J. L. Cordeiro, R. Bauer and J. P. de Magalhães, BMC Biol., 2021, 19, 56 CrossRef PubMed.
- Y. Diao, T. Hao, X. Liu and H. Yang, Acta Biomater., 2024, 174, 49–68 CrossRef CAS PubMed.
- K. A. Murray, Y. Gao, C. A. Griffiths, N. Kinney, Q. Guo, M. I. Gibson and T. F. Whale, JACS Au, 2023, 3, 1314–1320 CrossRef CAS PubMed.
- M. L. Etheridge, Y. Xu, L. Rott, J. Choi, B. Glasmacher and J. C. Bischof, Technology, 2014, 02, 229–242 CrossRef.
- A. Lawson, I. N. Mukherjee and A. Sambanis, Cryobiology, 2012, 64, 1–11 CrossRef CAS PubMed.
- A. Abazari, N. M. Jomha, J. A. W. Elliott and L. E. McGann, Cryobiology, 2013, 66, 201–209 CrossRef CAS PubMed.
- A. M. Vian and A. Z. Higgins, Cryobiology, 2014, 68, 35–42 CrossRef CAS PubMed.
- A. Fatehullah, S. H. Tan and N. Barker, Nat. Cell Biol., 2016, 18, 246–254 CrossRef PubMed.
- J. Tan, J. Li, C. Lin, N. Ye, H. Zhang, C. Liu, S. Han, Z. Li and X. Zhou, Life Sci., 2024, 355, 122980 CrossRef CAS PubMed.
- T. F. Zur Bruegge, A. Liese, S. Donath, S. Kalies, M. Kosanke, O. Dittrich-Breiholz, S. Czech, V. N. Bauer, A. Bleich and M. Buettner, Stem Cells Int., 2021, 2021, 9041423 Search PubMed.
- Q. Liu, T. Zhao, X. Wang, Z. Chen, Y. Hu and X. Chen, Micromachines, 2021, 12, 624 CrossRef PubMed.
- Y. K. Chong, T. B. Toh, N. Zaiden, A. Poonepalli, S. H. Leong, C. E. Ong, Y. Yu, P. B. Tan, S. J. See, W. H. Ng, I. Ng, M. P. Hande, O. L. Kon, B. T. Ang and C. Tang, Stem Cells, 2009, 27, 29–39 CrossRef CAS PubMed.
- U. A. Okoli, M. T. Okafor, K. A. Agu, A. C. Ndubuisi, I. J. Nwigwe, E. O. Nna, O. C. Okafor, F. I. Ukekwe, T. U. Nwagha, V. C. Menkiti, C. O. Eze, K. C. Onyekwelu, J. E. Ikekpeazu, C. A. Anusiem, A. U. Mbah, C. P. Chijioke and I. J. Udeniya, Cryobiology, 2020, 97, 179–184 CrossRef CAS PubMed.
- R. E. Thompson, M. A. Meyers, C. Premanandan and F. K. Hollinshead, Theriogenology, 2023, 196, 167–173 CrossRef PubMed.
- Z. Liu, X. Zheng and J. Wang, J. Am. Chem. Soc., 2022, 144, 5685–5701 CrossRef CAS PubMed.
- H. Dong, X. Li, K. Chen, N. Li and H. Kagami, Tissue Eng., Part C, 2021, 27, 253–263 CrossRef CAS PubMed.
- D. Kaiser, N. M. Otto, O. McCallion, H. Hoffmann, G. Zarrinrad, M. Stein, C. Beier, I. Matz, M. Herschel, J. Hester, G. Moll, F. Issa, P. Reinke and A. Roemhild, Front. Cell Dev. Biol., 2021, 9, 750286 CrossRef PubMed.
- D. Diaz-Dussan, Y. Peng, J. Sengupta, R. Zabludowski, M. K. Adam, J. P. Acker, R. N. Ben, P. Kumar and R. Narain, Biomacromolecules, 2020, 21, 1264–1273 CrossRef CAS PubMed.
- R. Li, K. Hornberger, J. R. Dutton and A. Hubel, Front. Bioeng. Biotechnol., 2020, 8, 1 CrossRef PubMed.
- T. F. Whale, M. Rosillo-Lopez, B. J. Murray and C. G. Salzmann, J. Phys. Chem. Lett., 2015, 6, 3012–3016 CrossRef CAS PubMed.
