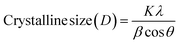Open Access Article
Open Access ArticleGreen synthesis of silver nanoparticles using the BT5 tea cultivar of Bangladesh: unveiling molecular mechanisms of anti-cancer activity in mice model†
Sk. Md. Atiqur
Rahman
 abef,
Rokshana Ara
Ruhi
abef,
Rokshana Ara
Ruhi
 abef,
Md. Mahmudul Hasan
Maruf
abef,
Md. Mahmudul Hasan
Maruf
 b,
Md. Ragib
Shariar
b,
Md. Ragib
Shariar
 b,
Mobasshir Noor
Shehab
b,
Khaled Mahmud
Sujon
b,
Mohammad Saiful
Islam
b,
Mobasshir Noor
Shehab
b,
Khaled Mahmud
Sujon
b,
Mohammad Saiful
Islam
 b,
Md. Abdul
Aziz
c,
Firoz
Ahmed
d,
Ananda Kumar
Saha
f,
Md. Anwarul Kabir
Bhuiya
*e and
Md. Abu
Reza
b,
Md. Abdul
Aziz
c,
Firoz
Ahmed
d,
Ananda Kumar
Saha
f,
Md. Anwarul Kabir
Bhuiya
*e and
Md. Abu
Reza
 *b
*b
aInstitute of Environmental Science, University of Rajshahi, Dr M A Wazed Miah Academic Building, Level # 1, Rajshahi-6205, Bangladesh
bMolecular Biology and Protein Science Laboratory, Department of Genetic Engineering and Biotechnology, University of Rajshahi, Sir Jagadish Chandra Bose Academic Building, Level # 4, Rajshahi-6205, Bangladesh. E-mail: reza.gen@ru.ac.bd
cBotany Division, Bangladesh Tea Research Institute, Sreemangal 3210, Moulvibazar, Bangladesh
dBCSIR Laboratories Rajshahi, Bangladesh Council of Scientific and Industrial Research (BCSIR), Rajshahi 6206, Bangladesh
eBio & Nanotechnology Research Lab, Department of Materials Science and Engineering, University of Rajshahi, DrM A Wazed Miah Academic Building, Level # 3, Rajshahi-6205, Bangladesh
fGenetics and Molecular Biology Lab, Department of Zoology, University of Rajshahi, Sir Jagadish Chandra Bose Academic Building, Level # 2, Rajshahi-6205, Bangladesh
First published on 1st April 2025
Abstract
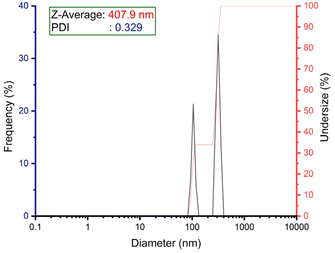
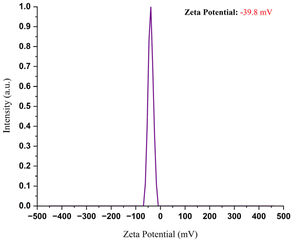
Silver nanoparticles (AgNPs) have sparked widespread interest due to their remarkable physiochemical capabilities, and they are now being used as a beneficial tool in the biomedical field. However, typical synthesis processes generate dangerous compounds that raise environmental and safety concerns. This is one of the main reasons for choosing a greener synthesis approach. The BT5 cultivar of green tea, which is high in phenolic and flavonoid compounds and has strong antioxidant activity (IC50 ≈ 97.8 μg mL−1), was used in an eco-friendly way to produce AgNPs. The green synthesis approach was confirmed by the rapid color change (light to dark brown) of AgNO3 solution upon the addition of the BT5 extract. Several methods were performed to characterize the synthesized BT5-AgNPs, using UV-vis spectroscopy (λmax = 424 nm), FTIR, DLS, zeta potential (−39.8 ± 0.45 mV), TGA, XRD (crystalline size = 11.25 nm), and FE-SEM (35 ± 8.36 nm). BT5-AgNPs showed strong antiproliferative properties inimical to Ehrlich-Lettre ascites carcinoma cells (EAC) of the Swiss albino variety of mice model, with low-concentration treatment (1 mg kg−1) resulting in 52.15% cell growth inhibition. Fluorescence microscopy using DAPI staining revealed morphological alterations via heterochromatization and karyorrhexis. Gene expression analysis revealed upregulation of hallmark genes p53 and BAX and Bcl2 downregulation in the low- and high-concentration groups, indicating activation of the apoptotic pathway. Most likely due to cytotoxic effects and altered cellular responses, a higher dosage (5 mg kg−1) resulted in slightly lower efficacy.
