 Open Access Article
Open Access ArticleBiological metasurfaces based on tailored Luria Bertani Agar growth medium formulations for photonic applications†
Francesca
Leone
ab,
Olga
Favale
ab,
Mauro Daniel Luigi
Bruno
ab,
Roberto
Bartolino
b,
Ferdinanda
Annesi
*b,
Vincenzo
Caligiuri
 *abcd and
Antonio
De Luca
*abcd and
Antonio
De Luca
 *ab
*ab
aDepartment of Physics, University of Calabria, via P. Bucci 31c, 87036, Rende (CS), Italy. E-mail: antonio.deluca@unical.it
bCNR Nanotec UOS Rende, via P. Bucci, 31c, 87036, Rende (CS), Italy. E-mail: ferdinanda.annesi@cnr.it
cOptoelectronics, Istituto Italiano di Tecnologia, via Morego 30, Genova, 16163, Italy
dLASCAMM – CR INSTM, Unità INSTM of Calabria Dipartimento di Chimica e Tecnologie Chimiche, Università della Calabria, via P. Bucci Cubo 14C, Arcavacata di Rende (CS), 87036, Italy. E-mail: vincenzo.caligiuri@unical.it
First published on 29th October 2024
Abstract
Biodegradable alternatives to classic solid-state components are rapidly taking place in front-end photonic systems like metamaterials, meta-surfaces and photonic crystals. From this point of view, numerous solutions have been proposed involving eco-friendly compounds. Among them, the Luria Bertani agar (LBA) growth medium has been recently proposed as a functional option with the remarkable advantage of allowing the growth of fluorescent protein expressing bacteria. Such a possibility promises to lead to development of a new generation of biological and eco-sustainable optical sources based on meta-surfaces. There is, however, still a main drawback to address, related to the highly scattering nature of these compounds. To ensure adequate nutritive elements for cell growth, LBA hosts several compounds like NaCl, yeast extracts and tryptone. The presence of these components leads to very scattering LBA films, thus hindering its performance as an optical polymer. A trade-off arises between nutritive capacity and optical performance. In this paper, we successfully address this trade-off, demonstrating that a reduction of the basic nutrients (net Agar concentration) of LBA largely enhances the optical properties of the film as a photonic polymer without compromising its cell-viability. We considered two new LBA formulations with two- (LB2A) and four-fold (LB4A) reduction of the nutrients and replicated a square-lattice meta-surface used as a benchmark architecture. We demonstrated that both the replica molding performances and the optical properties (absorption, scattering and diffraction efficiency) of LBA formulations increase with decreasing nutrient concentration, without losing their cell-growth capability. To demonstrate this fundamental aspect, we inoculated the most critical case of LB4A with green-fluorescent-protein-expressing E. coli bacteria, verifying both their vitality and good photoluminescence properties. These results overcome one of the main limitations of LBA as a functional biopolymer for optical applications, unlocking its use in a new generation of biological quantum optical frameworks for all-biological weak and strong light–matter interactions.
New conceptsIn this paper we introduce new formulations of the Luria Bertani Agar (LBA) growth medium as functional biopolymers for bio-photonic applications. The new formulations resolve the trade-off between cell-viability performances and optical scattering, which hinders the performance of LBA as a polymer for high-end photonics. We demonstrate that reducing the basic nutrients of the LBA can significantly enhance its optical properties without compromising its cell-viability. Our approach is substantially different not only from the classic paradigm of optoelectronics but also from the most broadly explored bio-inspired ones. Unlike the other available eco-friendly compounds, the LBA growth medium stands out for its ability to support the growth of fluorescent protein-expressing bacteria, paving the way for a new generation of bio-inspired, eco-sustainable optical sources based on meta-surfaces. Additionally, we demonstrate that new formulations of LBA are still suitable as growth media. Moreover, we demonstrate that the photonic structure is not affected by the presence of large bacteria, paving the way towards fully biological photonic technologies. Our findings also indicate the successive blending of the classic LBA with new polymers and elements to further customize its functional properties, for example, the fine tailoring of its refractive index and optical dispersion. |
1. Introduction
Biodegradable materials used to replace classic materials for photonic applications are emerging fast.1–7 The always increasing interest in these materials is due to the environmental advantage they possess over petroleum-based plastics. Their characteristics like large availability in nature and biodegradability lead to their exceptional performance in terms of transparency, Young's modulus and processability, making them a winning alternative to classic materials. Polynucleotides like RNA and DNA are typically used to decorate photonic structures like nanoparticles and to endow them with specific sensing capabilities.8–14 Polypeptides like silk and keratin15–17 or polysaccharides like cellulose,18,19 chitin,20,21 chitosan,22,23 and alginate24–26 are commonly used for applications in photonics. Great attention is paid to the optical functionality of the biopolymers but their capability to support living organisms is often neglected. Such a characteristic turns critical in a scenario where biological photonic structures are required to interact with bio-luminescent entities like fluorescent proteins expressed in competent bacteria towards all-biological optical sources.27 Fluorescent-protein-expressing bacteria constitute an exceptional alternative to harmful solid state semiconductors like perovskites or quantum dots.28–31 Bio-emitters such as fluorescent proteins can, indeed, easily reproduce the properties of their high-performing solid-state counterparts with the invaluable advantage of being eco-friendly. Bio-emitters like the green fluorescent protein show a naturally large transition dipole moment (TDM) that can compete and in some cases even outperform those of most recently developed Pb-based semiconductors like perovskites. For example, photoemissive proteins like phiYFP, EYFP and GFP-S65T (mutagenic version of the natural GFP) show a transition dipole moment of around 10.5 D,27,32 while recently calculated TDM values for perovskite quantum