Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Mass spectrometry-based profiling of phosphoinositide: advances, challenges, and future directions
Yuki
Ishino
,
Yuta
Shimanaka
 ,
Junken
Aoki
and
Nozomu
Kono
,
Junken
Aoki
and
Nozomu
Kono
 *
*
Laboratory of Health Chemistry, Graduate School of Pharmaceutical Sciences, the University of Tokyo, Hongo 7-3-1, Bunkyo-ku, Tokyo 113-0033, Japan. E-mail: nozomu@mol.f.u-tokyo.ac.jp; Fax: +81-3-3818-3173; Tel: +81-3-5841-1076
First published on 27th October 2025
Abstract
Phosphoinositides (PIPs), the phosphorylated derivatives of phosphatidylinositol (PI), are low-abundance yet critical components of eukaryotic membranes. They play pivotal roles in a wide array of cellular processes, including signal transduction, membrane trafficking, and cell motility. The seven PIP subclasses, generated by phosphorylation at the 3-, 4-, and 5-positions of the inositol ring, are tightly regulated in both spatial and temporal contexts. Dysregulation of PIP metabolism is associated with a range of diseases, including cancer, myopathy, and neurodegenerative and developmental disorders. While the importance of phosphorylation of the inositol ring is well established, recent studies have clarified the role of the fatty acyl chain composition of PIPs. This has resulted in a growing interest in analytical techniques that can determine fatty acyl chain profiles of PIPs. Over the past three decades, substantial advances have been made in mass spectrometry-based techniques, enabling detailed characterization of PIP molecular species, including their phosphate regioisomers. This review provides an overview of the development of mass spectrometric methods for analyzing PIPs, with a particular focus on those enabling the separation of PIP regioisomers and the profiling of their acyl chain composition.
Introduction
Phosphoinositides (PIPs), which are phosphorylated derivatives of phosphatidylinositol (PI), are minor yet crucial components of eukaryotic membranes, playing critical roles in regulating diverse cellular processes. Phosphorylation at the 3-, 4-, and 5-positions of the inositol ring generates seven distinct PIP subclasses: PI(3)P, PI(4)P, PI(5)P, PI(3,4)P2, PI(3,5)P2, PI(4,5)P2, and PI(3,4,5)P3 (Fig. 1A). Each subclass exhibits a unique subcellular distribution: PI(4,5)P2 is predominantly localized to the plasma membrane; PI(4)P to the Golgi apparatus and plasma membrane; PI(3,5)P2 to late endosomes and lysosomes; and PI(3)P to early and late endosomes1,2 (Fig. 1B). PI(3)P is also synthesized at the endoplasmic reticulum during autophagy initiation. PI(3,4)P2 and PI(3,4,5)P3 are produced transiently at the plasma membrane in response to extracellular stimuli. Although the precise localization of PI(5)P is unclear, it has been detected at the plasma membrane, early endosomes, and within the nucleus.3The spatial distribution of PIPs is tightly regulated by the coordinated actions of specific lipid kinases and phosphatases (Fig. 1A). At distinct subcellular locations, PIPs act as molecular landmarks that orchestrate the recruitment and activity of effector proteins involved in key cellular processes such as cell migration, adhesion, signal transduction, and membrane trafficking2 (Fig. 1B). Classically, PI(4,5)P2 serves as a precursor of the second messengers inositol-1,4,5-trisphosphate (IP3) and diacylglycerol (DAG), which are produced via phospholipase C-mediated hydrolysis of PI(4,5)P2 in response to extracellular stimuli.4 Recently, PI(4)P has also been recognized as a central regulator of intracellular lipid transport.2
Dysregulation of PIP metabolism has been increasingly recognized as a key contributor to a broad spectrum of human diseases.2,4 For instance, somatic activating mutations in PIK3CA, which encodes a class I PI3-kinase responsible for generating PI(3,4,5)P3, have been identified in breast, endometrial, colorectal, urinary tract, and ovarian cancers.5,6 PTEN, which encodes an inositol 3-phosphatase that dephosphorylates PI(3,4,5)P3, is frequently mutated in cancers such as glioblastoma, endometrial, and prostate cancers.6,7 Mutations in OCRL1, which encodes an inositol-5-phosphatase acting on PI(4,5)P2 and PI(3,4,5)P3, cause Lowe syndrome, an X-linked disorder characterized by congenital cataracts, intellectual disability, and renal failure.8 Mutations in MTM1, which affect an inositol 3-phosphatase that dephosphorylates PI(3)P and PI(3,5)P2, lead to X-linked myotubular myopathy.9 Mutations in MTMR2, another inositol 3-phosphatase targeting PI(3)P and PI(3,5)P2, are associated with Charcot-Marie-Tooth disease 4B1, a severe autosomal recessive demyelinating neuropathy.10
Increasing evidence indicates that PIP acyl chains influence the biological functions of PIPs. The acyl chain composition of PIPs affects Akt phosphorylation activity of PDK1,11 activation of potassium channel GIRK1/GIRK4,12 membrane binding of HIV-1 Gag protein,13 lipid transfer activity of Osh6p,14 and the coactivator peptide-binding affinity of SF-1.15 The acyl chain composition of PIPs also impacts the biophysical properties of membranes, including the formation of PIP-enriched nanodomains, which in turn influence membrane curvature16 and PIP metabolism.17,18 Moreover, PIP2 species with saturated acyl chains potentiate effector T cell signaling.19 Alterations in PI/PIP acyl chain profiles have been associated with tumorigenesis, raising the possibility that cancer cells exploit specific PIP molecular species to rewire signaling networks.20–22 These findings underscore the need for comprehensive PIP profiling that captures both head group phosphorylation and fatty acyl chain identity.
