 Open Access Article
Open Access ArticleSmall-molecule probes for imaging and impairing the Golgi apparatus in cancer
Phanindra Kumar
a,
Poulomi Sengupta
*b and
Sudipta Basu
 *a
*a
aDepartment of Chemistry, Indian Institute of Technology (IIT) Gandhinagar, Palaj, Gandhinagar, Gujarat 382355, India. E-mail: Sudipta.basu@iitgn.ac.in
bDepartment of Chemistry, Indrashil University, Rajpur, Kadi, Mehsana, Gujarat 382740, India. E-mail: poulomisg@gmail.com
First published on 16th June 2025
Abstract
The Golgi apparatus (GA) is one of the most important subcellular organelles controlling protein processing, post-translational modification and secretion. Dysregulation of the GA structure and function leads to multiple pathological states, including cancer development and metastasis. Consequently, visualizing GA dynamic structures and their impairment in cancer has emerged as a novel strategy for next-generation unorthodox cancer therapeutics. However, the major challenge in GA-mediated theranostic probe development is the specific targeting of the GA within the subcellular milieu due to the lack of GA-recognizing chemical entities. In this review, we delineated various chemical functionalities that are extensively used as GA-homing moieties. Moreover, we outlined GA imaging probes consisting of classical fluorophores as well as novel aggregation-induced emissive (AIE) probes tagged with GA-homing moieties. Furthermore, we described GA-impairing molecules that can damage GA morphology through chemotherapeutic and photodynamic therapy (PDT) in cancer. Finally, we addressed the current challenges in this emerging and underexplored field of GA-targeted theranostics and proposed potential solutions to guide future cancer therapeutics.
Introduction
The Golgi apparatus (GA) is a complex network of vesicles and folded membranes in the cytoplasm of eukaryotic cells. As a central organelle involved in cellular processing and trafficking, the GA plays a crucial role in the maturation, modification, sorting, and transportation of proteins and lipids.1–6 However, in cancer, where cellular homeostasis is disrupted, the GA undergoes significant structural and functional alterations that contribute to malignant transformation. These changes are associated with key cancer-related processes, including metastasis, invasion, and cell signalling, making the GA an emerging focus in cancer biology.7–12In cancer, the GA regulates glycosylation, a post-translational modification in which carbohydrates are added to proteins and lipid chains to alter their functions and stability.13 Abnormal glycosylation by the GA affects growth factor receptors, adhesion molecules, and immune checkpoint regulators, leading to tumor progression, immune evasion, and metastasis. Moreover, the GA regulates the secretion of matrix metalloproteinases (MMPs), which are enzymes that degrade the extracellular matrix (ECM) and facilitate cancer cell invasion. Dysregulation of this secretory process promotes the metastatic spread of cancer cells.14 A hallmark of many aggressive cancers is the extensive fragmentation of the GA, which enhances cell migration and invasion. Fragmented GA structures are often correlated with poor prognosis and aggressive tumor behaviour.15 Furthermore, the GA helps cancer cells survive in the harsh conditions of the tumor microenvironment, including hypoxia and nutrient deprivation.16 By modifying secretory pathways, the GA enables cancer cells to adapt and resist apoptosis, thereby contributing to tumor progression.17 The GA plays a vital role in modulating key cancer-related signalling pathways, including the epidermal growth factor receptor (EGFR) and transforming growth factor-beta (TGF-β) pathways. Dysregulation of these pathways leads to uncontrolled proliferation and therapy resistance.18,19
Given its pivotal role in tumor progression, visualization of the morphology and structural dynamics of GA in cancer cells has become an important strategy for developing anticancer therapeutics.20–24 Toward this end, small-molecule fluorescent probes and aggregation-induced emission (AIE) probes have gained immense importance in GA imaging. Moreover, the development of GA-targeted anticancer therapeutic strategies has remained elusive to date due to the lack of specificity in GA-homing moieties. To address this issue, in this review, we summarized (a) GA targeting chemical entities (b) GA imaging fluorescence probes and AIE probes and (c) GA-impairing probes in cancer treatment, including chemo and phototherapy. Furthermore, we highlighted the challenges of developing novel small-molecule probes for imaging and selectively impairing the GA in cancer cells compared with normal cells for next-generation chemophototherapy.