wells are around 9.5 D to 11 D.33 Fluorescent proteins also show a very large quantum yield (QY). For example, cyan fluorescent protein variants with QY from 0.84 to 0.87 were broadly documented,34,35 while a quantum yield as high as 0.93 was documented for the fluorescent protein mTurquoise2,36 revealing that the fluorescent protein can easily reproduce the emission properties of the most recently developed Pb-based semiconductors like perovskites.28,29,31 Moreover, the expression of photoluminescent proteins in living organisms like bacteria can lead to environmentally friendly production of a large number of proteins by simply leveraging the natural replication capabilities of these organisms, while classic semiconductors necessitate harmful synthesis procedures that pose serious issues for the disposal of synthesis byproducts like solvents. In the end, producing variants of the classic GFP with tailored emission wavelengths is a very easy, inexpensive and well-established routine that does not prevent the expression in competent bacteria,37 while tuning the emission wavelength of perovskites, for example, necessitates environmentally harmful processes.28 For all these reasons, the need for an all-biological photonic platform with performance comparable to the solid-state ones but with the advantage of being completely eco-friendly emerges. Bacteria, however, require a specific cell-viable scaffold to survive and keep producing fluorescent proteins through their metabolic functions. From this point of view, LBA is an unbeatable option but its glass-like refractive index necessitates its appropriate micro- or nano-structuring in order to manifest resonant properties and work as an in-plane optical cavity. This challenge has been recently addressed by our group, demonstrating that LBA can constitute a great alternative to common bio-polymers used for photonics, with the advantage of ensuring incomparable cell viability.38 However, there are two main drawbacks to overcome: (i) humidity/exsiccation and (ii) nutritive/scattering properties, which still hinder LBA from expressing its potentiality as an all-biological photonic structure. Certainly, these two concerns manifest as trade-offs and are intimately connected to LBA's dual nature of being a biological and an optical material. The former trade-off is brought about by the water content constraint in LBA, which must be drastically reduced with respect to the classic formulation to achieve a good replica molding, severely compromising the cell viability properties of the obtained LBA photonic structures. Such a challenge has, however, been adequately addressed in a previous publication, where water activity measurements allowed finding a compromise between humidity and replica-molding performances.38 The latter trade-off resides in the fact that the nutritive components of LBA, like NaCl, yeast extract and tryptone, are highly scattering in nature, leading to optical signals being affected by scattering-related speckles. This feature leads to poor optical properties, like blurred diffraction patterns.38 Techniques to reduce the scattering of a sample are several and sophisticated. For example, techniques based on statistical analysis and successive filtering have been revealed to be successful.39 Pump-detector depolarization or de-modulation techniques can also be very effective in scattering reduction.40,41 Signal chopping is another very efficient technique to reduce the scattering.42–45 Even though all these techniques may lead to exceptional results, they do not solve the problem from the root since they are external reduction methods of scattering of the sample. In our system, the photonic structure is directly replicated in the scattering polymeric matrix. Therefore, an external scattering reduction method would be useless. A different approach is needed that directly reduces the scattering of the polymeric matrix where the photonic structure is printed. The problem can, in principle, be faced following two different strategies: adding plasticizers like glycerol that could lead to smoother films or reducing the nutritive components. The first choice does not ensure an adequate reduction of the scattering parameters since, if on one hand it is true that smoother surfaces lead to less-scattering films, on the other, adding new components could hinder the functionality as a growth medium. The second strategy, reducing the nutritive components, is a more feasible choice that could result in significant scattering reduction and enable the new LBA composition to carry out its main biological task of keeping cells alive. In this work, we address this second trade-off by demonstrating that the composition of the LBA can be modified to accommodate the constraints posed by the photonic application, without losing its cell viability capabilities. We propose two additional formulations in which except for agar, whose amount is fixed, all the other components (NaCl, yeast extract and tryptone) are two- (LB2A) and four-fold (LB4A) decreased, respectively. We demonstrate that both scattering and absorbance are substantially decreased with respect to the classic LBA formulation. Regarding the cell growth properties, we found that those related to the LB2A formulation are comparable to classic LBA, while the LB4A composition is still adequate as a growth medium, despite its performances being slightly decreased. To prove the validity of both these new formulations as biological scaffolds for photonic applications, we replicated via a standard replica molding technique a common photonic crystal with a hexagonal unit cell, demonstrating through atomic force microscopy (AFM) measurements that reducing the scattering nutrients also leads to better replica-molding performance in terms of morphology and fidelity with respect to the original master mold. By collecting the diffraction pattern generated by each of the replicated samples, we demonstrate that the effect of the scattering-derived speckle is substantially reduced by decreasing the nutritive component percentage, with a related enhancement in the diffraction efficiency as well. In the end, we demonstrate the capability of the LB4A formulation (the most critical one for this task) to host and keep green-fluorescent-protein-expressing E. coli bacteria vital, without compromising the diffraction properties of the replicated structure. Our findings remove a fundamental obstacle towards the effective employment of growth media as high-end photonic platforms, paving the way towards much more sophisticated architectures to be replicated. In addition to the possibility of being functionalized with photo-emissive materials and/or inoculated with fluorescent-protein-expressing bacteria, the outstanding replica molding properties allow envisioning strong and weak light–matter interaction scenarios where these new LBA formulations can be involved.2. Results and discussion
