Open Access Article
Open Access ArticleDesign and synthesis of enantiopure NHC–silver(I) and NHC–gold(I) complexes with anticancer, anti-inflammatory and antioxidant properties
Domenico
Iacopetta
 a,
Assunta
D'Amato
a,
Assunta
D'Amato
 *b,
Jessica
Ceramella
a,
Annaluisa
Mariconda
*b,
Jessica
Ceramella
a,
Annaluisa
Mariconda
 c,
Camillo
Rosano
*d,
Maria
Marra
a,
Alessia
Catalano
e,
Pasquale
Longo†
b and
Maria Stefania
Sinicropi†
a
c,
Camillo
Rosano
*d,
Maria
Marra
a,
Alessia
Catalano
e,
Pasquale
Longo†
b and
Maria Stefania
Sinicropi†
a
aDepartment of Pharmacy, Health and Nutritional Sciences, University of Calabria, Via Pietro Bucci, 87036 Arcavacata di Rende, Italy
bDepartment of Chemistry and Biology “A. Zambelli”, University of Salerno, Via Giovanni Paolo II 132, 84084 Fisciano, Italy. E-mail: asdamato@unisa.it
cDepartment of Basic and Applied Sciences (DISBA), University of Basilicata, Via Dell'Ateneo Lucano 10, 85100, Potenza, Italy
dProteomics and Mass Spectrometry Unit, IRCCS Ospedale Policlinico San Martino, Largo Rosanna Benzi, 10, 16132 Genova, Italy. E-mail: camillo.rosano@hsanmartino.it
eDepartment of Pharmacy-Drug Sciences, University of Bari “Aldo Moro”, Via Orabona, 4, 70126 Bari, Italy
First published on 24th September 2025
Abstract
So far, several interesting reports dealing with N-heterocyclic carbene (NHC) complexes bearing silver and gold have been published, highlighting their versatility in several research fields and their various applications as well. However, most of the reported NHC complexes have been synthetically obtained and studied as racemates, whereas less is still known about the properties of enantiopure complexes. Aiming at contributing to fill this gap, herein a new series of enantiopure NHC complexes of silver(I) and gold(I) bearing an imidazole derivative, opportunely substituted, with one or two asymmetric carbons has been synthesized. These complexes have been characterized by 1H and 13C NMR, mass spectrometry, and elemental analysis and studied for their anticancer, anti-inflammatory and antioxidant properties. The most active complex was also further investigated for its ability in modulating two main enzymes involved in cancer and inflammatory diseases, viz. human topoisomerase I (hTopoI) and murine inducible nitric oxide synthase (iNOS). The outcomes highlight the role of the configuration and substituents in the regulation of the above-mentioned targets, strengthening the need to widen the studies on enantiopure NHC complexes, which may represent useful compounds to be further developed for the obtaining of tailored therapeutic regimens.
Introduction
Chirality plays a main role in drug affinity and interactions toward a given biological target, so that one enantiomer may become clinically more effective, possessing a safer and more selective profile compared to its racemate.1 Consequently, chiral drugs became a challenging issue in the design, synthesis and development of new drugs to be employed for treating different diseases. It is clear that the use of enantiopure drugs offers some advantages over the administration of racemic mixtures.2N-heterocyclic carbene (NHC) ligands and their metal complexes have gained the attention of many researchers because of their chemical and biological properties. In this scenario, several efforts have been made for the development of chiral NHC–metal complexes, which found several applications not only in asymmetric synthesis/catalysis but also in medicinal chemistry.3,4 Over time, several interesting results were disclosed in this area of research, and NHC–metal complexes have become powerful candidates for the treatment of various diseases.5–7 Different biological targets regulated by NHC–metal complexes were also investigated, corroborating their versatility and effectiveness as, for instance, thioredoxin reductase (TrxR), human topoisomerases, cyclooxygenases and so on.8,9 In the last decade, we focused our research activity on the design of gold and silver complexes stabilized by NHC ligands, in order to study the ability of these compounds in numerous catalytic and medicinal chemistry applications. To achieve these results, we developed tunable synthetic routes to obtain functionalized N-heterocyclic ligands containing methyl and alcohol side arms, starting from various imidazole derivatives with hydrogen, chloride, phenyl, xanthine, or a condensed benzene ring on the backbone. The substituents on the backbone of the imidazole ring strongly influence the σ-donation of the carbene to the metal and, therefore, the stability of the resulting complex, as well as its lipophilicity.The synthesized complexes have a substituent on one of the nitrogen atoms of the NHC ligand that possesses a stereogenic center (Fig. 1), but the used reagents produced a racemic mixture; thus, they were tested as racemates.10–12
However, since single enantiomers can exhibit significant different pharmacodynamic, pharmacokinetic, and toxicological properties, the resolution of racemic mixtures or enantiospecific synthesis are particularly important in developing new, more effective, and safer drugs.
Recently, we separately synthesized the R- and S-enantiomers of the silver and gold NHC complexes shown in Fig. 1 and studied their biological activity. In particular, we evaluated their anticancer, antibacterial, and anti-inflammatory properties. The obtained results, in silico and in vitro, showed that one of the two enantiomers was more active and selective.4
The previously obtained results have strengthened the idea that enantiopure NHC complexes based on gold and silver can be extremely useful for the development of increasingly effective multitarget drugs. Thus, in the present work, we synthesized and characterized by 1H and 13C NMR, mass spectrometry, and elemental analysis new enantiopure NHC complexes of silver and gold having an imidazole derivative containing substituents with an asymmetric carbon, namely the 2-phenylethyl group (see Scheme 1). The new series of enantiopure NHC–Ag(I) and NHC–Au(I) complexes were studied for their anticancer, anti-inflammatory and antioxidant properties. Particularly, the obtained outcomes highlighted that the gold-based complexes possessed a better anticancer activity than the silver-based ones and that it depends on the configuration and the nature of the NHC substituents. A similar trend was found for the anti-inflammatory and antioxidant properties. Additionally, two important targets, namely the human topoisomerase I and the inducible nitric oxide synthase (iNOS), implicated in the onset of cancer and inflammatory diseases, were found to be regulated by the most active member of this new series of enantiopure complexes. The obtained data revealed interesting differences related to the complexes' configuration and to the different substituents of the carbenic core, with the latter responsible for their different lipophilicity. These studies provide new insights into the importance of the design and synthesis of suitably substituted enantiopure molecules, and their relationships with biological effects which can lay the groundwork for further development in clinical applications.
Results and discussion
Chemistry
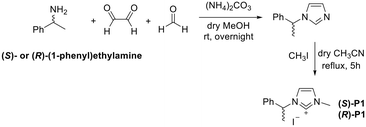
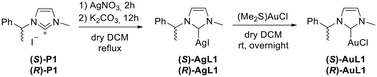
Similarly to the best performing typical N-heterocyclic carbene (NHC) ligand previously reported by us,10,13 we designed ligands (S)-P1/(R)-P1, bearing one methyl substituent on the N3 imidazole atom, and one enantiopure 2-phenylethyl moiety on the N1 atom. The synthesis was accomplished in an efficient one-pot procedure by reacting glyoxal with (S)- or (R)-2-phenylethylamine and achieving ring closure in the presence of p-formaldehyde (Scheme 1).14 Subsequent methylation with iodomethane under our optimized reaction conditions10 led to proligands (S)-P1/(R)-P1.N-heterocyclic carbene was achieved in an alkaline environment due to K2CO3, and metalation was performed in the presence of silver nitrate as the metal source to obtain complexes (S)-AgL1/(R)-AgL1, which were lastly subjected to transmetalation with (Me2S)AuCl to gain Au(I) analogous (S)-AuL1/(R)-AuL1 (Scheme 2).13
 | ||
| Scheme 2 Synthesis of enantiopure silver complexes (S)-AgL1/(R)-AgL1 and enantiopure gold complexes (S)-AuL1/(R)-AuL1. | ||
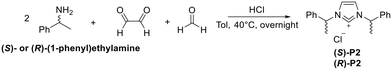
Then, we decided to investigate the influence of an increase in lipophilicity and, at the same time, the presence of a double stereogenic center on the NHC ligand, by switching from the methyl substituent on the N3 atom with an additional 2-phenylethyl unit. Proligands (S)-P2/(R)-P2 were synthesized according to literature procedures,15 by a convenient one-pot reaction, similarly to chiral mono-substituted imidazolium salts (S)-P2/(R)-P2, obviously employing two equivalents of the desired chiral amine, glyoxal and p-formaldehyde (Scheme 3).