Golgi apparatus targeting chemical functionalities
One of the major challenges in GA targeting is the lack of GA-selective chemical entities and knowledge about the surface functionality of GA membranes. To address this issue, over the years, several chemical moieties have been explored to route drugs and emissive molecules into GA for imaging and therapeutic purposes. During tumorigenesis, the GA apparatus undergoes changes that are linked to cyclooxygenase-2 (COX-2), an inducible enzyme that accumulates in the GA.25 Due to its overexpression in most cancer cells and relatively low expression in normal cells, COX-2 serves as an attractive target for molecular imaging. Additionally, COX-2 inhibitors, such as indomethacin (A) and celecoxib (B) bind selectively and tightly to the COX-2 enzyme side pocket.26 Hence, indomethacin and celecoxib (Fig. 1) can be used as targeting groups for COX-2 and be utilized for imaging the GA. Furthermore, recent studies have demonstrated that the selectivity of the COX-2 inhibitor SC-558 is attributed to its phenylsulphonamide moiety, which interacts with specific binding regions of COX-2 efficiently. Hence, the phenylsulphonamide moiety (C) is a promising GA-targeting unit for imaging and studying GA-associated biological processes. However, COX-2 is also housed in other organelles such as the endoplasmic reticulum (ER) and mitochondria. Hence, the use of phenylsulphonamide (C) for selective GA targeting is challenged by potential homing into the ER and mitochondria simultaneously.On the other hand, quinoline derivatives have been extensively employed in the development of fluorescent probes due to their superior photophysical properties. However, their potential for organelle-specific imaging has been largely underexplored. Quinoline naturally fluoresces in the near-ultraviolet region with tunable wavelengths through strategic structural modification. Furthermore, the pyridyl moiety in quinoline derivatives exhibits strong compatibility with the mildly acidic microenvironment (pH 6.0–6.7) of GA, establishing them as promising candidates for GA-targeted imaging and bioanalytical applications. Hence, 2-trifluoromethyl-7-aminoquinoline derivatives (D) have been successfully utilized for GA homing. Interestingly, galactosyltransferase and protein kinase D localize to the GA through cysteine residues or cysteine-rich domains, which is facilitated by the ability of cysteine thiol groups to form disulfide bonds with GA-associated thiols/disulphide moieties in the oxidizing environment of the GA lumen. Based on this mechanism, researchers have developed cysteine-tagged emissive probes (E) for early disease detection and GA-related therapeutics. Recently, peptides containing organelle-specific import or retention sequences have been shown to facilitate the targeted delivery of probes to subcellular structures such as the nucleus, trans-GA network (TGN), or ER when conjugated to fluorophores and drugs. Furthermore, substituting an oxygen atom with sulfur in the phosphodiester bond of phosphopeptide (F) triggered rapid dephosphorylation catalysed by alkaline phosphatase (ALP) and led to the formation of self-assembled thiopeptides.
This in situ self-assembly specifically targeted GA and selectively induced cancer cell death. On the other hand, ceramide serves as a precursor for complex sphingolipids, which are synthesized in the ER and transported to the GA via the ceramide transfer protein (CERT) in mammalian cells for further processing. Consequently, small-molecule fluorophores incorporating lipid-based tagging groups, such as ceramide (G), dodecanoic acid and tetradecyl chains with an amide unit, exhibit GA targeting ability in cell imaging. Despite having an elusive mechanism, probes with electron-withdrawing groups, such as cyano-stilbene (H and H′), were also explored for GA targeting. In addition, adenine nucleotide (I) was used to target and image GA.
Imaging Golgi apparatus
The GA is a highly dynamic organelle. Under diseased states like cancer, aberrant GA dynamics perturb the tumor microenvironment and immune responses, leading to enhanced invasion and metastasis.27 Moreover, dysregulation in GA dynamics includes altered GA orientation and morphology, which disturb GA-associated protein trafficking, post-translational modifications and cellular exocytosis. Hence, visualization of GA is of utmost importance for real-time monitoring of the structure–function relationship in tumor development and progression. Subsequently, several classical fluorescent probes as well as novel AIE probes tagged with GA-targeting moieties were explored to illuminate GA in cancer.28Classical fluorescent probes
 | ||
| Fig. 2 (a) Chemical structure of the indomethacin-tagged fluorophore. (b) Fluorescence emission spectra of 1 (2.0 μM) in the absence and presence of COX-2. (c) Fluorescence images of 1 (5.0 μM) (green) and BODIPY TR C5-ceramide (5.0 μM) (red) in HeLa cells. (d) Changes in GA morphology of cancer cells during apoptosis, stained with compound 1 in MCF-7 cells. (e) Chemical structure of nile blue-tagged indomethacin probe (2). (f) Confocal images of MCF-7 and HeLa cells stained with compound 2 (green) and NBD C6-ceramide (red). (g) Chemical structure of the BODIPY-tagged indomethacin-based viscosity rotor in GA (3). (h) Confocal fluorescence images of compound 3 (green) and GA tracker red in LO2 and HepG2 cells. (i) Confocal fluorescence imaging and FLIM imaging for LO2 and HepG2 cells incubated with compound 3 for 30 min. (j) Chemical structure of napthalimide–celecoxib-based GA imaging probe (4). (k) Fluorescence emission spectra of compound 4 in the presence of COX-2. (l) Confocal images of compound 4 (green) with commercial BODIPY FL-C5-ceramide (red) in MCF-7. (m) Chemical structure of celecoxib–nile blue conjugated probe (5) for GA imaging. Fig. 2b–d have been reproduced with permission from ref. 29. Copyright 2018 American Chemical Society. Fig. 2f has been reproduced with permission from ref. 30. Copyright 2015, The Royal Society of Chemistry. Fig. 2h and i have been reproduced with permission from ref. 31. Copyright 2022, The Royal Society of Chemistry. Fig. 2k and l have been reproduced with permission from ref. 32. Copyright 2018 American Chemical Society. | ||