We propose three different LBA compositions: (i) the standard LBA growth medium, (ii) an LB2A version, where the nutritive component concentration is half of the standard LBA, and (iii) LB4A, where the nutritive elements are four-fold reduced with respect to the standard LBA. The agar amount is equal for all three formulations. For each composition, a thin film has been fabricated by solvent casting (see the Experimental section). Its absorption and scattering properties have then been characterized through spectrophotometric measurements (Fig. 1a, absorption and Fig. 1b, scattering). Details are given in the Experimental section. Noticeably, the LB4A formulation resulted in the lowest absorption and scattering, serving as the best option for a replica-molding material for photonic applications. In turn, LB2A showed intermediate properties between LBA and LB4A, while, as expected, standard LBA was demonstrated to be the most absorptive and scattering one. In particular, the scattering value of the LB4A composition was two-fold decreased with respect to common LBA (Fig. 1b), thus demonstrating its suitability as a material for photonic applications. We attribute this achievement to the reduction of components like NaCl crystals and yeast extract powders that act as scattering centers. We notice that the absorption value of the LB4A formulation is still high in the spectral region below 450 nm, being significantly larger than those of conventional optical polymers like PDMS or biodegradable alternatives like cellulose.18,46,47 However, in the visible range (>450 nm), the absorption coefficient drastically reduces to around 30%, a value that, as will be demonstrated further, is suitable for optical applications. | ||
| Fig. 1 (a) Absorption and (b) scattering of the standard LBA (solid red curves), LB2A (dashed blue curves) and LB4A (dotted green curves). | ||
We then checked the viability of all three compositions to confirm that the enhancement of the optical performance did not affect their biological properties. Standard cell cultivation experiments were performed for each of the proposed formulations to obtain the related growth curves in terms of the concentration of colony-forming units per ml (CFU ml−1, log values) (Fig. 2a, see the Experimental section for details). In this case, as expected, the standard LBA showed the best performances (see Fig. 2a, the solid red curve and dots).
Noticeably, we found out that the LB2A formulation is almost as valid as the standard LBA growth medium (see Fig. 2a, the dashed blue curve and dots). The LB4A formulation, instead, presents the worst growth properties (see Fig. 2a, the dotted green curve and dots), but nonetheless it also proved to be valid as a growth medium. In addition, we estimated the effect of nutrient reduction in terms of the doubling time (g) and instantaneous growth rate (k), taking into account the exponential growth phase (for details, see the ESI,† Section S1). The doubling time for E. coli bacteria was estimated to be around 15 minutes for common LBA, ∼17 minutes for LB2A and ∼34 minutes for LB4A. Moreover, we found that the k value of LB4A is 1.20 h−1, thus confirming the slower replication rate of E. coli bacteria in this formulation with respect to LB2A and LBA, which present values of 2.43 and 2.73 h−1, respectively. To further confirm this slowdown, we determined CFU ml−1 (log values in Fig. 2b) as well as the optical density measured at λ = 600 nm (OD600, Fig. 2c) both as a function of time in the exponential growth phase (the first six hours), for all three formulations. The three systems start from almost the same values for both CFU ml−1 and OD600. Then, as expected, the evolution of the LB4A formulation (green dotted curves in Fig. 2b and c) is much slower than those of LB2A and LBA. To confirm this aspect, we performed a linear fit of all the measured CFU ml−1 concentrations (see curves in Fig. 2b) and demonstrated that the smallest slope is obtained for LB4A. This is evidenced by the fact that after 6 hours the logarithm of CFU ml−1 concentration is about 10 for the LB4A formulation, while it is around 12.5 for LB2A and 13 for LBA. This measurement also allows concluding that the LB2A formulation shows intermediate growth speed between LBA (the best one) and LB4A, as expected. The measurement of the OD600 quantity confirms the results obtained for the CFU ml−1 concentration: classic LBA shows a larger OD600 value (around 5, after 6 hours), LB2A shows intermediate OD600 (around 2.2, after 6 hours), while LB4A shows the lowest OD600 (around 1.2, after 6 hours). We underline that, despite, as expected, the viability properties of LB4A being the worst among the three formulations, it still shows cell-growth capability. Such a result was somehow expected since we are moving out of the optimal growth conditions for bacteria. On the other hand, the LB4A formulation is by far the best performing formulation from an optical point of view, showing an ideal trade-off between biological and optical properties towards bio-photonic applications. The validity of the three formulations as photonic platforms was proved by carrying out classic replica molding experiments, where a 2D square-lattice photonic crystal was used as a master mold. The technique is very similar to that shown in recent publications by our group.18,38 The performance of the replicas, which can act as biological metasurfaces, was characterized by atomic force microscopy (AFM) experiments (see the Experimental section for details). The AFM morphology of the master mold is shown in Fig. 3a, together with those of standard LBA (Fig. 3d), LB2A (Fig. 3g) and LB4A (Fig. 3j) replicas. The first qualitative analysis of the replicas is provided through a comparison of the AFM profiles collected along the white lines traced over each of the respective AFM topography panels (Fig. 3a, d, g and j), between the master mold (Fig. 3b) and LBA (Fig. 3e, orange profile), LB2A (Fig. 3h, orange profile) and LB4A (Fig. 3k, orange profile) replicas, respectively.