Traditional methods for PIP measurement have relied on metabolic labeling with [3H]inositol or [32P]phosphate, followed by deacylation and separation of PIP subclasses using high-performance liquid chromatography (HPLC) methods that separate by headgroup polarity.23 The radiolabeling methods revealed that the total PIP content is less than 10% of PI, which itself comprises 5–10% of the total phospholipids, with PI(4)P and PI(4,5)P2 being the major species detected.23 However, these methods do not allow for the quantification of endogenous PIP levels and thus are not applicable to tissue samples. Alternative non-radioactive methods, such as ELISA-based mass assays using PIP-binding domains and PIP kinase-based mass assays,24–26 can quantify endogenous PIP subclasses, but they do not provide information on fatty acyl chain composition.
Electrospray ionization mass spectrometry (ESI-MS) is a powerful and widely used technique for quantifying individual lipid species with high sensitivity.27,28 Over the past three decades, substantial methodological advances have enabled the precise quantification of PIP molecular species, which includes information about fatty acyl chains. More recently, innovations have enabled the discrimination of PIP1 and PIP2 regioisomers, along with their fatty acyl chain profiles. In this review, we provide an overview of historical and current mass spectrometry methods of PIP analysis (Table 1) and discuss where further technical innovations are needed.
| Authors | Year | Advances/novelty | Method | Derivatization | Regioisomer separation | Ref. |
|---|---|---|---|---|---|---|
| Michelsen et al. | 1995 | Application of ESI-MS to PIP1 and PIP2 analysis. | ESI-MS | No | — | 29 |
| Hsu et al. | 2000 | The detailed mechanism of fragmentation of PI(4)P and PI(4,5)P2 ions under CAD with ESI-MS/MS. | ESI-MS/MS | No | — | 30 |
| Wenk et al. | 2003 | Measurement of PIP1 and PIP2 acyl profiles in biological samples by ESI-MS/MS. | ESI-MS/MS | No | — | 31 |
| Enhancement of PIPs signal intensities by piperidine. | ||||||
| Milne et al. | 2005 | Measurement of PI(3,4,5)P3 molecular species in biological samples. | ESI-MS | No | — | 32 |
| Development of 2-step extraction method. | ||||||
| Pettitt et al. | 2006 | Measurement of PIP molecular species and PIP2 regioisomers by normal phase LC-MS. | NPLC-ESI-MS ESI-MSn | No | PIP2 and PI(4)P | 33 |
| Development of buffered citrate extraction method. | ||||||
| Use of ethylamine as a mobile phase modifier. | ||||||
| Distinct fragmentation pattern of PI(4)P from other PIP1s at MS3. | ||||||
| Ogiso et al. | 2008 | Measurement of PIP molecular species by reverse phase LC-MS. | RPLC-ESI-MS/MS | No | — | 34 |
| Use of DEAE column to obtain a PIP-rich fraction. | ||||||
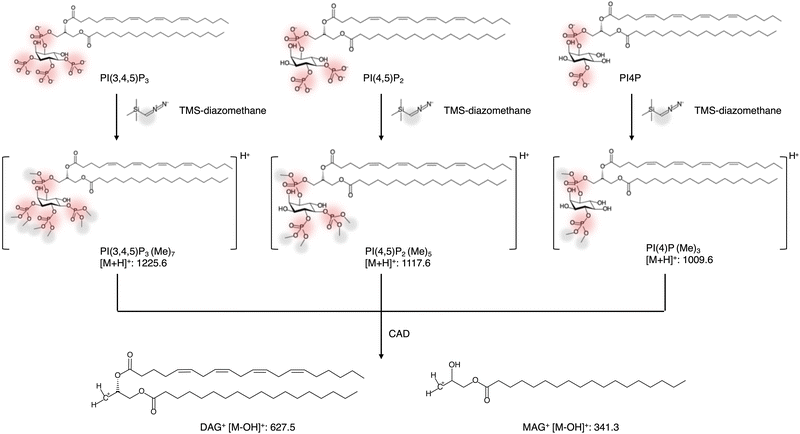
| Clark et al. | 2011 | Development of PIP derivatization method with TMS-diazomethane. | RPLC-ESI-MS/MS | Yes | — | 35 |
| Measurement of methylated PIPs by LC-MS/MS. | ||||||
| Wang et al. | 2016 | Development of a method for quantifying each PIP2 regioisomer and PI(3)P by simulation using the information of methylation patterns. | ESI-MS/MS | Yes | PIP2 and PI(3)P | 37 |
| Kim et al. | 2017 | Development of LC-MS/MS method for structural identification of sn-1 and sn-2 fatty acyl chains of PIPs. | RPLC-ESI-MS/MS | Yes | — | 36 |
| Enhancement of PIPs signal intensities by ammonium ion adduction. | ||||||
| Malek et al. | 2017 | Development of a method to distinguish between 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/20 0/20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 PI(4,5)P2 and PI(3,4)P2 in biological samples by shortening the acyl groups of PIPs by ozone cleavage. 4 PI(4,5)P2 and PI(3,4)P2 in biological samples by shortening the acyl groups of PIPs by ozone cleavage. |
RPLC-ESI-MS/MS | Yes | PI(4,5)P2 and PI(3,4)P2 | 39 |
| Bui et al. | 2018 | Development of a technique for measuring non-derivatized PIP2 regioisomers by RPLC-MS with the addition of the ion-pairing reagents. | RPLC-ESI-MS/MS | Yes | PIP2 | 38 |
| Li et al. | 2021 | Development of a method to separate all methylated PIP1 and PIP2 regioisomers by LC-MS using a cellulose-based chiral column. | RPLC-ESI-MS/MS | Yes | PIP1 and PIP2 | 40 |
| Use of SWATH to obtain a comprehensive PIP profile. | ||||||
| Morioka et al. | 2022 | Development of a method for simultaneously quantifying molecular species of all seven PIP classes by LC-ESI-MS/MS using a cellulose-based chiral column. | RPLC-ESI-MS/MS | Yes | PIP1 and PIP2 | 41 |
| Shimanaka et al. | 2022 | Development of a method for simultaneously quantifying molecular species of all seven PIP classes by SFC-ESI-MS/MS using a β-cyclodextrin column. | SFC-ESI-MS/MS | Yes | PIP1 and PIP2 | 42 |
ESI-MS analysis of intact PIPs