This time, the silver complexes were obtained by a reaction with silver oxide in an anhydrous environment.15 The analogous gold complexes were attained through transmetalation with (Me2S)AuCl (Scheme 4).
 | ||
| Scheme 4 Synthesis of enantiopure silver complexes (S)-AgL2/(R)-AgL2 and enantiopure gold complexes (S)-AuL2/(R)-AuL2. | ||
All the novel-synthesized molecules were thoroughly characterized via NMR spectroscopy and mass spectrometry. Moreover, polarimetric analysis confirmed the opposite configuration at the superimposed stereogenic centers (Table 1).
| Compound | [α]D25 (C = 0.5, CHCl3) |
|---|---|
| (S)-AgL1 | +98.6 |
| (R)-AgL1 | −101.3 |
| (S)-AuL1 | +109.5 |
| (R)-AuL1 | −108.3 |
| (S)-AgL2 | +88.6 |
| (R)-AgL2 | −87.2 |
| (S)-AuL2 | +123.7 |
| (R)-AuL2 | −121.2 |
Anticancer activity
The anticancer activity of the new series of enantiopure NHC complexes, bearing one or two chiral centers (L1 and L2 series, respectively), was determined adopting two breast cancer cell lines, which are the triple negative MDA-MB-231 and the ER-α positive MCF-7 cells, using the MTT assay. As a normal counterpart, the human mammary epithelial MCF-10A cells were also assayed, in order to determine the selectivity. Table 2 shows the calculated IC50 values and the relative selectivity index for each molecule.| IC50 (μM) | SI | ||||
|---|---|---|---|---|---|
| MDA-MB-231 | MCF-7 | MCF-10A | MDA-MB-231 | MCF-7 | |
| (S)-P1 | 86.95 ± 7.6 | >100 | >100 | >1.2 | >1 |
| (R)-P1 | 71.0 ± 6.8 | 34.0 ± 2.4 | 40.3 ± 3.6 | 0.6 | 1.2 |
| (S)-AgL1 | >100 | 7.5 ± 0.8 | 77.1 ± 6.8 | >0.8 | 10.3 |
| (R)-AgL1 | 20.6 ± 1.4 | 11.2 ± 0.6 | 72.8 ± 5.8 | 3.5 | 6.1 |
| (S)-AuL1 | 0.5 ± 0.1 | 1.7 ± 0.1 | 11.5 ± 0.5 | 21.2 | 6.5 |
| (R)-AuL1 | 5.8 ± 0.5 | 6.8 ± 0.4 | 2.1 ± 0.1 | 0.4 | 0.3 |
| (S)-P2 | 10.8 ± 1.1 | 16.3 ± 1.3 | 28.8 ± 1.9 | 2.7 | 1.8 |
| (R)-P2 | >100 | 58.0 ± 4.3 | 28.2 ± 1.9 | 0.3 | 0.5 |
| (S)-AgL2 | 14.3 ± 1.2 | 4.2 ± 0.3 | 24.0 ± 1.1 | 1.7 | 5.8 |
| (R)-AgL2 | 17.6 ± 1.7 | 9.7 ± 0.8 | 24.8 ± 1.7 | 1.4 | 2.6 |
| (S)-AuL2 | 2.3 ± 0.1 | 1.8 ± 0.1 | 5.3 ± 0.2 | 2.3 | 2.8 |
| (R)-AuL2 | 6.9 ± 0.3 | 7.0 ± 0.6 | 7.1 ± 0.5 | 1.0 | 1.0 |
| Cisplatin | 29.5 ± 0.9 | 37.3 ± 1.2 | 79.4 ± 1.6. | 2.7 | 2.1 |
At first sight, it is clearly evident that the most active enantiopure complexes were found to be those bearing gold as the metal, with respect to the silver ones. However, the configuration seems to strictly influence the activity and, mainly, the selectivity. Indeed, the most active candidate, in both the cell models adopted, was (S)-AuL1, with IC50 values of 0.5 ± 0.1 and 1.7 ± 0.1 μM for MDA-MB-231 and MCF-7 cells, respectively. This complex was also found high selective toward both the breast cancer cell lines, with respect to the normal counterpart, as shown in Table 2. However, a net drop in both anticancer activity and selectivity was observed for its enantiomer, (R)-AuL1. This is not surprising, since it is generally known that one enantiomer can be bioactive and the opposite inactive, less active or also responsible for unwanted effects.16 In the present case, this is a further validation of the importance to design and obtain enantiopure metal complexes for selective and safer drug development. Next, the corresponding silver bearing enantiopure complexes, (S)- and (R)-AgL1, were found to possess a lesser activity than the above-mentioned gold complexes, mostly against the MDA-MB-231 cells, where (S)-AgL1 was totally inactive and (R)-AgL1 exhibited an IC50 value of 20.6 ± 1.4 μM. A higher, but inverted, activity was found in MCF-7 cells, whereas a similar ability in impairing the MCF-10A cell viability was recorded (Table 2). Finally, both the pro-ligands, (S)- and (R)-P1, were tested and the obtained results clearly confirm that the configuration of the pro-ligands, and consequently of the relative complexes, is definitely determinant for the activity and selectivity, since (R)-P1 was found more cytotoxic toward all the adopted cell lines, opposite to (S)-P1. The L2 series structurally differentiate for the presence of two chiral carbons on the NHC ligand and for the substituent at the N in position 3, which increased the lipophilicity. These molecules produced different results, and indeed both the enantiopure pro-ligands (S)- and (R)-P2 affected the MCF-10A cell viability in a similar way and were already more cytotoxic than (S)- and (R)-P1 (Table 2). The above-mentioned pro-ligands influence, consequently, the behavior of the corresponding silver and gold complexes, as the selectivity lowered, overall, with respect to the L1 series, with the exception of (S)-AgL2 in MCF-7 cells and (R)-AuL2 in both cell lines. As the reference drug in these experiments, cisplatin was included. However, it is less effective and selective compared to the identified leads (Table 2). Summing up, the obtained outcomes suggest that the metal, the complexes' configuration, the presence of one or two chiral centers and the varied lipophilicity strongly influence the activity and selectivity of the enantiopure complexes, in the adopted cellular models.
Human topoisomerase I assay
It is known that NHC–metal complexes possess many pharmaceutical activities, exerted through the modulation of numerous biological pathways and key-point targets.17 However, the biological properties of enantiopure NHC–gold and NHC–silver complexes are quite an unexplored field, and especially the investigation of the possible interactions with biological targets involved in sundry diseases is a very interesting and growing scientific field. Considering the viability assay outcomes, we focused our attention on both the enantiomers (S)- and (R)-AuL1, aiming at further investigating the anticancer properties. With this purpose, both the enantiopure complexes were tested for their ability to modulate the hTopoI, an enzyme that our research group has studied for a long time and that was the target of different metal complexes, acting as inhibitors.18 Most importantly, these experiments were performed not only for investigating this cellular target that can be tuned by (S)- and (R)-AuL1, but also for studying the behavior of the enantiomers toward this enzyme, in order to establish a model to be used for obtaining new and selective anticancer drugs. Therefore, for determining the capability of (S)- and (R)-AuL1 to block the relaxation of supercoiled DNA activity of hTopoI, an in vitro assay was employed, exposing the enzyme to increasing concentrations of both the enantiopure complexes. The achieved outcomes, shown in Fig. 2, revealed that the enantiomers inhibited the hTopoI depending on the adopted concentration, suggesting a different affinity for the enzyme.Indeed, (R)-AuL1 had a better inhibitory activity than (S)-AuL1, considering that it was able to totally block the relaxation of the supercoil DNA substrate (pHOT1) already at the concentration of 0.01 μM (lane 6), as the evidenced by the net band of uncut DNA in the lowest area of the gel, and partially at 0.001 μM (lane 4). The inhibitory activity was, obviously, preserved at higher concentrations (0.1 and 1 μM, lanes 8 and 10 respectively). In contrast, (S)-AuL1 totally inhibited the enzyme only at the higher concentration (1 μM, lane 9), and partially and dose-dependently at the lower concentrations (0.01 and 0.1 μM, lanes 5 and 7, respectively), whereas no inhibition was observed at the lowest concentration (0.001 μM, lane 3). Indeed, in the latter case, the DNA was cut by the enzyme producing a pattern similar to that of the control reaction (lane 2), in which the fully active enzyme was exposed only to the vehicle (DMSO). Lane 1 shows the uncut supercoiled DNA plasmid, used as the negative control. It should be highlighted that (S)-AuL1 was found the most active and selective toward the breast cancer cells in the viability assay, in contrast to (R)-AuL1, even though the latter was found to inhibit the hTopoI activity at a concentration 100 times lower than that of (S)-AuL1. However, this behavior could explain the difference in selectivity amongst the two enantiomers, because a higher affinity toward hTopoI could also impair the basal activity of the enzyme, which is not overexpressed in normal breast cells used as a control in previous assays, or may also have some off-target cytotoxicity.19 Besides, the hTopoI is just one of the diverse targets that can be modulated by these complexes, and thus it is very likely that (S)-AuL1 can impact other targets involved in cancer cell pathways and the synergistic effects can be responsible for the found activity and selectivity. Indeed, as demonstrated in the next experiments, (S)-AuL1 has been found to also regulate the iNOS, in which abnormal activity has been strictly related to cancer onset and progression, affecting tumor heterogenicity, evolution and resistance to clinical treatment.20 Finally, other mechanisms, for instance membrane cell permeation and/or efflux pumps (e.g. PgP or other multidrug resistance (MDR) proteins)21 amongst all, may account for the diverse activity and selectivity exerted by the two complexes in the adopted cancer cell models.
