Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Unmasking the lipid landscape: carbamazepine induces alterations in Leydig cell lipidome†
Inês
Nobre
abc,
Inês M. S.
Guerra
ab,
Marisa
Pinho
a,
Ana D.
Martins
bd,
Laura
Goracci
 e,
Stefano
Bonciarelli
e,
Stefano
Bonciarelli
 f,
Tânia
Melo
ab,
Pedro
Domingues
b,
Artur
Paiva
ghi,
Pedro F.
Oliveira
bd and
M. Rosário
Domingues
f,
Tânia
Melo
ab,
Pedro
Domingues
b,
Artur
Paiva
ghi,
Pedro F.
Oliveira
bd and
M. Rosário
Domingues
 *ab
*ab
aCESAM-Centre for Environmental and Marine Studies, Department of Chemistry, University of Aveiro, Santiago University Campus, 3810-193 Aveiro, Portugal
bMass Spectrometry Centre, LAQV-REQUIMTE, Department of Chemistry, University of Aveiro, Santiago University Campus, 3810-193 Aveiro, Portugal
cServiço Patologia Clínica, ULS Coimbra, 3004-561 Coimbra, Portugal
dLAQV-REQUIMTE and Department of Chemistry, University of Aveiro, 3810-193 Aveiro, Portugal
eDepartment of Chemistry, Biology and Biotechnology, University of Perugia, 06123 Perugia, Italy
fMolecular Discovery Ltd., Hertfordshire WD6 4PJ, UK
gUnidade de Gestão Operacional em Citometria, Centro Hospitalar e Universitário de Coimbra (CHUC), 3004-561 Coimbra, Portugal
hFaculty of Medicine, Coimbra Institute for Clinical and Biomedical Research (iCBR), University of Coimbra, 3000-370 Coimbra, Portugal
iCiências Biomédicas Laboratoriais, ESTESC – Coimbra Health School, Instituto Politécnico de Coimbra, 3046-854 Coimbra, Portugal
First published on 4th February 2025
Abstract
Leydig cells rely on lipids and fatty acids (FA) for essential functions like maintaining structural integrity, energy metabolism, and steroid hormone synthesis, including testosterone production. Carbamazepine (CBZ), a common anticonvulsant medication, can influence lipid metabolism and profiles, potentially impacting Leydig cell function and testosterone levels. Understanding this interplay is crucial to optimize treatment strategies for individuals requiring CBZ therapy while mitigating any adverse effects on male reproductive health. This study focuses on evaluating the effects of selected CBZ concentrations on the lipid homeostasis of BLTK-1 murine Leydig cells. By employing liquid chromatography-mass spectrometry (LC-MS) and gas chromatography-mass spectrometry (GC-MS), we aimed to uncover the specific changes in lipid profiles induced by CBZ exposure (25 and 200 μM). FA analysis demonstrated a significant decrease in FA 22:6 n-3 with increasing CBZ concentration and an increase in the n-6/n-3 ratio. Furthermore, changes in the lipidome, particularly in lipid species belonging to phosphatidylethanolamine (PE), phosphatidylcholine (PC), phosphatidylglycerol (PG), and sphingomyelin (SM) classes were observed. PE and PC lipid species were significantly elevated in Leydig cells exposed to 200 μM CBZ, whereas PG and SM species were downregulated. CBZ treatment significantly altered the Leydig cell phospholipidome, suggesting specific phospholipids such as PG 40:4, PG 34:1, PC O-32:1, PC 32:2, and PE P-38:6, which exhibited the lowest p-values, as potential biomarkers for clinical assessment of CBZ's impact on Leydig cells. These findings underscore the intricate relationship between CBZ exposure and alterations in lipid profiles, offering potential insights for monitoring and mitigating the drug's effects on male reproductive health.
Introduction
Leydig cells are key testicular somatic cells with a key role in the production and secretion of testosterone, the primary male sex hormone.1 Testosterone is essential for the development and maintenance of male reproductive tissues and for spermatogenesis,2 and disruptions in Leydig cell homeodynamics can affect fertility. Additionally, Leydig cell dysfunction has been linked to changes in lipid and fatty acid (FA) metabolism within the testis.3,4 In fact, lipid droplet accumulation in these cells has been suggested to contribute to tumor growth, proliferation, and metastasis.5–7Alterations in Leydig cell function can have profound effects on male health and reproductive capabilities.8–10 These changes can occur due to various factors, including aging, hormonal imbalances, diseases, medications and environmental factors.11 Certain medications, such as glucocorticoids, opioids, chemotherapy drugs and antipsychotic medications can affect Leydig cells’ function and testosterone production.3 Among the former ones, carbamazepine (CBZ), an anticonvulsant and mood-stabilizing medication commonly used to treat epilepsy and bipolar disorder, has been associated with changes in lipid metabolism and lipid profiles, causing adverse effects on male reproductive health in some individuals.12,13 While CBZ primarily exerts its therapeutic effects by stabilizing neuronal membranes and reducing abnormal electrical activity in the brain, it can also have effects on the endocrine system, including Leydig cells’ function and testosterone production.14 Research in animals has shown that CBZ can affect Leydig cells’ morphology and reduce testosterone production and suggest that CBZ may negatively influence Leydig cells, which are responsible for the testosterone production in the testis and may be involved in the tumorigenesis of these cells.15,16
The use of CBZ may lead to changes in plasma lipid levels, including increases in total cholesterol, LDL (low-density lipoproteins) cholesterol, triglycerides and alterations in lipoprotein profiles.17 These effects are thought to be mediated by CBZ-induced hepatic enzymes (cytochrome P450), changes in insulin sensitivity among others.18,19 However, the precise mechanisms through which CBZ may influence Leydig cell function and potentially induce tumorigenesis remain unclear. It appears that the effects of CBZ can vary among individuals, influenced by factors such as dosage, treatment duration, individual metabolism, and concurrent medications.20,21 Understanding these mechanisms is important for developing therapeutic strategies aimed at restoring Leydig cell function and improving male reproductive health.
Therefore, in this work, we intend to study the variation in the lipid profile of BLTK-1 murine Leydig cells after chronic exposure to CBZ, as a model of Leydig cell tumor to induce dysfunction and infertility. BLTK-1 cells were selected for this study due to their stability in culture, even at passages higher than those utilized. Moreover, BLTK-1 cells are a well-established model for studying endocrine, paracrine, and autocrine regulation.22 Cell viability was evaluated after exposure to different concentrations of CBZ (25 and 200 μM), the FA profile analyzed by gas chromatography-mass spectrometry (GC-MS) and the lipid profile by liquid chromatography-mass spectrometry (LC-MS).
Experimental
Cell culture – BLTK-1 murine Leydig cells
BLTK-1 was first established in 1996 a murine Leydig cell line with the name BLT-1.23 This cell line was gently provided by Nafis Rahman, MD, PhD, Faculty of Medicine, Institute of Biomedicine, University of Turku (Finland). Cells were cultured in 75 cm2 T-flasks and kept at 37 °C with a 5% (v/v) CO2 humidified atmosphere in DMEM (Dulbecco's modified Eagle's medium):Ham's F12 culture medium supplemented with 10% heat-inactivated FBS (fetal bovine serum), 50 μmol mL−1 gentamicin, 50 U mL−1 penicillin, 50 mg mL−1 streptomycin sulfate, and 0.5 mg mL−1 fungizone. Cells from passages 23 to 28 were used, and their morphology and functionality were routinely accessed via optical microscopy.Experimental groups and design
For the cytotoxicity evaluation, CBZ was diluted in dimethyl sulfoxide (DMSO) to prepare a stock solution, which was then diluted and used to treat the seeded cells. BLTK-1 cells were divided into six groups, each treated with increasing concentrations of CBZ (0 (CTR), 5, 25, 50, 100, and 200 μM) in DMEM:Ham's F12 culture medium (as in 2.1.1) supplemented with only 5% FBS. The cells were exposed for 24 hours. The chosen CBZ concentrations were based on plasmatic levels reported in the literature,24,25 with additional intermediate concentrations included. A control group (CTR, 0 μM) was also included in which the cells were cultured with the culture medium previously described with no CBZ and the same concentration of DMSO as the other groups (0.5%).Sulforhodamine B (SRB) cytotoxicity assay
The sulforhodamine B (SRB) assay was performed as previously described.26 Briefly, cells were seeded in 48-well plates and exposed to increasing concentrations of CBZ after reaching approximately 80% confluence. After 24 h, the medium was removed and used for the lactate dehydrogenase (LDH) release assay. Cells were washed using phosphate buffered saline (PBS) and fixed with 1% acetic acid in methanol for at least 1 h at −20 °C. The cells were then incubated, for 1 hour, with a solution of 0.05% SRB in 1% acetic acid at 37 °C. After incubation, cells were again washed with 1% acetic acid. The SRB dye bound to the cells was removed, using a 10 mM Tris solution with pH 10, and 200 μL of the resulting solution was transferred to a 96-well plate in duplicates. The optical density was measured in a multiplate reader (MultiSkan Go, Thermo Fisher Scientific) at 510 nm. The obtained results were divided by the mean of the control group and expressed in fold variation to the control.Lactate dehydrogenase release assay
To perform the LDH release assay, the cell culture medium from the SRB assay was collected to a 96-well plate in duplicates. The LDH activity was measured using the LDH-CytoxTM Assay Kit (BioLegend®, USA), which was performed following manufacturer instructions. The optical density was measured in a multiplate reader (MultiSkan Go, Thermo Fisher Scientific) at 490 nm. The obtained results were divided by the mean of the control group and expressed in fold variation to the control.3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
The MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay was performed as described.27 Cells were seeded and exposed to respective doses of CBZ in a 48-well plate for 24 hours. Afterwards, the medium was removed, and replaced for DMEM:Ham's F12 medium containing 5 mg mL−1 of MTT. The cells were then incubated for 3 hours at 37 °C. Following incubation, the medium was removed, and DMSO was added to solubilize the formazan crystals. The solubilized formazan in DMSO was transferred to a 96-well plate in duplicates. Optical density was measured using a multiplate reader (MultiSkan Go, Thermo Fisher Scientific) at 570 nm and 655 nm. To analyse the results, the absorbance measured at 655 nm was subtracted from the absorbance measured at 570 nm. The resulting values were divided by the mean of the control group and expressed in fold variation to the control.Extraction of lipids
Lipids were extracted using the Bligh and Dyer method, with modifications.28,29 For this procedure, the cell pellets were resuspended in 1 mL of Milli-Q water and transferred from the microcentrifuge tube into the respective extraction tubes (Pyrex tubes). A volume of 3.75 mL of CH2Cl2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH mixture in a 1
MeOH mixture in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (v/v) ratio was added and vortexed for 1 minute. The mixture was incubated on ice for 30 minutes under agitation (75 rpm) using an orbital shaker (Stuart Reciprocating Shaker SSL2, Stuart, UK), with vortexing every 5 minutes. After 30 minutes, the tubes were vortexed again for 1 min. Following this, 1.25 mL of CH2Cl2 and 1.25 mL of Milli-Q water were sequentially added to each tube, with 1 minute of vortexing between each addition. The tubes were then placed in a centrifuge (Centurion Scientific, Pro-Analytical C4000R, Stoughton, UK) for 10 minutes at 2000 rpm to achieve phase separation. The organic phase from each tube was transferred to new tubes. To the remaining aqueous phase, 1.88 mL of CH2Cl2 was added, followed by vortexing for 1 minute and a new centrifugation step. The organic phase was collected into the previous glass tube, dried under a stream of nitrogen gas, and re-dissolved in CH2Cl2. The total lipid extracts were transferred to amber vials, dried, and stored at −80 °C until further analysis.