In 2022, Liu et al. designed a GA-targeting fluorescent probe 3 for detecting and diagnosing the early stages of alcoholic liver injury by monitoring GA viscosity (Fig. 2g). Compound 3 consists of boron dipyrromethene (BODIPY) as a fluorophore and indomethacin as a GA-targeting moiety, which showed efficient colocalization in the GA of LO2 and HepG2 cells which resemble liver disease models (Fig. 2h). Furthermore, compound 3 was used to map the viscosity of GA by utilizing the viscosity-dependent fluorescence lifetime under Fluorescence Lifetime Imaging Microscopy (FLIM) imaging (Fig. 2i) and to study the alcohol concentration-dependent cellular damage in hepatic models in zebrafish. Thus, compound 3 could be an available detection tool for alcohol-induced liver injury at the subcellular level.31 In 2018, Peng's group developed a GA-targeting fluorescent probe for COX-2, in which celecoxib is used as the GA-targeting moiety, conjugated with naphthalimide via a flexible six-carbon linker (4, Fig. 2j). Subsequently, after binding to COX-2, compound 3 exhibited an increase in fluorescence intensity (Fig. 2k). Compound 3 was selectively localized to the GA in cancer cells than normal cells with excellent efficiency in vitro (Fig. 2l), as well as in vivo models.32 Similarly, in 2020, Gurram et al. developed a GA-targeting fluorescent probe (5, Fig. 2m) by conjugating celecoxib with nile blue via a flexible six-carbon linker. Compound 5 binds to COX-2 increased the fluorescent intensity exhibited strong one- and two-photon fluorescence signals selectively in cancer cells by localizing in GA efficiently.33
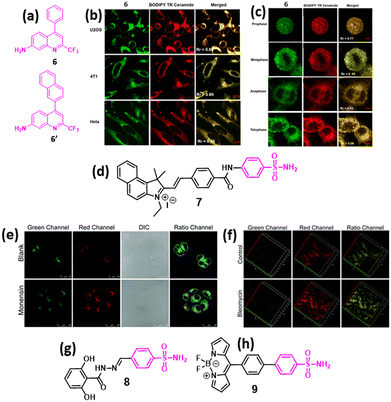 | ||
| Fig. 3 (a) Chemical structures of 2-trifluoromethyl-7-aminoquinoline fluorophores (6 and 6′) for GA imaging. (b) Confocal fluorescence micrographs showing GA stained by 6 (green), BODIPY TR ceramide (red), and the merged images in different cancer cells. (c) Subcellular localization in synchronously dividing HeLa cells of 6 (green), BODIPY TR ceramide (red) and the merged images. (d) Chemical structure of ratiometric GA polarity detection probe. (e) Confocal ratiometric fluorescence imaging of compound 7 in HL-7702 cells without or with monensin stimulation. (f) TP fluorescence imaging of kidney tissues in control mice and in LPS-induced AKI mice. (g and h) Chemical structures of 2,6-dihydroxybenzoyl–hydrazone-based (8) and BODIPY-based fluorescence probe (9) for imaging GA in cancer cells. Fig. 3b and c have been reproduced with permission from ref. 34. Copyright 2019 American Chemical Society. Fig. 3e and f have been reproduced with permission from ref. 37. Copyright 2021. The Royal Society of Chemistry. | ||
In recent years, the phenylsulfonamide group has become one of the most well-explored GA-targeting moieties. In 2021, Wang et al. developed a two-photon ratiometric fluorescent probe 7 (Fig. 3d) for monitoring GA polarity. Compound 7 exhibited higher fluorescence in normal cells than in cancer cells, indicating that cancer cells maintained lower GA polarity than normal cells. Moreover, compound 7 was further used to monitor GA polarity in HL-7702 cells before and after monensin treatment, which induced oxidative stress in GA leading to an increased internal polarity in GA (Fig. 3e). Compound 7-mediated polarity monitoring was also validated in the acute kidney injury (AKI) in Kunming mice model after treatment with bleomycin (Fig. 3f), confirming the utility of the phenylsulfonamide moiety for GA homing.37 In 2024, our group developed 2,6-dihydroxybenzoyl–hydrazone (8, Fig. 3g), which exhibited emissive properties due to long-range intramolecular charge transfer. Furthermore, compound 8 successfully homed into the GA of HCT-116 colon cancer cells very selectively compared with the breast (MCF7), lung (A549), and cervical (HeLa) cancer cells and non-cancerous RPE-1 cells without showing any toxicity.38
Very recently, Wang et al. developed a novel GA-targeting fluorescent probe (9, Fig. 3h) for real-time intracellular viscosity measurement, with potential applications in cancer diagnosis, as abnormal GA viscosity has been proposed as a biomarker for cancer detection. Compound 9 consists of a BODIPY-based viscosity rotor and a phenylsulfonamide moiety for GA-specific localization. Compound 9 showed fluorescence enhancement in response to viscosity changes and efficient colocalization into GA with minimal internalization in other organelles according to in vitro and in vivo models.39 Several other fluorescent probes conjugated with phenylsulfonamide were also developed for the sub-cellular detection of analytes such as H2O2, H2S and different ions in GA.28
 | ||