Nuclear DNA damage
In our experience, the impact on MDA-MB-231 breast cancer cell viability and the proved inhibition of hTopoI exerted by the lead enantiopure complex (S)-AuL1 could lead to the genomic DNA damage that, in turn, push cancer cells to die by the apoptotic mechanism. Indeed, a number of literature data inherent to racemic NHC–metal complexes, to which we contributed over the years, reported that the blockade of topoisomerases and nuclear DNA damage are strictly connected.22,23 Thus, aiming at investigating whether (S)-AuL1 could damage the nuclear DNA and induce cancer cell death by apoptosis, a TUNEL assay was performed on MDA-MB-231 cells. Briefly, cells were treated with (S)-AuL1, at a concentration equal to 1 μM, or DMSO (vehicle), for 24 h, then subjected to the TUNEL assay, as detailed in the experimental section. As shown in Fig. 3, MDA-MB-231 cells treated with (S)-AuL1 showed green fluorescence (panel B) that perfectly overlapped the blue fluorescence (panel A) from cell nuclei, which strongly indicate marked genomic DNA damage (panel C). In contrast, MDA-MB-231 cells exposed to the vehicle (DMSO, CTRL) did not show the green fluorescence (Fig. 2, panel B, CTRL), meaning that the nuclear DNA was undamaged (panels A and C, CTRL). The latter observations confirmed that the selective impact of (S)-AuL1 on MDA-MB-231 breast cancer cell viability depends on the cell death triggered by the nuclear DNA damage, due to the inhibition of hTopoI.Anti-inflammatory properties
The anti-inflammatory properties of the new series of synthesized enantiopure complexes were determined in murine macrophages RAW 264.7, measuring the NO production under bacterial lipopolysaccharide (LPS) stimulation by the means of the Griess reagent. A nonsteroidal anti-inflammatory drug (NSAID), i.e. indomethacin (Ind), was employed as the positive control. Two concentrations, 5 and 10 μM, chosen after testing the enantiopure pro-ligands and relative complexes for their cytotoxicity on the RAW264.7 murine macrophages via the MTT assay, were employed for the anti-inflammatory assay. The measurements were made after 24 hours of treatment. Ind was used at 5, 10 and 50 μM. As shown in Fig. 4, none of the assayed molecules produced a significant decrease of cell viability, with the exception of (R)-AuL1, (S)- and (R)-AuL2 at 10 μM, which reduced the viability by about 55–45%.Fig. 5 shows the results from the NO production measurements, where it is possible to see that none of the enantiopure pro-ligands P1 and P2 influenced the NO production under LPS stimulation.
Similar results were obtained from Ag-based complexes, whereas those bearing Au were found to be able to significantly decrease the induced NO production. Overall, (S)-AuL1 was found the most active, in reducing the NO production dose-dependently, of about 33 and 57% at 5 and 10 μM, respectively. It is important to emphasize that its enantiomer, (R)-AuL1, not only produced a lesser effect against the NO production (about 18%) at 5 μM, but it was also cytotoxic at the concentration of 10 μM (Fig. 4), which made the evaluation of NO production at the higher concentration misrepresented and useless (Fig. 5, not determined, n.d.). The other complexes (S)- and (R)-AuL2, bearing two stereocenters and an additional 2-phenylethyl substituent on the NHC core that raises the lipophilicity, demonstrated a low activity in decreasing the NO production with respect to (S)-AuL1, for both equal to about 18%, respectively, at the concentration of 5 μM, and the configuration seems not to play any role. Furthermore, they were found both cytotoxic at 10 μM (Fig. 4 and 5, not determined, n.d.). Notably, the complex (S)-AuL1 also exhibited a better activity than the reference molecule, Ind, which was able to reduce the NO stimulated production by only about 26% at 50 μM, namely ten- and five-fold higher concentrations than those adopted for (S)-AuL1 (Fig. 5). These outcomes suggest that only the complexes bearing gold inhibited the NO production in stimulated RAW 264.7 cells and that the configuration is absolutely responsible for the observed different activity in L1 series. Finally, the increase of the lipophilicity (see (S)-AuL1versus(S)-AuL2) is inversely proportional to the observed ability in inhibiting iNOS and, consequently, decreasing the NO production in the adopted cell context, confirming that for the most active complex (S)-AuL1, the lipophilicity tuning, due to the difference of substituents on the NHC core, contribute to the inhibitory activity.4 These outcomes are also in agreement with other published studies on structurally different iNOS inhibitors, even though little is still known about enantiopure inhibitors.24,25 It is accepted that iNOS modulation represents an important key-point in medicinal chemistry, since its overexpression and/or deregulation are linked to the onset and progression of several chronic diseases, included cancer.26 Additionally, drugs targeting iNOS may modulate its activity, as well as its expression, contributing to the varied amounts of NO production.27 Thus, the expression of iNOS in murine macrophages RAW 264.7 was assessed by the means of immunofluorescence assays, and the obtained results are shown in Fig. 6.
Particularly, LPS-stimulated macrophages revealed, as expected, an enhanced iNOS expression (red fluorescence, Fig. 6, panel 2A) with respect to cells treated with only the vehicle (DMSO) that showed basal levels (Fig. 6, panel 1A). The co-treatment with the enantiopure complexes (S)-AuL1 or (S)-AuL2 at 5 μM resulted in a net diminution of the intracellular iNOS expression, as shown in Fig. 6, panels 4A and 5A, with respect to the LPS-treated cells (panel 2A), mostly for (S)-AuL1 treatment. A similar behavior was found under ind co-treatment (Fig. 6, panel 3A), which was used at 50 μM (ten-fold higher than the concentration used for the complexes).
Overall, (S)-AuL1 was found to be able to reduce iNOS expression with a similar potency to ind, as it is easily deducible observing the fluorescence quantification graphic in Fig. 7, but at a ten-fold lower concentration.
This feature is also strictly connected with the previously shown results on the great impact of (S)-AuL1 on the viability of breast cancer cells, mostly for the highly aggressive and metastatic MDA-MB-231 cells. Indeed, the overexpression of iNOS is generally associated with poor outcomes in triple negative breast cancer (TNBC) patients;28 thus, the development of molecules able to modulate iNOS activity and expression represents a current and promising strategy in the anticancer therapy.
Docking simulations
Molecular docking simulations (Fig. 8) identified specific and well-defined binding modes for all tested compounds within the active site region of human iNOS, in close proximity to the heme prosthetic group. All four ligands exhibited interactions characteristic of metallodrug–protein complexes, involving both coordination bonds with the Au(I) center and non-covalent contacts, including hydrogen bonding and π-stacking.(S)-AuL1 was found to establish a direct coordination between its gold atom and the carboxylate group of Glu371, a key residue at the entrance of the heme pocket. The coordinated chlorine atom of (S)-AuL1 engaged in simultaneous interactions with the backbone amide nitrogen of Met368 and contributed to the stabilization of the complex. Furthermore, a π-stacking interaction was observed between a phenyl ring of the ligand and the porphyrin plane of the heme group, further anchoring the molecule in the binding pocket. (R)-AuL1 adopted a binding conformation characterized by strong coordination of the gold atom to a carboxylate oxygen atom (Cox) of the heme group, while the chlorine atom simultaneously interacted with a second Cox group on the same porphyrin structure. A phenyl ring of the ligand established a coplanar π-stacking interaction with the heme, stabilizing the compound in a conformation reminiscent of known iNOS inhibitors.