- Y. Hou, X. Sun, M. Dou, C. Lu, J. Liu and W. Rao, Nano Lett., 2021, 21, 4868–4877 CrossRef CAS PubMed.
- W. Han, L. Zhong, J. Zhang, Y. Li, N. Li, Z. Wang and J. Zhao, ACS Appl. Nano Mater., 2024, 7, 12783–12794 CrossRef CAS.
- Q. Lei, Y. Sun, J. Huang, W. Liu, X. Zhan, W. Yin, S. Guo, A. Sinelshchikova, C. J. Brinker, Z. He, J. Guo, S. Wuttke and W. Zhu, Angew. Chem., Int. Ed., 2023, 62, e202217374 CrossRef CAS PubMed.
- S. Ding, S. Ali, S. Zhang, J. Zhao, C. Liu, M. A. Aslam, X. Yu, M. Xi, L. Pan, N. Li and Z. Wang, ACS Omega, 2023, 8, 10466–10475 CrossRef CAS PubMed.
- M. Stefanic, K. Ward, H. Tawfik, R. Seemann, V. Baulin, Y. Guo, J. Fleury and C. Drouet, Biomaterials, 2017, 140, 138–149 CrossRef CAS PubMed.
- W. Rao, H. Huang, H. Wang, S. Zhao, J. Dumbleton, G. Zhao and X. He, ACS Appl. Mater. Interfaces, 2015, 7, 5017–5028 CrossRef CAS PubMed.
- Y. Zhang, H. Wang, S. Stewart, B. Jiang, W. Ou, G. Zhao and X. He, Nano Lett., 2019, 19, 9051–9061 CrossRef CAS PubMed.
- X. Yao, J. J. Jovevski, M. F. Todd, R. Xu, Y. Li, J. Wang and S. Matosevic, Adv. Sci., 2020, 7, 1902938 CrossRef CAS PubMed.
- A. Maruf, M. Milewska, K. Dudzisz, A. Lalik, S. Student, A. Salvati and I. Wandzik, Biomacromolecules, 2025, 26, 2835–2851 CrossRef CAS PubMed.
- Y. Cao, M. Hassan, Y. Cheng, Z. Chen, M. Wang, X. Zhang, Z. Haider and G. Zhao, ACS Appl. Mater. Interfaces, 2019, 11, 12379–12388 CrossRef CAS PubMed.
- G. Bai, Z. Song, H. Geng, D. Gao, K. Liu, S. Wu, W. Rao, L. Guo and J. Wang, Adv. Mater., 2017, 29, 1606843 CrossRef PubMed.
- C. Cline, H. Wang, J. Kong, T. Li, J. Liu and U. Wegst, Langmuir, 2022, 38, 15121–15131 CrossRef CAS PubMed.
- Z. Wang, B. Yang, Z. Chen, D. Liu, L. Jing, C. Gao, J. Li, Z. He and J. Wang, ACS Appl. Bio Mater., 2020, 3, 3785–3791 CrossRef CAS PubMed.
- F. Baniasadi, S. Hajiaghalou, A. Shahverdi, M. R. Ghalamboran, V. Pirhajati and R. Fathi, Reprod. Sci., 2023, 30, 2122–2136 CrossRef CAS PubMed.
- Y. Cao, T. Chang, C. Fang, Y. Zhang, H. Liu and G. Zhao, ACS Nano, 2022, 16, 8837–8850 CrossRef CAS PubMed.
- W. Zhu, J. Guo, J. O. Agola, J. G. Croissant, Z. Wang, J. Shang, E. Coker, B. Motevalli, A. Zimpel, S. Wuttke and C. J. Brinker, J. Am. Chem. Soc., 2019, 141, 7789–7796 CrossRef CAS PubMed.
- P. Zhang, R. Zou, S. Wu, L. Meyer, J. Wang and T. Kraus, Langmuir, 2022, 38, 2460–2466 CrossRef CAS PubMed.
- N. Jeon, I. Choi, P. C. W. Lee and E. Lee, ACS Appl. Nano Mater., 2022, 5, 15418–15428 CrossRef CAS.
- S. Liu, Z. Han, Z. Ye, M. Jiang, M. L. Etheridge, J. C. Bischof and Y. Yin, Nano Lett., 2024, 24, 11567–11572 CrossRef CAS PubMed.
- D. E. Mitchell, J. R. Lovett, S. P. Armes and M. I. Gibson, Angew. Chem., Int. Ed., 2016, 55, 2801–2804 CrossRef CAS PubMed.