Introduction
The use of nanoparticles is increasing exponentially worldwide because of their versatile use in industry, technology, and medicine. Their unique properties, including their greater surface-to-volume ratio, greater surface energy, and specific mechanical, thermal, electrical, magnetic, and optical characteristics, make them versatile for a wide range of applications and have enormous potential.1 They can be created through various techniques, primarily categorized into top-down approaches, which involve breaking down larger materials, and bottom-up methods, where nanoparticles are synthesized from atomic precursors. Synthesis techniques can also be categorized based on the methodology employed: physical, chemical, or biological.2 Although physiochemical reactions are the most efficient methods for nanoparticle synthesis, several other techniques like thermal, radiation/sono chemical, microwave-assisted, chemical reduction, and electrochemical processes are also used. However, numerous hazardous substances that are harmful to the environment and human health are regularly used in these procedures. Therefore, there is an increasing interest in biological approaches that do not involve the use of harmful chemicals or the formation of toxic byproducts. Among these, extracts prepared from plant leaves, seeds, roots, and stems are widely used in the production of green nanomaterials because they commonly contain polysaccharides, polyphenols, flavonoids, or alkaloids capable of functioning as agents of capping and reduction, facilitating the creation and preventing the agglomeration of nanoparticles. They are also usually harmless and biodegradable.3,4Among the numerous types of metal-based nanoparticles, silver nanoparticles (AgNPs) have garnered significant interest over the years because of their distinctive physicochemical characteristics, which include chemical stability, good conductivity and antifungal, antiviral, antibacterial, and catalytic activities.5 In recent years, the green synthesis of silver nanoparticles (AgNPs) has garnered noteworthy attention for its numerous advantages. Among the various biological methods, plant-based synthesis has emerged as a particularly promising approach that offers simplicity, scalability, and environmental sustainability. Plant extracts are rich in metabolites, e.g., sugars, proteins, and various phytochemicals like terpenoids, polyphenols, alkaloids, and phenolic acids aid greatly in silver ion reduction (Ag+) to metallic silver (Ag0). The diverse and abundant phytochemicals present in these extracts not only act as reducing agents but also stabilize the nanoparticles, making plant-based synthesis a highly effective and eco-friendly alternative.6–8 The tea plant, Camellia sinensis (L.) O. Kuntze has been a coveted source of non-alcoholic, caffeine-containing beverages since ancient times, with its origins tracing back to ancient China. Originally used as a medicinal drink, tea has evolved into a luxury beverage over the past 3000 years. Among the various tea varieties, green tea has gained considerable popularity in recent years because of its exceptional medicinal properties, including high antioxidant content. Green tea is known for its ability to promote intestinal health by supporting the propagation of beneficial microflora and safeguarding cellular and tissue systems from oxidative damage. Thus, green tea is an excellent oxygen-free radical scavenger. It also exhibits strong anti-inflammatory, antioxidant, and immune-boosting properties. From various notable epidemiological studies, the consumption of green tea can be further linked to a diminished risk of certain cancers, including those of the stomach, oral cavity, esophagus, and lungs.9,10 In favor of its production process, green tea is notable for its high concentration of tea polyphenols and other antioxidant compounds, including gallic acid, epigallocatechin gallate, and other catechins.11 Prior studies have emphasized the potential of green tea phytochemicals as highly effective reducing and stabilizing components in AgNP synthesis.12–14 The hydroxyl groups (–OH) and carboxyl (–COOH) present in these polyphenols and other phytochemical compounds facilitate the reduction of Ag+ ions, leading to AgNP synthesis.
In 1994, a remarkable breakthrough in nanoparticles was achieved with the advent of ‘Oncaspar’, a unique nanomedicine approved by the FDA for the treatment of Acute Lymphoblastic Leukemia by Servier Pharmaceuticals, followed by the approval of Doxil in 1995 by Janssen for the same indication. Since then, nanoparticles have gained importance in the development of anticancer drugs, enhancing the efficacy of existing chemotherapeutic agents, enabling targeted drug delivery, and overcoming drug resistance.15–18 In recent years, green tea has gained attention for its health-promoting properties, which are largely attributed to green tea polyphenols (GTPs), namely catechin, epicatechin, epigallocatechin, epicatechin-3-gallate (ECG), and epigallocatechin-3-gallate (EGCG). Several studies have demonstrated the potential of GTPs to prevent carcinogenesis and suppress the proliferation of various malignant cells.19–21
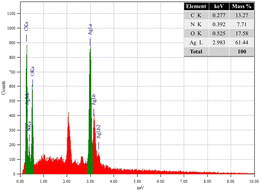
In this study, AgNPs were synthesized using BT5 extract (green tea) as both a reducing and capping agent. A comprehensive characterization of the synthesized AgNPs was performed using a broad spectrum of techniques, including UV-vis spectroscopy to confirm nanoparticle formation, Dynamic Light Scattering (DLS) and zeta potential analysis to evaluate particle surface charge and size, Fourier-Transform Infrared Spectroscopy (FTIR) to identify functional groups associated with synthesis, Thermogravimetric Analysis (TGA) to evaluate thermal stability, X-ray Diffraction (XRD) to evaluate crystal structure evaluation, Field Emission Scanning Electron Microscopy (FE-SEM) and Energy Dispersive X-ray (EDX) to evaluate elemental and morphological composition. To explore potential biomedical applications, the anti-cancer activity of AgNPs was evaluated in a mouse model bearing Ehrlich-Lettre ascites carcinoma (EAC) cells. Further studies will investigate the impact of AgNPs on apoptosis-related gene expression, specifically analyzing the levels of p53, Bax, and Bcl2 in cancer cells.
Materials and methods
Materials
Dried leaves of BT5, a green tea cultivar (Camellia sinensis L. O. Kuntze), were procured from the Bangladesh Tea Research Institute (BTRI), Srimangal, Bangladesh, which is located at an altitude of 21.95 meters above the mean sea level. The remaining chemicals, i.e., silver nitrate, ethanol, and methanol, were purchased from Sigma Aldrich, USA.Phytochemical and antioxidant analysis of BT5 cultivar
The plant material was analyzed to determine its bioactive compounds and antioxidant properties. The Total Phenolic Content (TPC) of the BT5 ethanolic extract was quantified by employing the Folin–Ciocalteu method22 with Gallic acid as the standard, while absorbance was recorded at 765 nm. A calibration curve (ESI, Fig. S1†) was generated using the equation y = 0.0039x + 0.0318 (R2 = 0.9986). Using the aluminium chloride colorimetric assay,23 the total flavonoid content (TFC) of the BT5 ethanolic extract was measured. Absorbance was detected at 415 nm using quercetin as the standard. The calibration curve was linear (ESI, Fig. S2†), with the equation y = 0.0061x + 0.13 (R2 = 0.9109). The free radical scavenging property was investigated via 1,1-diphenyl-2-picrylhydrazyl radical (DPPH) assay24 to quantify its ability to neutralize reactive oxygen species and calculated using the equation %RSA = ((Ac − As)/Ac) × 100. Using ascorbic acid as the standard, absorbance was recorded at 517 nm. The IC50 value, representing the concentration necessary to completely scavenge 50% of free radicals, was calculated from the linear regression equation y = 0.0945x + 40.755 (R2 = 0.8408).Further, specific phytochemical identification of the BT5 cultivar was performed via high-performance liquid chromatography (HPLC) (UltiMate 3000, DIONEX, Thermo SCIENTIFIC, Waltham, MA, USA) using a variable wavelength detector (VWD). As the non-polar stationary phase for HPLC, a C18 (Phenomenex, CA, USA) column was utilized (Particle size: 5 μm, pore size: 300 Å, dimension: 250 mm × 10 mm). Solvent A consisted of 0.1% trifluoroacetic acid (TFA) and 99.9% H2O, whereas solvent B contained 100% acetonitrile and 0.1% TFA in the mobile phase. Data processing was performed using Chromeleon™ 6.8 software.