The first application of ESI-MS to the analysis of PIP1 and PIP2 was demonstrated by Michelsen et al.29 They successfully quantified 1-stearoyl-2-arachidonoyl (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0_20
0_20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) species of PIP1 and PIP2 in bovine brain extracts. Using single-ion monitoring in negative-ion mode, they detected approximately 200 pmol of each species. Hsu et al.30 investigated the fragmentation mechanisms of PI(4)P and PI(4,5)P2 using collision-activated dissociation (CAD) in tandem ESI-MS (ESI-MS/MS), showing that ESI-MS/MS is a powerful tool for structural characterization of PIPs. Wenk et al.31 were the first to report comprehensive fatty acyl chain profiling of PIP1 and PIP2 molecular species in biological samples by ESI-MS/MS. They demonstrated that the addition of piperidine significantly enhanced the negative-mode ionization efficiency of PIP1 and PIP2. Using this approach, they found elevated levels of PIP2 species in fibroblasts derived from patients with Lowe syndrome. They also showed that deficiency of enzymes involved in PIP metabolism, such as Sac1, Pik1, and Vps34, in Saccharomyces cerevisiae affects not only the levels of PIPs but also their acyl chain profiles, suggesting that these enzymes exhibit substrate selectivity for specific acyl chains or that organelle-specific PI or PIP acyl chain composition may exist at the sites where these enzymes localize. Milne et al.32 further extended the scope of PIP profiling by enabling the quantification of PI(3,4,5)P3, which is the least abundant PIP subclass in vivo, along with PIP1 and PIP2. To minimize the interference of other phospholipids during analysis, they developed a selective two-step extraction protocol, in which an initial chloroform/methanol extraction removed unwanted non-polar lipids and a subsequent acidic extraction recovered PIPs from the residual pellet. Their analysis revealed that different profiles of PI(3,4,5)P3 were produced in macrophages stimulated by various agonists, such as lysophosphatidic acid and C5a, suggesting that the activation mechanisms of PI-3 kinase and/or substrate availability among PI(4,5)P2 species vary depending on the agonist.
4) species of PIP1 and PIP2 in bovine brain extracts. Using single-ion monitoring in negative-ion mode, they detected approximately 200 pmol of each species. Hsu et al.30 investigated the fragmentation mechanisms of PI(4)P and PI(4,5)P2 using collision-activated dissociation (CAD) in tandem ESI-MS (ESI-MS/MS), showing that ESI-MS/MS is a powerful tool for structural characterization of PIPs. Wenk et al.31 were the first to report comprehensive fatty acyl chain profiling of PIP1 and PIP2 molecular species in biological samples by ESI-MS/MS. They demonstrated that the addition of piperidine significantly enhanced the negative-mode ionization efficiency of PIP1 and PIP2. Using this approach, they found elevated levels of PIP2 species in fibroblasts derived from patients with Lowe syndrome. They also showed that deficiency of enzymes involved in PIP metabolism, such as Sac1, Pik1, and Vps34, in Saccharomyces cerevisiae affects not only the levels of PIPs but also their acyl chain profiles, suggesting that these enzymes exhibit substrate selectivity for specific acyl chains or that organelle-specific PI or PIP acyl chain composition may exist at the sites where these enzymes localize. Milne et al.32 further extended the scope of PIP profiling by enabling the quantification of PI(3,4,5)P3, which is the least abundant PIP subclass in vivo, along with PIP1 and PIP2. To minimize the interference of other phospholipids during analysis, they developed a selective two-step extraction protocol, in which an initial chloroform/methanol extraction removed unwanted non-polar lipids and a subsequent acidic extraction recovered PIPs from the residual pellet. Their analysis revealed that different profiles of PI(3,4,5)P3 were produced in macrophages stimulated by various agonists, such as lysophosphatidic acid and C5a, suggesting that the activation mechanisms of PI-3 kinase and/or substrate availability among PI(4,5)P2 species vary depending on the agonist.
Wenk et al.31 and Milne et al.32 employed direct infusion mass spectrometry, a technique susceptible to ion suppression effects, which could potentially compromise sensitivity. To address this limitation, Pettitt et al.33 developed a normal phase LC-MS method for PIP profiling. They thoroughly investigated each step, from extraction to final analysis of PIPs, and made several improvements. Especially, they developed a highly efficient buffered citrate extraction method to minimize acid-induced phosphoinositide degradation and a sensitive LC-MS method using ethylamine as a mobile phase modifier. Their approach enabled the detection of PIP1 in human platelets, with detection limits of 250 fmol for PIP1, 1 pmol for PIP2, and 5 pmol for PIP3.