(S)-AuL2 displayed a distinct binding pose in which the gold center coordinated the side chain of Gln257, while the chlorine atom formed a hydrogen bond with the hydroxyl group of Tyr367. The aromatic system of the ligand simultaneously engaged in dual π–π stacking interactions with both the heme prosthetic group and the aromatic ring of Tyr485, suggesting a favourable orientation for electronic delocalization and stabilization. (R)-AuL2 shared the same interaction profile as (S)-AuL2, with the coordination of Au to Gln257, hydrogen bonding between the chlorine atom and Tyr367, and π-stacking with both heme and Tyr485. However, the spatial arrangement of the aromatic groups differed slightly, potentially contributing to minor variations in the calculated binding affinities.
The calculated binding energies and predicted inhibition constants (Ki) for all four complexes are summarized in Table 3. These results support the hypothesis that both the Au–protein coordination and the geometry of π-stacking with the heme and adjacent residues are critical determinants of binding affinity.
| K i (μM) | E binding (kcal per mole) | |
|---|---|---|
| (S)-AuL1 | 2.3 | −8.59 |
| (R)-AuL1 | 2.24 | −8.6 |
| (S)-AuL2 | 0.55 | −10.03 |
| (R)-AuL2 | 0.33 | −10.33 |
In AutoDock, the inhibition constant (Ki) is calculated from the estimated binding free energy (ΔG) of the ligand–receptor complex using the following thermodynamic relationship:
| Ki = exp[(ΔG × 1000)/(R × T)] |
• ΔG is the predicted binding free energy (in kcal mol−1), returned by AutoDock for the best-ranked pose.
• R is the universal gas constant (1.987 cal mol−1 K−1).
• T is the temperature in Kelvin, typically set at 298.15 K (25 °C) by default in AutoDock.
It is important to highlight that NAC exhibited a reduction in ROS levels of about 25 and 40% at 10 × 103 and 20 × 103 μM, respectively, which are 1000 and 2000-fold higher than the (S)-AuL1 effective concentration (Fig. 9). Furthermore, a slight antioxidant activity was recorded for both the enantiopure complexes (S)-AgL2 and (R)-AgL2, about 15 and 12% ROS reduction, respectively, whereas the pro-ligands were found inactive. Finally, complexes (S)-AuL2 and (R)-AuL2 did not exert any antioxidant activity at the lower dosage (Fig. 9) and were found cytotoxic at the highest concentration (Fig. 4).
Summing up, (S)-AuL1 is the best antioxidant and anti-inflammatory complex, and was also found as the best and selective anticancer candidate. Meanwhile, its enantiomer (R)-AuL1 demonstrated a lesser activity in all the described experiments and also a dose dependent cytotoxicity toward the adopted cell models, both cancerous and normal.
Conclusions
In this paper, we reported the synthesis of a new series of enantiopure NHC–silver(I) and NHC–gold(I) complexes and disclosed some of their biological activities, such as anticancer, anti-inflammatory and antioxidant properties. We also individuated two important enzymes, namely hTopoI and iNOS, modulated by the most active complex and related to the above-mentioned activities. The reported results indicate that the gold(I)-based complexes are the most active and that the configuration and chemical substituents are fundamental for the modulatory properties exerted toward these two enzymes. Most interestingly, the stronger inhibitory activity exerted by (R)-AuL1 toward hTopoI with respect to (S)-AuL1, which is, instead, the best and more selective anticancer candidate, could be one of the main causes involved in the high cytotoxicity of (R)-AuL1 toward the normal breast cells used in these assays. (S)-AuL1 was also found to be the best iNOS inhibitor and antioxidant complex, and also the less cytotoxic toward the RAW 264.7 cells. Considering that there are only few literature reports in the field of NHC–gold and NHC–silver complexes, the present outcomes contribute to widen the knowledge about the design of more potent, selective and safer complexes to be further developed for the treatment of multifactorial diseases.Experimental section
All reagents were purchased from Merck Italy (Milan, Italy) and TCI Chemicals (Zwijndrecht, Belgium) and used without any purification. All solvents were bought from Carlo Erba Reagents srl (Milano, Italy) and were distilled over appropriate drying agents under nitrogen before use. The synthesis of silver(I) and gold(I) complexes was carried out under a nitrogen atmosphere by using Schlenk techniques in the dark. The glassware used was dried in an oven at 120 °C overnight. Deuterated solvents were dried on molecular sieves. 1H and 13C nuclear magnetic resonance spectra (NMR) were acquired on a Bruker Avance 300 spectrometer (300 MHz for 1H; 75 MHz for 13C), a Bruker AVANCE 400 spectrometer (400 MHz for 1H; 100 MHz for 13C) and a Bruker AVANCE 600 spectrometer (600.13 MHz for 1H, 150.90 MHz for 13C), operating at 298 K. NMR samples were prepared by dissolving about 15 mg of the compound in 0.5 mL of deuterated solvent (Eurisotop Cambridge Isotope Laboratories, Cambridge, UK). The chemical shifts of 1H-NMR and 13C-NMR spectra are referenced using the residual proton impurities of the deuterated solvents. 1H-NMR was reported relative to CDCl3δ 7.26 ppm. 13C-NMR was reported relative to CDCl3δ 77.16 ppm. The spectral multiplicities are indicated as follows: singlet (s), doublet (d), triplet (t), multiplet (m), and broad (br). MALDI-MS mass spectra were obtained using a Bruker SolariX XR Fourier transform ion cyclotron resonance mass spectrometer (Bruker Daltonik GmbH, Bremen, Germany) with a 7 T refrigerated actively shielded superconducting magnet (Bruker Biospin, Wissembourg, France). A MALDI ion source (Bruker Daltonik GmbH, Bremen, Germany) was used for the samples in positive ion mode. The mass range was set to m/z 200–3000. The laser power was 28% and 22 laser shots were utilized for each scan. The mass spectra were calibrated externally using a mix of peptide clusters in MALDI ionization positive ion mode. Ultraviolet-visible (UV-vis) measurements were performed with an Agilent Varian Cary 50 spectrophotometer (Agilent Technologies Inc., Santa Clara, California, USA). Optical rotations were measured on a digital polarimeter (JASCO P-2000) at a concentration of C = 0.50. A sodium lamp (λ = 589 nm) was used as a light source.Chemistry
Complexes (S)-AgL2/(S)-AuL2 (ref. 15) and (R)-AgL2/(R)-AuL2 were synthesized according to literature procedures,15 with slight modifications.(S)-1-(1-Phenylethyl)-1H-imidazole (0.596 g, 42%). 1 H-NMR (600 MHz, CDCl 3 , ppm) δ: 7.58 (1H, s, NC
![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) N), 7.33 (3H, m, Ar–
N), 7.33 (3H, m, Ar–![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) ), 7.16 (2H, d, J = 7.2 Hz, Ar–
), 7.16 (2H, d, J = 7.2 Hz, Ar–![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) ), 7.06 (1H, s, NC
), 7.06 (1H, s, NC![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) CHN), 6.92 (1H, s, NCHC
CHN), 6.92 (1H, s, NCHC![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) N), 5.33 (1H, q, J = 7.2 Hz, C
N), 5.33 (1H, q, J = 7.2 Hz, C![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) CH3), 1.85 (3H, d, J = 7.2 Hz, CHCH3).
CH3), 1.85 (3H, d, J = 7.2 Hz, CHCH3).