2 (v/v) ratio was added and vortexed for 1 minute. The mixture was incubated on ice for 30 minutes under agitation (75 rpm) using an orbital shaker (Stuart Reciprocating Shaker SSL2, Stuart, UK), with vortexing every 5 minutes. After 30 minutes, the tubes were vortexed again for 1 min. Following this, 1.25 mL of CH2Cl2 and 1.25 mL of Milli-Q water were sequentially added to each tube, with 1 minute of vortexing between each addition. The tubes were then placed in a centrifuge (Centurion Scientific, Pro-Analytical C4000R, Stoughton, UK) for 10 minutes at 2000 rpm to achieve phase separation. The organic phase from each tube was transferred to new tubes. To the remaining aqueous phase, 1.88 mL of CH2Cl2 was added, followed by vortexing for 1 minute and a new centrifugation step. The organic phase was collected into the previous glass tube, dried under a stream of nitrogen gas, and re-dissolved in CH2Cl2. The total lipid extracts were transferred to amber vials, dried, and stored at −80 °C until further analysis.
Phospholipid quantification
The quantification of the total phospholipids (PL) recovered after extraction was done using an adapted version of the Bartlett and Lewis method.30,31 Briefly, lipid extracts were dissolved in 200 μL of CH2Cl2, and 10 μL aliquots were transferred, in duplicate, to glass tubes previously washed with 5% nitric acid. The solvent was evaporated under a stream of nitrogen, followed by the addition of 125 μL of 70% perchloric acid to each tube. Samples were then incubated in a heating block (Stuart, UK) at 180 °C for 1 hour. After cooling to room temperature, 825 μL of Milli-Q water, 125 μL of 2.5% ammonium molybdate (25 μg μL−1 of Milli-Q water), and 125 μL of 10% ascorbic acid (100 μg μL−1 of Milli-Q water) were sequentially added to each sample, with vortexing between additions. The samples were further incubated in a water bath at 100 °C for 10 minutes and then immediately cooled in a cold-water bath. Phosphate standards ranging from 0.1 to 2 μg of phosphorus (P) were prepared using sodium dihydrogen phosphate dihydrate (NaH2PO4·2H2O, 0.1 μg μL−1 P). These standards underwent the same experimental procedure as the samples, excluding the heat block step. Absorbance was measured at 797 nm using a Multiskan GO 1.00.38 Microplate Spectrophotometer (Thermo Scientific, Hudson, NH, USA) controlled by SkanIT version 3.2 software (Thermo Scientific™). The amount of P present in each sample was determined by linear regression. For each lipid extract, the total PL amount was calculated by multiplying the phosphorus amount by 25.32Fatty acid analysis by gas chromatography coupled to mass spectrometry (GC-MS)
Fatty acids were analyzed by GC-MS using the transmethylation method described by Aued-Pimentel et al.,33 which is routinely employed in our laboratory.34 An aliquot of 15 μg of PL was transferred to a glass tube previously washed with 99% hexane, dried under a nitrogen stream, and dissolved in 1 mL of n-hexane containing the internal standard methyl-nonadecanoate (Sigma, St. Louis, MO, USA) at a concentration of 1.5 × 10−3 μg μL−1. Esterified fatty acids underwent conversion to fatty acid methyl esters (FAMEs) through the addition of 200 μL of a 2 M methanolic KOH solution, followed by vigorous vortexing for 2 minutes. Next, 2 mL of saturated NaCl solution (10 μg μL−1) was added. After centrifugation at 2000 rpm for 5 minutes, 600 μL of the organic phase was collected and dried using a nitrogen stream. The resulting FAME derivatives were dissolved in 100 μL of n-hexane, and 2 μL were injected in GC-MS for analysis utilizing an Agilent Technologies 8860 GC System (Santa Clara, CA, USA) equipped with a DB-FFAP column (Agilent Technologies). The GC system was connected to an Agilent 5977B Mass Selective Detector operating in electron impact mode at 70 eV, scanning the range m/z 50–550 in a 1s cycle using full scan mode acquisition. The oven temperature was programmed as follows: an initial temperature of 58 °C for 2 minutes, followed by a linear increase to 160 °C at 25 °C min−1, then a linear increase at 2 °C min−1 to 210 °C, further increased at 20 °C min−1 to 230 °C, and held at 230 °C for 15 minutes. The injector and detector temperatures were set at 220 °C and 230 °C, respectively. Helium served as the carrier gas at a flow rate of 1.4 × 103 μL min−1. The data acquisition software employed was GCMS5977B/Enhanced MassHunter, with data analysis performed using Agilent MassHunter Qualitative Analysis 10.0 software. Fatty acid identification was also confirmed by comparing the retention times and fragmentation patterns of the analytes with those of the Supelco 37 Component FAME Mix (Sigma-Aldrich), which includes a comprehensive range of fatty acid standards with chain lengths from C8 to C24 and saturation levels ranging from 0 to 6 double bonds. For fatty acids not included in the FAME mix, identification was further corroborated by comparing their mass spectra with entries in the NIST library database and the Lipid Map Web. The GC-MS approach utilized herein allows for the detection of fatty acids with chain lengths of C8 and higher, as confirmed by employing FAME standards. The GC-MS approach utilized herein allows for the detection of fatty acids with chain lengths of C8 and higher, as confirmed by employing FAME standards. Fatty acid quantification was executed utilizing calibration curves obtained from FAME standards under identical instrumental conditions.Characterization of the lipid profile by reverse phase liquid chromatography coupled to high-resolution tandem mass spectrometry (C18 LC-MS/MS)
Lipids were analyzed by C18 reverse-phase liquid chromatography in an HPLC system (Ultimate 3000 Dionex, Thermo Fisher Scientific, Bremen, Germany) coupled online to a Q-Exactive™ Hybrid Quadrupole-Orbitrap™ Mass Spectrometer (Thermo Fisher Scientific, Bremen, Germany). Lipid extracts were resuspended in CH2Cl2 to have a PL concentration of 1 μg PL μL−1. In a vial with a micro-insert, 10 μL of each sample, 8 μL of a mixture of internal standards and 82 μL of isopropanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (1
MeOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were mixed. A volume of 5 μL of this mixture was injected into the HPLC column (Ascentis® Express 90 Å C18 HPLC, Sigma-Aldrich®, 2.1 × 100 mm; 2.7 μm, Supelco®), at 50 °C and at a flow rate of 260 μL min−1. The internal standard mixture contained 0.04 μg of phosphatidylcholine (PC, 14:0/14:0), 0.04 μg of phosphatidylethanolamine (PE, 14:0/14:0), 0.024 μg of phosphatidylglycerol (PG, 14:0/14:0), 0.08 μg of phosphatidylinositol (PI, 16:0/16:0), 0.08 μg of phosphatidylserine (PS, 14:0/14:0), 0.16 μg of phosphatidic acid (PA, 14:0/14:0), 0.04 μg of lyso-PC (LPC, 19:0), 0.04 μg of sphingomyelin (SM, d18:1/17:0), 0.08 μg of ceramide (d18:1/17:0) and 0.16 μg of cardiolipin (CL, 14:0/14:0/14:0/14:0). Before lipidomic analysis, LC-MS data acquisition was conducted on a sample without internal standards to ensure that no lipid species corresponding to the selected internal standards were present in the samples. The elution started with 32% of mobile phase B, followed by the following gradient: 45% B (1.5 min), 52% B (4 min), 58% B (5 min), 66% B (8 min), 70% B (11 min), 85% B (14 min), 97% B (18 min, maintained for 7 min), and 32% B (25.01 min, followed by a re-equilibration period of 8 min prior next injection). Eluent A was composed of 60% acetonitrile, 40% water, 10 mM ammonium formate and 0.1% formic acid and eluent B was composed of 90% isopropanol, 10% acetonitrile, 10 mM ammonium formate and 0.1% formic acid. The Q-Exactive™ Orbitrap mass spectrometer, equipped with a heated electrospray ionization source, operated in both positive and negative modes. The electrospray voltage was set to 3.0 kV for the positive mode and −2.7 kV for the negative mode. Operational parameters included a sheath gas flow of 35 U, an auxiliary gas flow of 3 U, a capillary temperature of 320 °C, an S-lenses RF of 50 U, and a probe temperature of 300 °C. Full-scan MS spectra were acquired across an m/z range of 200–1600, with a resolution of 70