All three replicas showed good height fidelity with respect to the master mold. To further quantify the quality of the replicas, we calculated the average areas of the top (bottom) circular surfaces of the master mold's (replica's) pillars (holes). A very similar procedure has recently been proposed elsewhere by the authors.38 The average area of the top circular surface of the master mold is Sm = 2.3 μm2, while for the replicas we have: SLBA = 1.65 μm2 for the standard LBA formulation, SLB2A = 1.61 μm2 for the LB2A formulation, and SLB4A = 1.93 μm2 for the LB4A formulation. We define the figure of merit as F = Sreplica/Sm, which accounts for the accuracy with which the top surface of the pillars is converted into a hole. It turns out that for the LBA case F = 0.72, for LB2A F = 0.7 and for LB4A F = 0.84. Noticeably, the LB4A formulation shows the best surface fidelity during the replica molding process, confirming the beneficial role played by the nutrient reduction. The values obtained for the LBA and LB2A formulations are, however, very good as well, showing around 70% fidelity, which is very good compared to other results.38 The AFM characterization also provides rich information on the height fidelity of the fabricated replicas. According to a procedure reported in a previous publication,38 a statistical analysis is carried out over the z-coordinate (i.e., the heights) retrieved by AFM measurements. The frequency of occurrence C(zcoord) of a particular z-coordinate value can be fitted as a superposition of Gaussian distributions, as expressed in eqn (1):
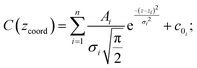 | (1) |
| Average surface area (μm2) | F (Srep/Smas) | 〈zg〉 (nm) | 〈zp〉 (nm) | 〈zh〉 (nm) | Δ p (nm) | Δ h (nm) | T (Δh/Δp) | |
|---|---|---|---|---|---|---|---|---|
| Master | 2.3 | — | 21.14 | 99.13 | — | 78 | — | — |
| LBA | 1.65 | 0.72 | −11 | — | −71 | — | 60 | 0.77 |
| LB2A | 1.61 | 0.7 | −10 | — | −73.2 | — | 63.2 | 0.81 |
| LB4A | 1.93 | 0.84 | −12 | — | −80.3 | — | 68.3 | 0.88 |
The diffractive properties of the proposed biological metasurfaces were analyzed by measuring their diffraction efficiency (see the Experimental section for details). All three proposed structures manifested good diffractive properties, in line with the expected diffraction pattern obtained for the master mold (Fig. 4a–c).38
It can be found that the pure LBA formulation shows a blurred diffraction pattern (Fig. 4a). This is mainly due to the effect of the speckle image generated by the large presence of scattering centers like NaCl crystals and yeast extracts. As demonstrated by the measurements reported in Fig. 1b, a reduction of the nutrients leads to a reduction of the scattering as well.
As a result, the diffraction pattern of the LB2A and LB4A replicas does not present any visible blurred contribution from the speckle. To better quantify such a beneficial effect, we measured the diffraction efficiency η1d, related to the first order (m = 1, white circles in Fig. 4a), for each of the three structures (see the Experimental section for details) and found that η1d is equal to 0.22 for the LBA structure, 0.32 for the LB2A structure and 0.67 for the LB4A structure. These results confirm the validity of LB4A as a biologically suitable platform for photonic applications.
To confirm the capability of the LB4A formulation to ensure cell viability, we inoculated green-fluorescent-protein-expressing E. coli bacteria into the LB4A solution before the replica-molding process. The produced micro-structured sample (Fig. 5a) shows very good diffraction properties, as demonstrated by the glaring presence of a multi-color diffraction pattern in Fig. 5a, confirming that the inoculation does not prevent the optical functionality of the LB4A growth medium. A fluorescence confocal microscopy analysis has been carried out within the area highlighted with a red dashed circle in Fig. 5a. This analysis reveals the presence of fluorescent bacteria agglomerates, as shown in Fig. 5b. To demonstrate that these bacteria are still alive, we cultivated the inoculated replica over a standard LBA plate to form new colonies (Fig. 5d). The growth colonies manifested the same photoluminescence properties as those of the original bacteria (reported in Fig. 5b), as detected via fluorescence confocal imaging analysis, thus demonstrating that the inoculated bacteria were successfully kept alive by the LB4A growth medium (Fig. 5e). This result confirms that the LB4A formulation constitutes an ideal platform to address the trade-off between nutritive properties and low optical scattering. Scanning electron microscopy (SEM) analysis has been performed to show the presence of these bacterial agglomerates over (but even included in) the obtained replicas (see Fig. 5c and f).