| Fig. 4 (a) Chemical structures of coumarin and napthalimide-based-lipid conjugated probes (10–12) for GA imaging. (b) Chemical structure of merocyanine–benzoyl difluoroboronate NIR probe (13) conjugated with cysteine for GA imaging. (c) Confocal fluorescence images of HL-7702 and SMMC-7721 cells stained with compound 13 (red). (d) Chemical structure of malononitrile-based GA imaging probe. (e) Confocal fluorescence images of compound 14 (red) in SH-SY5Y and LO2 cells stained with the GA tracker (green). Fig. 4c has been reproduced with permission from ref. 41. Copyright 2019 American Chemical Society. Fig. 4e has been reproduced with permission from ref. 42. Copyright 2024, Nature Publications. | ||
In 2019, Tang et al. developed a near-infrared (NIR) fluorescent probe 13 (Fig. 4b) for GA-targeted polarity visualization. Compound 13 exhibits aggregation within the GA, facilitated by cysteine, while its benzoyl difluoroboronate and merocyanine moieties enabled polarity detection by homing into the GA. Subsequently, compound 13 was used to identify polarity differences between normal (HL-7702) and cancerous cell lines (SMMC-7721), which showed high fluorescence in a cancerous cell line, indicating that the GA of the cancer cells has a low polar environment at the cellular level compared to the non-cancerous cells (Fig. 4c). Furthermore, compound 13 was used to study polarity changes in PC12 cells upon glutamate stimulation and increased polarity due to oxidative stress, leading to less fluorescence signal in in vitro and in vivo models. Moreover, compound 13 was used to study BDNF gene silencing in PC12 cells by using small interfering RNA (siRNA).41
In 2024, Zhao et al. developed a GA-targeting probe 14 (Fig. 4d), which can have an excellent ability to cross the brain barrier due to its lipophilicity and the presence of cyano groups. Compound 14 was specifically localized into the GA of SH-SY5Y and LO2 cells with higher efficiency (Fig. 4e). Furthermore, compound 14 was used to differentiate normal and cancerous cells using confocal imaging of LO2 and HepG2 cells, which showed higher fluorescence in HepG2 cells, indicating that GA has higher viscosity in liver cancer cells. Moreover, compound 14 was explored to study GA viscosity in Alzheimer's disease models, which showed higher fluorescence in Alzheimer's disease models compared to the control group, confirming that GA viscosity is significantly increased in Alzheimer's disease cells.42 However, the exact mechanism of the GA targeting moiety is elusive.
AIE-based probes
Most conventional organic dyes suffer from aggregation-caused quenching (ACQ) primarily due to the strong intermolecular π–π stacking. This effect severely limits their use in organic light-emitting diodes and organic nanodots for bioimaging because their emission is often quenched in solid films or aggregated states. Because traditional organic luminophores typically have planar aromatic cores that encourage π–π stacking upon aggregation, ACQ is a widespread phenomenon and has long been considered an inherent drawback. In 2001, Tang et al. introduced a new AIE phenomenon, which marked a significant breakthrough in luminescent materials.43 Unlike conventional ACQ dyes, which suffer from fluorescence quenching upon aggregation, AIEgen provides a wide avenue for exploring the potential scope for developing AIE-based probes that is useful for GA imaging. Hence, by fine-tuning the hydrophilic and hydrophobic moieties in the chemical structure of the probe and maintaining the required lipophilicity, the GA targeting property can be achieved.In 2021, Zhao et al. developed a GA-targeting AIE probe (15, Fig. 5a) containing tetraphenylethylene (TPE) as an AIE-inducing moiety and 2′,3′-O-isopropylideneadenosine (Ade) as a GA-targeting moiety. Compound 15 exhibited a remarkable increase in fluorescence intensity in an aqueous solution owing to restricted intramolecular rotations, leading to the AIE property. Furthermore, compound 15 efficiently stained GA in HL-7402 cells.44 In 2021, Tian et al. developed a “turn-on” AIEgen (16, Fig. 5b) consisting of 9,10-distyrylanthracene as the fluorophore and indomethacin as the GA-targeting group for detecting COX-2. In its native form in buffer solution, compound 16 exhibited a weak fluorescence. However, upon binding to COX-2, the fluorescence was significantly enhanced because of the restriction of intramolecular motion. Furthermore, compound 16 specifically stained GA in cancer cell lines (HeLa, MCF-7, and HepG2), producing strong fluorescence, whereas normal cell lines displayed only weak fluorescence. These findings confirmed that compound 16 selectively binds to COX-2, making it a potent and highly selective fluorescent probe for COX-2 detection (Fig. 5c).45 In 2021, Tang et al. explored an AIE-based fluorescence platform (17, Fig. 5d) having a phenylsulfonamide group to target GA and a triphenylamine (TPA) unit as a rotor and electron donor to induce AIE in THF/water binary solvent. Compound 17 homed into the GA with excellent photostability, biocompatibility and higher brightness compared to the commercially available GA tracker (Fig. 5e).46
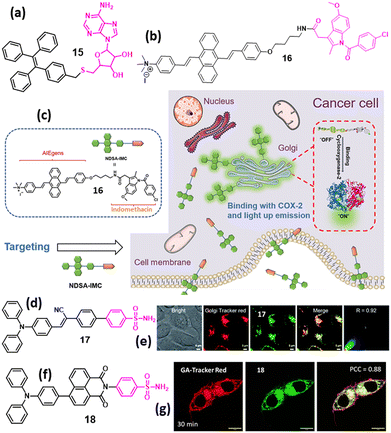 | ||