13 C-NMR (150 MHz, CDCl 3 , ppm) δ: 141.1, 135.6, 128.8, 128.4, 127.6, 125.5, 117.5, 56.1, 21.6.
(R)-1-(1-Phenylethyl)-1H-imidazole (0.738 g, 52%). 1 H-NMR (600 MHz, CDCl 3 , ppm) δ: 7.58 (1H, s, NC
![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) N), 7.33 (3H, m, Ar–
N), 7.33 (3H, m, Ar–![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) ), 7.15 (2H, d, J = 7.2 Hz, Ar–
), 7.15 (2H, d, J = 7.2 Hz, Ar–![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) ), 7.06 (1H, s, NC
), 7.06 (1H, s, NC![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) CHN), 6.92 (1H, s, NCHC
CHN), 6.92 (1H, s, NCHC![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) N), 5.33 (1H, q, J = 7.2 Hz, C
N), 5.33 (1H, q, J = 7.2 Hz, C![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) CH3), 1.84 (3H, d, J = 7.2 Hz, CHCH3).
CH3), 1.84 (3H, d, J = 7.2 Hz, CHCH3).
13 C-NMR (150 MHz, CDCl 3 , ppm) δ: 141.1, 135.6, 128.8, 128.4, 127.6, 125.5, 117.5, 56.1, 21.6.
The resulting crude intermediate (S/R)-1-(1-phenylethyl)-1H-imidazole (0.500 g, 2.91 mmol) was dissolved in acetonitrile (10 mL), and iodomethane (1.27 mL, 20.4 mmol) was added. The resulting mixture was stirred at reflux for 5 hours, then brought to room temperature and evaporated to dryness. Precipitation from acetone gave resulting salts (S)-P1/(R)-P1 as white solids.
(S)-3-Methyl-1-(1-phenylethyl)-1H-imidazol-3-ium iodide ((S)-P1, 0.874 g, 96%). 1 H-NMR (600 MHz, CDCl 3 , ppm) δ: 10.0 (1H, s, NC
![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) N), 7.48 (1H, s, NC
N), 7.48 (1H, s, NC![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) CHN), 7.42 (2H, d, J = 7.8 Hz Ar–
CHN), 7.42 (2H, d, J = 7.8 Hz Ar–![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) ), 7.38–7.34 (3H, overlapping signals, Ar–
), 7.38–7.34 (3H, overlapping signals, Ar–![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) ), 7.26 (1H, br s, NCHC
), 7.26 (1H, br s, NCHC![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) N), 5.85 (1H, q, J = 7.2 Hz, C
N), 5.85 (1H, q, J = 7.2 Hz, C![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) CH3), 4.08 (3H, s, NCH3), 2.00 (3H, d, J = 7.2 Hz, CHCH3).
CH3), 4.08 (3H, s, NCH3), 2.00 (3H, d, J = 7.2 Hz, CHCH3).
13 C-NMR (150 MHz, CDCl 3 , ppm) δ: 137.7, 136.1, 129.5, 127.0, 123.9, 120.9, 60.1, 37.3, 21.5.
(MALDI-ICR FTMS) m/z: HRMS calcd for C12H15N2+: 187.1230, found: 187.1225.
(R)-3-Methyl-1-(1-phenylethyl)-1H-imidazol-3-ium iodide ((R)-P1, 0.847 g, 93%). 1 H-NMR (600 MHz, CDCl 3 , ppm) δ: 10.0 (1H, s, NC
![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) N), 7.48 (1H, s, NC
N), 7.48 (1H, s, NC![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) CHN), 7.42 (2H, d, J = 7.8 Hz Ar–
CHN), 7.42 (2H, d, J = 7.8 Hz Ar–![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) ), 7.38–7.34 (3H, overlapping signals, Ar–
), 7.38–7.34 (3H, overlapping signals, Ar–![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) ), 7.26 (1H, br s, NCHC
), 7.26 (1H, br s, NCHC![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) N), 5.85 (1H, q, J = 7.2 Hz, C
N), 5.85 (1H, q, J = 7.2 Hz, C![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) CH3), 4.08 (3H, s, NCH3), 2.00 (3H, d, J = 7.2 Hz, CHCH3).
CH3), 4.08 (3H, s, NCH3), 2.00 (3H, d, J = 7.2 Hz, CHCH3).
13 C-NMR (150 MHz, CDCl 3 , ppm) δ: 137.7, 136.2, 129.5, 127.1, 123.8, 120.8, 60.1, 37.3, 21.5.
(MALDI-ICR FTMS) m/z: HRMS calcd for C12H15N2+: 187.1230, found: 187.1223.
To a stirred solution of (S)- or (R)-α-methylbenzylamine (2.02 g, 16.6 mmol) in toluene (18 mL), paraformaldehyde (0.50 g, 16.6 mmol) was added. After 30 min, the mixture was cooled to 0 °C and another equivalent of (S)- or (R)-α-methylbenzylamine (2.02 g, 16.6 mmol) was added, followed by 3 M HCl (4.97 mL, 16.6 mmol). After 15 min, the reaction was brought to room temperature, and glyoxal (1.89 mL, 16.6 mmol, 40% aqueous solution) was slowly added. The mixture was stirred for 12 h at 40 °C and then cooled to room temperature. The reaction was quenched with 2 mL of sat. solution of Na2CO3, then 10 mL of H2O and 10 mL of Et2O were added, the phases separated and the aqueous layer was washed with CH2Cl2 (3 × 10 mL). The organic phases were collected and dried over MgSO4, concentrated in vacuo. The residue was triturated with Et2O to give 1,3-bis((S)- or (R)-1′-phenylethyl)imidazolium chloride ((S)-P2/(R)-P2) as off-white powders.
1,3-Bis((S)-1′-phenylethyl)imidazolium chloride ((S)-P2, 3.58 g, 69%)15. NMR and HRMS characterization results were consistent with what already reported in the literature.
1,3-Bis((R)-1′-phenylethyl)imidazolium chloride ((R)-P2, 3.17 g, 61%). 1 H NMR (600 MHz, CDCl 3 , ppm) δ: 11.59 (1H, s, NC
![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) N), 7.45 (2H, br d, J 7.5 Hz, Ar–
N), 7.45 (2H, br d, J 7.5 Hz, Ar–![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) ), 7.40–7.35 (6H, overlapping, Ar–
), 7.40–7.35 (6H, overlapping, Ar–![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) ), 7.04 (2H, s, NC
), 7.04 (2H, s, NC![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) C
C![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) N), 6.05 (2H, q, J 7.2 Hz, NC
N), 6.05 (2H, q, J 7.2 Hz, NC![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) CH3), 2.03 (6H, d, J 7.2 Hz, NCHCH3).
CH3), 2.03 (6H, d, J 7.2 Hz, NCHCH3).
13 C NMR (150 MHz, CDCl 3 , ppm) δ: 137.8, 137.3, 129.4, 127.1, 119.9, 59.9, 21.2.
(MALDI-ICR FTMS) m/z: HRMS calculated for C19H21N2+: 277.3905, found: 277.3901.
(S)-3-Methyl-1-(1-phenylethyl)-1H-imidazol-3-ium silver(I) iodide ((S)-AgL1, 0.440 g, 81%). 1 H NMR (600 MHz, CDCl 3 , ppm) δ: 7.32–7.28 (5H, overlapping signals, Ar–
![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) ), 6.97 (1H, d, J = 1.2 Hz, NC
), 6.97 (1H, d, J = 1.2 Hz, NC![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) CHN), 6.93 (1H, d, J = 1.8 Hz, NCHC
CHN), 6.93 (1H, d, J = 1.8 Hz, NCHC![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) N), 5.89 (1H, q, J = 7.2 Hz, C
N), 5.89 (1H, q, J = 7.2 Hz, C![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) CH3), 3.82 (3H, s, NCH3), 1.82 (3H, d, J = 7.2 Hz, CHCH3).
CH3), 3.82 (3H, s, NCH3), 1.82 (3H, d, J = 7.2 Hz, CHCH3).
13 C-NMR (150 MHz, CDCl 3 , ppm) δ: 183.2, 140.6, 128.8, 128.1, 126.6, 122.3, 118.6, 60.0, 39.0, 21.6.
(MALDI-ICR FTMS) m/z: HRMS calculated for C12H15AgIN2+: 422.0381, found: 422.0379.
Elemental analysis: calculated for C12H14AgIN2 C, 34.23; H, 3.35; Ag, 25.62; I, 30.14; N, 6.65; found C, 34.12; H, 3.40; Ag, 25.68; I, 30.20; N, 6.70.