3. Conclusions
In conclusion, in this work we successfully addressed the trade-off between the viability and optical suitability of the LBA growth medium. In particular, we gradually reduced the concentration of nutritive elements like NaCl, yeast extracts and tryptone that made LBA highly scattering, verifying that not only the new formulations (two- and four-fold reduced nutrients) are still valid as growth media, but also, by reducing the scattering components, the optical properties are enhanced. We demonstrated this aspect by measuring the scattering and absorption of the three LBA versions and found that, as expected, both these parameters decrease with decreasing nutrient concentration. Moreover, we recorded the growth curves of the three new formulations and found out that LB2A shows a practically comparable growth capability relative to the standard LBA, while LB4A is still valid for cell growth, serving as the best option for bio-photonic applications where both good optical properties and cell viability are required. The replica molding performances are very good for all three compositions, while the diffraction efficiency increases when reducing the nutrients as a consequence of the reduced scattering. We also verified the capability of the LB4A-based metasurface to incorporate fluorescent bacteria keeping them vital. This result is of great interest, since it confirms the validity of the LB4A-inoculated replica as both a photonic platform and a growth medium. Our biological metasurfaces hold great promise in both optical and biological frameworks. From the optical point of view, the spotlight is pointed over new, fully biological planar and photoemissive systems. Structures like meta-lenses can be readily replicated in our new LBA formulations to produce biological and biodegradable meta-lenses. Planar optics like meta-lenses are gradually winning the competition with their classic, bulk counterparts. The proposed low-scattering LBA formulation serves as an option for these kinds of applications with the advantage that biological emitters can be readily incorporated within to endow the system with photoluminescence properties. These considerations offer the possibility of envisioning a biological quantum electrodynamical scenario, as described elsewhere, where the system introduced in this manuscript represents the very fundamental brick. In this bio-photonic study, the two-level emitters are photoemissive proteins expressed by competent bacteria, while cavities are represented by planar resonant structures like metasurfaces. The proposed systems offer a fundamental example of how photoemissive bacteria can be incorporated into low-scattering photonically structured growth media, thus demonstrating their feasibility. The results we found, therefore, pave the way towards a biological quantum optics scenario where strong light–matter interactions between resonant metasurfaces and emitters expressed by bacteria lead to the generation of polaritons, thus enabling the realization of polaritonic technologies and optical sources. From the biological point of view, a photonically structured growth medium opens up new opportunities for photoluminescent based assays where the photonic structuration of the growth medium could enhance the PL signal of the labels, thus increasing the sensitivity of the technique without compromising the viability of the culture medium.4. Experimental section
Materials
LBA, LB2A and LB4A formulations were prepared using the following reagents: tryptone (oxoid LP0042, lot 1900868); yeast extract (oxoid LP0021, lot 1444496-02); sodium chloride (Carlo Erba Reagents, catalog number 479687); bacteriological agar (oxoid LP0011, lot 1449443-02); double-distilled water; ampicillin sodium salt (Sigma-Aldrich, lot 0000208318); and chloramphenicol (Sigma-Aldrich, lot SLCJ7035).Average top circular surface area calculation
Starting from the AFM images shown in Fig. 1a, d, g and j, a thresholding procedure carried out through common routines in ImageJ software allowed isolating only the top circular surfaces of each of the cylinders included in the AFM image. This procedure provides the measure of the surface area of each of the individual circular surface. The average surface area is then calculated from the obtained values.Film preparation and replica molding
The film forming LBA, LB2A and LB4A composites are obtained using a standard stock solution of the Luria Bertani (LB) medium composed of tryptone (10 g L−1), NaCl (10 g L−1) and yeast extract (5 g L−1) powders with agar powder at a fixed concentration of 2.8% w/v. The LB solution was prepared by dissolving tryptone, NaCl and yeast extract powders in double-distilled water and magnetically stirred (200 rpm) at room temperature until complete dissolution was achieved. The LB2A and LB4A solutions were prepared by two- and four-fold diluting the original LB stock solution in double-distilled water. The solutions were autoclaved for 15 minutes at 15 psi and 121 °C and then poured (3 g of each solution) over the master mold at T = 50 °C. The drying process of all the formulations involves solvent evaporation through a multistep procedure consisting of a temperature ramp down (1 °C min−1 from 47 °C to 39 °C) and a soaking step at 38 °C for 12 hours. In the end, the replica is separated from the master mold by a peel-off procedure.38Bacterial culture and growth curve