Preparation of the BT5 extracts
Dried BT5 leaves were pulverized, and 5 g of the resulting powder was boiled in deionized water (100 mL) at 60 °C for 30 minutes. After cooling, the concoction was passed through a filter paper (Whatman no. 1). A rotary evaporator (SENCO Technology Co., Ltd, Shanghai, China) was used for condensation of the collected filtrate and a vacuum oven for subsequent drying at 60 °C. The dried extract was ground using a mortar and pestle and stored in a freezer for future use as an agent to stabilize and reduce AgNP synthesis.Green synthesis of BT5-AgNPs
A 450 mL solution of 5 mM silver nitrate (AgNO3) was prepared in a beaker and placed on a hot plate magnetic stirrer (XMTD-702, China). Simultaneously, 50 mL of BT5 (green tea) extract was dissolved in deionized water (at 1 mg mL−1 concentration). Once the AgNO3 solution reached 70 °C, the BT5 extract was gradually added to the beaker. The addition resulted in a notable color change from clear to brown, indicating that the silver ions were reduced to silver nanoparticles (Fig. 1). The mixture was stirred for an extended period of 20 minutes before being covered and stored in a dark place for 24 hours to stabilize the nanoparticles.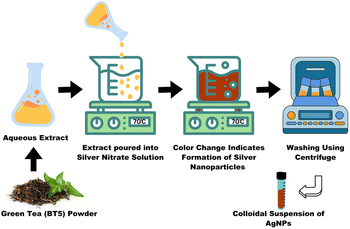 | ||
| Fig. 1 Synthesis of AgNPs using green tea extract from BT5 cultivar. 5 mM AgNO3 reduced using BT5 green tea extract and the color changes into dark brown indicating the formation of BT5-AgNPs. | ||
Centrifugation of the colloidal suspension was performed at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4 °C for 20 minutes; the pellet obtained was washed thrice with ddH2O and once with 100% ethyl alcohol using the centrifugation technique (Z36 HK, HERMLE Labortechnik GmbH, Wehingen, Germany). The purified nanoparticles were dispersed in deionized water (50 mL). A portion of the colloidal suspension was stored at 4 °C to assess its biological activity, whereas the rest of the suspension was vacuum-dried for biophysical characterization.
000 rpm at 4 °C for 20 minutes; the pellet obtained was washed thrice with ddH2O and once with 100% ethyl alcohol using the centrifugation technique (Z36 HK, HERMLE Labortechnik GmbH, Wehingen, Germany). The purified nanoparticles were dispersed in deionized water (50 mL). A portion of the colloidal suspension was stored at 4 °C to assess its biological activity, whereas the rest of the suspension was vacuum-dried for biophysical characterization.
Characterization of the synthesized BT5-AgNPs
In vivo experimental model and cell growth inhibition
A group of 24 male Swiss albino mice (8 weeks old, 25 ± 2 g) reared in the animal holding unit of the Molecular Biology and Protein Science Lab, Department of Genetic Engineering and Biotechnology, University of Rajshahi were used as in vivo experimental model animals. All mice were reared in cages maintained under ideal experimental conditions (25 °C ± 2 °C) with a 12 ± 1 hour dark/light cycle. Sufficient water and food (containing standard nutrients) were provided twice daily.EAC cells were obtained from the Protein and Enzyme Laboratory, Department of Biochemistry and Molecular Biology, University of Rajshahi, Bangladesh.
With a slight modification, the previously reported methodology (Sur & Ganguly) was used to perform an in vivo EAC cell growth inhibition experiment. For the investigation, four mice groups (n = 6 for each group) were used. 5 × 106 EAC cells were injected intraperitoneally into each mouse. The treatments were initiated 24 hours after the injection of EAC cells and administered every day for five days. BT5-AgNPs were administered intraperitoneally to Groups 1 and 2 at doses of 1 mg kg−1 (low-concentration) and 5 mg kg−1 (high-concentration), respectively. Group 4 was the untreated control and was administered 0.9% NaCl, whereas bleomycin (0.5 mg kg−1) was administered to Group 3. A 200 μL injection volume was used for all treatments and controls. On the sixth day after injecting EAC cells, the experimental subjects were euthanised, and the peritoneal fluid containing the cells was collected by washing the abdominal space with 1 mL of 0.9% salt solution. The collected cells were manually counted using a Neubauer haemocytometer (HBG, Germany) and a laboratory microscope (Optika, Italy). The efficacy of the treatments was assessed by calculating the inhibition of cell growth.
Ethical clearance
The Institutional Animal, Medical Ethics, Bio-safety and Bio-Security Committee (IAMEBBC) for Experimentation on Animal, Human, Microbes, and Living Natural Sources, memo no. 415 (27)/320/IAMEBBC/IBSc, Institute of Biological Sciences, University of Rajshahi, Bangladesh, granted permission to conduct this investigation.Determination of EAC cell growth inhibition
Cell growth inhibition was calculated using the following formula previously described by Sur and Ganguly:25where, Tw = mean number of EAC cells in the treated group. Cw = mean number of EAC cells in the control group of mice.
Assessment of apoptosis by DAPI staining
Morphological and nuclear alterations within EAC cells harvested from the control and treatment recipient sets of mice were examined using a fluorescence microscope (Olympus iX71, Hamburg, Germany). The extracted EAC cells (1 mL) were centrifuged at 4500 rpm (at 4 °C) for 5 minutes in a refrigerated centrifuge (Z36 HK, HERMLE Labortechnik GmbH, Wehingen, Germany). The resulting pellet was washed three times with normal saline (NS), with each wash at 4500 rpm (at 4 °C) for 2 minutes. The washed cells were subsequently stained using 5 μL of DAPI working solution (prepared from 10 mg mL−1 stock solution) for 15 minutes at 37 °C temperature in dark conditions, followed by a second centrifugation for 2 minutes at 4500 rpm. The DAPI-stained pellet was resuspended in 200 μL PBS. Finally, the cells were observed for morphological and nuclear alterations under a fluorescence microscope.Gene expression analysis
The total RNA content of harvested EAC cells from the treated and placebo (control) groups was extracted using the FavorPrep™ Total RNA Purification Mini kit (Favorgen, Ping Tung, Taiwan) following the instructions provided by the manufacturer. Nanodrop (Thermo Fisher, Madison, USA) was used for the quality assessment and quantification of RNA.First-strand complementary DNA (cDNA) synthesis was performed using the TIANScript M-MLV (TiangenR, Beijing, China) protocol. Two steps were involved in the reaction. In 15 μL of total reaction volume, RNA was first combined with oligo(dT) primers and dNTPs (ESI, Table ST1†). The reaction mixture (3–5 μL) was then incubated in 20 μL of final reaction volume with M-MLV Reverse Transcriptase (RT) and 5× First-Strand Buffer (ESI, Table ST2†). The reaction mixture was electrophoresed using 1% agarose gel and stained with ethidium bromide to ensure the synthesis of cDNA.
Primers for the target genes (p53, BAX, BCl2) and the reference gene (GAPDH) were designed using Primer3 software, ensuring optimal length (18–30 nucleotides), melting temperature (Tm) between 50 °C and 60 °C and GC content (40–60%). The primers were cross-checked for specificity using an Oligo analyzer to avoid non-specific binding. The primer sequences are presented in Table 1.
| Primer name | Primer sequence |
|---|---|
| GAPDH-forward | 5′-TCATCTCCGCCCCTTCTGC-3′ |
| GAPDH-reverse | 5′-TGCCTGCTTCACCACCTTCTT-3′ |
| p53-forward | 5′-GCGTCTTAGAGACAGTTGACT-3′ |
| p53-reverse | 5′-GGATAGGTCGGCGGTTCATCC-3′ |
| BCl2-forward | 5′-CAGCGGAGGAGGAGAAAGG-3′ |
| BCl2-reverse | 5′-GAATCGGGAGTTGGGGTCTG-3′ |
| BAX-forward | 5′-GCCCACCAGCTCTGAACAGATC-3′ |
| BAX-reverse | 5′-AAGTCCAGTGTCCAGCCCATG-3′ |
Real-time polymerase chain reaction (RT-PCR) was conducted using GoTaq qPCR Master Mix (Promega Corporation, USA) on a qTower3 G real-time PCR system (Analytik Jena, Germany). The reaction mixtures had a final volume of 20 μL, which contained 1 μL of cDNA template (10 times diluted), 10 μL of GoTaq qPCR Master Mix, 1 μL of forward primer (10 μM), 1 μL of reverse primer (10 μM), and 7 μL of nuclease-free water. A thermal cycler equipped with real-time detection was used for the amplification under the following conditions: initial denaturation for 3 minutes at 95 °C, 45 denaturation cycles for 15 seconds at 95 °C, annealing for 15 seconds at the primer-specific temperature, and extension for 20 seconds at 60 °C. The 2−(ΔΔCt) method26 was employed to calculate relative gene expression levels using Microsoft Excel.
Statistical analysis
A mean ± SD has been used to express the experimental results. To determine statistical significance, data were calculated using the ANOVA method in Microsoft Excel and GraphPad Prism 8. The p-value was required to be lower than 0.05 for a difference to be considered statistically significant. GraphPad Prism 8 and Origin2024b were used for the graphical presentation of data.Result and discussion
Phytochemical and antioxidant properties of BT5 cultivar
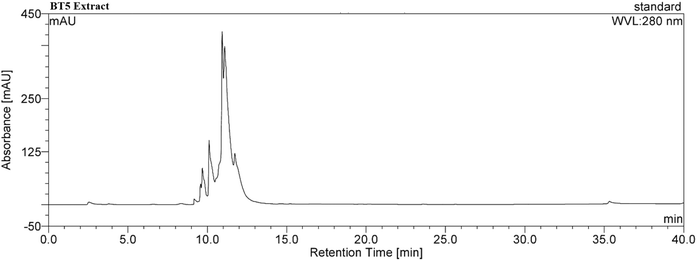
The BT5 cultivar of green tea exhibited significant phytochemical and antioxidant properties, highlighting its potential as a viable nanoparticle synthesis agent. The BT5 extract had a TPC of 133.73 μg mL−1 (133.73 mg per g dry weight) and a TFC of 102.19 μg mL−1 (102.19 mg per g dry weight). The extract exhibited a concentration-dependent increase in antioxidant activity. At 12.5 μg mL−1, the scavenging activity was 34.31%, which increased to 73.86% at 400 μg mL−1. The estimated IC50 was approximately 97.8 μg mL−1, suggesting moderate antioxidant activity (ESI, Fig. S3 and S4†). The HPLC analysis of the BT5 extract revealed significant peaks within the 9 to 12 minute retention time window, suggesting the presence of key polyphenolic compounds (Fig. 2). These peaks are linked to EGC and catechin, based on comparisons with recognized retention times.27Preparation of BT5-AgNPs
The AgNO3 solution was initially colorless, but upon gradual addition of the light brown BT5 green tea extract, the solution drastically turned dark brown, indicating the formation of colloidal AgNPs (Fig. 3). The compounds, which are mostly polyphenols present in the BT5 extract, help to form AgNPs from silver ions by donating electrons and acting as a stabilizing agent. Following visual confirmation, UV-vis, DLS, zeta, FT-IR, TGA, XRD, FE-SEM, and EDX were used to characterize BT5-AgNPS.13 | ||
| Fig. 3 Preparation of BT5-AgNPs. (A) Preheated AgNO3 solution before adding BT5 extract and (B) drastic colour change into dark brown after adding BT5 water extract. | ||
Characterization of BT5-AgNPss
 | ||
| Fig. 4 Prominent peak between 400–500 nm on the UV-visible spectrum indicates BT5-AgNPs formation (green line) and compares with BT5 plant extract (blue line) and AgNO3 solution (green line). | ||
| λ max (nm) | FT-IR peak frequencies (cm−1) | References |
|---|---|---|
| 435 | 3440.90, 2362.488, 1641.11, 2085.86, 466.88 | 29 |
| 420 | 3439, 1630,1384 | 30 |
| 447 | 3280, 2912, 1630, 1533, 1373, 1248, 1016, 775 | 31 |
| 410 | 3440, 2913, 1630, 1392, 1044 | 32 |
| 424 | 3425.58, 2360.87, 1631.78, 1382.96, 1072.42, 829.39, 596 | 33 |
| 479 | 3434, 1630, 1021 | 34 |
| 470 | 3315, 1634 | 13 |
| 410 | 3689.8, 2924.09, 1634, 1238.9, 1701, 1043, 761.8 | 12 |
| 434 | 3333, 1634, 1403, 1247.9, 1058.3, 555.12 | 35 |
| 415 | 3411, 2916, 1743, 1107, 800, 406 | 36 |
| 263 | 3421.72, 2937.32, 2358.94, 1643.35, 1369.46, 1037.70 | 37 |
| 421 | 3455, 1638 | 38 |
| 420 | 3435, 2923, 1040, 825, 610 | 39 |
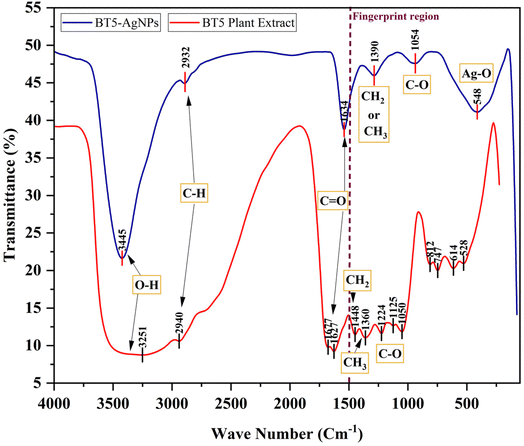
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) and alkene or aromatic (C
O) and alkene or aromatic (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) groups, respectively, which are characteristic of flavonoids and polyphenols present in the green tea extract. Additionally, peaks at 1224 cm−1, 1125 cm−1, and 1050 cm−1 correspond to the C–O stretching vibrations of ether (–C–O–C–) or alcohol (–C–OH) groups, while peaks at 1448 cm−1 and 1360 cm−1 are related to the CH2 and CH3 bending vibrations of aliphatic compounds. These functional groups collectively confirm the bioactive molecules present in the BT5 extract and act as reducing and capping agents to convert Ag+ ions to silver nanoparticles and stabilize them, thereby ensuring their colloidal stability.
C) groups, respectively, which are characteristic of flavonoids and polyphenols present in the green tea extract. Additionally, peaks at 1224 cm−1, 1125 cm−1, and 1050 cm−1 correspond to the C–O stretching vibrations of ether (–C–O–C–) or alcohol (–C–OH) groups, while peaks at 1448 cm−1 and 1360 cm−1 are related to the CH2 and CH3 bending vibrations of aliphatic compounds. These functional groups collectively confirm the bioactive molecules present in the BT5 extract and act as reducing and capping agents to convert Ag+ ions to silver nanoparticles and stabilize them, thereby ensuring their colloidal stability.
The FTIR spectrum of the BT5-AgNPs (Fig. 7, blue line) revealed shifts in several key vibrational peaks compared with the BT5 extract (Fig. 7, red line), confirming the involvement of these functional groups in nanoparticle synthesis. The broad O–H stretching peak at 3251 cm−1 in the extract deviated to 3445 cm−1 in the BT5-AgNPs, consistent with the detected phenolic compounds and aromatic alcohols. Aliphatic C–H stretching was observed at 2932 cm−1. Aromatic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching vibrations were evident at 1634 cm−1. Further peaks at 1390 cm−1 and 1054 cm−1 were assigned to aromatic C–H bending and C–O stretching (ether linkage), respectively. The emergence of a new peak at 548 cm−1 is accredited to Ag–O stretching vibrations,29,33,35,39 providing strong evidence for the interaction between the silver nanoparticles and the functional groups of the BT5 extract. These findings are consistent with previous reports on silver nanoparticle synthesis using green tea extract (Table 2).12,13,29–39
C stretching vibrations were evident at 1634 cm−1. Further peaks at 1390 cm−1 and 1054 cm−1 were assigned to aromatic C–H bending and C–O stretching (ether linkage), respectively. The emergence of a new peak at 548 cm−1 is accredited to Ag–O stretching vibrations,29,33,35,39 providing strong evidence for the interaction between the silver nanoparticles and the functional groups of the BT5 extract. These findings are consistent with previous reports on silver nanoparticle synthesis using green tea extract (Table 2).12,13,29–39
where K denotes the shape factor (0.9 for spherical particles), λ denotes the X-ray wavelength (0.1546 nm), while β denotes the full width at half maximum (FWHM) of the diffraction peak in radians, and θ is Bragg's angle. The crystalline sizes of all planes are given in Table 3.
 | ||
| Fig. 9 XRD peak of BT5-AgNPs corresponding to the silver lattice planes (111), (200), (220) and (311), indicating that the synthesized particles are cubic-shaped, single-phase BT5-AgNPs. | ||
| Sl. no. | Planes (hkl) | 2θ | Sin(θ) | Cos(θ) | FWHM (β) radians | Inter-planar spacing (d) | Lattice constant (a) | Cell volume (a3) | Crystalline size (D) (nm) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 111 | 37.94733 | 0.325134 | 0.945668 | 0.011402 | 2.36917 | 4.103523 | 69.098808 | 12.86 |
| 2 | 200 | 44.25451 | 0.376665 | 0.92635 | 0.023651 | 2.04504 | 4.09008 | 68.421944 | 6.33 |
| 3 | 220 | 64.34297 | 0.532455 | 0.846458 | 0.013251 | 1.44669 | 4.091857 | 68.511175 | 12.36 |
| 4 | 311 | 77.27808 | 0.624412 | 0.781095 | 0.013207 | 1.23364 | 4.091521 | 68.494288 | 13.44 |
Anti-proliferative activity of synthesized BT5-AgNPs on EAC cells
Based on these findings, the antiproliferative activity of BT5-AgNPs was determined. The synthesized BT5-AgNPs were subjected to experimentation to determine their capacity to reduce cancer cell proliferation using EAC cells induced in Swiss albino mice as a cancer model. Following six days of therapy, both the low-concentration dosage (1 mg kg−1 of body weight per day) and high-concentration dosage (5 mg kg−1 of body weight per day) treatment groups inhibited EAC cell development, with the low dose showing significant outcomes. The concentrations (1 mg kg−1 and 5 mg kg−1 body weight per day) reported for BT5-AgNPs represent the concentration of the entire nanoparticle formulation (i.e., the total weight of nanoparticles). Specifically, the low-dose group achieved 52.15% inhibition of cancer cell proliferation compared with 22.91% inhibition observed in the high-dose group (Fig. 12). The chemotherapeutic drug bleomycin was deliberately selected as a positive control at a standard therapeutic dosage of 0.5 mg kg−1 body weight per day based on established literature and clinical usage guidelines. Although this dosage differs from the nanoparticle concentrations tested, bleomycin's purpose was solely to serve as a validated reference point for its effective antiproliferative activity.44 The standard drug exhibited 81.59% suppression of EAC cell growth. The reduced efficacy observed at higher BT5-AgNP concentrations may be attributed to cytotoxic effects or altered cellular responses, highlighting the need for further optimization.Our findings align with earlier research, which demonstrated the anticancer efficacy of green tea extract-mediated AgNPs against EAC cells in vivo and enhanced therapeutic potential with minimum toxicity.45,46 Many other studies have highlighted the broad-spectrum chemotherapeutic activity of green tea-mediated AgNPs against MCF-7, HeLa, Hep-2, A549, KB3, AGS, and NTERA-2.31,47,48
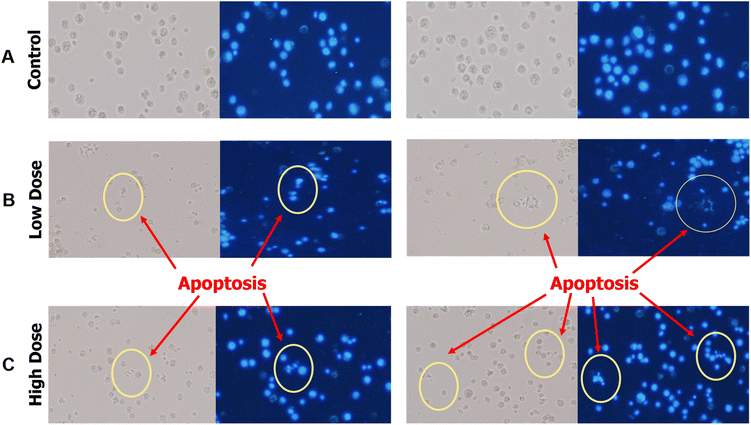
Detection of apoptosis EAC cell by DAPI staining
To further investigate the mechanism underlying the antiproliferative activity of BT5-AgNPs, DAPI staining was employed to determine whether EAC cells are undergoing apoptotic transformation or not. Fluorescence microscopy revealed notable differences between untreated control and treated cells. The control cells had a normal, rounded appearance with homogeneous nuclear staining (Fig. 13A). Cells treated with BT5-AgNPs (low-dose group and high-dose group) exhibited apoptotic characteristics such as heterochromatization, Karyorrhexis, bleb formation on the membrane, and shrinkage of cells (Fig. 13B and C). These structural alterations represent signs of apoptosis, a process of controlled cell death that rids the body of undesired or damaged cells while sparing healthy tissue.Expression of p53, BAX and Bcl2 gene analysis
The total RNA content extracted from the recipient EAC cells of treatment and placebo showed high purity and integrity (ESI, Fig. S5 and Table ST5†). cDNA synthesis (ESI, Table ST6†) and RT-qPCR were conducted according to the protocol presented in the methodology. The 2−(ΔΔCt) method26 was used to quantify relative gene expression by normalizing data to a reference gene (GAPDH in this study).The gene expression analysis revealed a dose-dependent modulation of the apoptotic pathway. p53 expression was upregulated 10 times in the low-dosage recipient group and 8 times in the high-dosage recipient group, suggesting activation of the intrinsic apoptotic pathway. Similarly, BAX, a pro-apoptotic gene, was upregulated 3 times in the low-dosage recipient group and 2.5 times in the high-dosage recipient group, thereby enhancing pro-apoptotic signalling. The anti-apoptotic gene BCl2 was downregulated 0.53 times in the low-dosage recipient group and 0.84 times in the high-dosage recipient group, reflecting reduced anti-apoptotic defences (Fig. 14). These results demonstrate that low-dose therapy promotes apoptosis better than high-dose therapy through the upregulation of p53 and BAX and downregulation of BCl2. This supports the notion of dose-dependent effects of nanoparticles in cancer therapy, as observed in previous studies.47
Conclusion