Reversed-phase (RP) LC is highly effective for separating phospholipid molecular species and detecting low-abundance components. Ogiso et al.34 developed a sensitive RPLC-MS method for profiling the acyl chain composition of PIP1, PIP2, and PIP3. They enriched PIP fractions using (diethylamino)ethyl (DEAE)-cellulose columns and utilized an alkaline-resistant C8 column for separation. Applying this method to epidermal growth factor (EGF)-stimulated A431 cells, they quantified 17 distinct PIP1 and PIP2 molecular species, demonstrating that EGF stimulation increased the levels of almost all PIP1 molecular species without affecting the levels of PIP2 molecular species. They also showed that levels of 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 PIP3, which appears to be the most abundant PIP3 species, increased in response to EGF stimulation. They achieved a detection limit of 25 fmol for each PIP molecular species.
2 PIP3, which appears to be the most abundant PIP3 species, increased in response to EGF stimulation. They achieved a detection limit of 25 fmol for each PIP molecular species.
Improving detection sensitivity of PIPs via derivatization
The difficulty in quantifying PIPs arises from (i) their low abundance in vivo, (ii) poor ionization efficiency by ESI, and (iii) low extraction efficiency. Notably, the low recovery efficiency and poor quantification reproducibility of PIPs are largely due to the adsorption of acidic phosphate head groups on glass and metal surfaces and their susceptibility to hydrolysis.33,34 To address these issues, Clark et al.35 developed a derivatization method in which phosphate groups are methylated using trimethylsilyl (TMS)-diazomethane (Fig. 2). Derivatization can be performed directly in the acidified chloroform-methanol extracts from cells or tissues and is completed within 10 minutes. This reaction introduces three, five, and seven methyl groups to PIP1, PIP2, and PIP3, respectively (Fig. 2). Methylation improves the recovery and chemical stability of PIPs and reduces their net negative charge. The methylated PIPs can be detected as positive ions via ESI-MS, and CAD of these methylated ions induces neutral loss of the methylated inositol phosphate headgroup, yielding charged diacylglycerol fragments. To further improve sensitivity for the low-abundance PIP3, Clark et al. concentrated methylated PIP3 using a C4 RPLC column and detected it via neutral loss scan or multiple reaction monitoring (MRM), targeting the diacylglycerol fragment ions as Q3 masses. Utilizing this approach, they were able to quantify PI(3,4,5)P3 species in fMLP-stimulated human neutrophils, glucose-stimulated human adipose tissue, and insulin-stimulated mouse liver. Notably, they demonstrated selective accumulation of 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/20
0/20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 PI(3,4,5)P3 in EGF-stimulated MCF10a breast cancer cells even though its precursor, 18
4 PI(3,4,5)P3 in EGF-stimulated MCF10a breast cancer cells even though its precursor, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/20
0/20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 PI(4,5)P2, is the least abundant PI(4,5)P2 species.
4 PI(4,5)P2, is the least abundant PI(4,5)P2 species.
Kim et al.36 developed an LC-MS/MS method for structurally characterizing the sn-1 and sn-2 fatty acyl chains of PIPs. They discovered that CAD of methylated PIPs generates sn-1 monoacylglycerol ions. Then, using positive ion mode, they performed precursor ion scans targeting these fragments, and neutral loss scans of the inositol headgroup. By comparing the LC retention times obtained from the two scan modes, they were able to determine the fatty acyl chains at the sn-1 and sn-2 positions. Furthermore, they found that ammonium adduction enhanced PIP signal intensities approximately twofold. Using this technique, they were able to quantitatively profile PIP species in insulin-treated AML12 hepatocytes and mouse liver with structural information of the sn-1 and sn-2 fatty acyl chains.
Development of chromatographic separation of PIP1 and PIP2 regioisomers
Since each PIP1 and PIP2 regioisomer plays a distinct role in cellular functions, the separation and quantification of individual regioisomers are crucial for understanding their specific biological roles. However, since PIP regioisomers have identical masses, they cannot be distinguished by mass spectrometry alone. Pettitt et al. addressed this issue by developing a method to analyze non-derivatized PIP regioisomers using normal-phase LC coupled with ion trap mass spectrometry.33 They demonstrated that the three PIP2 regioisomers could be separated using a silica column under normal-phase conditions. While the three PIP1 regioisomers could not be completely resolved by LC alone, PI(5)P eluted slightly earlier than PI(3)P and PI(4)P. Furthermore, they found that the MS3 fragmentation pattern of PI(4)P differed from those of PI(3)P and PI(5)P, enabling estimation of the relative abundance of PI(4)P among the PIP1 isomers. By combining chromatographic separation and diagnostic MS fragmentation, they successfully quantified each PIP1 regioisomer.Wang et al.37 found that the percentage of overmethylated PIPs produced in the reaction with TMS-diazomethane varied among PIP2 regioisomers and used the values to quantify each PIP2 regioisomer. For PIP1, only PI(3)P could be distinguished from PI(4)P and PI(5)P, as the latter two shared the same methylation pattern. Using this technique, they revealed alterations in PIP profiles in the cerebral cortex of db/db mice. Bui et al.38 reported a method for separating non-derivatized PIP2 regioisomers using C18 RPLC with diisopropylethanolamine (DiiPEA) and EDTA as ion-pairing agents. Melek et al.39 used an innovative approach to distinguish between 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/20
0/20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 PI(4,5)P2 and PI(3,4)P2 in biological samples by shortening the acyl chain through ozone-induced cleavage. Thus, while various efforts have been made to analyze individual PIP regioisomers, there remains a strong interest in developing a simple and versatile method capable of separating and simultaneously quantifying all PIP regioisomers.
4 PI(4,5)P2 and PI(3,4)P2 in biological samples by shortening the acyl chain through ozone-induced cleavage. Thus, while various efforts have been made to analyze individual PIP regioisomers, there remains a strong interest in developing a simple and versatile method capable of separating and simultaneously quantifying all PIP regioisomers.