The E. coli Rosetta (DE3) pLysS strain (Merck Life Science) and a psGFP-CVX plasmid (Voden) were used to achieve the expression of the fluorescent protein using a bacterial culture. The standard CaCl2 method48 was used to obtain competent cells for DNA uptake. The psGFP-CVX plasmid was acquired by the competent cells through heat-shock treatment. The transformed colonies were selected through antibiotic resistance analysis (the ampicillin-resistant gene of the plasmid and the chloramphenicol-resistant gene of the strain). Cell-growth on the two new compositions was tested over the standard LBA formulation. The standard colony forming unit (CFU) method was used (see Section S1 of the ESI† for details) to calculate the number (N) of formed colonies as a function of time.49 DF is the dilution factor and PV is the solution's plated volume. In the case of the reduced nutrient conditions, the corresponding reduced fresh media (LB/2 and LB/4) and agar plates (LB2-agar and LB4-agar plates) have been used.Spectrophotometric measurements
A UV-vis-NIR spectrophotometer Varian Cary-5E equipped with the integrating sphere DRA-CA-50 was used to measure the optical absorption and scattering of non-patterned films obtained as described in the section on film preparation. The measurement was conducted in a spectroscopic range spanning from 350 nm to 750 nm.Optical density (OD)
The optical density of cells at 600 nm (OD600) is a measurement used to monitor bacterial growth, based on the light scattering by cells in suspension (see the ESI†). As described above, for each time point (0, 1, 3, 6 and 24 hours) 1 ml of aliquot was taken to obtain information regarding the quantity of cells present in LB, LB2 and LB4 solutions during the growth. The value reported in Fig. 2c was evaluated by comparing it to the appropriate blank sample.GFP-expressing E. coli replica inclusion procedure
The GFP-expressing E. coli was cultivated for inoculation of LB4A. A bacterial aliquot is cultivated by following the method explained before and the culture was stopped after two hours of growth (OD600 ∼ 0.3). The culture was centrifuged and the residual pellet, containing only bacterial cells, was resuspended using phosphate buffered saline (PBS). The bacteria were added to the LB4A solution, to which 100 μg ml−1 ampicillin and 34 μg ml−1 chloramphenicol were added previously. The solution was slightly stirred and lastly 3 g of the solution was poured on the master mold. The inoculated LB4A solution was exsiccated using a thermal profile of standard replica, and lastly through peeling from the master mold, the replicas, including GFP-expressing bacteria cells, were achieved.Atomic force microscopy (AFM)
Topographic images of the samples were acquired using a Bruker Catalyst atomic force microscope (AFM) equipped with a Nanoscope V controller in tapping mode. The silicon cantilever probe (RTESPA-150, Bruker) with an elastic constant of 5 N m−1 and a tip radius of 8 nm is used. Depending on the size of the scan area, the acquisition speed and samples per line were set. Height profiles and height distribution data are extrapolated from topographic images acquired at 0.150–0.250 Hz and 512 samples per line. The topographic images were analysed using Nanoscope Analysis software (Bruker) and WSxM software.Diffraction efficiency measurement
A He–Ne laser source (λHe–Ne ∼ 633 nm, Melles Griot) has been used to acquire the far-field diffraction pattern of the obtained replicas. Before impinging on the sample, the laser beam crosses a filtering system composed of a zero-order half wave plate and a linear polarizer. The diffraction efficiency of the first order was calculated according to eqn (2): | (2) |
Confocal microscopy
The samples were observed with a laser-scanning confocal microscope (CLSM, TCS-SP8, Leica). The analysis was carried out within the spectral range from 515 nm to 615 nm using a 20× water-immersion objective and the excitation wavelength λex = 488 nm was tuned to the absorbance peak of the green fluorescent protein expressed by the inoculated bacteria.Scanning electron microscopy
Scanning electron microscopy images were recorded on a Quanta FEG 400 (FEI – Netherlands) scanning electron microscope (SEM) by using an electron beam of energy 10 keV. All acquired images show the typical morphology of the samples deposited on carbon tape. Every sample was sputter-covered with a thin graphite layer before analysis.Author contributions
Conceptualization: VC, FA, ADL; data curation: all the authors; formal analysis: VC, FL, ADL, FA; funding acquisition: VC, ADL, FA, RB; investigation: all the authors; methodology: FL, OF, VC; project administration: VC, FA, ADL; software: VC, FL; supervision and validation: VC, FA, RB, ADL; visualization: ADL, VC; writing – original draft: VC; writing – reviewing and editing: all the authors.Data availability
The data supporting this article, inherent to Fig. 1 and 2, have been included as part of the ESI.† Data produced for Fig. 3 are directly available within the main manuscript both in-line and in Table 1.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The present work was partially financed by “Progetto STAR 2 – PIR01_00008” – Ministero dell’Università e Ricerca/Italian Ministry of University and Research and supported by the project “STILE – Strategie nano-Tecnologiche anticontraffazione per il made-in-ItaLy Eco-sostenibile”, funded by PNRR – Missione 4, “Istruzione e ricerca” – Componente 2 “Dalla ricerca all’impresa” – Investimento 1.3 and European Union – NextGenerationEU, Partenariato Esteso PE0000004 – CUP D43C22003120001, title “MICS – Made in Italy Circolare e Sostenibile”. V. C. thanks the research project “Componenti Optoelettronici Biodegradabili ed Eco-Sostenibili verso la nano-fotonica “green”” (D. M. no. 1062, 10.08.2021, PON “Ricerca e Innovazione” 2014-2020), contract identification code 1062_R17_GREEN.Notes and references
- G. Jacucci, L. Schertel, Y. Zhang, H. Yang and S. Vignolini, Adv. Mater., 2020, 2001215 Search PubMed.