- P. G. Georgiou, I. Kontopoulou, T. R. Congdon and M. I. Gibson, Mater. Horiz., 2020, 7, 1883–1887 RSC.
- R. Gao, W. Wang, Z. Wang, Y. Fan, L. Zhang, J. Sun, M. Hong, M. Pan, J. Wu, Q. Mei, Y. Wang, L. Qiao, J. Liu and F. Tong, Adv. Healthcare Mater., 2024, 13, e2401833 CrossRef PubMed.
- L. E. Ehrlich, Z. Gao, J. C. Bischof and Y. Rabin, PLoS One, 2020, 15, e0238941 CrossRef CAS PubMed.
- M. E. Mamprin, E. E. Guibert and J. V. Rodriguez, Cryobiology, 2000, 40, 270–276 CrossRef CAS PubMed.
- K. Benhenia, H. Rahab, M. A. Smadi, H. Benmakhlouf, A. Lamara, T. Idres and M. Iguer-Ouada, Anim. Reprod. Sci., 2018, 195, 266–273 CrossRef CAS PubMed.
- J. P. Morrow, D. Pizzi, Z. A. I. Mazrad, A. I. Bush and K. Kempe, Biomater. Sci., 2023, 11, 3159–3171 RSC.
- A. A. Timralieva, I. V. Moskalenko, P. V. Nesterov, V. V. Shilovskikh, A. S. Novikov, E. A. Konstantinova, A. I. Kokorin and E. V. Skorb, ACS Omega, 2023, 8, 8276–8284 CrossRef CAS PubMed.
- T. Shimizu, R. Kishi, T. Yamada and K. Hata, RSC Adv., 2020, 10, 29419–29423 RSC.
- L. Rodriguez-Arco, I. A. Rodriguez, V. Carriel, A. B. Bonhome-Espinosa, F. Campos, P. Kuzhir, J. D. G. Duran and M. T. Lopez-Lopez, Nanoscale, 2016, 8, 8138–8150 RSC.
- W. A. Khalil, M. A. El-Harairy, A. E. B. Zeidan and M. A. E. Hassan, Theriogenology, 2019, 126, 121–127 CrossRef CAS PubMed.
- J. H. Crowe, L. M. Crowe, W. F. Wolkers, A. E. Oliver, X. Ma, J. Auh, M. Tang, S. Zhu, J. Norris and F. Tablin, Integr. Comp. Biol., 2005, 45, 810–820 CrossRef CAS PubMed.
- M. Wang, B. Zhang, C. Chen, Q. Gao, P. Zhou and G. Zhao, Langmuir, 2025, 41, 12264–12275 CrossRef CAS PubMed.
- D. S. Moore and S. C. Hand, Cryobiology, 2016, 73, 240–247 CrossRef CAS PubMed.
- T. Uchida, M. Furukawa, T. Kikawada, K. Yamazaki and K. Gohara, Cryobiology, 2017, 77, 50–57 CrossRef CAS PubMed.
- Y. Hu, X. Liu, F. Liu, J. Xie, Q. Zhu and S. Tan, ACS Biomater. Sci. Eng., 2023, 9, 1190–1204 CrossRef CAS PubMed.
- W. Zhang, J. Rong, Q. Wang and X. He, Nanotechnology, 2009, 20, 275101 CrossRef PubMed.
- M. Xu, Y. Song, J. Zhang, Y. Zhao, H. Geng, X. Weng, Z. Liu, J. Cheng and T. Yan, Chin. J. Biotechnol., 2024, 40, 2294–2307 CAS.
- K. A. Murray, Y. Gao, C. A. Griffiths, N. Kinney, Q. Guo, M. I. Gibson and T. F. Whale, JACS Au, 2023, 3, 1314–1320 CrossRef CAS PubMed.
- N. G. Lamson, A. Berger, K. C. Fein and K. A. Whitehead, Nat. Biomed. Eng., 2020, 4, 84–96 CrossRef CAS PubMed.
- H. Ruan, T. Wang and C. Gao, CryoLetters, 2020, 41, 26–30 Search PubMed.
- Y. XU, N. GUO, G. YANG, T. ZHAN, H. HAN, Y. CHENG, G. ZHAO, Q. WEI, X. ZHOU and B. LIU, Sci. Sin. Vitae, 2023, 53, 1021–1034 Search PubMed.