1) were mixed. A volume of 5 μL of this mixture was injected into the HPLC column (Ascentis® Express 90 Å C18 HPLC, Sigma-Aldrich®, 2.1 × 100 mm; 2.7 μm, Supelco®), at 50 °C and at a flow rate of 260 μL min−1. The internal standard mixture contained 0.04 μg of phosphatidylcholine (PC, 14:0/14:0), 0.04 μg of phosphatidylethanolamine (PE, 14:0/14:0), 0.024 μg of phosphatidylglycerol (PG, 14:0/14:0), 0.08 μg of phosphatidylinositol (PI, 16:0/16:0), 0.08 μg of phosphatidylserine (PS, 14:0/14:0), 0.16 μg of phosphatidic acid (PA, 14:0/14:0), 0.04 μg of lyso-PC (LPC, 19:0), 0.04 μg of sphingomyelin (SM, d18:1/17:0), 0.08 μg of ceramide (d18:1/17:0) and 0.16 μg of cardiolipin (CL, 14:0/14:0/14:0/14:0). Before lipidomic analysis, LC-MS data acquisition was conducted on a sample without internal standards to ensure that no lipid species corresponding to the selected internal standards were present in the samples. The elution started with 32% of mobile phase B, followed by the following gradient: 45% B (1.5 min), 52% B (4 min), 58% B (5 min), 66% B (8 min), 70% B (11 min), 85% B (14 min), 97% B (18 min, maintained for 7 min), and 32% B (25.01 min, followed by a re-equilibration period of 8 min prior next injection). Eluent A was composed of 60% acetonitrile, 40% water, 10 mM ammonium formate and 0.1% formic acid and eluent B was composed of 90% isopropanol, 10% acetonitrile, 10 mM ammonium formate and 0.1% formic acid. The Q-Exactive™ Orbitrap mass spectrometer, equipped with a heated electrospray ionization source, operated in both positive and negative modes. The electrospray voltage was set to 3.0 kV for the positive mode and −2.7 kV for the negative mode. Operational parameters included a sheath gas flow of 35 U, an auxiliary gas flow of 3 U, a capillary temperature of 320 °C, an S-lenses RF of 50 U, and a probe temperature of 300 °C. Full-scan MS spectra were acquired across an m/z range of 200–1600, with a resolution of 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, an automatic gain control (AGC) target of 3 × 106, and a maximum injection time (IT) of 100 ms. For MS/MS experiments, a top-10 data-dependent method was employed, wherein the top 10 most abundant precursor ions in the full MS were selected for fragmentation in the HCD collision cell. A stepped normalized collision energy scheme was applied, ranging between 25 and 30 eV for the positive ion mode and between 20, 24, and 28 for the negative ion mode. MS/MS spectra were acquired with a resolution of 17
000, an automatic gain control (AGC) target of 3 × 106, and a maximum injection time (IT) of 100 ms. For MS/MS experiments, a top-10 data-dependent method was employed, wherein the top 10 most abundant precursor ions in the full MS were selected for fragmentation in the HCD collision cell. A stepped normalized collision energy scheme was applied, ranging between 25 and 30 eV for the positive ion mode and between 20, 24, and 28 for the negative ion mode. MS/MS spectra were acquired with a resolution of 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, an AGC target of 1 × 105, an isolation window of 1 m/z, and a maximum IT of 100 ms. The acquisition cycle comprised one full-scan mass spectrum and ten data-dependent MS/MS scans, continuously repeated throughout the experiments, with the dynamic exclusion set to 30 s and an intensity threshold of 8 × 104. Data acquisition was managed using the Xcalibur data system (V3.3, Thermo Fisher Scientific, USA).
500, an AGC target of 1 × 105, an isolation window of 1 m/z, and a maximum IT of 100 ms. The acquisition cycle comprised one full-scan mass spectrum and ten data-dependent MS/MS scans, continuously repeated throughout the experiments, with the dynamic exclusion set to 30 s and an intensity threshold of 8 × 104. Data acquisition was managed using the Xcalibur data system (V3.3, Thermo Fisher Scientific, USA).
The LC-MS data were processed using the Lipostar software (Molecular Discovery Ltd, version 2.1.5 × 64).35 This software was used for raw data import, peak detection, alignment, noise reduction, and identification. Lipid assignment and identification were performed using a database created from the LIPID MAPS Structure Database (downloaded in February 2024).36 The database was fragmented using the DB Manager Module of Lipostar, following Lipostar fragmentation rules. The raw files were directly imported and aligned using the settings described by Lange et al.37 Automatic peak picking was carried out with the SDA smoothing level set to low and a minimum signal-to-noise ratio (S/N) of 3. Automatic isotope clustering was performed setting an m/z tolerance of 7 ppm and a retention time tolerance of 0.2 min. After data processing, t an automatic filtering was applied to retain only features with MS/MS spectra for identification. Lipid identification was based on the following parameters: a 5 ppm precursor ion mass tolerance and a 10 ppm product ion mass tolerance. An automatic approval process was applied to retain only identifications with a confidence level of 3–4 stars.35 The putative annotations from Lipostar were confirmed, considering the typical fragmentation pattern of each class lipid class.38
Statistical analysis
Multivariate and univariate statistical analyses were conducted using R version 4.3.139 in RStudio version 2024.04.2.40 Lipid species areas obtained from LC-MS were normalized by sum, log-transformed (base 10),41 and further normalized using EigenMS.42 Principal component analysis (PCA) was performed with the R packages FactoMineR43 and factoextra.44 Heatmaps were generated from autoscaled data using the R package heatmap, with “Euclidean” as the clustering distance and “ward.D” as the clustering method.45Normality and variance homogeneity of the data were assessed using the Shapiro-Wilk and Levene's tests, respectively. Based on these assessments, ANOVA was used if the assumptions were satisfied otherwise, the Kruskal–Wallis test was applied. Significant differences identified by ANOVA were further analyzed using Tukey's HSD test, while Dunn's test was used for post hoc comparisons following the Kruskal–Wallis test. P-Values were adjusted for multiple testing using the Benjamini–Hochberg method for the false discovery rate (FDR, q-values). All univariate analyses were conducted using the R package rstatix with a significance threshold of p < 0.05.46 All graphics and boxplots were created using the R package ggplot2.47
Results
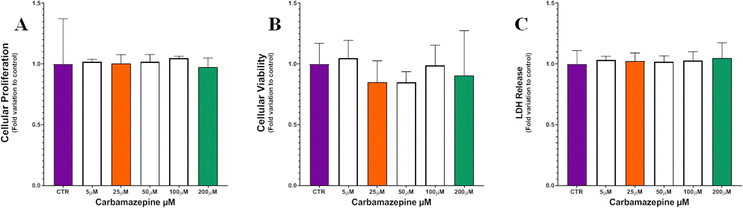
CBZ did not impact Leydig cell viability
The cellular proliferation was evaluated using the SRB assay and no significant differences were observed after cell exposure to the CBZ concentrations tested including 25 μM (1.00 ± 0.07-fold variation to control) and 200 μM (0.98 ± 0.07-fold variation to control), when compared to controls (1.00 ± 0.37-fold variation to control) (Fig. 1A). In what concerns the evaluation of the metabolic viability, using the MTT assay, the Leydig cells exposed to the highest concentrations of CBZ showed a tendency for reduction in the metabolic activity, particularly the group of Leydig cells exposed to 25 μM of CBZ (0.85 ± 0.18-fold variation to control) and to 200 μM of CBZ (0.91 ± 0.37-fold variation to control) when compared with the control group (1.00 ± 0.17-fold variation to control) (Fig. 1B). The results gathered from the LDH release assay, used to evaluate membrane damage, although without statistical significance, showed that the cells exposed to 200 μM of CBZ (1.05 ± 0.12-fold variation to control) exhibited a slight higher tendency for LDH release to the extracellular medium when compared to the Leydig cells from the control group (1.00 ± 0.11-fold variation to control) and from the group exposed to 25 μM (1.02 ± 0.07-fold variation to control) of CBZ (Fig. 1C).Fatty acid analysis by GC-MS
The profile of the esterified fatty acids of the Leydig cells was determined by GC-MS after alkaline transmethylation. A total of 12 FA were identified in Leydig cells from both CTR and CBZ groups (Table 1 and Fig. S1, ESI†). The palmitic acid (FA 16:0) was the most abundant FA in all groups followed by the oleic acid (FA 18:1 n-9), vaccenic acid (FA 18:1 n-7) and stearic acid (FA 18:0). These results are in line with the FA profile of Leydig cells previously described.7,48| Fatty acids | CTR | CBZ 25 μM | CBZ 200 μM |
|---|---|---|---|
| FA 14:0 | 0.8 ± 0.2 | 0.9 ± 0.2 | 1.0 ± 0.4 |
| FA 16:0 | 21.8 ± 1.3 | 21.7 ± 1.2 | 21.7 ± 1.0 |
| FA 16:1 n-9 | 1.0 ± 0.3 | 1.0 ± 0.2 | 0.7 ± 0.3 |
| FA 16:1 n-7 | 6.5 ± 1.0 | 7.1 ± 1.6 | 7.4 ± 1.6 |
| FA 18:0 | 13.6 ± 0.8 | 13.6 ± 0.9 | 14.2 ± 3.4 |
| FA 18:1 n-9 | 20.3 ± 1.3 | 19.9 ± 1.0 | 20.0 ± 1.7 |
| FA 18:1 n-7 | 18.5 ± 2.2 | 18.4 ± 1.8 | 18.3 ± 2.3 |
| FA 18:2 n-6 | 1.7 ± 0.3 | 1.9 ± 0.3 | 1.9 ± 0.3 |
| FA 20:4 n-6 | 5.3 ± 1.0 | 5.8 ± 1.1 | 5.4 ± 1.3 |
| FA 22:4 n-6 | 1.1 ± 0.4 | 1.0 ± 0.4 | 1.1 ± 0.5 |
| FA 22:5 n-3 | 1.8 ± 0.3 | 1.7 ± 0.4 | 1.8 ± 0.4 |
| FA 22:6 n-3 | 2.5 ± 1.1 | 2.5 ± 0.9 | 1.9 ± 1.0 |
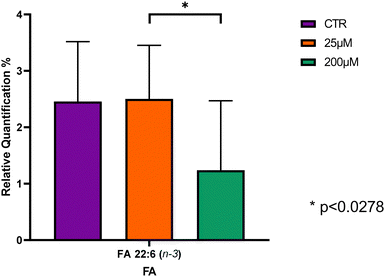
Statistical analysis showed that of the 12 FA identified, only the FA 22:6 n-3 revealed significant differences between the CBZ 25 μM and CBZ 200 μM groups, with Leydig cells treated with CBZ 200 μM showing the lowest content in this n-3 FA in comparison to CTR and CBZ 25 μM, however, no significant changes were observed when comparing controls and CBZ 200 μM groups (Fig. 2 and Table S1, ESI†).