| Fig. 5 (a) Chemical structure of the TPE-2′,3′-O-isopropylideneadenosine (Ade)-based AIEgen (15) for GA imaging. (b) Chemical structure of 9,10-distyrylanthracene–indomethacin-based AIEgen (16) (c) discrimination of COX-2 enzyme via specific compound 16 probe. (d) Chemical structure of phenylsulfonamide-based AIEgen (17) for GA imaging. (e) Confocal images of HeLa cells stained with compound 17 (green) and GA tracker (red). (f) Chemical structure of triphenylamine–naphthalimide-based “on–off–on” AIEgen (18) for GA imaging. (g) Confocal laser scanning microscopy images of HCT-116 cells incubated with compound 18 (green) for 30 min, followed by staining GA with GA-tracker red dye. Fig. 5c has been reproduced with permission from ref. 45. Copyright 2022, Chinese Chemical Society. Fig. 5e has been reproduced with permission from ref. 46. Copyright 2021, Royal Society of Chemistry. Fig. 5g has been reproduced with permission from ref. 47. Copyright 2025, American Chemical Society. | ||
In 2025, our group developed an “on–off–on” AIE probe 18 (Fig. 5f) with phenylsulfonamide as the GA homing and triphenylamine-napthalimide as the AIE-triggering moieties. Compound 18 efficiently localized to GA within 30 min in HCT-116 cells (Fig. 5g). Moreover, we showed that compound 18 also efficiently homed into the ER within 30 min in HCT-116 cells, indicating that the phenylsulfonamide moiety is not very specific for GA and can also home the tagged molecule into the ER. Hence, thorough exploration is needed to evaluate the homing of phenylsufonamide-tagged molecules into other subcellular organelles, especially the ER. Furthermore, we observed that compound 18 can also home into the GA of non-cancerous human retinal pigment epithelial (RPE-1) cells, indicating that the phenylsulfonamide moiety is not highly specific for cancerous cells alone.47
Impairing Golgi apparatus
The pivotal role of GA in tumorigenesis has been highlighted in recent years, which showed that the GA remains an attractive target in cancer biology. However, the precise role and mechanisms through which the GA is involved in the development of cancers are yet to be fully understood. Subsequently, scientists have explored various drug candidates targeting GA as potential cancer treatments. These drugs act through various mechanisms, such as disrupting glycosylation, targeting key proteins and oncogenes linked to cancer progression, or directly damaging GA to trigger malignant cell death.48 Despite decades of research, no GA-targeting drug has received clinical approval for cancer therapy. Several well-known compounds, such as brefeldin A, monensin, swainsonine, and retinoic acid, have been extensively studied, and innovative strategies, such as nano-formulations and phototherapy, are being used to enhance their effectiveness. However, major hurdles remain, (a) specific drug localization into GA, (b) treatment-related side effects, and (c) a lack of comprehensive efficacy data. Despite the current challenges, GA and related processes have untapped potential as therapeutic targets.AIE-based probes for phototherapy
In 2022, Guo et al. developed an AIEgen-based photosensitizer (19, Fig. 6a) with TPE as AIE and cyanovinyl–methylpyridinium moiety as a GA targeting moiety, which can effectively home into the GA through caveoline/raft-mediated endocytosis. After irradiation under 532 nm laser light, compound 19 generated ROS, fragmented the GA morphology, triggered GA oxidative stress, followed by mitochondrial outer membrane permeabilization, leading to apoptosis in vitro and in vivo model (Fig. 6b). Interestingly, without having a designated GA-targeting moiety, compound 19 efficiently homed into the GA due to the presence of a cyano-group, which induced molecular rod-like assemblies for specific GA targeting compared with the other organelles. This study opened a new avenue towards GA-targeted effective photodynamic therapy (PDT).49 This study also exhibited the importance of GA-targeted phototherapy in impairing GA in cancer therapeutics. In 2024, Qi et al. developed an AIEgen-based photosensitizer having indomethacin (NSAID) and triphenylamine–thiophene–vinyl–pyridinium cation (20, Fig. 6c), which can specifically target GA and generate ROS, leading to severe damage to the morphology and function of GA to trigger apoptosis.50 | ||
| Fig. 6 (a) Chemical structures of cyanovinyl–methylpyridinium-based photosensitizer AIEgens for GA targeting. (b) Schematic of AIEgen-induced GA stress and the crosstalk between GA and mitochondria during cell apoptosis upon PDT. (c) Chemical structure of AIEgen-based photosensitizer containing indomethacin and triphenylamine–thiophene–vinyl–pyridinium cation. Fig. 6a and b have been reproduced with permission from ref. 49. Copyright 2022, Nature Publications. | ||
Self-assembled thiopeptide
In 2021, Xu et al. developed a phosphopeptide-based GA-targeting probe (21, Fig. 7a), which can impair cancer cells through enzymatic self-assembly. Peptide 21 efficiently enters HeLa cervical cancer cells via caveolin-mediated endocytosis and micropinocytosis and reacts with ALP to change the self-assembled morphology from a micellar structure to nanofibers, followed by homing into the GA by disulfide bond formation with GA proteins. Furthermore, compound 21 selectively killed HeLa cells more efficiently than the parent oxygenated phosphopeptide.51 Similarly, in 2022, Xu et al. engineered a short peptide chain-based GA-targeting fluorescent probe (22, Fig. 7b) containing nitrobenzoxadiazole (NBD) as the fluorophore and an enzyme-responsive aminoethyl thioester locking group attached to the self-assembling D-diphenylalanine, for targeting GA in different cell lines. Peptide 22 was internalized into the HeLa cell through caveolin-mediated endocytosis or macropinocytosis, hydrolysed by GA-associated thioesterase, dimerized and homed into the GA as well as into the ER, leading to disruption of protein trafficking followed by cell death through multiple pathways (Fig. 7c). Both peptides 21 and 22 demonstrated that the enzyme-responsive self-assembly of peptides could be interesting platforms for GA-targeting and controlling cell fate towards novel anti-cancer therapeutics.52 | ||