- A. G. Dumanli, G. Kamita, J. Landman, H. van der Kooij, B. J. Glover, J. J. Baumberg, U. Steiner and S. Vignolini, Adv. Opt. Mater., 2014, 2, 646–650 CrossRef CAS.
- B. Frka-Petesic, G. Guidetti, G. Kamita and S. Vignolini, Adv. Mater., 2017, 29, 1701469 CrossRef.
- M. Jacobs, M. Lopez-Garcia, O. Phrathep, T. Lawson, R. Oulton and H. M. Whitney, Nat. Plants, 2016, 2, 1–6 Search PubMed.
- M. Lopez-Garcia, N. Masters, H. E. O’Brien, J. Lennon, G. Atkinson, M. J. Cryan, R. Oulton and H. M. Whitney, Sci. Adv., 2018, 4, eaan8917 CrossRef PubMed.
- J. Jaramillo-Fernandez, H. Yang, L. Schertel, G. L. Whitworth, P. D. Garcia, S. Vignolini and C. M. Sotomayor-Torres, Adv. Sci., 2022, 9, 2104758 CrossRef CAS PubMed.
- Priyanka, S. K. Saini, S. Sharma, N. Singh, M. Khokhar and R. V. Nair, J. Opt. Soc. Am. B, 2021, 38, 2297–2305 CrossRef.
- J. Chao, Y. Lin, H. Liu, L. Wang and C. Fan, Mater. Today, 2015, 18, 326–335 CAS.
- A. P. Alivisatos, K. P. Johnsson, X. Peng, T. E. Wilson, C. J. Loweth, M. P. Bruchez and P. G. Schultz, Nature, 1996, 382, 609–611 CAS.
- A. Crameri, E. A. Whitehorn, E. Tate and W. P. C. Stemmer, Nat. Biotechnol., 1996, 14, 315–319 CAS.
- V. Di Meo, M. Moccia, G. Sanità, A. Crescitelli, A. Lamberti, V. Galdi, I. Rendina and E. Esposito, Front. Bioeng. Biotechnol., 2021, 9, 666121 Search PubMed.
- J. Heintz, N. Markešević, E. Y. Gayet, N. Bonod and S. Bidault, ACS Nano, 2021, 15, 14732–14743 CAS.
- J. Yoon, M. Shin, J.-Y. Lee, S.-N. Lee, J.-H. Choi and J.-W. Choi, J. Controlled Release, 2022, 342, 228–240 CAS.
- T. Pylaev, E. Avdeeva and N. Khlebtsov, J. Innovative Opt. Health Sci., 2021, 14, 2130003 CAS.
- G. Guidetti, Y. Wang and F. G. Omenetto, Nanophotonics, 2021, 10, 137–148 Search PubMed.
- B. Marelli and F. G. Omenetto, J. Mater. Chem. C, 2015, 3, 2783–2787 CAS.
- G. Cavallaro, S. Milioto, S. Konnova, G. Fakhrullina, F. Akhatova, G. Lazzara, R. Fakhrullin and Y. Lvov, ACS Appl. Mater. Interfaces, 2020, 12, 24348–24362 CAS.
- V. Caligiuri, G. Tedeschi, M. Palei, M. Miscuglio, B. Martin-Garcia, S. Guzman-Puyol, M. K. Hedayati, A. Kristensen, A. Athanassiou, R. Cingolani, V. J. Sorger, M. Salerno, F. Bonaccorso, R. Krahne and J. A. Heredia-Guerrero, ACS Nano, 2020, 14, 9502–9511 CAS.
- A. Espinha, C. Dore, C. Matricardi, M. I. Alonso, A. R. Goñi and A. Mihi, Nat. Photonics, 2018, 12, 343–348 CrossRef CAS PubMed.
- E. Lizundia, T.-D. Nguyen, R. J. Winnick and M. J. MacLachlan, J. Mater. Chem. C, 2021, 9, 796–817 RSC.
- J. Hou, B. E. Aydemir and A. G. Dumanli, Philos. Trans. R. Soc., A, 2021, 379, 20200331 CrossRef CAS.
- G. Huang, Y. Yin, Z. Pan, M. Chen, L. Zhang, Y. Liu, Y. Zhang and J. Gao, Biomacromolecules, 2014, 15, 4396–4402 CrossRef CAS PubMed.
- S. B. Marpu and E. N. Benton, Int. J. Mol. Sci., 2018, 19, 1795 CrossRef.
- A. Dodero, P. Lova, S. Vicini, M. Castellano and D. Comoretto, Chemosensors, 2020, 8, 37 CrossRef CAS.
- B. D. D. Cruz, D. M. Correia, R. Polícia, N. Pereira, P. Nunes, M. Fernandes, C. R. Tubio, G. Botelho, S. Lanceros-Méndez and V. de Zea-Bermudez, Chem. Eng. J., 2023, 472, 144813 CrossRef CAS.
- C. Santos, M. de Araújo Gonçalves, L. F. de Macedo, A. H. F. Torres, G. D. Marena, M. Chorilli and E. Trovatti, Int. J. Biol. Macromol., 2023, 232, 123351 CrossRef CAS.