The sum of saturated (SFA), monounsaturated (MUFA) and polyunsaturated fatty acids (PUFA) were calculated as well as the ratios of n-6/n-3 (Table 2). MUFA was the group of FA most abundant in all conditions followed by SFA. The results further demonstrate that the most abundant FAs are MUFA (FA 18:1) and SFA (FA 16:0). The results are in accordance with a previous study on lipid metabolism of the M5480 murine Leydig cell tumor,7 which states that in Leydig cell tumors, SFA and MUFA are higher in comparison with PUFA.
| Σ fatty acids | CTR | CBZ 25 μM | CBZ 200 μM |
|---|---|---|---|
| Σ n-3 | 4.3 ± 1.3 | 4.2 ± 1.3 | 3.1 ± 1.5 |
| Σ n-6 | 8.2 ± 1.7 | 7.5 ± 3.7 | 8.4 ± 2.0 |
| Σ SFA | 36.3 ± 2.1 | 36.2 ± 1.8 | 36.9 ± 2.7 |
| Σ MUFA | 47.6 ± 4.0 | 47.7 ± 4.4 | 47.7 ± 5.5 |
| Σ PUFA | 12.4 ± 2.9 | 11.7 ± 4.8 | 11.5 ± 3.1 |
| n-6/n-3 | 2.0 ± 0.3 | 1.7 ± 0.8 | 3.2 ± 1.2 |
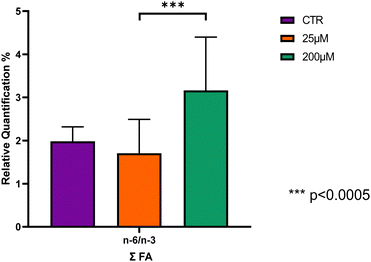
Only the n-6/n-3 ratio revealed significant differences, being significantly higher in Leydig cells treated with CBZ 200 μM compared with Leydig cells treated with CBZ 25 μM, p < 0.0005, however, no significant changes were observed when comparing controls and CBZ 200 μM groups (Fig. 3 and Table S2, ESI†).
Identification of the lipid profile by LC-MS/MS
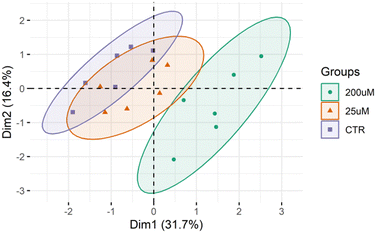
The lipid profile of the Leydig cells was analyzed by high-resolution LC-MS and MS/MS (Fig. S3–S9, ESI†). This lipidomic analysis allowed the identification of 367 different lipid molecular species (m/z values of molecular ions) belonging to 13 different classes, namely 146 phosphatidylcholines (PC) including diacyl, lyso PC (LPC), alkyl–acyl and alkenyl–acyl species, 68 phosphatidylethanolamines (PE) including diacyl, lyso PE (LPE), alkyl–acyl and alkenyl–acyl species, 15 phosphatidylinositols (PI), 8 phosphatidylserines (PS), 21 sphingomyelins (SMs), 14 phosphatidylglycerol (PG), 6 cardiolipin (CL), 12 ceramides (Cer), 4 Hexosylceramides (HexCer), 7 sphingosine (SPB), 1 coenzimeQ10, 8 diacylglycerols (DG) and 57 triacylglycerols (TG) (Table S3, ESI†).The differences between the lipid profiles of Leydig cells treated with CBZ 25 μM, with CBZ 200 μM, and CTR were assessed using multivariate statistical analysis. The PCA score plot of the two first dimensions (Dim) showed that the three groups were separated into three different clusters (90% confidence interval), with the model capturing 48.1% of the total variance in the data set (Dim 1:31.7%; and Dim 2:16.4%) (Fig. 4). The samples from the CTR group were scattered in the left region of the PCA plot while the group treated with 200 μM CBZ were scattered in the right region. Leydig cells treated with 25 μM of CBZ are clustered in the middle scattering of the PCA plot, with some overlap with the control group.
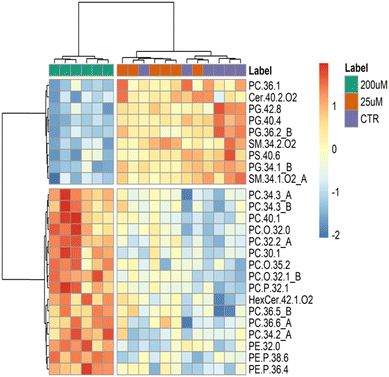
Additionally, we performed a hierarchical clustering analysis on the lipid data sets of the three groups (Fig. 5). A heatmap (double dendrograms) with two-dimensional hierarchical clustering was created using the top 25 q-values ANOVA test. The resulting hierarchical clustering (Fig. 5) showed a noticeable separation in the first dimension (upper hierarchical dendrogram) where samples are independently clustered into two groups, one for the Leydig cells treated with CBZ 200 μM (green), and a second group clustering Leydig cells treated with 25 μM (orange) and CTR (purple). Moreover, the second dimension shows two principal clusters: the first group (top in the dendrogram) includes 9 lipid species, namely 1 PC, 4 PG, 1 Cer, 2 SM and 1 PS, which are lower in the Leydig cells with the treated CBZ 200 μM group; while the second group (bottom on the dendrogram) with 16 different lipid species including 12 PC, 1 HexCer and 3 PE species with higher abundance in Leydig cells with the treated CBZ 200 μM group.
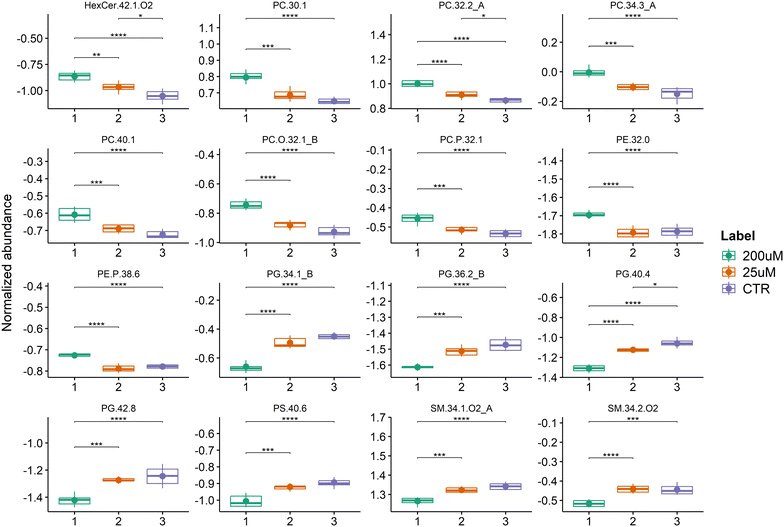
Univariate analysis of the LC-MS data from the three groups was performed to test for significant differences between the three groups (Table S4, ESI†). The lipid analysis is the total 66 PC, 34 ether-linked PC, 11 PG, 33 PE, 22 ether-linked PE, 4 HexCer and 7 PS different lipid species (m/z values of molecular ions). The boxplots of the 16 main species with the lowest q-values (q-value < 0.05) are shown in Fig. 6 and the major contributors correspond to 4 PC, 4 PG, 2 SM, 2 ether-linked PC, 1 PS, 1 PE, 1 ether-linked PC and 1 HexCer species. All of these PC species (PC 32:2_A that corresponds to PC 16:1_16:1, PC 40:1, PC 30:1, and PC 34:3_A that corresponds to PC 12:0_22:3), PC O-32:1_B (PC-O 16:0_16:1), PC P-32:1, PE 32:0, PE P-38:6 and HexCer 42:1;O2 were significantly increased in Leydig cells treated with 200 μM of CBZ compared to controls and 25 μM of CBZ. The HexCer 42:1;O2, PC 32:2 (PC 16:1_16:1), and PG 40:4 were also significantly increased in Leydig cells treated with 25 μM of CBZ compared to controls. In contrast, PG species (PC 34:1_B corresponding to PG 16:0_18:1, PG 36:2_B corresponding to PG 18:1_18:1, PG 40:4 and PG 42:8), PS 40:6 and SM species (SM 34:1;O2_A that corresponds to SM 18:1;O2_16:0 and SM 34:2;O2) were significantly reduced in Leydig cells treated with 200 μM.