(R)-3-Methyl-1-(1-phenylethyl)-1H-imidazol-3-ium silver(I) iodide ((R)-AgL1, 0.430 g, 79%). 1 H NMR (600 MHz, CDCl 3 , ppm) δ: 7.32–7.28 (5H, overlapping signals, Ar–
![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) ), 6.97 (1H, d, J = 1.2 Hz, NC
), 6.97 (1H, d, J = 1.2 Hz, NC![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) CHN), 6.92 (1H, d, J = 1.8 Hz, NCHC
CHN), 6.92 (1H, d, J = 1.8 Hz, NCHC![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) N), 5.89 (1H, q, J = 7.1 Hz, C
N), 5.89 (1H, q, J = 7.1 Hz, C![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) CH3), 3.82 (3H, s, NCH3), 1.81 (3H, d, J = 7.2 Hz, CHCH3).
CH3), 3.82 (3H, s, NCH3), 1.81 (3H, d, J = 7.2 Hz, CHCH3).
13 C-NMR (150 MHz, CDCl 3 , ppm) δ: 183.2, 140.6, 128.8, 128.1, 126.6, 122.3, 118.6, 60.0, 39.0, 21.6.
(MALDI-ICR FTMS) m/z: HRMS calculated for C12H15AgIN2+: 422.0381, found: 422.0384.
Elemental analysis: calculated for C12H14AgIN2 C, 34.23; H, 3.35; Ag, 25.62; I, 30.14; N, 6.65; found C, 34.25; H, 3.36; Ag, 25.65; I, 30.11; N, 6.68.
(S)-3-Methyl-1-(1-phenylethyl)-1H-imidazol-3-ium gold(I) chloride ((S)-AuL1, 0.183 g, 90%). 1 H NMR (600 MHz, CDCl 3 , ppm) δ: 7.36–7.31 (5H, overlapping signals, Ar–
![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) ), 6.91 (1H, d, J = 1.8 Hz, NC
), 6.91 (1H, d, J = 1.8 Hz, NC![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) CHN), 6.85 (1H, d, J = 1.8 Hz, NCHC
CHN), 6.85 (1H, d, J = 1.8 Hz, NCHC![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) N), 6.14 (1H, q, J = 7.2 Hz, C
N), 6.14 (1H, q, J = 7.2 Hz, C![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) CH3), 3.82 (3H, s, NCH3), 1.84 (3H, d, J = 7.2 Hz, CHCH3).
CH3), 3.82 (3H, s, NCH3), 1.84 (3H, d, J = 7.2 Hz, CHCH3).
13 C-NMR (150 MHz, CDCl 3 , ppm) δ: 170.8, 138.2, 129.0, 128.6, 126.8, 122.2, 117.8, 59.8, 38.3, 20.7.
(MALDI-ICR FTMS) m/z: HRMS calculated for C12H15AuClN2+: 419.6820, found: 419.6818.
Elemental analysis: calculated for C12H14AuClN2 C, 34.43; H, 3.37; Au, 47.05; Cl, 8.47; N, 6.69; found C, 34.50; H, 3.25; Au, 47.03; Cl, 8.41; N, 6.73.
(R)-3-Methyl-1-(1-phenylethyl)-1H-imidazol-3-ium gold(I) chloride ((R)-AuL1, 0.183 g, 90%). 1 H NMR (600 MHz, CDCl 3 , ppm) δ: 7.37–7.32 (5H, overlapping signals, Ar–
![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) ), 6.87 (1H, d, J = 1.8 Hz, NC
), 6.87 (1H, d, J = 1.8 Hz, NC![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) CHN), 6.85 (1H, d, J = 1.8 Hz, NCHC
CHN), 6.85 (1H, d, J = 1.8 Hz, NCHC![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) N), 6.13 (1H, q, J = 7.2 Hz, C
N), 6.13 (1H, q, J = 7.2 Hz, C![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) CH3), 3.83 (3H, s, NCH3), 1.84 (3H, d, J = 7.2 Hz, CHCH3).
CH3), 3.83 (3H, s, NCH3), 1.84 (3H, d, J = 7.2 Hz, CHCH3).
13 C-NMR (150 MHz, CDCl 3 , ppm) δ: 169.9, 138.2, 128.0, 127.6, 125.8, 121.1, 116.8, 58.8, 37.3, 19.7.
(MALDI-ICR FTMS) m/z: HRMS calculated for C12H15AuClN2+: 419.6820, found: 419.6819.
Elemental analysis: calculated for C12H14AuClN2 C, 34.43; H, 3.37; Au, 47.05; Cl, 8.47; N, 6.69; found C, 34.37; H, 3.34; Au, 47.09; Cl, 8.51; N, 6.72.
To a stirred solution of chiral proligand (S)-P2/(R)-P2 (0.500 g, 1.60 mmol) in dry dichloromethane (53 mL), under inert atmosphere, 4 Å molecular sieves were added, followed by silver oxide (0.444 g, 1.92 mmol). The suspension was stirred at reflux overnight, then cooled to room temperature and filtered through a celite plug. The solvent was evaporated to complete dryness, and the resulting powder was washed with hexane (3 × 10 mL) to obtain the off-white powders of complexes (S)-AgL2/(R)-AgL2.
1,3-Bis((S)-1′-phenylethyl)imidazolium silver(I) chloride ((S)-AgL2, 0.463 g, 69%)15. NMR and HRMS characterization results were consistent with what already reported in the literature.
1,3-Bis((R)-1′-phenylethyl)imidazolium silver(I) chloride ((R)-AgL2, 0.335 g, 50%). 1 H NMR (600 MHz, CDCl 3 , ppm) δ: 7.37–7.26 (10H, overlapping signals, Ar–
![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) ), 6.92 (2H, s, NC
), 6.92 (2H, s, NC![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) C
C![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) N), 5.76 (2H, q, J = 7.0 Hz, C
N), 5.76 (2H, q, J = 7.0 Hz, C![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) CH3), 1.84 (6H, d, J = 7.2 Hz, CHCH3).
CH3), 1.84 (6H, d, J = 7.2 Hz, CHCH3).
13 C-NMR (150 MHz, CDCl 3 , ppm) δ: 139.7, 129.1, 128.6, 126.6, 118.7, 61.0, 21.4.
(MALDI-ICR FTMS) m/z: HRMS calculated for C19H22AgClN2+: 421.7167, found: 421.7168.
Elemental analysis: calculated for C19H20AgClN2 C, 54.37; H, 4.80; Ag, 25.70; Cl, 8.45; N, 6.67; found C, 54.33; H, 4.75; Ag, 25.65; Cl, 8.41; N, 6.71.
Silver complexes (S)-AgL2/(R)-AgL2 (0.150 g, 0.317 mmol) were dissolved in dry dichloromethane (18 mL) under an inert atmosphere; (CH3)2SAuCl (0.105 g, 0.317 mmol) was added, and the resulting mixture was stirred at room temperature overnight. After completion, the reaction mixture was cooled to room temperature and filtered through a celite plug. The solvent was evaporated to complete dryness, and the resulting powder was washed with hexane (3 × 10 mL) to obtain the off-yellow powders of complexes (S)-AuL2/(R)-AuL2.
1,3-Bis((S)-1′-phenylethyl)imidazolium gold(I) chloride ((S)-AuL2, 0.077 g, 48%)15. NMR and HRMS characterization was consistent with the previously reported literature.
1,3-Bis((R)-1′-phenylethyl)imidazolium gold(I) chloride ((R)-AuL2, 0.089 g, 55%). 1 H NMR (600 MHz, CDCl 3 , ppm) δ: 7.37–7.34 (10H, overlapping signals, Ar–
![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) ), 6.82 (2H, s, NC
), 6.82 (2H, s, NC![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) C
C![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) N), 6.17 (2H, q, J = 7.2 Hz, C
N), 6.17 (2H, q, J = 7.2 Hz, C![[H with combining low line]](https://www.rsc.org/images/entities/i_char_0048_0332.gif) CH3), 1.84 (6H, d, J = 7.1 Hz, CHCH3).
CH3), 1.84 (6H, d, J = 7.1 Hz, CHCH3).
13 C-NMR (150 MHz, CDCl 3 , ppm) δ: 170.0, 139.2, 129.0, 128.6, 126.8, 118.1, 60.1, 20.8.
(MALDI-ICR FTMS) m/z: HRMS calculated for C19H22AuClN2+: 510.8150, found: 510.8148.
Elemental analysis: calculated for C19H20AuClN2 C, 44.85; H, 3.96; Au, 38.71; Cl, 6.97; N, 5.51; found C, 44.79; H, 3.99; Au, 38.67; Cl, 6.97; N, 5.49.