| Fig. 7 (a and b) Chemical structures of phospho and thioacetate peptides containing nitrobenzoxadiazole (NBD) for targeting GA. (c) Confocal images of the HeLa cells having peptide 22 homing into the GA. (d) Chemical structure of zinc phthalocyanines–indomethacin conjugate photosensitizer for GA targeting. Fig. 7c has been reproduced with permission from ref. 52. Copyright 2022, American Chemical Society. | ||
In 2022, the Chen group developed an ultra-efficient photosensitizer (23, Fig. 7d) by conjugating GA-targeting indomethacin along with zinc phthalocyanines as a photosensitizer unit to improve the therapeutic efficacy. Excitingly, compound 23 aggregated in the aqueous solution, causing fluorescence quenching. However, when it was bound to COX-2 after localization in GA, fluorescence enhancement was observed along with the generation of ROS, leading to excellent HepG2 cancer cell killing.53
Conclusions
In summary, we outlined small-molecule probes for imaging and impairing GA in cancer cells. We described the commonly used GA targeting chemical entities followed by imaging probes tagged with the GA-homing moieties. The imaging probes were subdivided into classical fluorophores and novel AIE probes. Furthermore, we outlined small-molecule probes tagged with GA-homing moieties for GA impairment in cancer cells via chemotherapy and PDT for cancer treatment.Outlook
In the last decade, GA has emerged as a key component in metastasis-causing cancer development because of its central role in protein processing and secretion. Therefore, in recent years, GA has gained immense attention as a novel target for cancer therapeutics. Subsequently, novel imaging agents (fluorophores and AIE probes) and GA-impairing agents have been developed for cancer treatment. Despite their tremendous potential, the development of novel GA-targeting agents has remained in its infancy because of the following challenges. Firstly, GA membrane biomarkers for the development of novel GA-homing chemical moieties have remained elusive. Although several GA-homing moieties were explored, none of the GA homing probes were thoroughly evaluated to understand their specificity towards GA and cross-homing to other subcellular organelles. Our recent work demonstrated that the widely used phenylsulfonamide moiety for GA homing routed the tagged AIEgen into the ER with equal efficiency.47 Hence, a novel GA-specific homing moiety is in high demand. Secondly, until now, very few examples of GA-induced impairment of molecules for chemotherapy have been reported. Hence, there is an immense uncharted territory to explore the target proteins inside GA and develop novel small molecules to specifically target them to understand their location–function relationship. Moreover, recent GA-targeted photosensitizers have been explored for noninvasive photodynamic therapy. However, there are no examples of GA-targeted chemo-photothermal agents for cancer therapy. Finally, one of the major challenges in GA-targeted therapy is to impair GA selectively in cancer tissues without causing any collateral damage to non-cancerous tissues, leading to toxic side effects for the patients. Towards this end, nanoscale materials have emerged as interesting platforms to deliver therapeutics into GA, specifically to cancer tissues.54–66 However, this field is also largely unexplored. Consequently, the development of novel biocompatible nanomaterials is needed for the GA-targeted combination of chemophototherapy. Furthermore, for the successful translation of GA-targeted theranostic probes from bench to bedside, it is important to develop in vivo disease models to understand the gap between cellular-level efficacy and animal animal-level potency. Toward this end, multiple in vivo mouse and zebrafish models for anticancer studies were explored, which showed promising results for further translation.30–33,37,39,42,49,50 We anticipate that focusing on the above-mentioned challenges would lead the way forward for the advancement of GA-targeted cancer theranostic applications.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Conflicts of interest
There are no conflicts to declare.Acknowledgements
PK acknowledges CSIR-UGC-JRF for doctoral fellowship. PS is thankful to the Science and Engineering Research Board (SRG/2021/000969) for funding. SB acknowledges IIT Gandhinagar, Gujarat Council on Science and Technology (GUJCOST/STI/R&D/2020-21/1302) and the Science and Engineering Research Board (CRG/2020/001127) for funding.Notes and references
- J. R. Baker, Nature, 1951, 168, 1089–1090 CrossRef CAS PubMed.
- C. Featherstone, Science, 1998, 282, 2172–2174 CrossRef CAS PubMed.
- J. Li, E. Ahat and Y. Wang, Results Probl. Cell Differ., 2019, 67, 441–485 CAS.
- F. Zappa, M. Failli and M. A. De Matteis, Curr. Opin. Cell Biol., 2018, 50, 102–116 CrossRef CAS PubMed.
- A. Eisenberg-Lerner, A. Benyair, S. Hizkiahou, T. N. Nudel, H. Maor-Nof, T. Kramer, N. Yayon, Y. Ulman, E. G. Yarden and Y. Elazar, Nat. Commun., 2020, 11, 409 CrossRef CAS PubMed.
- J. Gao, A. Gao, H. Zhou and L. Chen, Cell Biol. Int., 2022, 46, 1309–1319 CrossRef CAS PubMed.