Recently, three independent groups, including ours, have reported methods for the simultaneous analysis of all PIP regioisomers using chromatography-coupled tandem mass spectrometry.40–42 Li et al.40 successfully separated all methylated PIP1 and PIP2 regioisomers using HPLC with a cellulose-based chiral column. They also employed SWATH (sequential window acquisition of all theoretical mass spectra), a data-independent acquisition technique, to obtain a comprehensive PIP profile. Since SWATH comprehensively acquires MS2 spectra of PIPs, including DAG and MAG ions, it enables determination of the fatty acyl chain combinations of PIPs. Using their method, they reported complete PIP profiles of human plasma, Pichia pastoris, and HeLa cells, as well as changes in PIP profiles in HeLa cells upon treatment with PI3K inhibitor wortmannin. Morioka et al.41 also developed a method to simultaneously quantify the molecular species of all seven PIP subclasses by HPLC-ESI-MS/MS using a cellulose-based chiral column, employing MRM for sensitive and specific detection. Their method revealed (i) profound changes in acyl chain composition of PI(3,4)P2 and PI(3,4,5)P3 in Pten-deficient prostate cancerous tissues, (ii) extracellular mobilization of PIPs upon expression of oncogenic PIK3CA, and (iii) the PIP profile of exosomes. Although both methods employed cellulose-based chiral columns, the elution order of the PIP1 regioisomers differed: the orders were PI(3)P, PI(5)P, and PI(4)P in the method of Li et al., and PI(3)P, PI(4)P, and PI(5)P in the method of Morioka et al. In both methods, PIP2 regioisomers eluted in the order PI(3,5)P2, PI(3,4)P2, and PI(4,5)P2.
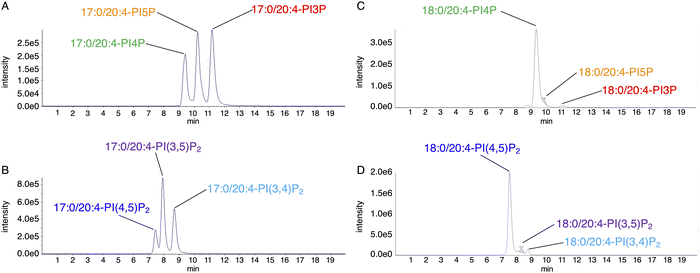
As an alternative chromatographic approach, we developed a method to separate methylated PIP1 and PIP2 regioisomers using supercritical fluid chromatography (SFC).42 A supercritical fluid is a substance maintained at a temperature and pressure above its critical point. It exhibits gas-like diffusivity and liquid-like solubility, enabling rapid and efficient separation. Importantly, SFC and HPLC have distinct separation behaviors, which makes it possible for SFC to separate compounds that HPLC cannot. We found that SFC using a β-cyclodextrin column effectively separated methylated PIP1 and PIP2 regioisomers (Fig. 3). Our SFC-based method provided a distinct elution profile compared to HPLC-based methods: PIP1 regioisomers eluted in the order PI(4)P, PI(5)P, and PI(3)P, while PIP2 regioisomers eluted in the order PI(4,5)P2, PI(3,5)P2, and PI(3,4)P2. Furthermore, SFC achieved broader separation windows, enhancing the resolution between regioisomers. Coupling SFC to ESI-MS/MS in MRM mode enabled the measurement of the molecular species of all seven PIP subclasses in biological samples. Using MRM transitions with the monoacylglycerol ion set as Q3, we were also able to detect PIPs with information on their fatty acyl chain combinations. Applying this method to mouse tissues, we discovered that PIPs containing two saturated fatty acids, which were hardly detected in most tissues or cultured cells, were abundant in the testis. We also found that less unsaturated PI(3)P molecular species, such as 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 34
1, 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 36
2, 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 36
1, and 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 PI(3)P, selectively increased during autophagy induction.
2 PI(3)P, selectively increased during autophagy induction.
Very recently, SFC-MS-based PIP profiling revealed that impaired production of PI(4,5)P2 in neural stem cells underlies the microcephaly in Mboat7 deficiency.43 LPLAT11, encoded by Mboat7, is a lysophospholipid acyltransferase that selectively incorporates arachidonic acid into phosphatidylinositol (PI).44 In humans, loss-of-function mutations in MBOAT7 are associated with neurodevelopmental disorders characterized by microcephaly and intellectual disability.45 Consistently, Mboat7-deficient mice exhibit microcephaly.45–47 We found that Mboat7 deficiency compromised the integrity of radial glial cells, which are the neural stem cells responsible for cortical development, resulting in reduced proliferation, impaired differentiation into intermediate progenitor cells, and increased apoptosis. These cellular defects were preceded by morphological abnormalities of the Golgi apparatus and reduced E-cadherin expression at the ventricular surface. SFC-MS analysis revealed that a reduction in arachidonic acid-containing species and a reciprocal increase in non-arachidonic acid-containing species occurred across PI, PI(4)P, and PI(4,5)P2 in Mboat7 KO cortices. In addition to changes in acyl chain composition, total PI(4,5)P2 levels were significantly reduced, whereas total PI and PI(4)P levels remained unchanged. This selective decrease is likely due to the acyl chain preference of PIP5Kγ,48 a key PI(4,5)P2-producing enzyme in the developing cortex. Furthermore, pharmacological inhibition of PIP5Kγ in wild-type embryonic cortices recapitulated the Golgi abnormalities and E-cadherin reduction observed in Mboat7 KO cortices. These findings show that PI(4,5)P2 has a critical role in maintaining radial glial cell integrity during cortical development and underscore the utility of SFC-MS in elucidating PIP-mediated mechanisms in physiology and pathology.