Discussion
In vivo, the primary role of Leydig cells in the testis is the synthesis and secretion of testosterone or androstenedione in response to luteinizing hormone (LH). However, the mechanisms by which carbamazepine acts as an endocrine disruptor in the testis remain unclear. It is well established that most endocrine imbalances directly or indirectly impact male fertility. Notably, several studies have reported alterations in male fertility associated with the use of antiepileptic medications. For instance, carbamazepine has been linked to asthenozoospermia.49 High doses of valproic acid have been associated with oligoasthenoteratozoospermia, a condition that improved upon reducing the dosage. Antiepileptic drugs, including carbamazepine, have also been reported to decrease sperm motility.50 Additionally, another study observed that men undergoing carbamazepine treatment exhibited lower sperm concentrations while maintaining normal morphology and motility.51CBZ is an anticonvulsant and mood-stabilizing drug commonly used to treat epilepsy, neuropathic pain, and bipolar disorder.52–54 Some studies have suggested that CBZ might disrupt the endocrine system, impacting testicular Leydig cells, hindering hormone production, and potentially leading to male infertility.55–57 CBZ may disrupt the normal function of these cells and can also have a direct inhibitory effect on their function.58 It can impair testosterone production, causing changes in androgen-dependent processes, such as male sexual differentiation, spermatogenic cycle establishment and maintenance.59–61 CBZ is known to depress spermatogenesis in mammals due to its cytotoxicity which results in the death of immature germ cells present in the seminiferous epithelium.62
In the present work, to assess the cytotoxic effect of CBZ in Leydig cells, we evaluated the cell (proliferation and membrane integrity) and the metabolic viability. Interestingly, none of the concentrations chosen altered cell proliferation, but treatment with 25 μM and 200 μM CBZ resulted in a tendency to decrease the metabolic viability. Results from the LDH assay show that the cells exposed to 200 μM of CBZ had significantly higher LDH release to the extracellular medium when compared to the Leydig cells from the control group, without statistical significance. These findings indicate that high concentrations of CBZ have a detrimental impact on Leydig cells, affecting the viability of the cells. Hence, we used 25 and 200 μM concentrations of CBZ for the subsequent stages of our study, being that these concentrations were also chosen to mimic the reported plasmatic levels of CBZ and a possible acute exposure, respectively.58
The effect of CBZ on the FA profile of Leydig cells was evaluated and a total of 12 FA were identified with FA 16:0, FA 18:0 and FA 18:1 as the most abundant FA, which is consistent with the profile previously reported in the literature in Leydig cells isolated from rats.7,51 Significant differences were only seen when Leydig cells were exposed to higher concentrations of CBZ with a significant decrease of FA 22:6 n-3. Reduced levels of FA 22:6 n-3 can result in less fluid cell membranes, impairing the function of membrane-bound proteins such as receptors, enzymes, and ion channels,63 which then may affect Leydig cell function. This reduction can potentially decrease testosterone production, vital for male reproductive health, cause inflammation, possibly leading to hypogonadism or other reproductive issues, and increase oxidative stress, damaging cellular components like DNA, proteins, and lipids.63–65 It was reported that in testis and Leydig cells, a decrease in this FA may lead to altered spermatogenesis, sperm quality, increased oxidative stress and hormonal changes, decreasing the production of male sex hormones, particularly testosterone.64,66,67
The alteration of the n-6/n-3 ratio was also observed, with a significant increase noted. The n-6/n-3 ratio was significantly higher in Leydig cells treated with CBZ at a concentration of 200 μM. Studies have shown that men experiencing fertility problems tend to have higher n-6/n-3 ratios in their blood and seminal plasma, as well as lower overall levels of n-3 FA.68 This imbalance has been associated with reduced sperm quality, including decreased sperm motility, lower sperm count, and increased DNA fragmentation.69 Furthermore, the elevated n-6/n-3 ratio may lead to increased oxidative stress and lipid peroxidation in sperm, potentially damaging sperm structure and function.70 These findings suggest that an increase in this ratio could have a significant impact on male reproductive potential and may play a role in the development of idiopathic oligoasthenoteratozoospermia, a common cause of male infertility.69
The plasticity of the Leydig cell lipidome was also evaluated. Lipidomics data sets were analyzed by multivariate PCA, hierarchical cluster analysis and univariate analysis for a comparison between the three groups to assess the differences between increasing concentrations of CBZ. Analysis of the PCA score plot (Fig. 4) showed that the lipid profile of Leydig cells was different among the CBZ treatments, particularly for the highest concentration. The results from hierarchical and univariate analysis (Fig. 5 and 6) revealed a significant increase in lipid species of PC, PE and HexCer and a decrease in lipid species of PG, PS and SM, which were predominantly observed in Leydig cells treated with 200 μM of CBZ.
In our study, 13 PC species were in the top 25 lipid species with a major variation observed in Leydig cells after exposure to CBZ (25 and 200 μM). The PC species which showed a significant increase in Leydig cells exposed to 200 μM CBZ are PC 34:3 (PC 12:0_22:3), PC 40:1, PC 32:2_A (PC 16:1_16:1), PC 30:1, PC 36:5, PC 36:6 and PC 34:2 and the alkyl-acyl PC O-32:0, PC O-35:2, PC O-32:1_B (O-16:0_16:1) and the alkenyl-acyl PC P-32:1. PC is the major class of phospholipids in cell membranes, being crucial for maintaining membrane structure and fluidity.5,6 So, changes in the profile and levels of this major phospholipid class may affect membrane and cell properties and functions. Increased PC levels may be an adaptive response of Leydig cells to CBZ-induced stress, helping to improve stress resistance and cell survival.71
The increased relative abundance of PC lipid species after exposure to CBZ is also an interesting feature observed in our results. The increase in PC has been associated and involved in carcinogenesis and tumor progression namely by supporting cell growth and proliferation.72 Interestingly, previous studies have reported that CBZ is associated with tumorigenesis in Leydig cells.21 In tumorigenesis, enzymes involved in the PC synthesis, such as choline kinase (CK), phosphocholine cytidylyltransferase (CCT) and choline phosphotransferase (CPT), exhibit increased activity.73 These enzymes are part of the Kennedy pathway, which is crucial for PC biosynthesis and can alter the lipid process, leading to increased conversion of other phospholipids into PC to support rapid cell division and growth.74,75
Few PE species were increased in Leydig cells treated with 200 μM of CBZ compared with cells under control conditions and the ones that contributed the most to the differentiation were the diacyl species PE 32:0 and the alkenyl-acyl PE P-38:6 and PE P-36:4. PE is the second most abundant class of PL in cells and plays an essential role in the membrane fusion, cell division and intracellular hierarchical structure organization, which also heavily affects testosterone synthesis.76,77 The increase in diacyl PE species may be caused by upregulated pathways for PE biosynthesis.78 Also the increase of the plasmanyl PE, well known endogenous antioxidant molecules may be part of a cellular adaptation, influencing the lipid metabolism, stress oxidative and inflammatory cytokines, leading to cell lysis and hypoxia.79
The decreased levels of PS species in cells exposed to the highest concentration of CBZ (200 μM), namely PS 40:6, could be attributed to the increased biosynthesis of PE and PC species, as reported previously.80 However, no significant variation in the PC or PE class level was observed. But changes in PS levels or membrane distribution might influence cholesterol transport within Leydig cells, affecting steroidogenesis.6 The PS class, at the cellular level, has been associated with anti-inflammatory roles and the loss of PS is an early indicator of apoptosis.80,81 Decreased PS levels can affect signaling pathways that promote cell survival, such PI3K/AKT pathway, potentially leading to enhanced tumor growth and enzymes like phosphatidylserine synthase (PSS) 1 and 2, which regulate PS levels, can be dysregulated in situations of tumorigenesis.78
Other PL and sphingolipid classes were downregulated in Leydig cells after exposure to CBZ. This is the case of PG and SM. Our results showed a significant decrease of 4 PG, namely of PG 42:8, PG 40:4, PG 36:2_B (PG 18:0_18:2 and PG 16:0_20:2) and PG 34:1_B (PG 16:0_18:1) in Leydig cells treated with 200 μM of CBZ, compared with those of the control group. PG is synthesized by the CDP-DAG pathway in the endoplasmic reticulum.82 PG plays several important roles in DNA replication, modification of cellular lipoprotein and cell death.83 The enzyme CDP-diacylglycerol synthetase plays a crucial role in regulating PG, with its decreased activity will cause the reduction of PG.84 Decrease of PG levels may be due to impairment of enzymes involved in PG synthesis and degradation, such phosphatidylglycerol phosphate synthase (PGPS) and phospholipase a2 (PLA2), that may have altered activities in tumorigenesis.85 Increased degradation or reduced synthesis of PG could lead to decreased levels. Tumor cells may divert precursors of PG, like phosphatidic acid (PA), towards the synthesis of other lipids that are more crucial for rapid cell proliferation and survival, such as PC or PE.79 Another reason is that PG is a precursor for cardiolipin, a lipid that is critical for mitochondrial function (including the structure, function, and stability of mitochondrial membranes).86 A decrease in PG might impact cardiolipin synthesis, with far-reaching effects on mitochondrial dynamics and function, potentially resulting in energy deficiency, increased oxidative stress, and cell death.87
Our study further reports a significant decrease of two SMs (SM 34:2;O2 and SM 34:1;O2_A corresponding to SM 18:1;O2_16:0) in Leydig cells with 200 μM of CBZ, compared to the control group. SMs contribute to membrane rigidity and stability, and hydrolysis of SMs by sphingomyelinases (SMases) produces ceramide, a bioactive lipid involved in regulating cell growth, differentiation and apoptosis thus important for cell function and survival,88,89 in other words, the decreased SM could be due to an increase of SMases that are enzymes that hydrolyse SM to produce Cer.90,91 Cellular stress can lead to the activation of SMase and the subsequent reduction of SM.88 Forced breakdown of SM by SMase has been demonstrated to decrease the cholesterol-holding capacity of the membranes and increase steroid hormone production.92 Conversely, SMase activity was associated with impaired Leydig cell function via degradation of SM and accumulation of the proapoptotic sphingolipid Cer.88,93
Conclusions
Our research demonstrates that CBZ significantly alters the lipid profile of Leydig cells at higher concentrations (200 μM), while lower doses (25 μM) have no discernible effect. This aligns with existing knowledge that high CBZ doses can impair spermatogenesis in mammals due to its cytotoxic impact on immature germ cells. The significant differences observed in the lipid molecular species highlight the complex interplay between CBZ dosage, exposure duration, and individual characteristics in influencing Leydig cell function, with an expected impact on hormone production and overall metabolic regulation. These findings contribute valuable insights into the potential mechanisms by which CBZ may disrupt male reproductive health and emphasize the importance of careful dosage consideration when using this medication.Author contributions
Conceptualization: I. N. and M. R. D.; methodology: A. D. M., I. M. S. G., M. P., T. M. and P. D.; validation: M. R. D. and T. M.; writing – original draft preparation: I. N, M. R. D and P. D. O; writing – review and editing: I. N., A. D. M., I. M. S. G., M. P., L. G., S. B., T. M., P. D., A. P., P. F. O.; visualization: M. R. D., I. N. and T. M.; supervision: R. D. and T. M. All authors have read and agreed to the published version of the manuscript.Data availability
The datasets supporting this article have been uploaded as part of the ESI.† Data for this paper, including mzML data files are available at Science Data Bank, 2024 [2024-11-08] at: https://doi.org/10.57760/sciencedb.16458.Conflicts of interest
The authors declare that there is no conflict of interests regarding the publication of this paper.Acknowledgements
The authors thank the University of Aveiro, Fundação para a Ciência e Tecnologia (FCT), and Ministério da Ciência Tecnologia e Ensino Superior (MCTES) for the financial support to the research units CESAM (UIDB/50017/2020 + UIDP/50017/2020 + LA/P/0094/2020) and LAQV-REQUIMTE (UIDB/50006/2020) through national funds and co-funded by ERDF, within Portugal 2020 Partnership Agreement and Compete 2020, and to the Portuguese Mass Spectrometry Network (RNEM, LISBOA-01-0145-FEDER-402-022125). The authors also acknowledge the COST Action EpiLipidNET, CA19105-Pan-European Network in Lipidomics and EpiLipidomics. Tânia Melo thanks the junior research contract in the scope of the Individual Call to Scientific Employment Stimulus 2020 [CEECIND/01578/2020, https://doi.org/10.54499/2020.01578.CEECIND/CP1589/CT0010]. Marisa Pinho (UI/BD/153346/2022) and Inês M. S. Guerra (2021.04754.BD, https://doi.org/10.54499/2021.04754.BD) are grateful to FCT for their grants.References
- B. R. Zirkin and V. Papadopoulos, Leydig cells: formation, function, and regulation, Biol. Reprod., 2018, 99, 101–111 CrossRef PubMed.
- R. P. Amann, Structure and Function of the Normal Testis and Epididymis, J. Am. Coll. Toxicol., 1989, 8(3), 457–471 CrossRef.
- U. M. Bello, M. Cathrine, M. Hermanus, B. G. Augustine and T. A. Aire, Changes in testicular histomorphometry and ultrastructure of Leydig cells in adult male Japanese quail exposed to di(n-butyl) phthalate (DBP) during the prepubertal period, Environ. Sci. Pollut. Res., 2023, 30, 55402–55413, DOI:10.1007/s11356-023-25767-2.
- H. G. Van Der, S. H. Kotzé, M. J. O. Riain, L. Maree and W. G. Breed, Testicular Structure and Spermatogenesis in the Naked Mole-Rat Is Unique (Degenerate) and Atypical Compared to Other Mammals, Front. Cell Dev. Biol., 2019, 7, 1–13 CrossRef PubMed.
- P. P. Koganti, L. N. Tu and V. Selvaraj, Functional metabolite reserves and lipid homeostasis revealed by the MA-10 Leydig cell metabolome, PNAS Nexus, 2022, 1, 1–14 CrossRef CAS PubMed.
- S. Venugopal, M. Galano, R. Chan, E. Sanyal, L. Issop and S. Lee, et al., Dynamic Remodeling of Membranes and Their Lipids during Acute Hormone-Induced Steroidogenesis in MA-10 Mouse Leydig Tumor Cells, Int. J. Mol. Sci., 2021, 22(2554), 1–24 Search PubMed.
- D. H. Albert, A. Mario and J. G. Coniglio, Lipid composition and gonadotropin-mediated lipid metabolism of the M5480 murine Leydig cell tumor, J. Lipid Res., 1980, 21(7), 862–867, DOI:10.1016/S0022-2275(20)34782-9.
- S. Dutta, P. Sengupta and P. Slama, Oxidative Stress, Testicular Inflammatory Pathways, and Male Reproduction, Int. J. Mol. Sci., 2021, 22(10043), 1–20 Search PubMed.
- A. Vermeulen, Effects of drugs on Leydig cell function, Int. J. Androl., 1982, 5, 163–182 CrossRef CAS.
- J. Chung, S. Brown, H. Chen, J. Liu and B. Zirkin, Effects of pharmacologically induced Leydig cell testosterone production on intratesticular testosterone and spermatogenesis, Biol. Reprod., 2020, 102, 489–498 CrossRef PubMed.
- K. Svechnikov, L. Landreh, J. Weisser, G. Izzo, E. Colón, I. Svechnikova and O. Soder, Origin, Development and Regulation of Human Leydig Cells, Horm. Res. Pediatr., 2010, 73, 93–101 CrossRef CAS PubMed.
- R. R. Andretta, F. K. Okada, C. C. Paccola, T. Stumpp, S. U. De Oliva and S. M. Miraglia, Carbamazepine-exposure during gestation and lactation affects pubertal onset and spermatic parameters in male pubertal offspring, Reprod. Toxicol., 2014, 44, 52–62, DOI:10.1016/j.reprotox.2013.09.009.
- J. Gierbolini, M. Giarratano, S. R. Benbadis, J. Gierbolini, M. Giarratano and S. R. Benbadis, Carbamazepine-related antiepileptic drugs for the treatment of epilepsy – a comparative review, Expert Opin. Pharmacother., 2016, 17(7), 885–888, DOI:10.1517/14656566.2016.1168399.
- S. U. De Oliva and S. M. Miraglia, Carbamazepine damage to rat spermatogenesis in different sexual developmental phases, Int. J. Androl., 2008, 563–574 Search PubMed.
- A. Blondet, G. Martin, P. Durand and M.-H. Perrard, A comparative study of the effects of 3 testicular toxicants in cultures of seminiferous tubules of rats or of domestic cats (veterinary waste): an alternative method for reprotoxicology, Toxicol. In Vitro, 2022, 83, 105397 CrossRef PubMed.
- H. Chen, J. Guo, R. Ge, Q. Lian, V. Papadopoulos and B. R. Zirkin, Steroidogenic Fate of the Leydig Cells that Repopulate the Testes of Young and Aged Brown Norway Rats after Elimination of the Preexisting Leydig Cells, Exp. Gerontol., 2015, 72, 8–15 CrossRef CAS PubMed.
- S. Bramswig, A. Kerksiek, T. Sudhop, C. Luers, K. V. O. N. Bergmann and H. K. Berthold, et al., Carbamazepine increases atherogenic lipoproteins: mechanism of action in male adults, Am. J. Physiol.: Heart Circ. Physiol., 2001, 282, 704–716 CrossRef PubMed.
- T. Ohnishi and Y. Ichikawa, Direct inhibitions of the activities of steroidogenic cytochrome P-450 mono-oxygenase systems by anticonvulsants, J. Steroid Biochem. Mol. Biol., 1997, 60, 77–85 CrossRef CAS PubMed.
- R. Lingamaneni and H. C. Hemmings Jr., Differential interaction of anaesthetics and antiepileptic drugs with neuronal Na + channels, Ca 2+ channels, and GABA A receptors, Br. J. Anaesth., 2003, 90(2), 199–211, DOI:10.1093/bja/aeg040.
- A. G. Herzog, F. W. Drislane, D. L. Schomer, P. B. Pennell, E. B. Bromfield and K. M. Kelly, et al., Differential Effects of Antiepileptic Drugs on Sexual Function and Reproductive Hormones in Men with Epilepsy: Interim Analysis of a Comparison between Lamotrigine and Enzyme-inducing Antiepileptic Drugs, Epilepsia, 2004, 45(7), 764–768 CrossRef CAS PubMed.
- D. E. Prentice, A review of drug-induced Leydig cell some comparisons with man, Hum. Exp. Toxicol., 1995, 14, 562–572 CrossRef CAS PubMed.
- C. A. Alvarez-gonza, I. S. Sandbakken, H. Su, L. Johansen, Y. Zhang and E. Ringø, et al., Replacing fishmeal with salmon hydrolysate reduces the expression of intestinal in fl ammatory markers and modulates the gut microbiota in Atlantic salmon (Salmo salar), Front. Biosci.-Landmark, 2024, 1–15 Search PubMed.
- K. Kananen, et al., The mouse inhibin α-subunit promoter directs SV40 T-antigen to Leydig cells in transgenic mice, Mol. Cell. Endocrinol., 1996, 135–146 CrossRef CAS PubMed.
- K. Rootwelt, T. Ganes and S. I. Johannessen, Effect of carbamazepine, phenytoin and phenobarbitone on serum levels of thyroid hormones and thyrotropin in humans, Scand. J. Clin. Lab. Invest., 1978, 38, 731–736 CrossRef CAS PubMed.
- J. I. Isojärvi and A. J. M. V. Pakarinen, Serum lipid levels during carbamazepine medication. A prospective study, Arch. Neurol., 1993, 50(6), 590–593 CrossRef PubMed.
- A. D. Martins and P. F. A. M. Oliveira, Assessment of Sertoli Cell Proliferation by 3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide and Sulforhodamine B Assays, Curr. Protoc. Toxicol., 2019, 81(1), e85 CrossRef PubMed.
- A. Martins, P. Oliveira and M. Alves, Assessment of Sertoli Cell Proliferation by 3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide and Sulforhodamine B Assays, Curr. Protoc. Toxicol., 2019, 81(7), e85 CrossRef PubMed.
- B. Sousa, T. Melo, A. Campos, A. S. Moreira, E. Maciel, P. Domingues, R. P. Carvalho, T. R. Rodrigues, H. Girão and M. Domingues, Alteration in Phospholipidome Profile of Myoblast H9c2 Cell Line in a Model of Myocardium Starvation and Ischemia, J. Cell. Physiol., 2016, 231(10), 2266–2274 CrossRef CAS PubMed.
- E. G. Bligh and W. J. Dyer, A rapid method of total lipid extraction and purification, Can. J. Biochem. Physiol., 1959, 37(1), 911–917 CrossRef CAS PubMed.
- E. M. Bartlett and D. H. Lewis, Spectrophotometric determination of phosphate esters in the presence and absence of orthophosphate, Anal. Biochem., 1970, 36(1), 159–167 CrossRef CAS PubMed.
- H. Ferreira, T. Melo, A. Monteiro, A. Paiva, P. Domingues and M. R. Domingues, Serum phospholipidomics reveals altered lipid profile and promising biomarkers in multiple sclerosis, Arch. Biochem. Biophys., 2021, 697, 108672 CrossRef CAS PubMed.
- G. W. Chapman and R. B. Russell, A Conversion Factor to Determine Phospholipid Content in Soybean and Sunflower Crude Oils, JAOCS, 1980, 299–302 CrossRef CAS.
- S. Aued-pimentel, G. Lago, M. Helena and E. Emy, Evaluation of a methylation procedure to determine cyclopropenoids fatty acids from Sterculia striata St. Hil. Et Nauds seed oil, J. Chromatogr. A, 2004, 1054, 235–239 CrossRef CAS PubMed.
- I. Guerra, M. Pinho, P. Domingues, R. Domingues and A. Moreira, Plasma Phospholipidomic Profile Differs between Children with Phenylketonuria and Healthy Children, J. Proteome Res., 2021, 20, 2651–2661 CrossRef CAS PubMed.
- L. Goracci, S. Tortorella, P. Tiberi, R. M. Pellegrino, A. Di Veroli and A. Valeri, et al., Lipostar, a Comprehensive Platform-Neutral Cheminformatics Tool for Lipidomics, Anal. Chem., 2017, 89, 6257–6264 CrossRef CAS PubMed.
- J. Conr, R. M. Andrews, S. Andr, A. Dennis, F. Eoin and C. Gaud, et al., LIPID MAPS: update to databases and tools for the lipidomics community, Nucleic Acids Res., 2024, 52, D1677–D1682 CrossRef PubMed.
- M. Lange, G. Angelidou, Z. Ni, M. Fedorova and M. Bl, Article AdipoAtlas: a reference lipidome for human white adipose tissue, Cell Rep. Med., 2021, 1–26 Search PubMed.
- T. Pluskal, S. Castillo, A. Villar-briones and M. Ore, MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data, BMC Bioinf., 2010, 11(395), 1–11 Search PubMed.
- The R project for statistical computing [Internet]. [cited 2024 Feb 21]. Available from: https://www.r-project.org/.
- RStudio|Open source & professional software for data scienceteams [Internet]. [cited 2024 Feb 21]. Available from: https://rstudio.com/.
- J. Xia and D. S. Wishart, MetaboAnalyst 3.0 for comprehensive metabolomics data analysis, Curr. Protoc. Bioinf., 2016, 55(1), 14.10.1–14.10.91 CrossRef PubMed.
- Y. V. Karpievitch, et al., Metabolomics Data Normalization with EigenMS, PLoS One, 2014, 9(12), e116221 CrossRef PubMed.
- FactoMineR: Multivariate Exploratory Data Analysis and Data Mining [Internet]. [cited 2024 Feb 21]. Available from: https://rdrr.io/cran/FactoMineR/.
- factoextra: Extract and Visualize the Results of Multivariate Data Analyses [Internet]. [cited 2024 Feb 21]. Available from: https://rdrr.io/cran/factoextra/.
- heatmap: Pretty Heatmaps, version 1.0.12 from CRAN.
- rstatix: Pipe-Friendly Framework for Basic Statistical Tests [Internet]. [cited 2024 Feb 21]. Available from: https://rdrr.io/cran/rstatix/.
- Create Elegant Data Visualisations Using the Grammar of Graphics [Internet]. [cited 2024 Feb 21]. Available from: https://ggplot2.tidyverse.org/.
- G. E. H. De Catalfo and I. N. T. De Go, Lipid dismetabolism in Leydig and Sertoli cells isolated from streptozotocin-diabetic rats, Int. J. Biochem. Cell Biol., 1998, 30, 1001–1010 CrossRef CAS PubMed.
- T. Hayashi, A. Yoshinaga, R. Ohno, N. Ishii, S. Kamata and T. Y. T. Watanabe, Asthenozoospermia: possible association with long-term exposure to an anti-epileptic drug of carbamazepine, Int. J. Urol., 2005, 12(1), 113–114 CrossRef PubMed.
- S.-S. Chen, M.-R. Shen, T.-J. Chen and S.-L. Lai, Effects of Antiepileptic Drugs on Sperm Motility of Normal Controls and Epileptic Patients with Long-Term Therapy, Epilepsia, 1992, 33(1), 149–153 CrossRef CAS PubMed.
- J. I. Isojärvi, E. Löfgren, K. S. Juntunen, A. J. Pakarinen, M. Päivänsalo and I. T. L. Rautakorpi, Effect of epilepsy and antiepileptic drugs on male reproductive health, Neurology, 2024, 62(2), 247–253 CrossRef PubMed.
- R. R. Andretta, F. K. Okada, C. C. Paccola, T. Stumpp, S. U. De Oliva and S. M. Miraglia, Carbamazepine-exposure during gestation and lactation affects pubertal onset and spermatic parameters in male pubertal offspring, Reprod. Toxicol., 2014, 44, 52–62, DOI:10.1016/j.reprotox.2013.09.009.
- E. D. Clegg, E. Chapin, P. M. D. Foster and G. P. Dasto, Leydig Cell Hyperplasia And Adenoma Formation: Mechanisms And Relevance To Humans, Reprod. Toxicol., 1997, 11(1), 107–121 CrossRef CAS PubMed.
- J. C. Cook, G. R. Klinefelter, J. F. Hardisty, M. Richard and P. M. D. Foster, Rodent Leydig Cell Tumorigenesis: A Review of the Physiology, Pathology, Mechanisms, and Relevance to Humans, Crit. Rev. Toxicol., 1999, 29(2), 169–261 CrossRef CAS PubMed.
- T. Hayashi, A. Yoshinaga, R. Ohno, N. Ishii, S. Kamata and T. Y. T. Watanabe, Asthenozoospermia: possible association with long-term exposure to an anti-epileptic drug of carbamazepine, Int. J. Urol., 2005, 12(1), 113–114 CrossRef PubMed.
- J. J. Clare, S. N. Tate, M. Nobbs and M. A. Romanos, Voltage-gated sodium channels as therapeutic targets, Drug Discovery Today, 2000, 5(11), 506–520 CrossRef CAS PubMed.
- P. K. Panda, I. K. Sharawat, I. K. Sharawat and P. N. Division, Impact of carbamazepine and lacosamide on serum lipid levels, Epilepsia, 2021, 1034–1035 CrossRef CAS PubMed.
- S. De Oliva, R. Engelbrecht and S. Miraglia, Maternal CBZ exposure impairs testicular development, spermatogenesis and sperm parameters in male offspring at puberty, Andrologia, 2020, 52(9), e13657 CrossRef CAS PubMed.
- A. L. Forgacs, Q. Ding, R. G. Jaremba, I. T. Huhtaniemi and N. A. Rahman, BLTK1 Murine Leydig Cells: A Novel Steroidogenic Model for Evaluating the Effects of Reproductive and Developmental Toxicants, Toxicol. Sci., 2012, 127(2), 391–402 CrossRef CAS PubMed.
- M. Ceylan, A. Yalcin, O. Faruk, I. Karabulut and A. Rıza, Effects of levetiracetam monotherapy on sperm parameters and sex hormones: data from newly diagnosed patients with epilepsy, Seizure, 2016, 41, 70–74, DOI:10.1016/j.seizure.2016.06.001.
- S. A. Rgberts, T. M. Nett, H. A. Hartman, E. Thomas and R. E. Stoll, SDZ 200-1 10 Induces Leydig Cell Tumors By Increasing Gonadotropins in Rats, J. Am. Coll. Toxicol., 1989, 8(3), 487–505 CrossRef.
- A. J. Shetty, K. Narayana, K. L. Bairy, P. Bhat, A. Dill and L. Eberlin, et al., The effect of carbamazepine on sperm counts in Wistar rats – reflecting upon its mitogenic, Reprod. Biol., 2007, 7(2), 177–181 Search PubMed.
- C. Huang, H. J. Hsu, M. E. Wang, M. C. Hsu and L. S. Wu, Fatty acids suppress the steroidogenesis of the MA – 10 mouse Leydig cell line by downregulating CYP11A1 and inhibiting late – stage autophagy, Sci. Rep., 2021, 1–12, DOI:10.1038/s41598-021-92008-2.
- K. E. Hopperton, R. E. Duncan, R. P. Bazinet and M. C. Archer, Fatty acid synthase plays a role in cancer metabolism beyond providing fatty acids for phospholipid synthesis or sustaining elevations in glycolytic activity, Exp. Cell Res., 2014, 320(2), 302–310, DOI:10.1016/j.yexcr.2013.10.016.
- C. C. C. R. De Carvalho and M. Jose, The Various Roles of Fatty Acids, Molecules, 2018, 23(2583), 1–36 CAS.
- K. Ri and T. Yokomizo, Omega-6 highly unsaturated fatty acids in Leydig cells facilitate male sex hormone production, Commun. Biol., 2022, 5(1001) CAS.
- J. G. Coniglio, A. R. Whorton and J. K. Beckman, Essencial fatty acids in testes, Funct. Biosynth. Lipids, 1977, 1965, 575–576 Search PubMed.
- S. Abdollahzadeh, A. Riasi, M. Tavalaee, F. Jafarpour and M. H. N. Esfahani, Omega 6/Omega 3 Ratio Is High in Individuals with Increased Sperm DNA fragmentation, Reprod. Sci., 2023, 30(12), 3469–3479, DOI:10.1007/s43032-023-01313-w.
- M. R. Safarinejad and S. Safarinejad, The roles of omega-3 and omega-6 fatty acids in idiopathic male infertility, Asian J. Androl., 2012, 14(4), 514–515, DOI:10.1038/aja.2012.46.
- L. Yan, X. Bai, Z. Fang, L. Che, S. Xu and D. Wu, Effect of different dietary omega-3/omega-6 fatty acid ratios on reproduction in male rats, Lipids Health Dis., 2013, 12(33), 1–9 Search PubMed.
- M. Galano, Y. Li, L. Li, C. Sottas and V. Papadopoulos, Role of Constitutive STAR in Leydig Cells, Int. J. Mol. Sci., 2021, 22, 1–17 Search PubMed.
- M. Cheng, Z. M. Bhuajwalla and K. Glunde, Targeting Phospholipid Metabolism in Cancer, Front. Oncol., 2016, 6, 1–17 Search PubMed.
- R. D. F. Saito, L. Nogueira, D. S. Andrade and S. O. Bustos, Phosphatidylcholine-Derived Lipid Mediators: The Crosstalk Between Cancer Cells and Immune Cells, Front. Immunol., 2022, 13, 1–24 Search PubMed.
- P. Fagone and S. Jackoeski, Phosphatidylcholine and the CDP-Choline Cycle, Biochim. Biophys. Acta, 2013, 1831(3), 523–532 CrossRef CAS PubMed.
- M. Tavasoli, S. Lahire, T. Reid, M. Brodovsky, C. R. Mcmaster and N. Scotia, Genetic diseases of the Kennedy pathways for membrane synthesis, J. Biol. Chem., 2020, 295(51), 17877–17886 CrossRef CAS PubMed.
- J. N. Van Der Veen, J. P. Kennelly, S. Wan, J. E. Vance, D. E. Vance and R. L. Jacobs, The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease, Biochim. Biophys. Acta, Biomembr., 2017, 1859(9), 1558–1572, DOI:10.1016/j.bbamem.2017.04.006.
- P. J. Quinn and L. G. White, Phospholipid And Cholesterol Content Of Epididymal And Ejaculated Ram Spermatozoa And Seminal Plasma In Relation To Cold Shock, Aust. J. Biol. Sci., 1967, 20(6), 1205–1216 CrossRef CAS PubMed.
- C. Stoica, A. K. Ferreira, K. Hannan and M. Bakovic, Bilayer Forming Phospholipids as Targets for Cancer Therapy, Int. J. Mol. Sci., 2022, 23(9), 1–17 Search PubMed.
- W. Szlasa, Lipid composition of the cancer cell membrane, J. Bioenerg. Biomembr., 2020, 52, 321–342 CrossRef CAS PubMed.
- P. A. Leventis and S. Grinstein, The Distribution and Function of Phosphatidylserine in Cellular Membranes, Annu. Rev. Biophys., 2010, 39(1), 407–427 CrossRef CAS PubMed.
- H. A. Majeed, Histopathological Effects of Carbamazepine on The Reproductive System of Male Rats, Al-Qadisiyah J. Pure Sci., 2021, 26, 39–43 CrossRef.
- C. Kent, Phospholipid Metabolism in Mammals, Encycl. Biol. Chem., 2004, 314–320 CAS.
- I. Domonkos, H. Laczkó-dobos and Z. Gombos, Progress in Lipid Research Lipid-assisted protein – protein interactions that support photosynthetic and other cellular activities, Prog. Lipid Res., 2008, 47(6), 422–435, DOI:10.1016/j.plipres.2008.05.003.
- S. Jian, Y. Li, S. Lin, H. Yang, X. Guan and H. Zhou, et al., Mass spectrometry-based lipidomics analysis using methyl tert-butyl ether extraction in human hepatocellular carcinoma tissues, Anal. Methods, 2015, 7, 8466–8471 RSC.
- O. Bogojevica, Y. Zhanga, C. D. Wolffa, J. V. Nygaarda, L. Wikingb, C. Arevångc and Z. Guo, Phospholipase D catalyzed transphosphatidylation for synthesis of rare complex phospholipid species – Hemi-bis(monoacylglycero)phosphate & Bis(diacylglycero)phosphate, ACS Sustainable Chem. Eng., 2023, 11(8), 3506–3516 CrossRef.
- S. T. Ahmadpour, K. Mah and L. Brisson, Cardiolipin, the Mitochondrial Signature Lipid: Implication in Cancer, Int. J. Mol. Sci., 2020, 21(8031), 1–16 Search PubMed.
- E. M. Mejia, V. W. Dolinsky and G. M. Hatch, Role of Phospholipases in Regulation of Cardiolipin Biosynthesis and Remodeling in the Heart and Mammalian Cells, Biomed. Life Sci., 2014, 39–53 Search PubMed.
- D. Wang, Y. Tang and Z. Wang, Role of sphingolipid metabolites in the homeostasis of steroid hormones and the maintenance of testicular functions, Front. Endocrinol., 2023, 1–8 Search PubMed.
- N. Mohammadzadeh, M. Zarezadeh, S. A. Molsberry and A. Ascherio, Changes in plasma phospholipids and sphingomyelins with aging in men and women: a comprehensive systematic review of longitudinal cohort studies, Ageing Res. Rev., 2021, 68, 101340, DOI:10.1016/j.arr.2021.101340.
- K. Bienias, A. Fiedorowicz, A. Sadowska, S. Prokopiuk and H. Car, Regulation of sphingomyelin metabolism, Pharmacol. Rep., 2016, 68(3), 570–581 CrossRef PubMed.
- X. Li, T. Luo, H. Li and N. Yan, Sphingomyelin Synthase 2 Participate in the Regulation of Sperm Motility and Apoptosis, Molecules, 2020, 25(4231), 1–13 Search PubMed.
- Y. Chen and Y. Cao, The sphingomyelin synthase family: proteins, diseases, and inhibitors, Biol. Chem., 2017, 398(12), 1319–1325 CAS.
- N. C. Lucki and M. B. Sewer, Multiple Roles for Sphingolipids in Steroid Hormone Biosynthesis, Subcell. Biochem., 2008, 49, 387–412 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4mo00221k |
| This journal is © The Royal Society of Chemistry 2025 |