- S. Bui, I. Mejia, B. Díaz and Y. Wang, Front. Cell Dev. Biol., 2021, 9, 806482 CrossRef PubMed.
- R. Bajaj, A. N. Warner, J. F. Fradette and D. L. Gibbons, Cells, 2022, 11, 1484 CrossRef CAS PubMed.
- J. Liu, Y. Huang, T. Li, Z. Jiang, L. Zeng and Z. Hu, Int. J. Mol. Med., 2021, 47, 38 Search PubMed.
- Y. Li, L. Mu, Y. Li, Y. Mi, Y. Hu, X. Li, D. Tao and J. Qin, Cell Death Dis., 2024, 15, 417 Search PubMed.
- M. Martins, A. S. Fernandes and N. Saraiva, Int. J. Biochem. Cell Biol., 2022, 145, 106174 CrossRef CAS PubMed.
- M. Martins, J. Vieira, C. Pereira-Leite, N. Saraiva and A. S. Fernandes, Biology, 2024, 13, 1 CrossRef CAS PubMed.
- X. Zhang, Front. Cell Dev. Biol., 2021, 9, 665289 Search PubMed.
- V. Millarte and H. Farhan, Sci. World J., 2012, 2012, 498278 Search PubMed.
- A. Petrosyan, Biochem. Mol. Biol. J., 2015, 1, 16 Search PubMed.
- D. Mennerich, S. Kellokumpu and T. Kietzmann, Antioxid. Redox Signaling, 2019, 30, 113–137 CrossRef CAS PubMed.
- D. Spano and A. Colanzi, Cells, 2022, 11, 1990 CrossRef CAS PubMed.
- J. Wang, P. K. Lau, C. W. Li and Y. Guo, J. Biol. Chem., 2023, 299, 102979 CrossRef CAS PubMed.
- H. J. S. Stewart, R. Curtis, K. R. Jessen and R. Mirsky, Eur. J. Neurosci., 1995, 7, 1761–1772 CrossRef CAS PubMed.
- C. Preisinger, B. Short, V. De Corte, E. Bruyneel, A. Haas, R. Kopajtich, J. Gettemans and F. A. Barr, J. Cell Biol., 2004, 164, 1009–1020 CrossRef CAS PubMed.
- J. I. Sbodio, B. D. Paul, C. E. Machamer and S. H. Snyder, Cell Rep., 2013, 4, 890–897 CrossRef CAS PubMed.
- L. Lu, Q. Zhou, Z. Chen and L. Chen, J. Cell. Physiol., 2018, 233, 2911–2919 CrossRef CAS PubMed.
- J. I. Sbodio, S. H. Snyder and B. D. Paul, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 780–785 CrossRef CAS PubMed.
- C. Kienzle and J. von Blume, Trends Cell Biol., 2014, 24, 584–593 Search PubMed.
- C. Yuan and W. L. Smith, J. Biol. Chem., 2015, 290, 5606–5620 CrossRef CAS PubMed.
- S. Chennamaneni, B. Zhong, R. Lama and B. Su, Eur. J. Med. Chem., 2012, 56, 17–29 CrossRef CAS PubMed.
- B. V. Howley and P. H. Howe, Oncoscience, 2018, 5, 142–143 CrossRef PubMed.
- S. Xu, K.-C. Yan, Z.-H. Xu and X.-P. He, Chem. Soc. Rev., 2024, 53, 7590–7631 RSC.
- H. Zhang, J. Fan, J. Wang, S. Zhang, B. Dou and X. Peng, J. Am. Chem. Soc., 2013, 135, 11663–11669 CrossRef CAS PubMed.
- B. Wang, J. Fan, X. Wang, H. Zhu, J. Wang, H. Mu and X. Peng, Chem. Commun., 2015, 51, 792–795 RSC.
- C. Liu, L. Zhou, Y. Zheng, H. Man, Z. Ye, X. Zhang, L. Xie and Y. Xiao, Chem. Commun., 2022, 58, 10052–10055 RSC.
- B. Gurram, S. Zhang, M. Li, H. Li, Y. Xie, H. Cui, J. Du, J. Fan, J. Wang and X. Peng, Anal. Chem., 2018, 90, 5187–5193 CrossRef CAS PubMed.
- B. Gurram, M. Li, J. Fan, J. Wang, S. Zhang and X. Peng, Front. Chem. Sci. Eng., 2020, 14, 41–52 CrossRef CAS.
- J. Chen, H. Liu, L. Yang, J. Wang and Y. Yang, ACS Med. Chem. Lett., 2019, 10, 954–959 CrossRef CAS PubMed.
- J. Li and Y. Wang, Cells, 2022, 11, 289 CrossRef CAS PubMed.
- S. Kellokumpu, Front. Cell Dev. Biol., 2019, 7, 93 Search PubMed.
- H. Wang, M. Dong, H. Wang, F. Huang, P. Li, W. Zhang, W. Zhang and B. Tang, Chem. Commun., 2021, 57, 5838–5841 RSC.
- X. Wang, J. Fan, J. Wang and X. Peng, ChemBioChem, 2024, 25, e202400507 Search PubMed.
- X. Wang, X. Li, Z. Liu, Y. Meng, X. Fan, H. Wang, J. Nie and B. Xue, Sens. Actuators, B, 2024, 278, 126497 Search PubMed.