- R. Chen, B. Wang, Y. Liu, R. Lin, J. He and D. Li, Cryobiology, 2018, 82, 1–7 CrossRef CAS PubMed.
- L. Eini, M. Naseri, F. Karimi-Busheri, M. Bozorgmehr, R. Ghods and Z. Madjd, J. Cell. Physiol., 2020, 235, 2452–2463 CrossRef CAS PubMed.
- G. Dunaevskiy, E. Gavrilin, A. Pomytkin, R. Sorokin, A. Kuznetsov, V. Antipov and A. Nechaev, Sci. Rep., 2023, 13, 1362 CrossRef CAS PubMed.
- L. Cirino, S. Tsai, L. Wang, C. Chen, W. Hsieh, C. Huang, Z. Wen and C. Lin, Cryobiology, 2021, 98, 80–86 CrossRef CAS PubMed.
- N. E. Estrin, A. Lesniewski, S. McClain, W. Hou and G. E. Romanos, Photobiomodulation, Photomed., Laser Surg., 2022, 40, 410–416 CrossRef CAS PubMed.
- Y. Hou, C. Lu, M. Dou, C. Zhang, H. Chang, J. Liu and W. Rao, Acta Biomater., 2020, 102, 403–415 CrossRef CAS PubMed.
- C. Alvarez, C. Berrospe-Rodriguez, C. Wu, J. Pasek-Allen, K. Khosla, J. Bischof, L. Mangolini and G. Aguilar, Front. Bioeng. Biotechnol., 2022, 10, 957481 CrossRef PubMed.
- A. Chiu-Lam and C. Rinaldi, Adv. Funct. Mater., 2016, 26, 3933–3941 CrossRef CAS PubMed.
- J. Wang, G. Zhao, Z. Zhang, X. Xu and X. He, Acta Biomater., 2016, 33, 264–274 CrossRef CAS PubMed.
- M. L. Etheridge, Y. Xu, J. Choi and J. C. Bischof, Cryobiology, 2013, 67, 398–399 CrossRef.
- T. Wakabayashi, M. Kaneko, T. Nakai, M. Horie, H. Fujimoto, M. Takahashi, S. Tanoue and A. Ito, Bioeng. Transl. Med., 2022, 8, e10416 CrossRef PubMed.
- P. Singh, K. Duraisamy, C. Raitmayr, K. S. Sharma, T. Korzun, K. Singh, A. S. Moses, K. Yamada, V. Grigoriev, A. A. Demessie, Y. Park, Y. T. Goo, B. Mamnoon, A. P. M. Souza, K. Michimoto, K. Farsad, A. Jaiswal, O. R. Taratula and O. Taratula, Adv. Funct. Mater., 2025, 35, 2414719 CrossRef CAS PubMed.
- Z. Ye, S. Liu and Y. Yin, Mater. Chem. Front., 2023, 7, 3427–3433 RSC.
- P. Joshi and Y. Rabin, PLoS One, 2023, 18, e0290063 CrossRef CAS PubMed.
- N. Manuchehrabadi, Z. Gao, J. Zhang, H. L. Ring, Q. Shao, F. Liu, M. McDermott, A. Fok, Y. Rabin, K. G. M. Brockbank, M. Garwood, C. L. Haynes and J. C. Bischof, Sci. Transl. Med., 2017, 9, eaah4586 CrossRef PubMed.
- S. Karimi, S. N. Tabatabaei, M. G. Novin, M. Kazemi, Z. S. Mofarahe and A. Ebrahimzadeh-Bideskan, Heliyon, 2023, 9, e18828 CrossRef CAS PubMed.
- D. Ivanov, A. Hoeppel, T. Weigel, R. Ossikovski, S. Dembski and T. Novikova, in Photonics, 2023, vol. 10 Search PubMed.
- X. Liu, G. Zhao, Z. Chen, F. Panhwar and X. He, ACS Appl. Mater. Interfaces, 2018, 10, 16822–16835 CrossRef CAS PubMed.
- H. L. Ring, Z. Gao, A. Sharma, Z. Han, C. Lee, K. G. M. Brockbank, E. D. Greene, K. L. Helke, Z. Chen, L. H. Campbell, B. Weegman, M. Davis, M. Taylor, S. Giwa, G. M. Fahy, B. Wowk, R. Pagotan, J. C. Bischof and M. Garwood, Magn. Reson. Med., 2020, 83, 1750–1759 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |