 Open Access Article
Open Access ArticleEvaluation of the antimicrobial activity of chitosan- and curcumin-capped copper oxide nanostructures against multi-drug-resistant microorganisms†
Noura
El-Kattan
a,
Mostafa A.
Ibrahim
b,
Ahmed N.
Emam
 *cd,
Khaled
Metwally
*cd,
Khaled
Metwally
 e,
Fady Sayed
Youssef
f,
Nourelhuda Ahmed
Nassar
g and
Ahmed S.
Mansour
hi
e,
Fady Sayed
Youssef
f,
Nourelhuda Ahmed
Nassar
g and
Ahmed S.
Mansour
hi
aDepartment of Microbiology, Research Institute of Medical Entomology, General Organization for Teaching Hospitals and Institutes, Giza, Egypt
bProduction and R&D Unit, NanoFab Technology Company, 6th October City, Giza, Egypt
cRefractories, Ceramics and Building Materials Department, Advanced Materials Technology & Mineral Resources Research Institute, National Research Centre (NRC), El Bohouth St., Dokki, 12622 Cairo, Egypt. E-mail: ahmed.gsc.ndp@gmail.com; an.emam@nrc.sci.eg
dNanomedicine & Tissue Engineering Research Lab, Medical Research Centre of Excellence, National Research Centre (NRC), El Bohouth St., Dokki, 12622 Cairo, Egypt
eGenetics Department, Faculty of Agriculture, Ain Shams University, P.O. Box 68, Hadayek Shoubra, 11241, Cairo, Egypt
fDepartment of Pharmacology Faculty of Veterinary Medicine, Cairo University, 12211 Giza, Egypt
gClinical Pathology Department, Al-Sahel Teaching Hospital, Cairo, Egypt
hDepartment of Laser Applications in Meteorology, Chemistry and Agriculture, National Institute of Laser Enhanced Sciences (NILES), Cairo University, Cairo, Egypt
iFaculty of Postgraduate Studies for Nanotechnology, Cairo University, Zayed City, Giza, Egypt
First published on 19th March 2025
Abstract
The emergence of multi-drug-resistant microorganisms presents a serious threat to infection control, for which new antimicrobial strategies are urgently needed. Herein, the antimicrobial activities of copper oxide nanoparticles capped with curcumin (Cur-CuO NPs) and copper oxide nanoparticles capped with chitosan (CS-CuO NPs) were investigated. They were prepared via the co-precipitation method. A total of 180 clinical ICU patients were found to have 70% Gram-negative and 30% Gram-positive isolates. Antimicrobial susceptibility testing indicated resistance of these isolates to 14 among the 21 tested antibiotics. Physicochemical properties of the curcumin-capped (Cur-CuO NPs) and chitosan-capped (CS-CuO NPs) copper oxide nanoparticles were identified using UV-vis spectroscopy, transmission electron microscopy (TEM), dynamic light scattering (DLS), zeta-potential (ζ), and Fourier transform infrared (FT-IR) spectroscopy. Cur-CuO- and CS-CuO-NPs exhibited potent antimicrobial efficacy, wherein CS-CuO NPs were found to possess a lower minimum inhibitory concentration (MIC) (3.9–15.6 μg mL−1) than Cur-CuO NPs (14.5–31.2 μg mL−1). Biocompatibility assay showed that Cur-CuO NPs were safer with an IC50 dose of 74.17 μg mL−1 than CS-CuO NPs with an IC50 dose of 41.01 μg mL−1. Results revealed that the Cur-CuO- and CS-CuO-NPs have the potential to be safely used as effective antimicrobial agents in clinical applications at low concentrations (6.25–12.5 μg mL−1).
1. Introduction
Nosocomial infections are among the most significant global health care problems and have an unfavorable impact on clinical outcomes.1,2 The rise of multidrug-resistant bacteria poses a critical challenge to medical treatment, as these organisms can withstand multiple antimicrobial interventions, severely limiting therapeutic options.3 The pathogeny of antibiotic-resistant genes in diverse bacterial species has emerged owing to the misuse of antibiotics. According to researchers, antimicrobial nano-formulation is a promising avenue of future research owing to the significant antimicrobial activities exhibited by nanomaterials.4 Nanoparticles with a size of less than 100 nm are increasingly replacing the application of traditional antibiotics in medicine. Nanobiotechnology in the future is expected to bring the development of new antibacterial agents via the production of important size- and form-specific metal oxide nanoparticles. Metal oxide nanoparticles are of great interest owing to their functionalities in electric-, electrochemical-, paint/ink materials, catalysis, and magnetism.5–7Copper oxide nanoparticles and other metal oxide nanoparticles are now the focus of research because of their antibacterial and biocidal properties, which are employed in many biological applications.5 Silver, zinc oxide, titanium oxide, copper oxide, and iron oxide nanoparticles are the most widely used in antimicrobial research. However, silver is expensive, and thus, there is a need for inexpensive materials that can yield equivalent efficacy. Copper presents antibacterial possibilities in making antimicrobial textiles. Antibacterial action can cause ROS generation, destruction of the cell membrane through electrostatic interactions, disruption in metal/metal ion homeostasis, and dysfunction of proteins and enzymes.8 The antibacterial activities of copper and copper oxide nanoparticles are well documented and are said to work against bacteria by puncturing their cell membranes and impairing their vital enzymes. Nevertheless, in the case of Gram-positive and Gram-negative bacteria, which are generally negatively charged, Gram-positive bacteria with a thick peptidoglycan cover will be influenced more than Gram-negative bacteria with a sophisticated structure on metal uptake.9,10 Electrostatic interactions will attract the positively charged nanoparticles and disrupt the cell wall with increased permeability. The nanoparticles release the metal ions from their extracellular environment, inducing a biological response and generation of reactive oxygen species (ROS) using the above-mentioned pathways. Metal ions will bind to the cellular constituents, thereby disrupting cell activities and creating very strong coordination bonds with organic and biomolecular fragments.11,12
CuO NPs possess better antibacterial properties than silver NPs.13 Throughout history, copper has been recognized for its powerful antimicrobial properties, demonstrating the ability to eliminate up to 99.9% of microorganisms through its metal oxide interactions. Copper/copper oxide NPs exhibit extensive antimicrobial action against Gram-positive as well as Gram-negative bacteria, thereby eradicating these pathogens, which are responsible for hospital-acquired infections.14 The antibacterial effect by CuO NPs is determined regarding bacterial cell properties, which is especially significant in terms of cell wall structure and Gram character. CuO NPs destroyed 100% of Gram-negative E. coli at concentrations greater than 9.5%, while the same concentration was less effective against Gram-positive S. aureus.15,16 Antibacterial activity is affected by particle size and surface properties, where smaller particles have more antibacterial power because of their higher surface area.17,18 Although limited studies have been done on CuO NPs, they present good potential bactericidal activity for a variety of infectious organisms including E. coli, B. subtilis, V. cholera, P. aeruginosa, S. typhus, and S. aureus.19–21
Turmeric curcumin extract (Curcuma longa Linns/Curcuma domestica Valeton) can be used as a biocompatible reducing and capping agent during the synthesis of nanoparticles. This compound has been used as a spice, food color,22 and Chinese medicine for thousands of years and possesses multiple therapeutic activities such as antioxidant, anti-inflammatory, antiseptic, and anticancer activities.23–28 FDA has approved the safety of curcumin in a dose of up to 12 g per day.29 Similar to antibiotics, curcumin possesses more than one way of killing bacteria including causing membrane damage, production of reactive oxygen species (ROS), inhibition of efflux pumps, and inhibition of cell division. The abundant hydroxyl groups in the phenolic molecules in curcumin interact with the bacterial cell membrane in a specific manner. This results in the loss of permeability and alteration in fatty acid and phospholipid profiles, hence inhibiting energy metabolism and de novo synthesis of its genetic material. Curcumin has been shown experimentally to cause high dose rumpling of Gram-positive (S. aureus and Enterococcus faecalis) and Gram-negative (E. coli and Pseudomonas aeruginosa) bacteria membranes, cause apoptosis of bacterial cells, and markedly suppresses the efflux pump resistance mechanisms of bacteria such as P. aeruginosa and S. aureus.30,31
Chitosan is a shellfish and crustacean-derived polysaccharide from chitin.32 It can be used in tissue engineering for treating hypertension and high cholesterol, and wound healing due to its antioxidant and antibacterial properties.33 Chitosan possesses an advantageous property of compatibility with metals, metal oxide nanoparticles, and polymers. Its antimicrobial effect is through the disruption of microbial cell membranes through electrostatic interaction between its positively charged amino groups and the negatively charged cell surface components;32 inhibition of nutrient transfer in Gram-negative bacteria (high-molecular-weight chitosan); and inhibition of DNA/RNA and protein synthesis (low-molecular-weight chitosan).34,35 Chitosan also serves as a template in the synthesis of metal oxide nanoparticles, which can alter the surface properties of the resulting particles.36–38
To the best of our knowledge, the antimicrobial efficacy of chitosan-capped CuO NPs and curcumin-capped CuO NPs against multi-drug-resistant microbes has yet to be studied except in a few reports.39–45 The present study aimed to evaluate the antibacterial activity of green-synthesized copper oxide nanoparticles (CuO NPs) in the presence of curcumin against the multi-drug-resistant (MDR) bacteria, which existed in the turmeric ethanolic extract and chitosan extracts. In addition, efficient hybrid nanocomposites were developed based on the formation of chitosan-capped CuO (CS-CuO NPs) and curcumin-capped CuO (Cur-CuO NPs) nanoparticles. The morphological, optical, surface and colloidal properties of the as-prepared nanoparticles were investigated using TEM, UV-vis absorption spectroscopy, FT-IR, DLS and zeta-potential measurements. In addition, the antimicrobial activity of both CS-CuO and Cur-CuO NPs was tested against the most popular MDR microbes in Egyptian hospitals, especially in the intensive care unit (ICU). Our results revealed that the chitosan-capped CuO NPs have higher antimicrobial efficacy with a lower minimum inhibition concentration (MIC) than previously reported values in the literature. Also, the curcumin-capped CuO NPs exhibited significant antimicrobial activity against MDR microbes with a lower MIC than that reported in the previous study by Varaprasad et al.45 Finally, Cur-CuO NPs showed remarkable biocompatibility, which was higher than that previously reported by Varaprasad et al.45 All these results indicate that Cur-CuO and CS-CuO NPs can be used as active ingredients in antimicrobial coating paint applications.
2. Experimental
2.1. Materials
Copper(II) sulfate pentahydrate LR (CuSO4·5H2O, 98.5%) was purchased from SD-Fine Chem Limited. Ethanol absolute (C2H5OH, EtOH 95%) was obtained from Central Drug House (P) LTD, chitosan medium molecular weight (CS M. wt. 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–350,000, 99%) was from Alpha Chemika, and turmeric powder (Curcuma longa Linns, “synonym; Curcuma domestica Valeton, Zingiberaceae”) was obtained from the local spice market as an imported product from India by Aava Ayurveda Private Limited, Ludhiana, India. Pure curcumin as the standard was purchased from Herbal House Centers, (Egypt – Lot No. HHC092021). Mueller–Hinton Agar (Oxoid Limited, Cat. No. CM0337), antibiotics including amikacin, amoxicillin/clavulanic acid, tigecycline, cefepime, cefotaxime, ceftazidime, cefaclor, ciprofloxacin, gentamicin, imipenem, meropenem, levofloxacin, tetracycline, tobramycin, cefazolin, and cefoxitin were purchased from Oxoid Limited. SRPMI-1640 medium, MTT, and DMSO were obtained from Sigma Co., St. Louis, USA, and fetal bovine serum was from GIBCO, UK.
000–350,000, 99%) was from Alpha Chemika, and turmeric powder (Curcuma longa Linns, “synonym; Curcuma domestica Valeton, Zingiberaceae”) was obtained from the local spice market as an imported product from India by Aava Ayurveda Private Limited, Ludhiana, India. Pure curcumin as the standard was purchased from Herbal House Centers, (Egypt – Lot No. HHC092021). Mueller–Hinton Agar (Oxoid Limited, Cat. No. CM0337), antibiotics including amikacin, amoxicillin/clavulanic acid, tigecycline, cefepime, cefotaxime, ceftazidime, cefaclor, ciprofloxacin, gentamicin, imipenem, meropenem, levofloxacin, tetracycline, tobramycin, cefazolin, and cefoxitin were purchased from Oxoid Limited. SRPMI-1640 medium, MTT, and DMSO were obtained from Sigma Co., St. Louis, USA, and fetal bovine serum was from GIBCO, UK.
2.2. Methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 silylation reagent and 50 μL pyridine to derivatize the functional groups in the samples to trimethylsilyl groups (abbreviated TMS) before GC measurement. Gas chromatography-mass spectrometry analysis (GC-MS). The Central Laboratories Network, National Research Centre, Cairo, Egypt, used an Agilent Technologies GC-MS system with a gas chromatograph (7890B) and a mass spectrometer detector (5977A). The GC had an HP-5MS column (30 mm × 0.25 mm internal diameter, 0.25 μm film thickness). Analyses were conducted using hydrogen as the carrier gas at a flow rate of 2.0 mL min−1, splitless injection volume of 2 μL, and the following temperature program: 50 °C for 5 min; increase at 5 °C min−1 to 100 °C and hold for 0 min; then increase at 10 °C min−1 to 320 °C and hold for 10 min. The injector and detector were kept at 280 °C and 320 °C, respectively. Mass spectra were acquired using electron ionization (EI) at 70 eV, in the spectral range of m/z 25–700 and a solvent delay of 6 min. The temperature of the source was 230 °C, while that of the quad was 150 °C. Various constituents were discovered by comparing the spectrum fragmentation pattern to that in the Wiley and NIST databases.
1 silylation reagent and 50 μL pyridine to derivatize the functional groups in the samples to trimethylsilyl groups (abbreviated TMS) before GC measurement. Gas chromatography-mass spectrometry analysis (GC-MS). The Central Laboratories Network, National Research Centre, Cairo, Egypt, used an Agilent Technologies GC-MS system with a gas chromatograph (7890B) and a mass spectrometer detector (5977A). The GC had an HP-5MS column (30 mm × 0.25 mm internal diameter, 0.25 μm film thickness). Analyses were conducted using hydrogen as the carrier gas at a flow rate of 2.0 mL min−1, splitless injection volume of 2 μL, and the following temperature program: 50 °C for 5 min; increase at 5 °C min−1 to 100 °C and hold for 0 min; then increase at 10 °C min−1 to 320 °C and hold for 10 min. The injector and detector were kept at 280 °C and 320 °C, respectively. Mass spectra were acquired using electron ionization (EI) at 70 eV, in the spectral range of m/z 25–700 and a solvent delay of 6 min. The temperature of the source was 230 °C, while that of the quad was 150 °C. Various constituents were discovered by comparing the spectrum fragmentation pattern to that in the Wiley and NIST databases.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min (SIGMA, Germany). The resulting pellet was dried in an oven at 100 °C. The collected precipitate was ground into a fine powder, followed by calcination for 2 h at 500 °C to get brownish-black powder.
000 rpm for 15 min (SIGMA, Germany). The resulting pellet was dried in an oven at 100 °C. The collected precipitate was ground into a fine powder, followed by calcination for 2 h at 500 °C to get brownish-black powder.
2.3. Characterization
The photophysical properties, including the UV-vis absorption spectra to provide insights into the electronic transitions, of the as-green synthesized curcumin (Cur-CuO) and chitosan-capped (CS-CuO) copper oxide nanoparticles were investigated using a TG-80 double beam spectrophotometer. The absorption spectra were recorded in the range of 200 to 900 nm, with an increment and wavelength step of about 0.2 and 5 nm, respectively.Transmission electron microscopy (TEM) JEOL, model JEM 2100F, was used to visualize the morphological properties of the obtained Cur-CuO and CS-CuO NPs at an operating voltage of 160 kV. Also, to confirm the elemental composition and presence of copper and oxygen in the nanoparticles, ensuring the successful synthesis of copper oxide nanoparticles, elemental X-ray (EDX) analysis was carried out on a TESCAN VEGA II SBU scanning electron microscope at an operating voltage in the range of 200 V to 30 kV.
Crystallographic structure was investigated by X-ray diffraction (XRD) measurements using a Bruker D8 advanced X-ray powder diffractometer operating with a Cu target with Kα1 = 1.54060 Å, Kα2 = 1.5444 Å, in the 2θ range of 10° to 50° at a step of 0.02°.
The critical information about the size distribution, surface charge (i.e. zeta potential), and colloidal stability for the as-prepared curcumin and chitosan-capped CuO NPs was measured using a Malvern Zetasizer Nano ZS Nano instrument with an He/Ne laser (i.e., λ = 633 nm) at an angle of 173° collecting backscatter optics.
Furthermore, FT-IR was used to identify the functional groups present in the synthesized curcumin and chitosan-capped CuO NPs and investigate the potential interactions among curcumin, chitosan, and copper oxide. FT-IR spectra were recorded in the range of 400 to 4000 cm−1 using a JASCO 6700 Fourier transform infrared spectrometer (FT-IR).
2.4. Microbiological investigation
| Source of microorganism isolates | S. aureus MSSA | S. aureus MRSA | P. aeruginosa | K. Pneumoniae | A. baumannii | P. mirabilis | P. vulgaris | E. coli | E. faecalis | S. Pyogenes | C. albicans |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sputum | 0 | 3 | 0 | 5 | 1 | 0 | 0 | 2 | 0 | 0 | 0 |
| Urine | 4 | 0 | 6 | 11 | 3 | 2 | 1 | 7 | 4 | 0 | 5 |
| Wound | 5 | 11 | 9 | 12 | 5 | 3 | 2 | 7 | 3 | 2 | 2 |
| Blood | 1 | 5 | 0 | 5 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
2.5. Biocompatibility assay
where A570 is the optical absorbance obtained at the wavelength of 570 nm for both the treated and untreated cell lines.58,59
2.6. Statistical analysis
The statistical analysis was performed using IBM SPSS (version 25) with a population at (P < 0.05) and (P < 0.01) compared to the control. The experiments were conducted in triplicate and expressed as mean ± SD. Significant differences among means were evaluated using Duncan's multiple range or one-way ANOVA test.3. Results and discussion
3.1. Identification of the ethanolic extract of turmeric (Curcuma longa Linn) powder
The validation of for the ethanolic extract of Curcuma longa Linn (i.e., C. longa) against pure (standard) curcumin was achieved as illustrated in our previously published work.4 The UV-vis spectra of C. longa ethanolic extract and pure (standard) curcumin showed a single broad band with the maximum absorbance peak at 426 nm, which is attributed to low energy π–π* excitation. In addition, pure (standard) curcumin showed a single narrow emission band at 539 nm, as shown in Fig. 1. Alternatively, the C. longa ethanolic extract also showed a single narrow emission band, but with a slight shift at 543 nm depending on the sample, which is consistent with previous reports (see Fig. 1).60–62 The TLC bands of both pure curcumin and the ethanolic extract of Curcuma longa Linn (i.e., C. longa) powder were observed after 1 h of exposure to mobile phase vapor. The standard pure curcumin showed a single TLC band after irradiation to UV light at a wavelength of 366 nm (see Fig. 2).63 Alternatively, the ethanolic extract of C. longa showed two bands, as shown in Fig. 2. The upper band is ascribed to curcumin, while the middle band is demethoxycurcumin, which agree with the study reported by Kamble and Dahake Pavan.64 | ||
| Fig. 1 Validation process using (a) UV-vis absorption and (b) emission spectra of C. longa ethanolic extract (black line) against standard pure curcumin (orange line). | ||
 | ||
| Fig. 2 TLC visual bands of pure standard curcumin (left side) and ethanolic extract of C. longa (right side). | ||
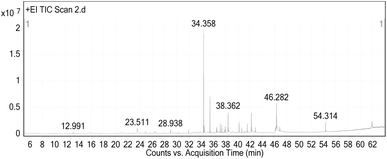
In addition, a study using gas chromatography-mass spectrometry (GC-MS) was conducted on the ethanolic extract of Curcuma longa Linn (turmeric) powder vs. pure curcumin to show their chemical composition. This study found ten different chemicals in the pure curcumin purchased from Herbal House Center, Egypt – (Lot No. HHC092021), and their bioactive chemicals, retention time (RT), peak areas (%), and molecular formulas are listed in Table 2. The main chemicals found were 9-octadecenoic acid, (E)-(37.51%), n-hexadecanoic acid (32.4%), and 3-buten-2-one, 4-(4-hydroxy-3-methoxyphenyl) (7.77%), which are consistent with previous findings, as shown in the chromatogram illustrated in Fig. 3 and S1–S3 in the ESI,† respectively.65,66
| Peak | RT | Name | Formula | Area | Area sum % |
|---|---|---|---|---|---|
| 1 | 23.519 | 2-Methoxy-4-vinylphenol | C9H10O2 | 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 289 289![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 710.7 710.7 |
1.85 |
| 2 | 38.371 | 3-Buten-2-one, 4-(4-hydroxy-3-methoxyphenyl)- | C11H12O3 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 954 954![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 073.27 073.27 |
7.77 |
| 3 | 42.263 | n-Hexadecanoic acid | C16H32O2 | 61![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 189 189![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 184.17 184.17 |
32.4 |
| 4 | 46.358 | 9-Octadecenoic acid, (E)- | C18H34O2 | 72![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 579 579![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 212.42 212.42 |
37.51 |
| 5 | 46.434 | Curlone | C15H22O | 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 528 528![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 061.47 061.47 |
2.12 |
| 6 | 46.85 | Oleic acid | C18H34O2 | 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 825 825![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 001.8 001.8 |
4.09 |
| 7 | 54.315 | Bis(2-ethylhexyl)phthalate | C24H38O4 | 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 344 344![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 755.35 755.35 |
3.7 |
| 8 | 57.68 | Ar-turmerone | C15H20O | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 724 724![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 070.17 070.17 |
6.23 |
| 9 | 61.96 | Borneol, pentafluoropropionate | C13H17F5O2 | 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 179 179![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 711.5 711.5 |
1.23 |
| 10 | 62.58 | (+)-Alpha-curcumene | C15H22 | 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 321 321![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 477.21 477.21 |
3.1 |
Alternatively, the gas chromatography-mass spectrometry chromatogram of the ethanolic extract from Curcuma longa Linn (turmeric) powder identified 26 compounds, as shown in Table 3, Fig. 4 and S4–S8 in the ESI.† The major molecules were Ar-turmerone (30.39%), followed by curlone (12.33%), 9-octadecenoic acid, (E)-(9.77%), n-hexadecanoic acid (7.62%), and 3-buten-2-one, 4-(4-hydroxy-3-methoxyphenyl)-(7.38%). Earlier studies also identified the majority of turmerones,67–71 which are terpenoid chemicals and are important components of the Curcuma species.72 These components have a variety of pharmacological activities, including antibacterial, antioxidant, and anti-inflammatory properties.72 The bioactive components of Curcuma longa are responsible for its medicinal properties, according to the findings of Anekwe et al.73 Furthermore, the chemical makeup of the turmeric rhizome is determined by its genotype, field circumstances, and postharvest processing.74
| Peak | RT | Name | Formula | Area | Area sum % |
|---|---|---|---|---|---|
| 1 | 12.991 | p-Cymene | C10H14 | 699![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 955.52 955.52 |
0.35% |
| 2 | 23.511 | 2-Methoxy-4-vinylphenol | C9H10O2 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 542 542![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 020.92 020.92 |
0.78% |
| 3 | 24.857 | Eugenol | C10H12O2 | 803![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 411.42 411.42 |
0.41% |
| 4 | 26.386 | Vanillin | C8H8O3 | 825![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 515.89 515.89 |
0.42% |
| 5 | 28.938 | (+)-Alpha-curcumene | C15H22 | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 517 517![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 379.14 379.14 |
1.27% |
| 6 | 30.244 | Cyclohexene, 3-(1,5-dimethyl-4-hexenyl)-6-methylene-, [S-(R*, S*)]- | C15H24 | 629![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 517.91 517.91 |
0.32% |
| 7 | 31.892 | Benzene, 1,1′-(1,1,2,2-tetramethyl-1,2-ethanediyl)bis- | C20H26 | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 054 054![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 993.87 993.87 |
1.04% |
| 8 | 34.358 | Ar-turmerone | C15H20O | 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 029 029![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 102.04 102.04 |
30.39% |
| 9 | 35.4 | Curlone | C15H22O | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 966 966![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 625.58 625.58 |
12.33% |
| 10 | 36.504 | 1-(4-Hydroxybenzylidene)acetone | 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 605 605![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 314 314 |
2.69% | |
| 11 | 37.09 | Dicumyl peroxide | C18H22O2 | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 622 622![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 002.45 002.45 |
2.85% |
| 12 | 37.338 | 6-Isopropenyl-4,8a-dimethyl-4a,5,6,7,8,8a-hexahydro-1H-naphthalen-2-one | C15H24O | 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 483 483![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 499.65 499.65 |
2.27% |
| 13 | 37.833 | 4,4-Diallyl-cyclohexanone | C16H24O6 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 845 845![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 258.02 258.02 |
0.93% |
| 14 | 37.897 | Borneol, pentafluoropropionate | C13H17F5O2 | 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 998 998![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 162.43 162.43 |
2.02% |
| 15 | 38.362 | 3-Buten-2-one, 4-(4-hydroxy-3-methoxyphenyl)- | C11H12O3 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 575 575![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 091.99 091.99 |
7.38% |
| 16 | 40.175 | 1-Adamantanecarboxylic acid, 3-phenylpropyl ester | C17H20O2 | 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 724 724![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 212.31 212.31 |
3.4% |
| 17 | 40.595 | 1-Methoxybicyclo[2,2,2]oct-5-en-2-yl methyl ketone | C11H16O2 | 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 381 381![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 998.35 998.35 |
1.71% |
| 18 | 41.075 | 2,2,6-Trimethyl-1-(3-methylbuta-1,3-dienyl)-7-oxabicyclo[4.1.0]heptan-3-ol | C14H22O2 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 040 040![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 626.31 626.31 |
0.53% |
| 19 | 41.518 | (E)-2-Isopropyl-5-methylphenyl 2-methylbut-2-enoate | C15H22O2 | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 069 069![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 205.11 205.11 |
2.57% |
| 20 | 42.176 | n-Hexadecanoic acid | C16H32O2 | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 062 062![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 805.96 805.96 |
7.62% |
| 21 | 42.82 | 3-Methyl-but-2-enoic acid, 1,7,7-trimethyl-bicyclo[2.2.1]hept-2-yl ester | C15H26O2 | 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 307 307![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 390.87 390.87 |
1.67% |
| 22 | 46.104 | 9,12-Octadecadienoic acid (Z,Z)- | C18H32O2 | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 799 799![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 112.78 112.78 |
1.42% |
| 23 | 46.282 | 9-Octadecenoic acid, (E)- | C18H34O2 | 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 306 306![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 924.05 924.05 |
9.77% |
| 24 | 46.812 | Oleic acid | C18H34O2 | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 710 710![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 804.34 804.34 |
1.37% |
| 25 | 54.314 | Bis(2-ethylhexyl) phthalate | C24H38O4 | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 609 609![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 218.7 218.7 |
2.84% |
| 26 | 61.9 | 1-Heptatriacotanol | C37H76O | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 829 829![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 870.02 870.02 |
1.65% |
 | ||
| Fig. 4 GC-MS/MS chromatogram of all the identified compounds in the ethanolic extract from C. longa (Turmeric) powder. | ||
3.2. Characterization of curcumin capped- and chitosan capped-copper nanoparticles
The photophysical properties of the as-prepared Cur-CuO NPs and CS-CuO NPs were investigated by monitoring their UV-vis absorption spectra, as shown in Fig. 5. Cur-CuO NPs exhibit a single broad absorption band at 400 nm and a noticeable shoulder at 460 nm, showing the existence of copper oxide nanoparticles capped with curcumin extract (Cur-CuO NPs) (see Fig. 5, black line). Alternatively, the chitosan-capped copper oxide nanoparticles (CS-CuO NPs) showed a characteristic feature at the shoulder at 370 nm, indicating the formation of CS-CuO NPs (see Fig. 5, red line), which agrees with previous studies.40,46 Moreover, the morphological properties, including the particle size and shape of Cur-CuO and CS-CuO NPs, are depicted in Fig. 6. Both the green-synthesized Cur-CuO and CS-CuO NPs show quasi-spherical-like particles with average sizes of 25 ± 10 nm (Fig. 6a and b) and 10 ± 5 nm (Fig. 6c and d), respectively.The EDX spectra show Cu and O as the main elemental composition, indicating the purity of the CS-CuO and Cur-CuO NPs samples, as shown in Fig. 7. The atomic percentage of copper (Cu) and oxygen (O) in CS-CuO NPs was about 28.51% and 51.07%, respectively. The atomic percentage of copper (Cu) and oxygen (O) in Cur-CuO NPs was about 41.26% and 36.96%, respectively. In addition, the presence of nitrogen and carbon in the EDX spectrum of CS-CuO NPs is attributed to the amino groups and carbon present in chitosan (see Fig. 7a). Besides, the EDX spectrum of Cur-CuO NPs displayed the presence of copper, oxygen and carbon elements, indicating the formation of Cur-CuO NPs (see Fig. 7b). Moreover, the EDX spectra of CS-CuO and Cur-CuO NPs clearly explained the fact that the amount of Cu decreased with a decrease in the CuO content in these hybrid nanocomposites. Finally, the presence of sulfur (S) element is due to the inorganic nature of the copper precursor (i.e., copper sulfate, CuSO4).
Fig. 8 presents the X-ray diffraction (XRD) patterns of the curcumin-capped CuO nanoparticles (Cur-CuO NPs) (see Fig. 8a) and chitosan-capped CuO nanoparticles (CS-CuO NPs), as shown in Fig. 8b.
The XRD pattern of Cur-CuO NPs shows the presence of both monoclinic CuO (ICCD 01-080-0076) and cubic Cu2O (ICCD 03-065-3288) calculated as diffraction peaks with a change in 2θ (see Fig. 8a). The strongest peaks are found at the 2θ values of 35.5° and 38.7°, which correspond to the (1 1 0) and (1 1 1) planes, respectively. The other peaks corresponding to the (1 1 0), (2 0 2), and (0 2 0) planes are located at around 32.5°, 48.7°, and 53.5°, respectively. In addition, another two diffraction patterns, specifically the (1 1 1) and (2 0 0) planes that characterize to Cu2O crystal phases, are distinctly visible in the XRD pattern of the CuO/Cu2O NPs sample. The average crystallite size was calculated to be 14.35 nm. The XRD pattern for CS-CuO NPs (see Fig. 8b) exhibits almost the same major peaks at around 35.5° and 38.7° for the (0 0 2) and (1 1 1) planes of a monoclinic CuO. A peak is observed also for the (2 0 2) plane at around 48.7°. CS-CuO NPs exhibits a stronger and broader diffraction pattern than curcumin-capped Cu2O, supporting the conclusion that they have a smaller particle size. The average crystallite size was about 4.59 nm, which is smaller than that of CS-CuO NPs, and in good agreement with the TEM data.
The large background observed in both patterns, especially in the low 2θ range (10–20°), is attributed to presence of amorphous constituents such amorphous capping ligands (curcumin and chitosan).
In addition, colloidal properties, including dynamic light scattering (DLS) and electrophoretic mobility based on zeta potential measurements, were investigated for Cur-CuO NPs and CS-CuO NPs in a vehicle solution, as shown in Fig. 9 and Table 4. The hydrodynamic diameter (HD) of Cur-CuO NPs was about 105.1 ± 36.58 nm with a polydispersity index (PDI) of 0.28, which is smaller than that of CS-CuO NPs. The average HD of CS-CuO NPs was about 1631 ± 205.5 nm with a more extensive polydispersity index (PDI) of 0.698 (Fig. 9a and b and Table 4). The zeta potential of Cur-CuO NPs was about −1.07 mV, which is lower than the zeta-potential of the CS-CuO NPs, with a value of about +7.1 mV, as shown in Fig. 9c and d, respectively, and Table 4. According to the previously mentioned colloidal properties based on the DLS data, the hydrodynamic particle size of the as-prepared nanoparticles is enlarged, consistent with their agglomeration. This agglomeration is because of their hydrophilicity. In addition, the intensity of the steric forces of the functional groups on the surface of the nanoparticles is generated by creating a layer of water around the material.
 | ||
| Fig. 9 DLS data of Cur-CuO NPs (a) and CS-CuO NPs (b). Zeta potential of Cur-CuO NPs (c) and CS-CuO NPs (d). | ||
| Sample | Dynamic light scattering (DLS) | Zeta potential (ζ, mV) | |
|---|---|---|---|
| Hydrodynamic diameter (HD, nm) | Polydispersity index (PDI) | ||
| Cur-CuO NPs (0 Time) | 105.1 ± 36.58 | 0.28 | −1.07 |
| Cur-CuO NPs (12 Months) | 107.7 ± 19.26 | 0.79 | −4.39 |
| CS-CuO NPs (0 Time) | 1631 ± 205.5 | 0.698 | +7.1 |
| CS-CuO NPs (12 Months) | 1319 ± 208 | 0.599 | +2.44 |
Furthermore, the surface properties of Cur-CuO NPs and CS-CuO NPs were investigated via Fourier-transform infrared (FT-IR) based on their transmittance as a function of wavenumber, as shown in Fig. 10. The FT-IR spectrum of Cur-CuO NPs showed a strong stretching band at a wavenumber of 3401 cm−1 due to the presence of –OH intermolecular bonded alcohol in the ethanolic solution or the phenolic components in the turmeric extract. Two stretching aliphatic –CH bands were observed at 2977 and 2896 cm−1, corresponding to the sp2 C–H bond existing in –OCH3 (i.e., methoxy) groups in the curcumin component.75–77 Also, the asymmetric stretching vibration at 2348 cm−1 indicates the presence of the O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C
C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group due to the decarboxylation of the phenolic compounds ferulic acid and its derivatives such as 3-buten-2-one, 4-(4-hydroxy-3-methoxyphenyl) (i.e., feruloylmethane) present in the turmeric extract and the –COOH attached to their aromatic ring (see Fig. 10, black line).78,79 In addition, the stretching strong vibration at 1635 cm−1 is assigned to C
O group due to the decarboxylation of the phenolic compounds ferulic acid and its derivatives such as 3-buten-2-one, 4-(4-hydroxy-3-methoxyphenyl) (i.e., feruloylmethane) present in the turmeric extract and the –COOH attached to their aromatic ring (see Fig. 10, black line).78,79 In addition, the stretching strong vibration at 1635 cm−1 is assigned to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O in the β-diketone moiety (O
O in the β-diketone moiety (O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH
C–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–C
CH–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) and C–H bending frequency of the aromatic overtone, which are present in turmeric extract flavonoids such as curcumin, and other curcuminoid compounds present in turmeric, such as demethoxycurcumin and bisdemethoxycurcumin.76,77 The symmetric bending band at 1379 cm−1 is due to the vibration of the –CH3 group present in the flavonoids in the turmeric ethanolic extract.80 The weak stretching band of aromatic C–O enol and bending phenolic –OH group of the curcuminoid components are assigned to the peak at 1272 cm−1. The sharp stretching band at 1052 cm−1 is assigned to the C–O–C stretching vibration of phenyl alkyl ether, which confirmed the molecular structure of curcumin and other curcuminoid components extracted from turmeric.81 The weak stretching band at 663 cm−1 corresponds to Cu–O as a result of the interaction between the turmeric extract components and copper oxide surface.81,82 Finally, the weak sharp band at 879 cm−1 is attributed to the aromatic C–H out-of-plane bending vibration in curcumin and other curcuminoid components.77
O) and C–H bending frequency of the aromatic overtone, which are present in turmeric extract flavonoids such as curcumin, and other curcuminoid compounds present in turmeric, such as demethoxycurcumin and bisdemethoxycurcumin.76,77 The symmetric bending band at 1379 cm−1 is due to the vibration of the –CH3 group present in the flavonoids in the turmeric ethanolic extract.80 The weak stretching band of aromatic C–O enol and bending phenolic –OH group of the curcuminoid components are assigned to the peak at 1272 cm−1. The sharp stretching band at 1052 cm−1 is assigned to the C–O–C stretching vibration of phenyl alkyl ether, which confirmed the molecular structure of curcumin and other curcuminoid components extracted from turmeric.81 The weak stretching band at 663 cm−1 corresponds to Cu–O as a result of the interaction between the turmeric extract components and copper oxide surface.81,82 Finally, the weak sharp band at 879 cm−1 is attributed to the aromatic C–H out-of-plane bending vibration in curcumin and other curcuminoid components.77
Alternatively, CS-CuO NPs, as shown in Fig. 10 (red line), showed the characteristic peaks for chitosan at 3500 and 1112 cm−1, corresponding to the –OH and –C–O–C– stretching vibrations, respectively. The –NH2 bending vibration peak was observed in chitosan at 1629 cm−1. The peaks at 2922 cm−1 correspond to the stretching vibration of –CH3 and –NH, and the peak at 1400 cm−1 belongs to the stretching vibration of the C–H bond. In addition, the stretching band of C–O in the spectrum of chitosan was observed at 894 cm−1 due to the conjugation with Cu–O. According to the comparison between the FTIR spectra of Cur-CuO and CS-CuO NPs, two characteristic features are observed at 2977 and 2896 cm−1 in Cur-CuO NPs than CS-CuO NPs due to the stretching vibration of –CH of alkyne in the curcumin molecule and intermolecular-bonded –OH alcohol that exists in the ethanolic solution, respectively. The second difference was observed at 2348, 1920, and 1052 cm−1 due to the medium stretching bands for the –O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C
C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O–, –CCC– and –C–O groups existing in curcumin and primary alcohol, respectively. The last difference observed at 3229, 2850 and 1725 cm−1 in CS-CuO compared to Cur-CuO NPs due to –NH2 of chitosan, –CH, C
O–, –CCC– and –C–O groups existing in curcumin and primary alcohol, respectively. The last difference observed at 3229, 2850 and 1725 cm−1 in CS-CuO compared to Cur-CuO NPs due to –NH2 of chitosan, –CH, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of aliphatic ketone, and –OH group in carboxylic acid.40 Furthermore, the sharp band at 1634 cm−1 and less intense band at 1377 cm−1 are attributed to the M–O out-of-plane rocking and in-plane rocking, respectively.40
O of aliphatic ketone, and –OH group in carboxylic acid.40 Furthermore, the sharp band at 1634 cm−1 and less intense band at 1377 cm−1 are attributed to the M–O out-of-plane rocking and in-plane rocking, respectively.40
3.3. Evaluation of the long-term stability and shelf-life of Cur-CuO and CS-CuO NPs
Stability is essential for determining the usefulness of nanoparticles in real-life applications such as biomedicine, catalysis, and environmental science. Thus, a 12-month stability study was conducted considering parameters such as hydrodynamic diameter (HD), polydispersity index (PDI), and zeta potential using DLS, as shown in Table 4. The findings reveal that Cur-CuO NPs only exhibited small variations in size, which slightly changed from 105.1 ± 36.58 nm to just 107.7 ± 19.26 nm. However, their PDI increased from 0.28 to 0.79, indicating that this system was becoming increasingly heterogeneous, which demonstrates the growth of some relatively small aggregates. Moreover, the zeta potential became more negative (−1.07 mV to −4.39 mV), suggesting that changes in surface charge interactions and colloidal stability occurred. However, despite the slight changes in size, the resulting data confirmed the sufficient size stability of the Cur-CuO NPs and proved their decent shelf-life (see Table 4).Alternatively, the CS-CuO NPs exhibited a rather substantial reduction in hydrodynamic diameter from 1631 ± 205.5 nm to 1319 ± 208 nm, which is probably due to the structural changes or partial sedimentation of the large nanoparticle aggregates over time. The PDI showed a decrease in value from 0.698 to 0.599, denoting that the size uniformity improved slightly. Nevertheless, the zeta potential decreased from the previous level of +7.1 mV to +2.44 mV, indicating that there was less repulsion between the particles, which might be one reason for their instability. On the negative side, the zeta potential was between −10 mV and +10 mV, which shows that CS-CuO NPs are more likely to form aggregates after some time. These results are consistent with former investigations showing that particles with a larger initial size and smaller surface charge present higher risks of sedimentation and aggregation, as shown in Table 4.83
The capping agents are responsible for the dissimilarity in the long-term stability of the Cur-CuO and CS-CuO NPs. Curcumin, a nanoparticle-stabilizing agent, is superior due to its stabilizing action, which allows the nanoparticles to stay dispersed and resistant to head-to-head aggregation.84 Alternatively, chitosan deposits at the beginning of the reaction, given that it is positively charged, showing stability for a short time. Nonetheless, it may be highly likely that its dispersion may be hampered or even lost because of its destruction due to structural degradation or deterioration of electrostatic interactions. Alternatively, PDI values below the threshold of 0.3 can be overtly stable nanoparticles, and numbers above 0.7 can mean aggregation. The increase in the PDI of Cur-CuO and the CS-CuO zeta decrease in potential are the main signs causing us to believe that they undergo degradation processes, but with Cur-CuO showing better long-term stability.85
3.4. Distribution and prevalence of clinically isolated pathogenic bacteria
One hundred eighty specimens were collected from different sources from patients diagnosed with diverse infections, as shown in Table 5. Among the sources sampled, 128 (71.1%) had pathogenic microbial growth. After the isolation and characterization, the results showed that the most frequent isolated pathogenic bacteria were Klebsiella pneumoniae accounting for 33 strains (25.8% of total isolated strains), followed by Staphylococcus aureus (MRSA), which gave a frequency of 19 strains (14.8%). Ten strains (7.8%) were Staphylococcus aureus (MSSA).| Category | Organisms | No. of strains | Frequency % |
|---|---|---|---|
| Gram-positive bacteria | Enterococcus faecalis | 7 | 5.5 |
| Staphylococcus aureus MSSA | 10 | 7.8 | |
| Staphylococcus aureus MRSA | 19 | 14.8 | |
| Streptococcus pyogenes | 2 | 1.6 | |
| Gram-negative bacteria | Klebsiella pneumoniae | 33 | 25.8 |
| Escherichia coli | 17 | 13.3 | |
| Pseudomonas aeruginosa | 15 | 11.7 | |
| Acinetobacter baumannii | 10 | 7.8 | |
| Proteus mirabilis | 5 | 3.9 | |
| Proteus vulgaris | 3 | 2.3 | |
| Fungi | Candida albicans | 7 | 5.5 |
| Total | 128 | 100 | |
Moreover, Escherichia coli and Pseudomonas aeruginosa were present with a frequency of 17 strains (13.3%) and 15 (11.7%), respectively. Acinetobacter baumannii was present, recording 10 strains (7.8%), whereas Enterococcus faecalis had 7 strains (5.5%). In the case of the Proteus genus, 5 strains (3.9%) were Proteus mirabilis, and 3 strains (2.3%) were Proteus vulgaris. Finally, the lowest recorded bacteria were Streptococcus pyogenes, accounting for 2 strains (1.6%). However, Candida albicans accounts for 7 strains (5.5%). Our findings agree with previous reports,84,86 stating that the most frequently distributed bacteria were Gram-negative bacteria compared with Gram-positive bacteria and the presence of Candida albicans in intensive care units.
3.5. Antibiotic inhibition capacity
The antibiotic resistance capacities of the isolated microorganisms were investigated against 21 antibiotic discs, as shown in Table 6. The results revealed that methicillin-sensitive S. aureus (MSSA) strains resisted cefotaxime and ofloxacin. Nevertheless, they showed strong resistance against ampicillin, ceftazidime, levofloxacin, doxycycline, and erythromycin with values of 90%, 80%, 80%, 80% and 70%, respectively. MSSA also showed moderate resistance, with 60% against amikacin and gentamicin and 50% against ampicillin/sulbactam, ciprofloxacin, tetracycline, and clindamycin. Moreover, MSSA showed moderate to weak resistance against amoxycillin/clavulanic acid, cefazolin, cefepime, and tigecycline with values of 40%, 40%, 30% and 20%, respectively. Conversely, MSSA was completely sensitive to vancomycin. The methicillin-resistant S. aureus (MRSA) strains showed 100% resistance against 14 out of the 21 tested antibiotics, which are amoxycillin/clavulanic acid, ampicillin/sulbactam, cefazolin, cefepime, cefotaxime, ceftazidime, cefoxitin, gentamicin, imipenem, meropenem, levofloxacin, ofloxacin, ampicillin, and erythromycin. However, MRSA showed strong to moderate resistance against clindamycin, tetracycline, doxycycline, amikacin, ciprofloxacin, and tigecycline, recording 84.2%, 68.4%, 63.2%, 52.6% and 36.8% resistance, respectively. Only vancomycin was shown to be effective against 100% of the MRSA strains.| Antibiotic discs | S. aureus MSSA | S. aureus MRSA | E. faecalis | S. pyogenes | A. baumannii | E. coli | K. pneumoniae | P. vulgaris | P. mirabilis | P. aeruginosa |
|---|---|---|---|---|---|---|---|---|---|---|
| Amikacin (30 μg) | 60 | 63.2 | 71.4 | 100 | 70 | 29.4 | 72.7 | 33.3 | 60 | 66.7 |
| Amoxycillin/clavulanic acid (30 μg) | 40 | 100 | 57.1 | 100 | 80 | 88.2 | 100 | 100 | 100 | 100 |
| Ampicillin/sulbactam (30 μg) | 50 | 100 | 85.7 | 100 | 80 | 64.7 | 100 | 100 | 80 | 80 |
| Cefazolin (30 μg) | 40 | 100 | 71.4 | 100 | 100 | 82.4 | 100 | 66.7 | 60 | 86.7 |
| Cefepime (30 μg) | 30 | 100 | 57.1 | 50 | 80 | 52.9 | 90.9 | 33.3 | 40 | 100 |
| Cefotaxime (10 μg) | 100 | 100 | 100 | 100 | 90 | 82.4 | 100 | 100 | 100 | 100 |
| Ceftazidime (30 μg) | 80 | 100 | 85.7 | 50 | 90 | 70.6 | 100 | 100 | 100 | 80 |
| Cefoxitin (30 μg) | 0 | 100 | 100 | 100 | 100 | 41.2 | 93.9 | 100 | 100 | 100 |
| Ciprofloxacin (5 μg) | 50 | 52.6 | 57.1 | 100 | 80 | 82.4 | 84.8 | 66.7 | 80 | 100 |
| Doxycycline (5 μg) | 80 | 63.2 | 85.7 | 100 | 70 | 64.7 | 60.6 | 100 | 100 | 93.3 |
| Gentamicin (10 μg) | 60 | 100 | 71.4 | 50 | 90 | 41.2 | 75.8 | 33.3 | 40 | 66.7 |
| Imipenem (10 μg) | 0 | 100 | 0 | 50 | 60 | 17.6 | 54.5 | 0 | 20 | 60 |
| Meropenem (10 μg) | 0 | 100 | 0 | 0 | 30 | 11.8 | 63.6 | 0 | 0 | 40 |
| Levofloxacin (5 μg) | 80 | 100 | 57.1 | 50 | 50 | 82.4 | 75.8 | 100 | 80 | 60 |
| Ofloxacin (5 μg) | 100 | 100 | 85.7 | 100 | 90 | 52.9 | 90.9 | 100 | 100 | 80 |
| Tetracycline (30 μg) | 50 | 68.4 | 28.9 | 50 | 50 | 88.2 | 66.7 | 66.7 | 60 | 80 |
| Tigecycline (15 μg) | 20 | 36.8 | 14.3 | 0 | 0 | 0 | 15.2 | 0 | 0 | 33.3 |
| Ampicillin (10 μg) | 90 | 100 | 100 | 100 | ||||||
| Erythromycin (15 μg) | 70 | 100 | 57.1 | 100 | ||||||
| Clindamycin (2 μg) | 50 | 84.2 | 71.4 | 50 | ||||||
| Vancomycin (30 μg) | 0 | 0 | 14.3 | 0 |
Our results are consistent with previous reports87,88 stating that S. aureus is one of the most prevalent pathogenic bacteria that frequently resist multiple drugs, contributing to various infections. Methicillin-resistant S. aureus (MRSA) is particularly concerning, which substantially impacts antimicrobial resistance levels across numerous countries. The S. aureus-acquired resistance against various antibiotics is attributed to genetic factors and mutations.89,90E. faecalis showed complete resistance (100%) against cefotaxime, cefoxitin, and ampicillin, as previously reported.4,91 Nevertheless, E. faecalis exhibited strong resistance, accounting for 85.7% against ampicillin/sulbactam, ceftazidime, ofloxacin, and doxycycline; 71.4% against amikacin, cefazolin, gentamicin, and clindamycin; and 57.1% against amoxycillin/clavulanic acid, cefepime, ciprofloxacin, levofloxacin, and erythromycin. Moreover, as previously stated, weak resistance was observed against tigecycline and vancomycin, recording 14.3% resistance.92 Imipenem and meropenem antibiotics efficiently killed 100% of E. faecalis. The obtained results are consistent with previous reports stating that E. faecalis is considered one of the most dangerous multi-drug resistant bacteria, and the acquired resistance is attributed to its high capacity in transferring and acquiring antibiotic resistance genes through mobile genetic elements such as plasmids.53,93
S. pyogenes demonstrated 100% resistance against 11 antibiotics (amikacin, amoxycillin/clavulanic acid, ampicillin/sulbactam, cefazolin, cefotaxime, cefoxitin, ciprofloxacin, doxycycline, ofloxacin, ampicillin, and erythromycin). The results agree with that obtained by Yu and co-workers,94 stating the strong resistance of S. pyogenes against β-lactam antibiotics.94 In contrast, the other antibiotics showed moderate efficiency, accounting for 50% against S. pyogenes, where only tigecycline and vancomycin effectively killed S. pyogenes. 100% of A. baumannii strains exhibited complete resistance against cefazolin and cefoxitin. However, A. baumannii displayed strong resistance ranging from 70% to 90% against several antibiotics, including amikacin, doxycycline, amoxycillin/clavulanic acid, cefepime, cefotaxime, gentamicin, and ofloxacin, where the same observations were reported in a previous study.95
Moreover, it exhibited moderate resistance, accounting for 50% against both levofloxacin and tetracycline and 60% against imipenem. A. baumannii showed 100% sensitivity against tigecycline.96 Alternatively, E. coli also showed 100% sensitivity against tigecycline, which elucidated the usability of this antibiotic in combating E. coli.97E. coli strains showed different antibiotic resistance capacities ranging between moderate as observed against amikacin, cefoxitin, gentamicin, cefepime, ofloxacin, ampicillin/sulbactam, and doxycycline accounting (29.4%, 41.2%, 41.2%, 52.9%, 52.9%, 64.7% and 64.7%, respectively) to strong resistance, as observed against ceftazidime, cefazolin, ciprofloxacin, cefotaxime, levofloxacin, amoxycillin/clavulanic acid, and tetracycline accounting for 70.6%, 82.4%, 82.4%, 82.4%, 82.4%, 88.2% and 88.2%, respectively. At the same time, it showed weak resistance, 11.8% and 17.6%, against meropenem and imipenem antibiotics, respectively, which agrees with previous reports.98
Furthermore, P. vulgaris exhibited complete resistance against amoxycillin/clavulanic acid, ampicillin/sulbactam, cefotaxime, ceftazidime, cefoxitin, doxycycline, levofloxacin, and ofloxacin, while moderate resistance of 33.3% was observed with amikacin, cefepime, and gentamicin, and 66.6% with cefazolin, ciprofloxacin, and tetracycline. Imipenem, meropenem, and tigecycline were shown to be effective in combating P. vulgaris, resulting in killing 100% of bacteria. Moving forward to the other strain for the Proteus genus P. mirabilis, nearly the same resistance patterns were observed, with a slight difference in amikacin accounting for 33.3% resistance compared with 60% observed with P. vulgaris. K. pneumoniae strains exhibited 100% resistance against 5 tested antibiotics, including amoxycillin/clavulanic acid, ampicillin/sulbactam, cefazolin, cefotaxime, and ceftazidime. Moreover, it showed strong resistance ranging from 60.6% to 93.9% against the other tested antibiotics, and the most interesting observed result was that none of the tested antibiotics showed 100% efficacy against K. pneumoniae; however, only tigecycline showed the highest killing capacity against K. pneumonia (killing around 84.8% of treated strains), which was the same as previously reported.99 The obtained results reflected the threats correlated with K. pneumoniae as a multi-drug resistant bacteria, agreeing with previous reports.100,101 Lastly, P. aeruginosa was shown to have complete resistance against five antibiotics including amoxycillin/clavulanic acid, cefepime, cefotaxime, cefoxitin, and ciprofloxacin. None of the tested antibiotics were effective against P. aeruginosa; however, the lowest recorded resistance obtained was 33.3% against tigecycline.
3.6. Antimicrobial activity of curcumin copper oxide nanoparticles (Cur-CuO NPs)
The antimicrobial activity of the curcumin-capped copper oxide nanoparticles (Cur-CuO NPs) was investigated against clinically isolated pathogens using the disc diffusion assay at three concentrations (i.e., 10 μL–5 μg mL−1; 25 μL–15 μg mL−1; 50 μL–25 μg mL−1, respectively), and the obtained results are presented in Table 7 and Fig. S9–S11 in the ESI.† The results revealed that the three tested concentrations resulted in zones of inhibition against all the investigated pathogenic bacteria together with pathogenic fungi. However, it was noticed that by increasing the concentration of nanoparticles, the observed zone of inhibition was increased, as shown in previously reported studies.4,45,102 The highest observed zone of inhibition was recorded in the treatment using 50 μL (25 μg mL−1) of Cur-CuO against Proteus vulgaris, accounting for 17.3 mm, followed by treatment using the same concentration against one of the most multi-drug resistant (MDR) pathogenic bacteria, Staphylococcus aureus (MRSA), which resulted in the appearance of a 16.6 mm inhibition zone, as previously reported.103 A slightly lower inhibition zone was obtained using 25 μL (15 μg mL−1) against MRSA, recording 14.2 mm. The lowest recorded zone of inhibition was observed against Staphylococcus aureus (MSSA) using 10 μL (5 μg mL−1) and 25 μL (15 μg mL−1) of 8.6 and 11 mm, respectively.| Microorganism | Mean of zone inhibition in mm (mean ± SD) | ||
|---|---|---|---|
| A | B | C | |
| a A: 10 μL–5 μg mL; B: 25 μL–15 μg mL; C: 50 μL–25 μg mL−1 of Cur-CuO NPs. The diameter of the inhibition zone expressed as mean ± SD (experiment conducted in triplicate), P < 0.05 or P < 0.01. | |||
| Staphylococcus aureus (MSSA) | 8.6 ± 0.52 | 11.0 ± 0.67 | 14.5 ± 0.53 |
| Staphylococcus aureus (MRSA) | 10.6 ± 0.58 | 14.2 ± 0.69 | 16.6 ± 0.68 |
| Enterococcus faecalis | 10.2 ± 1.07 | 12.8 ± 0.69 | 15.1 ± 0.69 |
| Streptococcus pyogenes | 10.0 ± 0.0 | 14.5 ± 0.7 | 16.5 ± 0.7 |
| Acinetobacter baumannii | 10.5 ± 0.7 | 13.5 ± 0.85 | 15.4 ± 0.7 |
| Escherichia coli | 9.0 ± 0.75 | 11.3 ± 0.47 | 14.3 ± 0.59 |
| Klebsiella pneumonia | 9.5 ± 0.8 | 12.2 ± 0.82 | 15.0 ± 0.91 |
| Proteus mirabilis | 10.4 ± 0.55 | 13.4 ± 0.55 | 16.0 ± 0.7 |
| Proteus vulgaris | 11.0 ± 0.0 | 14.0 ± 0.0 | 17.3 ± 0.58 |
| Pseudomonas aeruginosa | 10.4 ± 0.91 | 12.3 ± 0.9 | 16.4 ± 0.82 |
| Candia albicans | 10.4 ± 0.53 | 14.0 ± 1.0 | 16.4 ± 0.53 |
The antimicrobial activity of Cur-CuO NPs was investigated by Jayarambabu et al.104 In their study, Cur-CuO NPs were used as suspensions with a concentration of 100, 150, 200 and 250 μL against two bacterial isolates of Basilus subtilis (i.e., Gram-positive bacteria) and Escherichia coli (Gram-negative bacteria). In their study, the used concentrations were higher than the concentrations used in the current study (i.e., 10 μL–5 μg mL−1; B: 25 μL–15 μg mL−1; C: 50 μL–25 μg mL−1 of Cur-CuO NPs). In addition, the antimicrobial activity was tested against a broad range of bacterial isolates, either Gram-positive or Gram-negative bacteria.
Moreover, Cur-CuO NP discs showed efficiency in combating not only pathogenic bacteria but also pathogenic fungi such as Candida albicans, where the inhibition zones were monitored using three tested concentrations of Cur-CuO NPs (10.4, 14.0 and 16.4 mm for 10 μL–5 μg mL−1; 25 μL–15 μg mL−1; and 50 μL–25 μg mL−1, respectively), in agreement with previous studies.105,106 The obtained results give insights into the usability of Cur-CuO NPs against pathogenic bacteria and fungi.107–109
3.7. Antimicrobial activity of chitosan copper oxide nanoparticles (CS-CuO NPs)
The antimicrobial activity of the chitosan-capped copper oxide nanoparticles (CS-CuO NPs) was investigated against clinically isolated pathogens, and data are presented in Table 8 and Fig. S12 and S13 in the ESI.† The results showed a variation in the inhibition zones monitored according to the different concentrations of nanoparticles (i.e., 10 μL–5 μg mL−1; 25 μL–15 μg mL−1; 50 μL–25 μg mL−1) and the tested microorganisms. Generally, the antimicrobial effect was strengthened as the concentration of CS-CuO NPs increased in the same manner observed when investigating the Cur-CuO NPs. The highest monitored zone of inhibition of 24.4 mm was observed using 50 μL (25 μg mL−1) of CS-CuO NPs and against Enterococcus faecalis. In contrast, the lowest observed zone of inhibition of 10.5 mm was observed for the concentration of NPs of 10 μL (5 μg mL−1) against Escherichia coli. Furthermore, CS-CuO NPs were effective in combating the pathogenic fungi Candida albicans, resulting in the formation of inhibition zones (12.3, 16.0 and 20.1 mm) for the three concentrations (i.e., 10 μL–5 μg mL−1; 25 μL–15 μg mL−1; 50 μL–25 μg mL−1), respectively. Overall, the inhibition zones were slightly higher than that obtained by Cur-CuO NPs. Our results are consistent with that reported in previous studies.37,110–112 Furthermore, in the case of chitosan-capped copper oxide nanoparticles (CS-CuO NPs), only two studies have been devoted to investigating their antimicrobial efficacy against bacterial isolates.37,113 Sathiyavimal et al. investigated the antibacterial activity against Gram-positive (S. pneumoniae and S. epidermidis) and Gram-negative (E. coli and P. mirabilis) with different concentrations (20, 40, 60, and 80 μg mL−1), which were higher than the concentrations used in the current study (i.e., 5 μg mL−1, 15 μg mL−1 and 25 μg mL−1 of CS-CuO NPs).37| Microorganism | Mean of zone inhibition in mm (mean ± SD) | ||
|---|---|---|---|
| A | B | C | |
| a A: 10 μL–5 μg mL; B: 25 μL–15 μg mL; C: 50 μL–25 μg mL−1 of CS-CuO NPs. The diameter of the inhibition zone is expressed as mean ± SD (experiment conducted in triplicate), P < 0.05 or P < 0.01. | |||
| Staphylococcus aureus (MSSA) | 14.3 ± 0.48 | 17.1 ± 0.57 | 20.5 ± 1.08 |
| Staphylococcus aureus (MRSA) | 12.8 ± 1.17 | 16.4 ± 0.6 | 19.1 ± 0.94 |
| Enterococcus faecalis | 16.1 ± 0.9 | 20.3 ± 0.76 | 24.4 ± 0.53 |
| Streptococcus pyogenes | 14.0 ± 0.0 | 16.5 ± 0.7 | 19.0 ± 0.0 |
| Acinetobacter baumannii | 11.0 ± 0.82 | 14.2 ± 0.42 | 16.3 ± 0.48 |
| Escherichia coli | 10.5 ± 0.62 | 13.5 ± 0.87 | 16.2 ± 0.56 |
| Klebsiella pneumonia | 11.2 ± 0.85 | 15.2 ± 0.73 | 19.5 ± 0.87 |
| Proteus mirabilis | 11.6 ± 0.55 | 15.2 ± 0.45 | 18.4 ± 0.55 |
| Proteus vulgaris | 12.3 ± 0.7 | 15.0 ± 0.0 | 19.3 ± 0.58 |
| Pseudomonas aeruginosa | 14.4 ± 0.74 | 17.2 ± 0.94 | 21.4 ± 1.06 |
| Candia albicans | 12.3 ± 0.76 | 16.0 ± 0.82 | 20.1 ± 0.69 |
3.8. Plain curcumin and chitosan antimicrobial activity
Antimicrobial activity was investigated to determine whether the antimicrobial effects observed in Table 7 and 8 for Cur-CuO NPs and CS-CuO NPs, respectively, were solely attributed to the copper oxide nanoparticles or the capping materials (curcumin and chitosan), as shown in Table 9 and Fig. S14 and S15 in the ESI,† respectively. The results revealed that plain curcumin did not show any antimicrobial activity against the investigated pathogens, which aligns with the previous report.4 These results proved that the obtained antimicrobial capacity of Cur-CuO NPs presented in Table 7 was driven by the CuO NPs. However, curcumin can act as a helper in facilitating the diffusion of CuO NPs through the microbe cell membrane due to its hydrophobicity, as previously reported.114,115 Thus, using plain chitosan resulted in the appearance of inhibition zones for all the investigated pathogenic bacteria and fungi.| Microorganism | Mean of zone inhibition in mm (mean ± SD) | |
|---|---|---|
| A | B | |
| a 0 is no inhibition. A: plain curcumin and B: plain chitosan. The diameter of the inhibition zone is expressed as mean ± SD (experiment conducted in triplicate), P < 0.05 or P < 0.01. | ||
| Staphylococcus aureus (MSSA) | 0.0 ± 0.0 | 14.2 ± 0.63 |
| Staphylococcus aureus (MRSA) | 0.0 ± 0.0 | 15.0 ± 0.52 |
| Enterococcus faecalis | 0.0 ± 0.0 | 15.1 ± 0.38 |
| Streptococcus pyogenes | 0.0 ± 0.0 | 15.0 ± 0.0 |
| Acinetobacter baumannii | 0.0 ± 0.0 | 12.4 ± 0.84 |
| Escherichia coli | 0.0 ± 0.0 | 11.2 ± 0.9 |
| Klebsiella pneumonia | 0.0 ± 0.0 | 15.2 ± 0.84 |
| Proteus mirabilis | 0.0 ± 0.0 | 12.0 ± 0.7 |
| Proteus vulgaris | 0.0 ± 0.0 | 10.3 ± 0.58 |
| Pseudomonas aeruginosa | 0.0 ± 0.0 | 14.3 ± 0.72 |
| Candia albicans | 0.0 ± 0.0 | 12.3 ± 0.52 |
In contrast, the highest inhibition zone of 15.2 mm was observed against Klebsiella pneumonia, whereas the lowest inhibition zone of 10.3 mm was observed against Proteus vulgaris. The antimicrobial capacity of chitosan could result from its polycationic structure, thus electrostatically interacting with the anionic components of the microorganisms,116 in addition to its hydrophobic and chelating capacities.117
The antimicrobial mechanisms of the curcumin-capped copper oxide (Cur-CuO) nanoparticles and chitosan-capped copper oxide (CS-CuO) nanoparticles operate through distinct and complementary pathways, as follows.
Cur-CuO NPs exert their effect through their antimicrobial property by the generation of reactive oxygen species (ROS) primarily, damage to the membrane, inhibition of DNA replication, and inhibition of the efflux pump mechanism of bacteria. The as-generated ROS, such as hydroxyl radicals (OH˙), superoxide anions  , and hydrogen peroxide (H2O2), induce oxidative stress, which leads to lipid peroxidation, protein oxidation, and DNA damage.118 Additionally, membrane permeation is heightened by curcumin due to its polyphenolic nature, which alters the permeability of bacterial membranes and leads to intracellular leakage.119 Cur-CuO NPs bind with nucleic acid and transcriptional enzymes within the bacterial cells and hinder DNA replication, suppressing protein synthesis together with bacterial growth.120 Furthermore, curcumin was found to suppress bacterial efflux pumps, which are major contributors to antibiotic resistance, thus adding to the general antimicrobial effect of Cur-CuO NPs, especially against multidrug-resistant (MDR) strains.121
, and hydrogen peroxide (H2O2), induce oxidative stress, which leads to lipid peroxidation, protein oxidation, and DNA damage.118 Additionally, membrane permeation is heightened by curcumin due to its polyphenolic nature, which alters the permeability of bacterial membranes and leads to intracellular leakage.119 Cur-CuO NPs bind with nucleic acid and transcriptional enzymes within the bacterial cells and hinder DNA replication, suppressing protein synthesis together with bacterial growth.120 Furthermore, curcumin was found to suppress bacterial efflux pumps, which are major contributors to antibiotic resistance, thus adding to the general antimicrobial effect of Cur-CuO NPs, especially against multidrug-resistant (MDR) strains.121
CS-CuO NPs utilize electrostatic interactions, ROS-stimulated oxidative damage, and induce metabolic impairment to exert their antimicrobial activity. The effectiveness of the nanocomposite is due to the following processes: firstly, the electrostatic interaction between the positively charged amino (–NH3+) functional groups in chitosan and the negatively charged bacterial cell membrane lead to the destabilization of the cell membrane, thereby allowing increased permeability and leakage of the vital intracellular components.122,123 Similar to Cur-CuO NPs, CS-CuO NPs also yield ROS, which in turn causes oxidative damage to bacterial lipids, proteins, and DNA, thus leading to cell dysfunction and death. Contiguously, CS-CuO NPs also interact with bacterial nutrient uptake and the metabolic procedures that lead to ion exchange and energy generation disorders, the occurrence of which negatively affects the bacterial cell life cycle.34,124 The entry of chitosan in bacterial cells further inhibits the DNA replication and transcription process, and therefore no proteins are produced, preventing their growth. The combinatorial effect of the mechanisms makes CS-CuO NPs highly effective against various bacterial pathogens.125,126
Both Cur-CuO and CS-CuO NPs exhibit strong antimicrobial effects due to their unique but partly similar modes of action. Cur-CuO NPs make use of the bioactive properties of curcumin to inhibit efflux pumps and bring about oxidative stress, while CS-CuO NPs work mainly by the destabilization of the electrostatic membrane and metabolic disruption.
3.9. Antimicrobial mechanism based on generation of ROS in bacterial cell
Fig. 11 and Table 10 present the effects of Cur-capped and CS-capped CuO nanoparticles exposure to S. aureus and K. pneumoniae as models for Gram-positive and Gram-negative bacterial isolates, respectively, compared to the control. The CS-capped CuO NPs produced lower levels of ROS in bacterial cells than Cur-capped CuO NPs. Also, S. aureus Gram-positive bacterial isolates were more susceptible to antibacterial based on the induction of ROS than K. pneumoniae Gram-negative bacterial isolates using both Cur-capped and CS-capped CuO NPs, which may be due to the difference in the cell wall structure of these two classes and agrees with earlier reports.127,128 Reactive oxygen species (ROS)-induced oxidative stress plays a critical role in the antibacterial action of copper. ROS are oxygen-containing derivatives that are made up of highly unstable oxygen radicals such as superoxide , hydroxyl (OH˙), hydrogen peroxide (H2O2), and singlet oxygen (O2). The atomic or molecular orbitals of ROS contain one or more unpaired electrons, making them very reactive. In this regard, Cu2+ ions can cause oxidative damage to the unsaturated fatty acids of the phospholipids in the bacterial cell membrane by producing extracellular ROS, whereas OH can drive the non-enzymatic peroxidation of unsaturated double bonds of fatty acids, triggering a series of reactions and causing extensive changes in the structure of the phospholipid bilayer. This destroys the biophysical properties of the membrane, eventually leading to membrane loss.128,129
, hydroxyl (OH˙), hydrogen peroxide (H2O2), and singlet oxygen (O2). The atomic or molecular orbitals of ROS contain one or more unpaired electrons, making them very reactive. In this regard, Cu2+ ions can cause oxidative damage to the unsaturated fatty acids of the phospholipids in the bacterial cell membrane by producing extracellular ROS, whereas OH can drive the non-enzymatic peroxidation of unsaturated double bonds of fatty acids, triggering a series of reactions and causing extensive changes in the structure of the phospholipid bilayer. This destroys the biophysical properties of the membrane, eventually leading to membrane loss.128,129
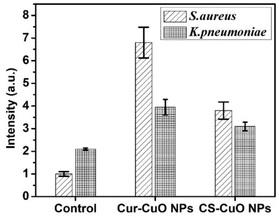 | ||
| Fig. 11 Antibacterial activity mechanism of Cur-CuO NPs and CS-CuO NPs against S. aureus (Gram-positive) and K. pneumoniae (Gram-negative). | ||
| Bacterial species | Control | Cur-CuO NPs | Cs–CuO NPs |
|---|---|---|---|
| +ve Gram bacteria | |||
| S. aureus | 1.0 ± 0.0 | 6.8 ± 0.66 | 3.95 ± 0.6 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| −ve Gram bacteria | |||
| K. pneumoniae | 2.1 ± 0.0 | 3.8 ± 0.55 | 3.1 ± 0.55 |
3.10. Determination of minimum inhibitory concentration (MIC)
The minimum inhibitory concentration (MIC) evaluation for the Cur-CuO NPs and CS-CuO NPs was carried out as shown in Table 11. Results revealed that all the MIC values observed in the case of CS-CuO NPs were lower than those surveyed in Cur-CuO NPs among the investigated microorganisms, where the lowest MIC recorded for Cur-CuO NPs was 14.5 μg mL−1 against Enterococcus faecalis, while it was 3.9 μg mL−1 for CS-CuO NPs against Streptococcus pyogenes and Proteus vulgaris. The MIC of Cur-CuO was 31.2 μg mL−1 for Staphylococcus aureus (MSSA) and Escherichia coli, whereas it was in the range of 14.5 to 15.6 μg mL−1 against the other investigated bacteria and fungi.130 Alternatively, the MIC of CS-CuO was 4.3 μg mL−1 for both Staphylococcus aureus (MSSA) and (MRSA) and 4.5, 5.1, 7 and 8.6 μg mL−1 for Enterococcus faecalis, Klebsiella pneumonia, Proteus mirabilis, and Acinetobacter baumannii, respectively.| Microorganism | Minimum inhibition concentration (μg mL−1) | |
|---|---|---|
| Cur-CuO NPs | CS-CuO NPs | |
| Staphylococcus aureus (MSSA) | 31.2 ± 0.0 | 4.3 ± 1.2 |
| Staphylococcus aureus (MRSA) | 15.2 ± 1.8 | 4.3 ± 1.4 |
| Enterococcus faecalis | 14.5 ± 2.9 | 4.5 ± 1.5 |
| Streptococcus pyogenes | 15.6 ± 0.0 | 3.9 ± 0.0 |
| Acinetobacter baumannii | 14.8 ± 2.4 | 8.6 ± 2.4 |
| Escherichia coli | 31.2 ± 0.0 | 15.6 ± 0.0 |
| Klebsiella pneumonia | 15.1 ± 1.9 | 5.1 ± 1.8 |
| Proteus mirabilis | 15.6 ± 0.0 | 7.0 ± 1.7 |
| Proteus vulgaris | 15.6 ± 0.0 | 3.9 ± 0.0 |
| Pseudomonas aeruginosa | 14.6 ± 2.7 | 5.2 ± 1.9 |
| Candia albicans | 15.6 ± 0.0 | 5.0 ± 1.9 |
Our findings demonstrated that the MIC of CS-CuO NPs against Acinetobacter baumannii was 8.6 μg mL−1, which is lower than that reported by Sarfraz and co-workers,43 stating that the MIC value for CS-CuO was 62.5 μg mL−1. In addition, the MIC (i.e., 14 to 31 μg mL−1) in the current study is lower than the MIC obtained by Sathiyavimal et al. (i.e., 25 to 100 μg mL−1).37 Also, Jayaramudu et al. explored the antibacterial activity against Gram-positive and Gram-negative bacterial isolates with a concertation of 1 mg mL−1, which is also higher than that used in the current study.113
3.11. Biocompatibility based on MTT assay
In the biocompatibility assays, WI38 cells, as an in vitro test model of a normal cell line, were exposed to both Cur-CuO and CS-CuO NPs, respectively, to determine their (see Fig. 12 and Table 12). In this study, various concentrations of 1.56, 3.125, 6.25, 12.5, 25, 50 and 100 μg mL−1 of Cur-CuO NPs, CS-CuO NPs, plain curcumin, plain chitosan, and DOX respectively, were subjected to MTT screening colorimetric assay at a 48 h interval, as shown in Fig. 12. Our results, as presented in Table 12, showed that Cur-CuO NPs have an IC50 dose of 74.17 μg mL−1, which is higher than that of CS-CuO NPs with IC50 of 41.01 μg mL−1. In addition, Cur-CuO NPs were found to be safer than both plain chitosan (IC50 of ∼65.8 μg mL−1) and DOX (IC50 of ∼6.72 μg mL−1). Moreover, the lowest cytotoxic effect was observed for plain curcumin with an IC50 of 92.17 μg mL−1 (see Table 12). In the case of Cur-CuO NPs, the highest dose could kill about 55% of cells, whereas when applied at low concentrations of 1.56, 3.125 and 6.25 μg mL−1, they only killed 0 and 1.8%, respectively, of cells after 48 h of cell exposure (Fig. 12). Treatment of cells with the highest concentration of 100 μg mL−1 CS-CuO NPs killed about 68% of the cells, whereas treatment of cells with low concentrations of 1.56, 3.125 and 6.25 μg mL−1 killed about 0 and 6.6% of the cells after 48 h of exposure (Fig. 12). | ||
| Fig. 12 Biocompatibility assay of Cur-CuO NPs, CS-CuO NPs, plain curcumin, plain chitosan and DOX at different levels of exposure doses. | ||
| No. | Sample | Lethal cytotoxic dose (IC50, μg mL−1) |
|---|---|---|
| a IC50 (ppm): 1–10 (very strong). 11–20 (strong). 21–50 (moderate). 51–100 (weak) and above 100 (non-cytotoxic). b DOX: doxorubicin data were measured after 48 h. Means a significant difference (P < 0.05) compared to the control. Mean ± SD (experiment conducted in triplicate). | ||
| Cont | DOX | 6.72 ± 0.5 |
| 1 | Cur-CuO NPs | 74.17 ± 3.7 |
| 2 | CS-CuO NPs | 41.01 ± 2.2 |
| 3 | Plain curcumin | 92.17 ± 4.2 |
| 4 | Plain chitosan | 65.80 ± 3.1 |
3.11. Comparison with other antimicrobial agents and traditional antibiotics
In this work, the effectiveness of Cur-CuO and CS-CuO nanoparticles was compared to our previous studies on the Cur-Ag and Cur-ZnO materials and traditional drugs such as ciprofloxacin, amoxicillin, gentamicin, and vancomycin.4 The outcomes revealed that Cur-Ag NPs possess the maximum antibacterial function, which are superior to the traditional antibiotics, particularly MRSA, Pseudomonas aeruginosa, and Candida albicans. Furthermore, CS-CuO NPs were found to demonstrate significant antibacterial activity and the results showed that they were as effective as gentamicin and vancomycin making them a suitable alternative. Although the traditional antibiotics, especially ciprofloxacin, have lower minimum inhibitory concentration (MIC) values, and thus they are proven to be more potent, the nanoparticles still remained quite efficient, especially against the drug-resistant bacterial strains. In comparison with other nanoparticles, Cur-Ag NPs had the largest zones of inhibition and lowest MIC values at every stage, whereas CS-CuO NPs were almost compatible showing their antimicrobial property, as shown in Table 13.| Microorganism | Cur-CuO NPs | CS-CuO NPs | Cur-Ag NPs | Cur-ZnO NPs | Ciprofloxacin | Amoxicillin | Gentamicin | Vancomycin |
|---|---|---|---|---|---|---|---|---|
| a The values are represented as zone of inhibition (mm)/MIC (μg mL−1). | ||||||||
| Staphylococcus aureus (MSSA) | 14.5/5.0 | 20.5/3.2 | 21.2/2.5 | 18.4/3.8 | 24.1/1.2 | 15.6/8.0 | 20.3/2.0 | 18.2/3.5 |
| Staphylococcus aureus (MRSA) | 16.6/4.2 | 19.1/3.0 | 22.5/2.1 | 19.2/3.5 | 21.3/1.5 | 12.4/9.3 | 18.6/2.3 | 16.8/4.0 |
| Enterococcus faecalis | 15.1/6.0 | 24.4/3.5 | 20.8/3.0 | 18.9/4.0 | 23.0/2.0 | 14.2/10.5 | 19.0/3.5 | 21.6/3.8 |
| Streptococcus pyogenes | 16.5/5.8 | 19.0/3.3 | 21.3/2.8 | 19.0/3.9 | 25.3/1.8 | 15.8/7.8 | 20.5/2.8 | 18.7/3.6 |
| Acinetobacter baumannii | 15.4/6.5 | 16.3/4.0 | 18.9/3.5 | 16.2/4.2 | 20.4/2.3 | 11.7/12.0 | 16.5/4.0 | 14.3/4.8 |
| Escherichia coli | 14.3/6.8 | 16.2/4.2 | 20.0/3.2 | 17.4/4.5 | 23.5/1.7 | 13.5/11.2 | 19.8/3.3 | 15.9/4.6 |
| Klebsiella pneumoniae | 15.0/6.3 | 19.5/4.0 | 20.6/3.3 | 18.1/4.4 | 22.1/2.0 | 14.0/10.8 | 18.9/3.4 | 16.5/4.2 |
| Proteus mirabilis | 16.0/6.1 | 18.4/3.8 | 19.5/3.4 | 17.5/4.2 | 21.7/2.1 | 12.9/9.7 | 17.6/3.6 | 15.8/4.0 |
| Proteus vulgaris | 17.3/5.9 | 19.3/3.6 | 21.8/2.9 | 19.0/3.8 | 22.5/1.9 | 14.8/8.9 | 19.2/3.1 | 16.4/3.7 |
| Pseudomonas aeruginosa | 16.4/6.7 | 21.4/4.5 | 22.1/3.8 | 18.7/4.6 | 23.0/2.5 | 13.9/10.2 | 20.1/3.7 | 16.9/4.5 |
| Candida albicans | 16.4/6.4 | 20.1/4.1 | 22.0/3.0 | 19.5/4.3 | 21.0/2.0 | 12.7/11.0 | 18.5/3.5 | 14.7/5.0 |
In contrast, Cur-CuO and CS-CuO NPs showed higher biocompatibility and safety than the Cur-Ag NPs and Cur-ZnO NPs mentioned in our previous study.4 Cur-CuO NPs and CS-CuO NPs have an IC50 dose of 74 and 41 μg mL−1, respectively, which is higher than the recorded IC50 of Cur-Ag and Cur-ZnO NPs of 30 μg mL−1 mentioned in our previous work.4
3.12. Potential applications in healthcare and antimicrobial strategies
These nanoparticles can be combined with another drug to make new antimicrobial compositions. In a treatment plan implementation, glass substrates and glasses are effective in the prevention of biofilm formation and device-associated infections to be used as coatings of medical devices, such as catheters, implants, and ventilators. In addition, they can be considered good alternatives to some hospital equipment such as filtration membranes, tubing, prosthesis, artificial lung devices, resistivity measurement, manufacturing processes, and air disinfection systems.Cur-CuO and CS-CuO NPs have been proven as promising materials that have the potential to replace the traditional antibiotics of the past. This is because of their broad-spectrum antimicrobial activity, which targets the whole spectrum of microorganisms. Furthermore, although the biocompatible chitosan coating ensures that the development of resistance is prevented, it is also beneficial. Thus, if the most efficient and effective building-related technologies are implemented, considerable energy reduction can be achieved.
Cur-CuO and CS-CuO nanoparticles are microorganisms that have broad variability to different drugs, which range from those which can kill microorganisms to other drugs with an inhibitory effect on growth. The structural and physical properties of these CuO nanomaterials include hardness, brittleness, and uniaxial orientation of their crystals.
Their main function is oxidative stress induction and bacterial membrane disruption, which make it hard for pathogens to develop resistance, thus overcoming this big issue when trying to fight infections. Another potential use is in inhalable aerosol formulations for respiratory infections, especially in ICUs where there are many cases of pneumonia that are resistant to antibiotics, and thus a very concerning development.
4. Conclusion
In the present study, curcumin- and chitosan-capped copper oxide nanoparticles (Cur-CuO and CS-CuO NPs) were successfully green synthesized in the presence of an ethanolic extract of Curcuma longa Linns (synonym; Curcuma domestica Valeton, Zingiberaceae) and 1% aqueous solution of acetic acid, respectively. The average particle sizes with spherical shape were 25 ± 10 (Cur-CuO NPs) and 10 ± 5 nm (CS-CuO NPs). In addition, their antimicrobial activity was investigated against several multi-drug resistance (MDR) bacterial and fungal isolates. The tested microorganisms were clinically isolated from patients suffering from different infections. Identification and frequency determination for these microorganisms revealed Klebsiella pneumoniae as the most frequent (25.8% of total isolated strains), followed by Staphylococcus aureus (MRSA) (14.8% of total isolated strains). Streptococcus pyogenes had the lowest representation, with only 1.6% of the strains. Candida albicans was the only recorded fungi, comprising 5.5% of the isolated strains. In addition, the isolated microorganisms showed complete resistance to about 14 antibiotics from 21 tested antibiotics. Cur-CuO and CS-CuO NPs were effective against all the clinically isolated microorganisms, even when using the lowest investigated concentration. In addition, the results for both Cur-CuO NPs and CS-CuO NPs revealed that overall, the minimum inhibitory concentration was lower for the chitosan-capped copper oxide nanoparticles compared with the curcumin-capped copper oxide nanoparticles, with values of 3.9 μg mL−1 to 15.6 μg mL−1 for CS-CuO NPs, while 14.5 to 31.2 μg mL−1 for Cur-CuO NPs. The antimicrobial activity for plain curcumin and plain chitosan was investigated, reflecting that plain curcumin did not show any antimicrobial activity; conversely, plain chitosan showed strong antimicrobial activity, giving zones of inhibition ranging in size from 10.3 to 15.2 mm against Proteus vulgaris and Klebsiella pneumonia, respectively. However, after capping CuO with curcumin, the complex has strong antimicrobial activity, which could be due to the binding of the curcumin to the bacterial or fungal cell membrane, thus facilitating the penetration of CuO into the interior of the cells, leading to the formation of ROS and DNA damage.131,132 The remarkable efficacy of the curcumin- and chitosan-capped copper oxide nanoparticles in effectively combating highly resistant multi-drug pathogens highlights their promising potential for future applications. Finally, the biocompatibility assay showed that Cur-CuO NPs have an IC50 dose of 74.17 μg mL−1, which is higher than that of CS-CuO NPs with an IC50 of 41.01 μg mL−1. In addition, Cur-CuO NPs exhibited higher safety than both plain chitosan (IC50 of ∼65.8 μg mL−1) and DOX (IC50 of ∼6.72 μg mL−1). The obtained results show that Cur-CuO NPs are safer to use as an antimicrobial agent compared to CS-CuO NPs. However, both Cur-CuO and CS-CuO NPs could be safely used at low concentrations of 6.25 to 12.5 μg mL−1. Our findings highlight the applicability of using Cur-CuO NPs and CS-CuO NPs and how the capping procedure could affect antimicrobial activity.Ethical approval
This study was performed in strict accordance with the GOTHI guidelines ethics regulations issued by the Minister of Health & Population Cairo, Egypt: no. 238/2003, Articles 52-6121, approved by the medical research ethics committee. In addition, the approval for performing this study has been registered under No IME 00069 in 2022.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
Noura El-Kattan; conceptualization, performed all the microbiological studies and contributed to manuscript writing. Mostafa Ahmed Ibrahim and Ahmed Sadek Mansour; synthesis of nanomaterials, performed TEM imaging and their analysis & discussion. Ahmed Nabile Emam (Corresponding author) performed characterization experiments, including UV-vis optical absorption spectroscopy, FTIR, dynamic light scattering (DLS), EDX elemental analysis, XRD and zeta potential. In addition, data analysis, writing/formulating, revision, and submission of the manuscript. Khaled Metwally: contributed to cytotoxicity assay, the discussion on the microbiological part and in writing of the manuscript. Fady Sayed Youssef: performed the gas chromatography-mass spectrometry (GC-MS) on the ethanolic extract of Curcuma longa Linn (turmeric) powder vs. the pure curcumin. Nourelhuda Ahmed; contributed to cytotoxicity assay. Documentation of patient's data and collection of the clinical samples, including blood, sputum, wound, and urine, from patients admitted to the intensive care unit (ICU) in El-Sahel Teaching Hospital, the general organization for teaching hospitals and institutes (GOTHI), Cairo, Egypt.Conflicts of interest
The authors have no relevant financial or non-financial interests to disclose.Acknowledgements
The authors are grateful to Prof. Ashraf Bakry Abd El-Razik and NanoFab Technology for funding the Chemicals used in the fabrication of nanomaterials. In addition, funding the TEM imaging investigation that was included and used in this study.Notes and references
- E. L. Tsalik, Y. Li, L. L. Hudson, V. H. Chu, T. Himmel, A. T. Limkakeng, J. N. Katz, S. W. Glickman, M. T. McClain and K. E. Welty-Wolf, Ann. Am. Thorac. Soc., 2016, 13, 401–413 Search PubMed.
- T. Gottlieb and G. R. Nimmo, Med. J. Aust., 2011, 194, 281–283 Search PubMed.
- M. S. Usman, M. E. E. Zowalaty, K. Shameli, N. Zainuddin, M. Salama and N. A. Ibrahim, Int. J. Nanomed., 2013, 4467–4479 Search PubMed.
- N. El-Kattan, A. N. Emam, A. S. Mansour, M. A. Ibrahim, A. B. Abd El-Razik, K. A. Allam, N. Y. Riad and S. A. Ibrahim, RSC Adv., 2022, 12, 18022–18038 Search PubMed.
- A. Rahman, A. Ismail, D. Jumbianti, S. Magdalena and H. Sudrajat, Indones. J. Chem., 2009, 9, 355–360 CrossRef.
- M. Ahamed, H. A. Alhadlaq, M. M. Khan, P. Karuppiah and N. A. Al-Dhabi, J. Nanomater., 2014, 2014, 17 CrossRef.
- K. Gopalakrishnan, C. Ramesh, V. Ragunathan and M. Thamilselvan, Digest J. Nanomater. Biostruct., 2012, 7, 833–839 Search PubMed.
- S. Jagadeeshan and R. Parsanathan, Advanced Nanostructured Materials for Environmental Remediation, 2019, pp. 59–90 Search PubMed.
- V. Stanić and S. B. Tanasković, in Nanotoxicity, Elsevier, 2020, pp. 241–274 Search PubMed.
- L. Bruslind, Microbiology, Open Oregon Stat, Corvallis, OR, USA, 2017 Search PubMed.
- E. Sánchez-López, D. Gomes, G. Esteruelas, L. Bonilla, A. L. Lopez-Machado, R. Galindo, A. Cano, M. Espina, M. Ettcheto and A. Camins, Nanomaterials, 2020, 10, 292 Search PubMed.
- M. C. Stensberg, Q. Wei, E. S. McLamore, D. M. Porterfield, A. Wei and M. S. Sepúlveda, Nanomedicine, 2011, 6, 879–898 CrossRef PubMed.
- S. Aruna, N. Vasugi Raaja and S. Sathiesh Kumar, Int. J. Innov. Res. Sci. Eng. Technol., 2016, 5, 2112–2119 Search PubMed.
- I. Subhankari and P. Nayak, World J. Nanosci. Nanotechnol., 2013, 2, 10–13 Search PubMed.
- A. M. Eid, A. Fouda, S. E.-D. Hassan, M. F. Hamza, N. K. Alharbi, A. Elkelish, A. Alharthi and W. M. Salem, Catalysts, 2023, 13, 348 CrossRef.
- U. T. Khatoon, A. Velidandi and G. N. Rao, Inorg. Chem. Commun., 2023, 110372 Search PubMed.
- G. Ungur and J. Hruza, Fibers Polym., 2015, 16, 621–628 CrossRef CAS.
- S. P. Selvaraj, Mater. Today: Proc., 2022, 50, 2865–2868 CAS.
- N. Y. Lee, W. C. Ko and P. R. Hsueh, Front. Pharmacol, 2019, 10, 1153 CrossRef CAS PubMed.
- O. O. Adeniji, N. Nontongana, J. C. Okoh and A. I. Okoh, Int. J. Mol. Sci., 2022, 23, 15038 CrossRef CAS PubMed.
- G. Waktole and B. Chala, J. Biomater. Nanobiotechnol., 2023, 14, 1–22 CrossRef CAS.
- A. Goel, A. B. Kunnumakkara and B. B. Aggarwal, Biochem. Pharmacol., 2008, 75, 787–809 CrossRef PubMed.
- S. J. Hewlings and D. S. Kalman, Foods, 2017, 6, 92 Search PubMed.
- M. Karandish, H. Mozaffari-Khosravi, S. M. Mohammadi, M. Azhdari and B. Cheraghian, Trials, 2020, 21, 1–11 CrossRef PubMed.
- I. Brouet and H. Ohshima, Biochem. Biophys. Res. Commun., 1995, 206, 533–540 CrossRef PubMed.
- S. Cikrikci, E. Mozioglu and H. Yilmaz, Rec. Nat. Prod., 2008, 2, 19 Search PubMed.
- P. Anand, A. B. Kunnumakkara, R. A. Newman and B. B. Aggarwal, Mol. Pharmaceutics, 2007, 4, 807–818 CrossRef PubMed.
- H. Elabd, H. H. Mahboub, S. M. R. Salem, A. M. Abdelwahab, K. M. Alwutayd, M. Shaalan, S. H. Ismail, A. M. Abdelfattah, A. Khalid, A. T. Mansour, H. S. Hamed and H. Youssuf, Fishes, 2023, 8, 333 Search PubMed.
- J. Sharifi-Rad, Y. El Rayess, A. Abi Rizk, C. Sadaka, R. Zgheib, W. Zam, S. Sestito, S. Rapposelli, K. Neffe-Skocińska and D. Zielińska, Front. Pharmacol., 2020, 11(2020), 550909 Search PubMed.
- M. Kumari and D. K. Nanda, Burns, 2022, 49(5), 1003–1016 CrossRef PubMed.
- D. Zheng, C. Huang, H. Huang, Y. Zhao, M. R. U. Khan, H. Zhao and L. Huang, Chem. Biodiversity, 2020, 17, e2000171 CrossRef PubMed.
- A. Guarnieri, M. Triunfo, C. Scieuzo, D. Ianniciello, E. Tafi, T. Hahn, S. Zibek, R. Salvia, A. De Bonis and P. Falabella, Sci. Rep., 2022, 12, 8084 CrossRef PubMed.
- S. K. Shukla, A. K. Mishra, O. A. Arotiba and B. B. Mamba, Int. J. Biol. Macromol., 2013, 59, 46–58 CrossRef CAS PubMed.
- D. Yan, Y. Li, Y. Liu, N. Li, X. Zhang and C. Yan, Molecules, 2021, 26, 7136 CrossRef CAS PubMed.
- C.-L. Ke, F.-S. Deng, C.-Y. Chuang and C.-H. Lin, Polymers, 2021, 13, 904 CrossRef CAS PubMed.
- D. Bharathi, R. Ranjithkumar, B. Chandarshekar and V. Bhuvaneshwari, Int. J. Biol. Macromol., 2019, 141, 476–483 CrossRef CAS PubMed.
- S. Sathiyavimal, S. Vasantharaj, T. Kaliannan and A. Pugazhendhi, Carbohydr. Polym., 2020, 241, 116243 CrossRef CAS PubMed.
- P. S. Umoren, D. Kavaz, A. Nzila, S. S. Sankaran and S. A. Umoren, Polymers, 2022, 14, 1832 CrossRef CAS.
- S. B. Ahmed, H. I. Mohamed, A. M. Al-Subaie, A. I. Al-Ohali and N. M. Mahmoud, Sci. Rep., 2021, 11, 9540 CrossRef CAS PubMed.
- A. R. Maheo, Results in Surf. and Interf., 2022, 6, 100048 Search PubMed.
- S. Sathiyavimal, S. Vasantharaj, T. Kaliannan, H. A. Garalleh, M. Garaleh, K. Brindhadevi, N. T. L. Chi, A. Sharma and A. Pugazhendhi, Environ. Res., 2023, 218, 114986 CrossRef PubMed.
- M. H. Sarfraz, S. Muzammil, S. Hayat, M. Khurshid and A. H. Sayyid, Int. J. Biol. Macromol., 2023, 242, 124954 CrossRef PubMed.
- M. H. Sarfraz, M. Zubair, B. Aslam, A. Ashraf, M. H. Siddique, S. Hayat, J. N. Cruz, S. Muzammil, M. Khurshid and M. F. Sarfraz, Front. Microbiol., 2023, 14, 1188743 CrossRef PubMed.
- Y. Haldorai and J.-J. Shim, Int. J. Photoenergy, 2013, 2013(1), 245646 Search PubMed.
- K. Varaprasad, M. López, D. Núñez, T. Jayaramudu, E. R. Sadiku, C. Karthikeyan and P. Oyarzúnc, J. Mol. Liq., 2020, 300, 112353 CrossRef.
- S. Faisal, N. S. Al-Radadi, H. Jan Abdullah, S. A. Shah, S. Shah, M. Rizwan, Z. Afsheen, Z. Hussain, M. N. Uddin, M. Idrees and N. Bibi, Coatings, 2021, 11, 849 Search PubMed.
- N. Rabiee, M. Bagherzadeh, M. Kiani, A. M. Ghadiri, F. Etessamifar, A. H. Jaberizadeh and A. Shakeri, Int. J. Nanomed., 2020, 15, 3983–3999 CrossRef PubMed.
- A. M. Beyene, M. Moniruzzaman, A. Karthikeyan and T. Min, Nanomaterials, 2021, 11(2), 460 Search PubMed.
- A. R. Maheo, B. S. M. Vithiya, T. A. A. Prasad, P. Tamizhdurai and V. Mangesh, Arabian J. Chem., 2022, 15, 103661 Search PubMed.
- W. WC, Koneman's Color Atlas and Textbook of Diagnostic Microbiology, 2006, pp. 67–110 Search PubMed.
- B. A. Forbes, D. F. Sahm and A. S. Weissfeld, Bailey & Scott's Diagnostic Microbiology, Mosby, St. Louis, 12th edn, 2007, pp. 187–214 Search PubMed.
- P. A. Wayne, Clinical and laboratory standards institute: performance standards for antimicrobial susceptibility testing: informational supplement, M100, Clinical and Laboratory Standards Institute (CLSI), 2018 Search PubMed.
- A.-P. Magiorakos, A. Srinivasan, R. B. Carey, Y. Carmeli, M. Falagas, C. Giske, S. Harbarth, J. Hindler, G. Kahlmeter and B. Olsson-Liljequist, Clin. Microbiol. Infect., 2012, 18, 268–281 CrossRef PubMed.
- F. R. Cockerill, M. Wikler, K. Bush, M. Dudley, G. Eliopoulos and D. Hardy, Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Second Informational Supplement, 2012 Search PubMed.
- C. Negrei, A. Hudita, O. Ginghina, B. Galateanu, S. N. Voicu, M. Stan, M. Costache, C. Fenga, N. Drakoulis and A. M. Tsatsakis, Front. Pharmacol., 2016, 7, 172 Search PubMed.