Future perspectives in phosphoinositide analysis
Recent advances have made it possible to measure molecular species in all seven PIP subclasses. This will greatly improve our understanding of the physiological and pathological roles of PIPs. Nevertheless, challenges remain in the absolute quantification of individual PIP species. Because PI(4)P and PI(4,5)P2 are far more abundant than other PIP classes in biological samples, minor PIP1s [PI(5)P and PI(3)P] and minor PIP2s [PI(3,4)P2 and PI(3,5)P2] often co-elute on the shoulders of the dominant PI(4)P and PI(4,5)P2 peaks, respectively42 (Fig. 3), hindering their accurate quantification. Indeed, the levels of these minor PIP species measured by HPLC-ESI-MS/MS or SFC-ESI-MS/MS were substantially higher than those determined using traditional radioisotope-labeling techniques40,42,49–51 (Table 2). While differences in culture conditions during radioisotope labeling may partially account for this discrepancy, it is also possible that minor PIP signals are artificially elevated due to overlap with the abundant PI(4)P and PI(4,5)P2 peaks. Although peak-integration methods, such as tangential skim, can partially address the overestimation caused by peak overlap, further improvements in the chromatographic separation of PIP regioisomers will be essential to achieve more accurate absolute quantification of PIP molecular species in biological matrices.| Method | Cell type | |||
|---|---|---|---|---|
| Mouse embryonic fibroblasts | HeLa cells | |||
| Methylation followed by SFC-MS | [3H]Inositol-labeling followed by HPLC | Methylation followed by HPLC-MS | [3H]Inositol-labeling followed by HPLC | |
| Values represent % of total PIPs. | ||||
| PI(3)P | 9.1 | 2.2 | 5.7 | 2.8 |
| PI(4)P | 36.2 | 37.2 | 36.1 | 45.3 |
| PI(5)P | 8.1 | 3.5 | 7.6 | 0.8 |
| PI(3,4)P2 | 2.6 | Not detected | 1.9 | Not detected |
| P(3,5)P2 | 3.7 | 0.5 | 2.7 | Not detected |
| PI(4,5)P2 | 40.4 | 56.6 | 46.1 | 51.0 |
| Ref. | Shimanaka et al.42 | Steinfeld et al.49 | Li et al.40 | Sarkes et al.50 & Fujii et al.51 |
Recent advances in mass spectrometry (MS)-based imaging techniques have enabled the visualization of diverse phospholipid species at single-cell resolution within tissues.52,53 However, there have been only a few reports on MS imaging of PIPs to date,54,55 and none of them are capable of distinguishing between PIP regioisomers. For MS imaging of PIP regioisomers, it is necessary to separate the regioisomers inside the mass spectrometer. One promising approach for isomer-specific imaging of PIPs is ion mobility spectrometry (IMS), a gas-phase separation technique in which ions are separated base on their collision cross-section (CCS), which reflects differences in molecular size and shape.56 Even when two isomers have the same mass-to-charge ratio, differences in their CCS values can result in distinct drift times, enabling their separation prior to mass analysis. Moreover, when integrated with LC-ESI-MS/MS or SFC-ESI-MS/MS workflows, IMS provides an additional orthogonal dimension of separation, enhancing the resolution of PIP regioisomers and thereby improving absolute quantification. Incorporating emerging technologies such as IMS into PIP analysis platforms will help overcome current limitations and further advance our understanding of the complex physiological and pathological functions of PIPs.
Conflicts of interest
YS and NK are co-inventors of Japanese Patent 7017704, titled “biological membrane phosphoinositide separation method.” All other authors declare no competing interests.Data availability
No primary research results, software or code have been included and no new data were generated or analyzed as part of this review.Acknowledgements
This work was supported by Grant-in-Aid for Early-Career Scientists to (18K14898 to YS); by Grants-in-Aid for Scientific Research (B) (23K27125 to NK); by AMED-CREST, AMED (JP 21gm1210013 to NK); and by Moonshot R&D Program (JPMJMS2024-11 and JPMJMS2024-15 to JA).References
- L. Picas, F. Gaits-Iacovoni and B. Goud, F1000 Res., 2016, 5, 422 Search PubMed.
- Y. Posor, W. Jang and V. Haucke, Nat. Rev. Mol. Cell Biol., 2022, 23, 797–816 CrossRef PubMed.
- J. Hasegawa, B. S. Strunk and L. S. Weisman, Cell Struct. Funct., 2017, 42, 49–60 CrossRef PubMed.
- T. Balla, Physiol. Rev., 2013, 93, 1019–1137 CrossRef PubMed.
- J. A. Engelman, J. Luo and L. C. Cantley, Nat. Rev. Genet., 2006, 7, 606–619 CrossRef.
- T. L. Yuan and L. C. Cantley, Oncogene, 2008, 27, 5497–5510 CrossRef PubMed.
- L. Salmena, A. Carracedo and P. P. Pandolfi, Cell, 2008, 133, 403–414 CrossRef PubMed.
- O. Attree, I. M. Olivos, I. Okabe, L. C. Bailey, D. L. Nelson, R. A. Lewis, R. R. McInnes and R. L. Nussbaum, Nature, 1992, 358, 239–242 CrossRef PubMed.
- J. Laporte, L. J. Hu, C. Kretz, J. L. Mandel, P. Kioschis, J. F. Coy, S. M. Klauck, A. Poustka and N. Dahl, Nat. Genet., 1996, 13, 175–182 CrossRef PubMed.