Biology
MTT assay
The 3-(4,5-dimethylthiazol-2-y1)-2,5-diphenyltetrazolium bromide (MTT, Sigma-Aldrich, Milan, Italy) assay was used for cell viability determination, as already published.31 Cells were seeded on 48 well plates, and then incubated for 24 h (for the anti-inflammatory/antioxidant activity studies) or 72 h (for the anticancer activity) with different concentrations of the tested compounds (dissolved in DMSO). The IC50 values, ± standard deviation (SD), were calculated from the percent (%) of control using the software GraphPad Prism 10 (GraphPad Software, La Jolla, CA, USA). For graphics, columns ± standard deviation (SD) represent the % of residual cell viability vs. the experimental control. Data are representative of three independent experiments performed in triplicate.Human topoisomerase I relaxation assay
Human topoisomerase I (hTopo I) relaxation assays were performed, as already described (https://doi.org/10.1016/j.ejmech.2024.116757), using 2 units of the recombinant hTopo I and 0.25 μg of supercoiled plasmid pHOT1, as substrates (TopoGEN, Port Orange, FL, USA). The tested complexes, dissolved in DMSO, were used at the indicated concentrations. The mixture was incubated for 1 h at 37 °C, then loaded onto an 1% agarose gel made with 1× TAE buffer without ethidium bromide (EB). After the run, the gel was stained with EB (0.5 mg mL−1 in 1× TAE buffer) for 30 min, then washed with distilled water thrice for 5 min and visualized using a UV transilluminator.TUNEL assay
TUNEL assay was used for nuclear DNA damage, according to the manufacturer instructions (One-step TUNEL Assay Kit (Green, FITC), MBS2567761, MyBioSource, Inc., San Diego, CA 92195-3308 USA). Briefly, cells were grown on glass cover slips, treated with (S)-AuL1 at 1 μM (in DMSO), then PBS washed thrice and methanol-fixed (−20 °C, 15 min). The enzyme was added and incubated for 35 minutes at 37 °C, protected from light. Next, they were counterstained using a DAPI solution (0.2 mg mL−1, 10 min, room temperature). After washing three times with cold PBS, one drop of mounting solution was added and cells were observed and imaged using a fluorescence microscope (Leica DM6000; 20× magnification) with excitation/emission wavelength maxima of 490 nm/515 nm (CFTM488A) or 350 nm/460 nm (DAPI). Representative fields of three independent experiments are shown.Anti-inflammatory and antioxidant activity
The anti-inflammatory and the antioxidant activities in terms of NO and ROS production, respectively, were determined on LPS stimulated RAW 264.7 murine macrophages.32 In detail, cells were plated on 48 multi-wells in a full medium, and after 24 hours, they were treated with the tested molecules (in DMSO), at the indicated concentrations, in a medium containing 1% of FBS, and lipopolysaccharide (LPS, Sigma Aldrich, Milan, Italy), at 1 μg mL−1, was used to induce NO production and oxidative stress. After 24 h, the cell medium was harvested and mixed with the Griess reagent (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio), then left under agitation for 30 min, at room temperature. The absorbance was measured by a multiplate reader, at 540 nm, and used for calculating the % of NO production compared to the control. For the antioxidant assays, after medium removal, cells were PBS-washed, added with 2′,7′-dichlorofluorescein diacetate (H2DCFDA, 25 μM) and incubated (40 min, 37 °C, 5% CO2). Then, cells were PBS-washed again and the fluorescence was quantified using a multiplate reader (λex = 485 nm, λem = 535 nm). The ROS production vs. LPS, expressed as percentage (%), was calculated using the following formula:
1 ratio), then left under agitation for 30 min, at room temperature. The absorbance was measured by a multiplate reader, at 540 nm, and used for calculating the % of NO production compared to the control. For the antioxidant assays, after medium removal, cells were PBS-washed, added with 2′,7′-dichlorofluorescein diacetate (H2DCFDA, 25 μM) and incubated (40 min, 37 °C, 5% CO2). Then, cells were PBS-washed again and the fluorescence was quantified using a multiplate reader (λex = 485 nm, λem = 535 nm). The ROS production vs. LPS, expressed as percentage (%), was calculated using the following formula:where FLPS represents the mean fluorescence obtained for the LPS alone treatment and Fsample represents the mean fluorescence obtained for the co-treatment with LPS + tested compound.
Immunofluorescence studies
For the immunofluorescence (IF) studies, cells were grown on glass cover slips in a full medium, then treated with the complexes or indomethacin at the indicated concentrations (dissolved in DMSO), in a medium containing 1% of FBS for 24 h. Next, cells were PBS-washed, fixed with cold methanol (15 min, −20 °C) and incubated overnight (4 °C) with the anti-iNOS, (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, diluted in BSA 2%, Santa Cruz Biotechnology Inc., CA, USA), as described.33 Alexa Fluor®568 conjugate goat-anti-mouse (1
100, diluted in BSA 2%, Santa Cruz Biotechnology Inc., CA, USA), as described.33 Alexa Fluor®568 conjugate goat-anti-mouse (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 diluted in BSA 2%, Thermo Fisher Scientific, MA, USA) was used as the secondary antibody. Nuclei were stained with 0.2 μg mL−1 of DAPI (10 min, Sigma Aldrich, Milan, Italy). Cells were visualized using a fluorescence microscope (Leica DM 6000) and LAS-X software (version 3.5.7.23225) was employed to acquire the images. Fluorescence quantification was done with Image J software (version 1.54d).
500 diluted in BSA 2%, Thermo Fisher Scientific, MA, USA) was used as the secondary antibody. Nuclei were stained with 0.2 μg mL−1 of DAPI (10 min, Sigma Aldrich, Milan, Italy). Cells were visualized using a fluorescence microscope (Leica DM 6000) and LAS-X software (version 3.5.7.23225) was employed to acquire the images. Fluorescence quantification was done with Image J software (version 1.54d).
We adopted a protocol that was previously widely used by our group, some examples are described in ref. 37. Docking grids were centered on the whole protein to identify the most likely binding pockets. A grid box of 126 × 126 × 126 points with a spacing of 0.375 Å was used. Docking poses were clustered using a root mean square deviation (RMSD) cutoff of 2.0 Å. The best binding mode for each compound was selected from the lowest energy conformation within the most populated cluster. Images of the ligand–protein interactions were generated with UCSF Chimera.38
Data analysis
All the experiments were repeated three times, each in triplicate and the results were expressed as mean ± standard deviation (SD). Differences were determined by one-way analysis of variance (ANOVA), followed by Dunnett's multiple comparison test, performed using GraphPad Prism 10. Differences were considered significant at p < 0.05.Conflicts of interest
The authors declare that they have no personal relationships that could have appeared to influence the work reported in this paper.Data availability
Supplementary information: supplementary information (SI) reporting NMR characterization of the compounds is available. See DOI: https://doi.org/10.1039/D5MD00651A.The data supporting this article have been included as part of the SI.
Acknowledgements
This work was supported by the Italian Ministry of Health (Ricerca Corrente) to Camillo Rosano, by PRIN 2022 PNRR, Code P20222BLAZ—Enhanced pharmacological activity of noble metal carbene-N-heterocyclic complexes by oligopeptide counterion (CUP MASTER: D53D23016900001)—and PRIN 2022, Code 2022HARH5W—HyMTA (Hybrid Multi-Target Agents) Synthesis and biological evaluation of chimeric hybrid molecules containing NHC-metal complexes and carbazole moieties, as innovative multi-target anticancer and antiviral agents (CUP MASTER: C53D23004490001). Mr Antonio Danubio is acknowledged for valuable experimental work.References
- N. Senkuttuvan, B. Komarasamy, R. Krishnamoorthy, S. Sarkar, S. Dhanasekaran and P. Anaikutti, RSC Adv., 2024, 14, 33429–33448 RSC.
- L. Song, M. Pan, R. Zhao, J. Deng and Y. Wu, J. Controlled Release, 2020, 324, 156–171 CrossRef CAS PubMed.
- C. Fliedel, A. Labande, E. Manoury and R. Poli, Coord. Chem. Rev., 2019, 394, 65–103 CrossRef CAS.
- M. Marra, A. Mariconda, D. Iacopetta, J. Ceramella, A. D'Amato, C. Rosano, K. Tkachenko, M. Pellegrino, S. Aquaro, M. S. Sinicropi and P. Longo, Molecules, 2024, 29, 5262 CrossRef CAS PubMed.