- N. J. Schmidt, Enteroviruses and Reoviruses, 1995, pp. 513–579 Search PubMed.
- A. Emam, S. Loutfy, A. Mostafa, H. Awad and M. Mohamed, RSC Adv., 2017, 7(38), 23502–23514 CAS.
- T. Mosmann, J. Immunol. Methods, 1983, 65, 55–63 CAS.
- F. Denizot and R. Lang, J. Immunol. Methods, 1986, 89, 271–277 CAS.
- F. Zsila, Z. Bikádi and M. Simonyi, Org. Biomol. Chem., 2004, 2, 2902–2910 CAS.
- C. F. Chignell, P. Bilskj, K. J. Reszka, A. G. Motten, R. H. Sik and T. A. Dahl, Photochem. Photobiol., 1994, 59, 295–302 CAS.
- G. Stati, F. Rossi, T. Trakoolwilaiwan, L. D. Tung, S. Mourdikoudis, N. T. K. Thanh and R. Di Pietro, Molecules, 2022, 27, 282 CAS.
- D. Setyaningsih, Y. B. Murti, A. Fudholi, W. L. Hinrichs, R. Mudjahid, S. Martono and T. Hertiani, Indones. J. Pharm. Sci., 2017, 14, 147–157 Search PubMed.
- S. Kamble and R. Dahake Pavan, Int. J. Contemp. Res. Multidiscipl., 2015, 3, 90–96 Search PubMed.
- I. I. Anekwe, C. I. Chikwendu, E. S. Amadi, N. U. Nwogwugwu and F. C. Ihenetu, Int. J. Biol. Chem. Sci., 2023, 17, 1199–1207 Search PubMed.
- N. K. M. Altir, A. M. A. Ali, A.-R. Z. Gaafar, A. A. Qahtan, E. M. Abdel-Salam, A. Alshameri, M. S. Hodhod and B. Almunqedhi, Open Chem., 2021, 19, 945–952 CAS.
- J. O. Momoh, A. A. Manuwa and Y. O. Bankole, J. Adv. Microbiol., 2022, 22, 116–131 Search PubMed.
- Y. Zheng, C. Pan, Z. Zhang, W. Luo, X. Liang, Y. Shi, L. Liang, X. Zheng, L. Zhang and Z. Du, Microchem. J., 2020, 154, 104608 CAS.
- A. F. Guimarães, A. C. A. Vinhas, A. F. Gomes, L. H. Souza and P. B. Krepsky, Quim. Nova, 2020, 43, 909–913 Search PubMed.
- G. B. Avanço, F. D. Ferreira, N. S. Bomfim, R. M. Peralta, T. Brugnari, C. A. Mallmann, B. A. de Abreu Filho, J. M. G. Mikcha and M. Machinski Jr, Food Control, 2017, 73, 806–813 Search PubMed.
- N. Motohashi, C. Yamagami, H. Tokuda, T. Konoshima, Y. Okuda, M. Okuda, T. Mukainaka, H. Nishino and Y. Saito, Cancer Lett., 1998, 134, 37–42 CAS.
- A. Gupta, S. Mahajan and R. Sharma, Biotechnol. Rep., 2015, 6, 51–55 Search PubMed.
- I. I. Anekwe, C. I. Chikwendu, E. S. Amadi, N. U. Nwogwugwu and F. C. Ihenetu, Int. J. Biol. Chem. Sci., 2023, 17(3), 1199–1207 Search PubMed.
- B. H. Kebede, S. F. Forsido, Y. B. Tola and T. Astatkie, Heliyon, 2021, 7(2), e06239 CAS.
- H. Van Nong, L. X. Hung, P. N. Thang, V. D. Chinh, L. V. Vu, P. T. Dung, T. Van Trung and P. T. Nga, SpringerPlus, 2016, 5, 1147 Search PubMed.
- S. S. Hettiarachchi, S. P. Dunuweera, A. N. Dunuweera and R. G. Rajapakse, ACS Omega, 2021, 6, 8246–8252 CAS.
- A. Putri, T. Octavianty, N. Wahyudi and A. Safitri, J. Phys.: Conf. Ser., 2019, 1374(1), 012027 CAS.
- R. S. Nunavath, K. C. Bhadram and K. Nagappan, J. Pharm. Biomed. Anal., 2023, 235, 115614 CrossRef CAS.
- O. N. Gordon and C. Schneider, Trends Mol. Med., 2012, 18, 361–364 CrossRef CAS PubMed.
- A. Rohman, D. Ramadhani and A. Nugroho, Res. J. Med. Plant, 2015, 9, 179–186 Search PubMed.
- M. Qasem, R. El Kurdi and D. Patra, ChemistrySelect, 2020, 5, 1694–1704 CrossRef CAS.
- N. E. Safie, N. A. Ludin, M. S. Su’ait, N. H. Hamid, S. Sepeai, M. A. Ibrahim and M. A. M. Teridi, Malay. J. Anal. Sci., 2015, 19, 1243–1249 Search PubMed.
- S. Bhattacharjee, J. Controlled Release, 2016, 235, 337–351 CAS.
- D. Patra and R. El Kurdi, Green Chem. Lett. Rev., 2021, 14, 474–487 CrossRef.
- M. Danaei, M. Dehghankhold, S. Ataei, F. Hasanzadeh Davarani, R. Javanmard, A. Dokhani, S. Khorasani and M. Mozafari, Pharmaceutics, 2018, 10, 57 Search PubMed.
- S. Dasgupta, S. Das, N. S. Chawan and A. Hazra, Indian J. Crit. Care Med., 2015, 19, 14 Search PubMed.
- S. Stefani, D. R. Chung, J. A. Lindsay, A. W. Friedrich, A. M. Kearns, H. Westh and F. M. MacKenzie, Int. J. Antimicrob. Agents, 2012, 39, 273–282 Search PubMed.
- S. Y. Tong, J. S. Davis, E. Eichenberger, T. L. Holland and V. G. Fowler Jr, Clin. Microbiol. Rev., 2015, 28, 603–661 CrossRef PubMed.
- J. A. Lindsay, Int. J. Med. Microbiol., 2014, 304, 103–109 Search PubMed.
- B. Mlynarczyk-Bonikowska, C. Kowalewski, A. Krolak-Ulinska and W. Marusza, Int. J. Mol. Sci., 2022, 23, 8088 Search PubMed.
- G. Sakoulas, A. S. Bayer, J. Pogliano, B. T. Tsuji, S.-J. Yang, N. N. Mishra, V. Nizet, M. R. Yeaman and P. A. Moise, Antimicrob. Agents Chemother., 2012, 56, 838–844 CrossRef PubMed.
- J. Top, R. Willems and M. Bonten, FEMS Immunol. Med. Microbiol., 2008, 52, 297–308 CrossRef PubMed.
- A. R. Freitas, C. Novais, A. P. Tedim, M. V. Francia, F. Baquero, L. Peixe and T. M. Coque, PLoS One, 2013, 8, e60589 Search PubMed.
- D. Yu, Y. Zheng and Y. Yang, Infect. Drug Resist., 2020, 2323–2327 CrossRef PubMed.
- I. Kyriakidis, E. Vasileiou, Z. D. Pana and A. Tragiannidis, Pathogens, 2021, 10, 373 CrossRef PubMed.
- C.-R. Lee, J. H. Lee, M. Park, K. S. Park, I. K. Bae, Y. B. Kim, C.-J. Cha, B. C. Jeong and S. H. Lee, Front. Cell. Infect. Microbiol., 2017, 7, 55 Search PubMed.
- Y.-F. Zhou, P. Liu, C.-J. Zhang, X.-P. Liao, J. Sun and Y.-H. Liu, Front. Microbiol., 2020, 10, 2957 CrossRef PubMed.
- Y. Paitan, Escherichia coli, a Versatile Pathogen, 2018, pp. 181–211 Search PubMed.
- G. Granata and N. Petrosillo, Infect. Dis. Rep., 2017, 9, 7104 Search PubMed.
- M. Bassetti, E. Righi, A. Carnelutti, E. Graziano and A. Russo, Expert Rev. Anti-Infect. Ther., 2018, 16, 749–761 CrossRef CAS PubMed.
- B. Jana, A. K. Cain, W. T. Doerrler, C. J. Boinett, M. C. Fookes, J. Parkhill and L. Guardabassi, Sci. Rep., 2017, 7, 42483 CAS.
- S. V. Chakka, N. Thanjavur, S. Lee and S. Kim, J. Rare Earths, 2022, 41(10), 1606–1615 Search PubMed.
- C. Karthikeyan, K. Varaprasad, A. Akbari-Fakhrabadi, A. S. H. Hameed and R. Sadiku, Carbohydr. Polym., 2020, 249, 116825 Search PubMed.
- N. Jayarambabu, A. Akshaykranth, T. V. Rao, K. V. Rao and R. R. Kumar, Mater. Lett., 2020, 259, 126813 CAS.
- M. Jayandran, M. M. Haneefa and V. Balasubramanian, Indian J. Sci. Technol., 2016, 9, 1–9 CAS.
- K. Cheraghipour, B. Ezatpour, L. Masoori, A. Marzban, A. Sepahvand, A. K. Rouzbahani, A. Moridnia, S. Khanizadeh and H. Mahmoudvand, Curr. Drug Discovery Technol., 2021, 18, 379–390 CAS.
- I. M. El-Nahhal, J. Salem, F. S. Kodeh, A. Elmanama and R. Anbar, Mater. Chem. Phys., 2022, 285, 126099 CAS.
- J. K. Trigo-Gutierrez, Y. Vega-Chacón, A. B. Soares and E. G. D. O. Mima, Int. J. Mol. Sci., 2021, 22, 7130 CAS.
- N. P. Rajkumari, P. Roy, S. Siddika, K. Adhikary and P. Goswami, Mater. Sci. Eng. B., 2023, 292, 116416 CAS.
- C. D. Tran, J. Makuvaza, E. Munson and B. Bennett, ACS Appl. Mater. Interfaces, 2017, 9, 42503–42515 CAS.
- S. Logpriya, V. Bhuvaneshwari, D. Vaidehi, R. SenthilKumar, R. Nithya Malar, B. Pavithra Sheetal, R. Amsaveni and M. Kalaiselvi, J. Nanostruct. Chem., 2018, 8, 301–309 CAS.
- J. Li and S. Zhuang, Eur. Polym. J., 2020, 138, 109984 CAS.
- T. Jayaramudu, K. Varaprasad, R. D. Pyarasani, K. K. Reddy, K. D. Kumar, A. Akbari-Fakhrabadi, R. Mangalaraja and J. Amalraj, Int. J. Biol. Macromol., 2019, 128, 499–508 CAS.
- B. Vishalakshi, B. Umakanth, A. P. Shanbhag, A. Ghatak, N. Sathyanarayanan, M. Madhav, G. G. Krishna and H. Yadla, 3 Biotech, 2017, 7, 1–6 CAS.
- N. Zainuddin, I. Ahmad, H. Kargarzadeh and S. Ramli, Carbohydr. Polym., 2017, 163, 261–269 CrossRef CAS PubMed.
- H. Tan, R. Ma, C. Lin, Z. Liu and T. Tang, Int. J. Mol. Sci., 2013, 14, 1854–1869 CrossRef CAS PubMed.
- M. Kong, X. G. Chen, K. Xing and H. J. Park, Int. J. Food Microbiol., 2010, 144, 51–63 CrossRef CAS PubMed.
- Z. Song, Y. Wu, H. Wang and H. Han, Mater. Sci. Eng. C, 2019, 99, 255–263 CrossRef CAS PubMed.
- L. Wang, C. Hu and L. Shao, Int. J. Nanomed., 2017, 1227–1249 CrossRef PubMed.
- C. Gunawan, W. Y. Teoh, C. P. Marquis and R. Amal, ACS Nano, 2011, 5, 7214–7225 CAS.
- S. Zorofchian Moghadamtousi, H. Abdul Kadir, P. Hassandarvish, H. Tajik, S. Abubakar and K. Zandi, BioMed Res. Int., 2014, 2014(1), 186864 Search PubMed.
- D. Raafat and H. G. Sahl, Microb. Biotechnol., 2009, 2, 186–201 CrossRef CAS PubMed.
- W. Sajomsang, P. Gonil and S. Tantayanon, Int. J. Biol. Macromol., 2009, 44, 419–427 Search PubMed.
- G. Kravanja, M. Primožič, Ž. Knez and M. Leitgeb, Molecules, 2019, 24, 1960 CrossRef CAS PubMed.
- J. Vinsova and E. Vavrikova, Curr. Pharm. Des., 2011, 17, 3596–3607 CrossRef CAS PubMed.
- S. Paul, D. MK and S. Peter, J. Pharm. Innovation, 2022, 1–10 Search PubMed.
- G. Applerot, J. Lellouche, A. Lipovsky, Y. Nitzan, R. Lubart, A. Gedanken and E. Banin, Small, 2012, 8, 3326–3337 Search PubMed.
- X. Ma, S. Zhou, X. Xu and Q. Du, Front. Surg., 2022, 9, 905892 CrossRef PubMed.
- M. Tiwari, K. Narayanan, M. B. Thakar, H. V. Jagani and J. Venkata Rao, IET Nanobiotechnol., 2014, 8, 230–237 Search PubMed.
- S.-Y. Teow, S. A. Ali and P. J. Pharm, Sci, 2015, 28, 2109–2114 Search PubMed.
- A. A. Targhi, A. Moammeri, E. Jamshidifar, K. Abbaspour, S. Sadeghi, L. Lamakani and I. Akbarzadeh, Bioorg. Chem., 2021, 115, 105116 Search PubMed.
- P. Bhavyasree and T. Xavier, Curr. Res. Green Sustainable Chem., 2022, 5, 100249 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4na00955j |
| This journal is © The Royal Society of Chemistry 2025 |