This study used BT5 extract, a commercial green tea cultivar from Bangladesh, to synthesize silver nanoparticles (BT5-AgNPs) using an eco-friendly, green approach. Before synthesis, the BT5 extract was analysed, revealing significant amounts of bioactive compounds, such as total phenolics, flavonoids, and demonstrating strong antioxidant activity. These bioactive components are believed to contribute to the reduction and stabilization of silver ions during synthesis. The synthesized BT5-AgNPs were characterized using FE-SEM, EDX, XRD, FTIR, UV-vis spectroscopy, DLS, zeta potential, and TGA techniques to confirm their size, elemental composition, crystalline structure, PDI, hydrodynamic diameter, and surface charge. The FT-IR results indicated that the green-synthesized BT5-AgNPs displayed similar diffraction peaks and chemical functional groups as the BT5 extract, suggesting the involvement of phytochemicals in reducing Ag+ to AgNPs. Furthermore, BT5-AgNPs exhibit enhanced thermal stability compared with green tea extract. The antiproliferative activity of the synthesized BT5-AgNPs was assessed in an experimental tumor model. The results demonstrate significant inhibition of cell growth and apoptosis-inducing ability. Expression analysis revealed a dose-dependent modulation of key apoptotic genes, including downregulation of BCl2 and upregulation of p53 and BAX, further supporting their potential as natural therapeutic agents for cancer treatment.Data availability
The data supporting the findings of this article are included in the ESI.†Author contributions
Sk. Md. Atiqur Rahman: methodology, investigation, formal analysis, writing–original draft, writing–review & editing, visualization, project administration, and data curation. Rokshana Ara Ruhi: methodology, investigation, writing–original draft, and writing–review & editing. Md. Mahmudul Hasan Maruf: investigation. Md. Ragib Shariar: investigation. Mobasshir Noor Shehab: investigation and writing–review & editing. Khaled Mahmud Sujon: investigation. Mohammad Saiful Islam: investigation. Md. Abdul Aziz: investigation. Firoz Ahmed: investigation. Ananda Kumar Saha: writing–review & editing. Md. Anwarul Kabir Bhuiya: writing–review & editing, supervision, and funding acquisition. Md. Abu Reza: conceptualization, supervision, resources, writing–review & editing, validation, and funding acquisition.Conflicts of interest
The authors declare that there are no conflicts of interest.Acknowledgements
The current research was supported by a special research grant of Rajshahi University (A-701/6/109 (research), 2022) and the Ministry of Education's Grant for Advanced Research in Education (GARE) (Project ID: PS20201507 and ET20232558). The authors wish to extend their sincere gratitude to the Bangladesh Council of Scientific and Industrial Research for their support with DLS, Zetasizer, and FE-SEM with EDX instruments. Additionally, we acknowledge the Molecular Pathology Laboratory, Institute of Biological Science, R. U., and the Plant Breeding and Gene Engineering Lab, Department of Botany, R.U. for their assistance with HPLC and phytochemical analysis.References
- M. Xing, L. Ge, M. Wang, Q. Li, X. Li and J. Ouyang, Int. J. Nanomed., 2014, 2399, DOI:10.2147/ijn.s55015
.
- H. Chen, M. C. Roco, X. Li and Y. Lin, Nat. Nanotechnol., 2008, 3, 123–125 CrossRef CAS PubMed
.
- M. Govarthanan, Y.-S. Seo, K.-J. Lee, I.-B. Jung, H.-J. Ju, J. S. Kim, M. Cho, S. Kamala-Kannan and B.-T. Oh, Artif. Cells, Nanomed., Biotechnol., 2016, 44, 1878–1882 CrossRef CAS PubMed
.
- O. V. Kharissova, H. V. R. Dias, B. I. Kharisov, B. O. Pérez and V. M. J. Pérez, Trends Biotechnol., 2013, 31, 240–248 CrossRef CAS PubMed
.
- M. Baghayeri, B. Mahdavi, Z. Hosseinpor-Mohsen Abadi and S. Farhadi, Appl. Organomet. Chem., 2018, 32, 2 CrossRef
.
- K.-J. Lee, S.-H. Park, M. Govarthanan, P.-H. Hwang, Y.-S. Seo, M. Cho, W.-H. Lee, J.-Y. Lee, S. Kamala-Kannan and B.-T. Oh, Mater. Lett., 2013, 105, 128–131 CrossRef CAS
.
- V. V. Makarov, A. J. Love, O. V. Sinitsyna, S. S. Makarova, I. V. Yaminsky, M. E. Taliansky and N. O. Kalinina, Acta Naturae, 2014, 6, 35–44 CrossRef CAS PubMed
.
- K. Parveen, V. Banse and L. Ledwani, AIP Conf. Proc., 2016, 1724, 1 Search PubMed
.
- A. Rietveld and S. Wiseman, J. Nutr., 2003, 133, 3285S–3292S Search PubMed
.
- T. Kondo, T. Ohta, K. Igura, Y. Hara and K. Kaji, Cancer Lett., 2002, 180, 139–144 CrossRef CAS PubMed
.
- S. Wang, N. Moustaid-Moussa, L. Chen, H. Mo, A. Shastri, R. Su, P. Bapat, I. Kwun and C.-L. Shen, J. Nutr. Biochem., 2014, 25, 1–18 CrossRef CAS PubMed
.
- H. A. Widatalla, L. F. Yassin, A. A. Alrasheid, S. A. Rahman Ahmed, M. O. Widdatallah, S. H. Eltilib and A. A. Mohamed, Nanoscale Adv., 2022, 4, 911–915 RSC
.
- K. Parvathalu, D. N. Kumar, K. Rajitha, M. G. Kishan, B. N. Kumar, J. Bhemarajam, S. R. Naidu, G. L. Merlinsheeba, P. Mandal, S. Banne, A. Dayanand, V. Morampudi, B. Murali, S. E. N. Vinodini, Y. V. Reddy and P. B. Bhaskar, Plasmonics, 2023, 18, 1837–1845 CrossRef CAS
.
- M. Golpour, P. Ebrahimnejad, Z. R. Gatabi, A. Najafi, A. Davoodi, R. Khajavi, M. Alimohammadi and T. Mousavi, Inorg. Chem. Commun., 2024, 161, 111989 CrossRef CAS
.
- M. J. Mitchell, M. M. Billingsley, R. M. Haley, M. E. Wechsler, N. A. Peppas and R. Langer, Nat. Rev. Drug Discovery, 2021, 20, 101–124 CrossRef CAS PubMed
.
- H. Sabit, M. Abdel-Hakeem, T. Shoala, S. Abdel-Ghany, M. M. Abdel-Latif, J. Almulhim and M. Mansy, Pharmaceutics, 2022, 14, 1566 CrossRef CAS PubMed
.