- A. Bolino, M. Muglia, F. L. Conforti, E. LeGuern, M. A. Salih, D. M. Georgiou, K. Christodoulou, I. Hausmanowa-Petrusewicz, P. Mandich, A. Schenone, A. Gambardella, F. Bono, A. Quattrone, M. Devoto and A. P. Monaco, Nat. Genet., 2000, 25, 17–19 CrossRef PubMed.
- L. Stephens, K. Anderson, D. Stokoe, H. Erdjument-Bromage, G. F. Painter, A. B. Holmes, P. R. Gaffney, C. B. Reese, F. McCormick, P. Tempst, J. Coadwell and P. T. Hawkins, Science, 1998, 279, 710–714 CrossRef PubMed.
- T. Rohács, J. Chen, G. D. Prestwich and D. E. Logothetis, J. Biol. Chem., 1999, 274, 36065–36072 CrossRef PubMed.
- B. Olety, S. L. Veatch and A. Ono, J. Virol., 2015, 89, 7861–7873 CrossRef PubMed.
- R. Ghai, X. Du, H. Wang, J. Dong, C. Ferguson, A. J. Brown, R. G. Parton, J. W. Wu and H. Yang, Nat. Commun., 2017, 8, 757 CrossRef PubMed.
- J. M. Bryant, M. M. Malabanan, B. H. Vanderloop, C. M. Nichols, Z. Haratipour, K. T. Poon, S. D. Sherrod, J. A. McLean and R. D. Blind, J. Lipid Res., 2021, 62, 100081 CrossRef PubMed.
- L. Borges-Araújo, M. M. Domingues, A. Fedorov, N. C. Santos, M. N. Melo and F. Fernandes, Commun. Chem., 2021, 4, 164 CrossRef.
- T. Nishimura, M. Gecht, R. Covino, G. Hummer, M. A. Surma, C. Klose, H. Arai, N. Kono and C. J. Stefan, Mol. Cell, 2019, 75, 1043–1057.e8 CrossRef CAS PubMed.
- V. Chan, C. Camardi, K. Zhang, L. A. Orofiamma, K. E. Anderson, J. Hoque, L. N. Bone, Y. Awadeh, D. K. C. Lee, N. J. Fu, J. T. S. Chow, L. Salmena, L. R. Stephens, P. T. Hawkins, C. N. Antonescu and R. J. Botelho, Mol. Biol. Cell, 2024, 35, ar118 CrossRef CAS PubMed.
- J. Edwards-Hicks, P. Apostolova, J. M. Buescher, H. Maib, M. A. Stanczak, M. Corrado, R. I. Klein Geltink, M. E. Maccari, M. Villa, G. E. Carrizo, D. E. Sanin, F. Baixauli, B. Kelly, J. D. Curtis, F. Haessler, A. Patterson, C. S. Field, G. Caputa, R. L. Kyle, M. Soballa, M. Cha, H. Paul, J. Martin, K. M. Grzes, L. Flachsmann, M. Mitterer, L. Zhao, F. Winkler, D. A. Rafei-Shamsabadi, F. Meiss, B. Bengsch, R. Zeiser, D. J. Puleston, D. O'Sullivan, E. J. Pearce and E. L. Pearce, Nat. Immunol., 2023, 24, 516–530 CrossRef CAS.
- A. Naguib, G. Bencze, D. D. Engle, I. I. Chio, T. Herzka, K. Watrud, S. Bencze, D. A. Tuveson, D. J. Pappin and L. C. Trotman, Cell Rep., 2015, 10, 8–19 CrossRef CAS PubMed.
- A. Koizumi, S. Narita, H. Nakanishi, M. Ishikawa, S. Eguchi, H. Kimura, S. Takasuga, M. Huang, T. Inoue, J. Sasaki, T. Yoshioka, T. Habuchi and T. Sasaki, Sci. Rep., 2019, 9, 13257 CrossRef.
- R. M. Epand, J. Membr. Biol., 2017, 250, 353–366 CrossRef CAS.
- H. Guillou, L. R. Stephens and P. T. Hawkins, Methods Enzymol., 2007, 434, 117–130 CAS.
- Y. Irino, E. Tokuda, J. Hasegawa, T. Itoh and T. Takenawa, J. Lipid Res., 2012, 53, 810–819 CrossRef CAS.
- J. B. Morris, K. A. Hinchliffe, A. Ciruela, A. J. Letcher and R. F. Irvine, FEBS Lett., 2000, 475, 57–60 CrossRef CAS.
- G. Chicanne, S. Severin, C. Boscheron, A. D. Terrisse, M. P. Gratacap, F. Gaits-Iacovoni, H. Tronchère and B. Payrastre, Biochem. J., 2012, 447, 17–23 CrossRef CAS.
- M. Pulfer and R. C. Murphy, Mass Spectrom. Rev., 2003, 22, 332–364 CrossRef CAS.
- M. R. Wenk, Cell, 2010, 143, 888–895 CrossRef PubMed.
- P. Michelsen, B. Jergil and G. Odham, Rapid Commun. Mass Spectrom., 1995, 9, 1109–1114 CrossRef.
- F. F. Hsu and J. Turk, J. Am. Soc. Mass Spectrom., 2000, 11, 986–999 CrossRef.
- M. R. Wenk, L. Lucast, G. Di Paolo, A. J. Romanelli, S. F. Suchy, R. L. Nussbaum, G. W. Cline, G. I. Shulman, W. McMurray and P. De Camilli, Nat. Biotechnol., 2003, 21, 813–817 CrossRef.