- V. Caligiuri, F. Leone, F. Annesi, A. Pane, R. Bartolino and A. De Luca, Photonics, 2021, 8, 470 CrossRef CAS.
- M. Imran, V. Caligiuri, M. Wang, L. Goldoni, M. Prato, R. Krahne, L. De Trizio and L. Manna, J. Am. Chem. Soc., 2018, 140, 2656–2664 CrossRef CAS PubMed.
- Q. A. Akkerman, G. Rainò, M. V. Kovalenko and L. Manna, Nat. Mater., 2018, 17, 394–405 CrossRef CAS.
- A. Castelli, B. Dhanabalan, A. Polovitsyn, V. Caligiuri, F. Di Stasio, A. Scarpellini, R. Brescia, M. Palei, B. Martín-García, M. Prato, L. Manna, I. Moreels, R. Krahne and M. P. Arciniegas, Adv. Opt. Mater., 2019, 1901463 Search PubMed.
- B. Dhanabalan, G. Biffi, A. Moliterni, V. Olieric, C. Giannini, G. Saleh, L. Ponet, M. Prato, M. Imran, L. Manna, R. Krahne, S. Artyukhin and M. P. Arciniegas, Adv. Mater., 2021, 33, 2008004 CrossRef CAS PubMed.
- R. Nifosì, B. Mennucci and C. Filippi, Phys. Chem. Chem. Phys., 2019, 21, 18988–18998 RSC.
- A. H. Proppe, G. W. Walters, A. Y. Alsalloum, A. A. Zhumekenov, E. Mosconi, S. O. Kelley, F. De Angelis, L. Adamska, P. Umari, O. M. Bakr and E. H. Sargent, J. Phys. Chem. Lett., 2020, 11, 716–723 CrossRef CAS PubMed.
- J. Goedhart, L. Van Weeren, M. A. Hink, N. O. E. Vischer, K. Jalink and T. W. J. Gadella, Nat. Methods, 2010, 7, 137–139 CrossRef CAS PubMed.
- M. L. Markwardt, G. J. Kremers, C. A. Kraft, K. Ray, P. J. C. Cranfill, K. A. Wilson, R. N. Day, R. M. Wachter, M. W. Davidson and M. A. Rizzo, PLoS One, 2011, 6, 17896 CrossRef.
- J. Goedhart, D. Von Stetten, M. Noirclerc-Savoye, M. Lelimousin, L. Joosen, M. A. Hink, L. Van Weeren, T. W. J. Gadella and A. Royant, Nat. Commun., 2012, 3, 1–9 CrossRef.
- R. Y. Tsien, Annu. Rev. Biochem., 1998, 67, 509–544 CrossRef CAS.
- V. Caligiuri, F. Leone, O. Favale, M. De Santo, M. D. L. Bruno, O. Mileti, A. Pane, A. Patra, L. Petti, S. Guzman-Puyol, J. A. Heredia-Guerrero, R. Krahne, N. Baldino, R. Bartolino, M. Galluccio, F. Annesi and A. De Luca, Adv. Opt. Mater., 2024, 12, 2301236 CrossRef CAS.
- K. E. H. Anderson, S. L. Sewall, R. R. Cooney and P. Kambhampati, Rev. Sci. Instrum., 2007, 78, 073101 CrossRef.
- P. Malý, J. Ravensbergen, J. T. M. Kennis, R. van Grondelle, R. Croce, T. Mančal and B. van Oort, Sci. Rep., 2017, 7, 43484 CrossRef.
- P. Bado, S. B. Wilson and K. R. Wilson, Rev. Sci. Instrum., 1982, 53, 706–707 CrossRef CAS.
- J. Yue, L. Zhou, P. Su and W. Zhang, Chem. Phys. Lett., 2022, 802, 139766 CrossRef CAS.
- I. A. Heisler, R. Moca, F. V. A. Camargo and S. R. Meech, Rev. Sci. Instrum., 2014, 85, 063103 CrossRef PubMed.
- R. Augulis and D. Zigmantas, Opt. Express, 2011, 19, 13126–13133 CrossRef CAS PubMed.
- L. Guo, D. M. Monahan and G. Fleming, Opt. Express, 2016, 24, 18126–18132 CrossRef CAS PubMed.
- E. D. Palik, Handbook of Optical Constants of Solids, Academic Press, 1998 Search PubMed.
- F. Schneider, J. Draheim, R. Kamberger and U. Wallrabe, Sens. Actuators, A, 2009, 151, 95–99 CrossRef CAS.
- S. Panja, S. Saha, B. Jana and T. Basu, J. Biotechnol., 2006, 127, 14–20 CrossRef CAS PubMed.
- H. L. Green and E. Goldman, Practical Handbook of Microbiology, CRC Press, Taylor & Francis Group, 4th edn, 2021 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Section 1 – colony forming units, along with the details of the calculation of the unit forming colonies; Section 2 – scattering contribution of each single nutrient of LB4A formulation, along with the evaluation of the contribution of each single component to the overall scattering; and Section 3 – tabulated data for Fig. 1 and 2. See DOI: https://doi.org/10.1039/d4mh00861h |
| This journal is © The Royal Society of Chemistry 2025 |