- A. Rodzinski, R. Guduru, P. Liang, A. Hadjikhani, T. Stewart, E. Stimphil, C. Runowicz, R. Cote, N. Altman, R. Datar and S. Khizroev, Sci. Rep., 2016, 6, 20867 CrossRef CAS PubMed
.
- M. M. Safhi, S. M. Sivakumar, A. Jabeen, F. Zakir, F. Islam, T. Anwer, U. S. Bagul, M. E. Elmobark, G. Khan, R. Siddiqui, A. Hussien and M. F. Alam, Elsevier, 2017, 159–169, DOI:10.1016/b978-0-323-52725-5.00008-3
.
- C. S. Yang, X. Wang, G. Lu and S. C. Picinich, Nat. Rev. Cancer, 2009, 9, 429–439 Search PubMed
.
- A. Bag and N. Bag, J. Nat. Sci. Biol. Med., 2018, 9, 2 CrossRef CAS PubMed
.
- Y. Miyata, T. Matsuo, K. Araki, Y. Nakamura, Y. Sagara, K. Ohba and H. Sakai, Medicines, 2018, 5, 87 CrossRef CAS PubMed
.
- M. Nasiruddin and S. N. Sinha, Asian J. Med. Biol. Res., 2020, 6, 187–195 CrossRef CAS
.
- C. Quettier-Deleu, B. Gressier, J. Vasseur, T. Dine, C. Brunet, M. Luyckx, M. Cazin, J.-C. Cazin, F. Bailleul and F. Trotin, J. Ethnopharmacol., 2000, 72, 35–42 CrossRef CAS PubMed
.
- İ. Gulcin and S. H. Alwasel, Processes, 2023, 11, 2248 CrossRef
.
- P. Sur and D. Ganguly, Planta Med., 1994, 60, 106–109 CrossRef CAS PubMed
.
- K. J. Livak and T. D. Schmittgen, Methods, 2001, 25, 402–408 CrossRef CAS PubMed
.
- P. L. Fernandez, M. J. Martin, A. G. Gonzalez and F. Pablos, Analyst, 2000, 125, 421–425 Search PubMed
.
- D. Paramelle, A. Sadovoy, S. Gorelik, P. Free, J. Hobley and D. G. Fernig, Analyst, 2014, 139, 4855 Search PubMed
.
- A. Kharabi Masooleh, A. Ahmadikhah and A. Saidi, IET Nanobiotechnol., 2019, 13, 183–188 CrossRef PubMed
.
- W. Xu, Y. Fan, X. Liu, D. Luo, H. Liu and N. Yang, Mater. Res. Express, 2018, 5, 045029 CrossRef
.
- D. Arumai Selvan, D. Mahendiran, R. Senthil Kumar and A. Kalilur Rahiman, J. Photochem. Photobiol., B, 2018, 180, 243–252 Search PubMed
.
- W. R. Rolim, M. T. Pelegrino, B. De Araújo Lima, L. S. Ferraz, F. N. Costa, J. S. Bernardes, T. Rodigues, M. Brocchi and A. B. Seabra, Appl. Surf. Sci., 2019, 463, 66–74 Search PubMed
.
- P. Prema, V. Veeramanikandan, K. Rameshkumar, M. K. Gatasheh, A. A. Hatamleh, R. Balasubramani and P. Balaji, Environ. Res., 2022, 204, 111915 CrossRef CAS PubMed
.
- Y. Fang, C.-Q. Hong, F.-R. Chen, F.-Z. Gui, Y.-X. You, X. Guan and X.-H. Pan, Inorg. Chem. Commun., 2021, 132, 108808 CrossRef CAS
.
- A. Bergal, G. H. Matar and M. Andaç, BioNanoScience, 2022, 12, 307–321 CrossRef
.
- M. A. Taleb Safa and H. Koohestani, Results Eng., 2024, 21, 101808 CrossRef CAS
.
- H. H. Salih, Baghdad Sci. J., 2024, 21, 1470 CrossRef
.
- D. Hermanto, N. Ismillayli, D. H. Fatwa, U. K. Zuryati, H. Muliasari, R. Wirawan, D. Prasetyoko and S. Suprapto, S. Afr. J. Chem. Eng., 2024, 47, 136–141 Search PubMed
.
- A. Wirwis and Z. Sadowski, ACS Omega, 2023, 8, 30532–30549 Search PubMed
.
- P. Mukhopadhyay and P. P. Kundu, RSC Adv., 2015, 5, 93995–94007 RSC
.
- M. Singh, A. Mallick, M. Banerjee and R. Kumar, Bull. Mater. Sci., 2016, 39, 1871–1878 CrossRef CAS
.
- M. Flores-González, M. Talavera-Rojas, E. Soriano-Vargas and V. Rodríguez-González, New J. Chem., 2018, 42, 2133–2139 RSC
.
- Q. Sun, X. Cai, J. Li, M. Zheng, Z. Chen and C.-P. Yu, Colloids Surf., A, 2014, 444, 226–231 CrossRef CAS
.
- Pfizer, bleomycin for Injection, USP, https://www.pfizermedicalinformation.com/patient/bleomycin Search PubMed.
- M. E. Hosen, M. A. Rahman, M. S. Rahman, S. Akash, M. Khalekuzzaman, A. A. Alsahli, M. Bourhia, H. A. Nafidi, M. A. Islam and R. Zaman, Chem. Biodiversity, 2024, 21, 3 CrossRef PubMed
.
- A. Magdy, E. Sadaka, N. Hanafy, M. A. El-Magd, N. Allahloubi and M. El Kemary, Mol. Cell. Toxicol., 2020, 16, 271–282 CrossRef CAS
.
- F. Mobaraki, M. Momeni, M. Jahromi, F. M. Kasmaie, M. Barghbani, M. E. T. Yazdi, Z. Meshkat, F. H. Shandiz and S. M. Hosseini, Process Biochem., 2022, 119, 106–118 CrossRef CAS
.
- S. K. Sahu, A. Mansoori, S. K. Jana, A. Kumar and T. K. Ghorai, J. Mol. Struct., 2025, 1320, 139690 CrossRef CAS
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5na00115c |
| This journal is © The Royal Society of Chemistry 2025 |