- S. B. Milne, P. T. Ivanova, D. DeCamp, R. C. Hsueh and H. A. Brown, J. Lipid Res., 2005, 46, 1796–1802 CrossRef.
- T. R. Pettitt, S. K. Dove, A. Lubben, S. D. Calaminus and M. J. Wakelam, J. Lipid Res., 2006, 47, 1588–1596 CrossRef PubMed.
- H. Ogiso and R. Taguchi, Anal. Chem., 2008, 80, 9226–9232 CrossRef PubMed.
- J. Clark, K. E. Anderson, V. Juvin, T. S. Smith, F. Karpe, M. J. Wakelam, L. R. Stephens and P. T. Hawkins, Nat. Methods, 2011, 8, 267–272 CrossRef PubMed.
- S. H. Kim, H. E. Song, S. J. Kim, D. C. Woo, S. Chang, W. G. Choi, M. J. Kim, S. H. Back and H. J. Yoo, J. Lipid Res., 2017, 58, 469–478 CrossRef PubMed.
- C. Wang, J. P. Palavicini, M. Wang, L. Chen, K. Yang, P. A. Crawford and X. Han, Anal. Chem., 2016, 88, 12137–12144 CrossRef PubMed.
- H. H. Bui, P. E. Sanders, D. Bodenmiller, M. S. Kuo, G. P. Donoho and A. S. Fischl, Anal. Biochem., 2018, 547, 66–76 CrossRef PubMed.
- M. Malek, A. Kielkowska, T. Chessa, K. E. Anderson, D. Barneda, P. Pir, H. Nakanishi, S. Eguchi, A. Koizumi, J. Sasaki, V. Juvin, V. Y. Kiselev, I. Niewczas, A. Gray, A. Valayer, D. Spensberger, M. Imbert, S. Felisbino, T. Habuchi, S. Beinke, S. Cosulich, N. Le Novère, T. Sasaki, J. Clark, P. T. Hawkins and L. R. Stephens, Mol. Cell, 2017, 68, 566–580.e10 CrossRef.
- P. Li and M. Lämmerhofer, Anal. Chem., 2021, 93, 9583–9592 CrossRef PubMed.
- S. Morioka, H. Nakanishi, T. Yamamoto, J. Hasegawa, E. Tokuda, T. Hikita, T. Sakihara, Y. Kugii, C. Oneyama, M. Yamazaki, A. Suzuki, J. Sasaki and T. Sasaki, Nat. Commun., 2022, 13, 83 CrossRef PubMed.
- Y. Shimanaka, K. Matsumoto, Y. Tanaka, Y. Ishino, S. Ni, J. Guan, H. Arai and N. Kono, Commun. Chem., 2022, 5, 61 CrossRef CAS.
- Y. Ishino, Y. Kishi, T. Iwama, N. Kuwayama, H. Arai, Y. Gotoh, J. Aoki and N. Kono, bioRxiv, 2024, preprint, 2024.04.30.588048 DOI:10.1101/2024.04.30.588048.
- H. C. Lee, T. Inoue, R. Imae, N. Kono, S. Shirae, S. Matsuda, K. Gengyo-Ando, S. Mitani and H. Arai, Mol. Biol. Cell, 2008, 19, 1174–1184 CrossRef CAS.
- I. Tang, A. Nisal, A. Reed, T. B. Ware, A. Johansen, M. S. Zaki, B. F. Cravatt and J. G. Gleeson, Sci. Transl. Med., 2025, 17, eadp5247 CrossRef CAS PubMed.
- H. C. Lee, T. Inoue, J. Sasaki, T. Kubo, S. Matsuda, Y. Nakasaki, M. Hattori, F. Tanaka, O. Udagawa, N. Kono, T. Itoh, H. Ogiso, R. Taguchi, M. Arita, T. Sasaki and H. Arai, Mol. Biol. Cell, 2012, 23, 4689–4700 CrossRef CAS PubMed.
- K. E. Anderson, A. Kielkowska, T. N. Durrant, V. Juvin, J. Clark, L. R. Stephens and P. T. Hawkins, PLoS One, 2013, 8, e58425 CrossRef CAS PubMed.
- Y. V. Shulga, R. A. Anderson, M. K. Topham and R. M. Epand, J. Biol. Chem., 2012, 287, 35953–35963 CrossRef CAS.
- N. Steinfeld, S. S. P. Giridharan, E. J. Kauffman and L. S. Weisman, Methods Mol. Biol., 2021, 2251, 1–17 CrossRef CAS.
- D. Sarkes and L. E. Rameh, Biochem. J., 2010, 428, 375–384 CrossRef CAS.
- M. Fujii, S. Kita, M. Hirata and T. Iwamoto, Nihon Yakurigaku Zasshi, 2013, 142, 236–240 CrossRef CAS PubMed.
- D. Gode and D. A. Volmer, Analyst, 2013, 138, 1289–1315 RSC.
- M. K. Passarelli and A. G. Ewing, Curr. Opin. Chem. Biol., 2013, 17, 854–859 CrossRef CAS.
- N. Li, P. Wang, X. Liu, C. Han, W. Ren, T. Li, X. Li, F. Tao and Z. Zhao, Anal. Chem., 2019, 91, 15873–15882 CrossRef CAS.
- K. C. Pathmasiri, M. R. Pergande, F. Tobias, R. Rebiai, A. Rosenhouse-Dantsker, E. R. Bongarzone and S. M. Cologna, J. Lipid Res., 2020, 61, 1004–1013 CrossRef CAS.
- F. Lanucara, S. W. Holman, C. J. Gray and C. E. Eyers, Nat. Chem., 2014, 6, 281–294 CrossRef.
| This journal is © The Royal Society of Chemistry 2025 |