- T. J. Siciliano, M. C. Deblock, K. M. Hindi, S. Durmus, M. J. Panzner, C. A. Tessier and W. J. Youngs, J. Organomet. Chem., 2011, 696, 1066–1071 CrossRef CAS.
- S. J. Berners-Price and A. Filipovska, Metallomics, 2011, 3, 863–873 CrossRef CAS PubMed.
- S. A. Patil, S. A. Patil, R. Patil, R. S. Keri, S. Budagumpi, G. R. Balakrishna and M. Tacke, Future Med. Chem., 2015, 7, 1305–1333 CrossRef CAS PubMed.
- Y.-F. Zhang, Y.-K. Yin, H. Zhang and Y.-F. Han, Coord. Chem. Rev., 2024, 514, 215941 CrossRef CAS.
- I. Ott, Coord. Chem. Rev., 2009, 253, 1670–1681 CrossRef CAS.
- A. Mariconda, M. Sirignano, C. Costabile and P. Longo, Mol. Catal., 2020, 480, 110570 Search PubMed.
- C. Saturnino, I. Barone, D. Iacopetta, A. Mariconda, M. S. Sinicropi, C. Rosano, A. Campana, S. Catalano, P. Longo and S. Andò, Future Med. Chem., 2016, 8, 2213–2229 CrossRef CAS PubMed.
- M. Napoli, C. Saturnino, E. I. Cianciulli, M. Varcamonti, A. Zanfardino, G. Tommonaro and P. Longo, J. Organomet. Chem., 2013, 725, 46–53 CrossRef CAS.
- M. Sirignano, A. D'Amato, C. Costabile, A. Mariconda, A. Crispini, F. Scarpelli and P. Longo, Front. Chem., 2023, 11, 1260726 CrossRef CAS PubMed.
- A. P. da Costa, M. Sanaú, E. Peris and B. Royo, Dalton Trans., 2009, 6960–6966 RSC.
- G. A. Price, A. K. Brisdon, S. Randall, E. Lewis, D. M. Whittaker, R. G. Pritchard, C. A. Muryn, K. R. Flower and P. Quayle, J. Organomet. Chem., 2017, 846, 251–262 CrossRef CAS.
- E. V. Semenova, E. V. Belova, A. V. Sulimov and V. B. Sulimov, Chirality, 2024, 36, e23712 CrossRef CAS PubMed.
- P. O. Wagers, M. J. Panzner, M. R. Southerland, M. A. DeBord, M. C. Deblock, C. A. Tessier, C. L. Cannon and W. J. Youngs, in N-Heterocyclic Carbenes: From Laboratory Curiosities to Efficient Synthetic Tools, ed. S. Diez-Gonzalez, The Royal Society of Chemistry, 2016, pp. 567–595 Search PubMed.
- A. Chimento, C. Saturnino, D. Iacopetta, R. Mazzotta, A. Caruso, M. R. Plutino, A. Mariconda, A. Ramunno, M. S. Sinicropi, V. Pezzi and P. Longo, Bioorg. Med. Chem., 2015, 23, 7302–7312 CrossRef CAS PubMed.
- A. Lin, C. J. Giuliano, A. Palladino, K. M. John, C. Abramowicz, M. L. Yuan, E. L. Sausville, D. A. Lukow, L. Liu, A. R. Chait, Z. C. Galluzzo, C. Tucker and J. M. Sheltzer, Sci. Transl. Med., 2019, 11, eaaw8412 CrossRef CAS PubMed.
- T. Chen, Cancer Lett., 2024, 592, 216931 CrossRef CAS PubMed.
- N. K. Sharma, A. Bahot, G. Sekar, M. Bansode, K. Khunteta, P. V. Sonar, A. Hebale, V. Salokhe and B. K. Sinha, Cancers, 2024, 16, 680 CrossRef CAS PubMed.
- Q. Zhao, B. Han, C. Peng, N. Zhang, W. Huang, G. He and J.-L. Li, Med. Res. Rev., 2024, 44, 2194–2235 CrossRef CAS PubMed.
- J. Ceramella, A. Mariconda, D. Iacopetta, C. Saturnino, A. Barbarossa, A. Caruso, C. Rosano, M. S. Sinicropi and P. Longo, Bioorg. Med. Chem. Lett., 2020, 30, 126905 CrossRef CAS PubMed.
- C. Bonnefous, J. E. Payne, J. Roppe, H. Zhuang, X. Chen, K. T. Symons, P. M. Nguyen, M. Sablad, N. Rozenkrants, Y. Zhang, L. Wang, D. Severance, J. P. Walsh, N. Yazdani, A. K. Shiau, S. A. Noble, P. Rix, T. S. Rao, C. A. Hassig and N. D. Smith, J. Med. Chem., 2009, 52, 3047–3062 CrossRef CAS PubMed.
- J. Zuo, T.-H. Zhang, L. Xiong, L. Huang, C. Peng, Q.-M. Zhou and O. Dai, Molecules, 2022, 27, 6102 CrossRef CAS PubMed.
- A. Inkanuwat, R. Sukaboon, O. Reamtong, P. Asawanonda, A. Pattaratanakun, T. Saisavoey, P. Sangtanoo and A. Karnchanatat, Food Technol. Biotechnol., 2019, 57, 200–212 CrossRef CAS PubMed.
- B. Hinz, K. Brune, T. Rau and A. Pahl, Pharm. Res., 2001, 18, 151–156 CrossRef CAS PubMed.
- M. D. Carrión, B. Rubio-Ruiz, F. Franco-Montalban, P. Amoia, M. C. Zuccarini, C. De Simone, M. E. Camacho, R. Amoroso and C. Maccallini, Eur. J. Med. Chem., 2023, 248, 115112 CrossRef PubMed.
- A. J. Didier, J. Stiene, L. Fang, D. Watkins, L. D. Dworkin and J. F. Creeden, Antioxidants, 2023, 12, 632 CrossRef CAS PubMed.
- Y. J. Oh, S. E. Jin, H.-K. Shin and H. Ha, Sci. Rep., 2023, 13, 18891 CrossRef CAS PubMed.
- D. Iacopetta, C. Costabile, M. La Chimia, A. Mariconda, J. Ceramella, D. Scumaci, A. Catalano, C. Rosano, G. Cuda, M. S. Sinicropi and P. Longo, ACS Med. Chem. Lett., 2023, 14, 1567–1575 CrossRef CAS PubMed.
- Y. Tian, S. Zhou, R. Takeda, K. Okazaki, M. Sekita and K. Sakamoto, Biomed. Pharmacother., 2021, 141, 111854 CrossRef CAS PubMed.
- A. Mariconda, D. Iacopetta, M. Sirignano, J. Ceramella, A. D'Amato, M. Marra, M. Pellegrino, M. S. Sinicropi, S. Aquaro and P. Longo, Int. J. Mol. Sci., 2024, 25, 2599 CrossRef CAS PubMed.
- D. R. Cheshire, A. Åberg, G. M. K. Andersson, G. Andrews, H. G. Beaton, T. N. Birkinshaw, N. Boughton-Smith, S. Connolly, T. R. Cook, A. Cooper, S. L. Cooper, D. Cox, J. Dixon, N. Gensmantel, P. J. Hamley, R. Harrison, P. Hartopp, H. Käck, P. D. Leeson, T. Luker, A. Mete, I. Millichip, D. J. Nicholls, A. D. Pimm, S. A. St-Gallay and A. V. Wallace, Bioorg. Med. Chem. Lett., 2011, 21, 2468–2471 CrossRef CAS PubMed.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, J. Comput. Chem., 2009, 30, 2785–2791 CrossRef CAS PubMed.
- M. F. Sanner, B. S. Duncan, C. J. Carrillo and A. J. Olson, in Biocomputing '99, World Scientific, 1998, pp. 401–412 Search PubMed.
- C. Rosano, R. Lappano, M. F. Santolla, M. Ponassi, A. Donadini and M. Maggiolini, Curr. Med. Chem., 2012, 19, 6199–6206 CrossRef CAS PubMed.
- E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng and T. E. Ferrin, J. Comput. Chem., 2004, 25, 1605–1612 CrossRef CAS PubMed.
Footnote |
| † Co-senior authors. |
| This journal is © The Royal Society of Chemistry 2025 |