- X.-C. Feng, G. Zhang, R. Sun, Y.-J. Xu and J.-F. Ge, Sens. Actuators, B, 2023, 394, 134469 CrossRef CAS.
- P. Li, X. Guo, X. Bai, X. Wang, Q. Ding, W. Zhang, W. Zhang and B. Tang, Anal. Chem., 2019, 91, 3382–3388 CrossRef CAS PubMed.
- W. Wu, L. Zhao, Y. Zhang, J. Wei, J. Han, Y. Zhang and Z. Zhao, Sci. Rep., 2024, 14, 1336 CrossRef CAS PubMed.
- J. Luo, Z. Xie, J. W. Y. Lam, L. Cheng, H. Chen, C. Qiu, H. S. Kwok, X. Zhan, Y. Liu, D. Zhu and B. Z. Tang, Chem. Commun., 2001, 1740–1741 Search PubMed.
- X. Xing, Y. Jia, J. Zhang, Z. Wu, M. Qin, P. Li, X. Feng, Y. Sun and G. Zhao, Sens. Actuators, B, 2021, 329, 129245 CrossRef CAS.
- Y. Luo, S. Zhang, H. Wang, Q. Luo, Z. Xie, B. Xu and W. Tian, CCS Chem., 2022, 4, 456–463 CrossRef CAS.
- P. Xiao, K. Ma, M. Kang, L. Huang, Q. Wu, N. Song, J. Ge, D. Li, J. Dong, L. Wang, D. Wang and B. Z. Tang, Chem. Sci., 2021, 12, 13949–13957 RSC.
- P. Kumar, T. Mishra, Sanyam, A. Mondal and S. Basu, ACS Appl. Bio Mater., 2025, 8, 1524–1532 CrossRef CAS PubMed.
- Z. Y. Lee, W. H. Lee, J. S. Lim, A. A. A. Ali, J. S. E. Loo, A. Wibowo, M. F. Mommat and J. B. Foo, Life Sci., 2024, 352, 122868 CrossRef CAS PubMed.
- M. Liu, Y. Chen, Y. Guo, H. Yuan, T. Cui, S. Yao, S. Jin, H. Fan, C. Wang, R. Xie, W. He and Z. Guo, Nat. Commun., 2022, 13, 2179 CrossRef CAS PubMed.
- X. Wang, X. Wang, Y. Li and Z. Qi, Dyes Pigm., 2024, 222, 111897 CrossRef CAS.
- W. Tan, Q. Zhang, J. Wang, M. C. Quiñones-Frías, J. Wang and B. Xu, Angew. Chem., Int. Ed., 2021, 60, 12796–12801 CrossRef CAS PubMed.
- W. Tan, Q. Zhang, M. C. Quiñones-Frías, A. Y. Hsu, Y. Zhang, A. Rodal, P. Hong, H. R. Luo and B. Xu, J. Am. Chem. Soc., 2022, 144, 6709–6713 CrossRef CAS PubMed.
- J. Chen, K. Huang, J. Xue, M. Yan and H. Zhang, Dyes Pigm., 2022, 198, 109997 CrossRef.
- R.-Y. Yu, L. Xing, P.-F. Cui, J.-B. Qiao, Y.-J. He, X. Chang, T.-J. Zhou, Q.-R. Jin, H.-L. Jiang and Y. Xiao, Biomater. Sci., 2018, 6, 2144–2155 RSC.
- H. Li, P. Zhang, J. Luo, D. Hu, Y. Huang, Z.-R. Zhang, Y. Fu and T. Gong, ACS Nano, 2019, 13, 9386–9396 CrossRef CAS PubMed.
- X. Zhou, J. Xu, N. Ya, L. Deng, Y. Zeng and X. Zhou, ACS Appl. Nano Mater., 2024, 7, 7520–7532 CrossRef CAS.
- J. Luo, T. Gong and L. Ma, Carbohydr. Polym., 2020, 249, 116887 CrossRef CAS PubMed.
- L. Chen, P. Jiang, X. Shen, J. Lyu, C. Liu, L. Li and Y. Huang, Small, 2023, 19, 2204747 CrossRef CAS PubMed.
- H. Li, C. Deng, Y. Tan, J. Dong, Y. Zhao, X. Wang, X. Yang, J. Luo, H. Gao, Y. Huang, Z.-R. Zhang and T. Gong, Acta Biomater., 2022, 146, 357–369 CrossRef CAS PubMed.
- H. Li, P. Zhang, J. Luo, D. Hu, Y. Huang, Z.-R. Zhang, Y. Fu and T. Gong, Nano Lett., 2021, 21, 8455–8465 CrossRef PubMed.
- M. Zhang, N. Xu, W. Xu, G. Ling and P. Zhang, Pharmacol. Res., 2022, 175, 105861 CrossRef CAS PubMed.
- J. Chen, Y. Ma, W. Du, T. Dai, Y. Wang, W. Jiang, Y. Wan, Y. Wang, G. Liang and G. Wang, Adv. Funct. Mater., 2020, 30, 2001566 Search PubMed.
- J. Luo, P. Zhang, T. Zhao, M. Jia, P. Yin, W. Li, Z.-R. Zhang, Y. Fu and T. Gong, ACS Nano, 2019, 13, 3910–3923 Search PubMed.
- Y. Tang, X. Wang, G. Zhu, Z. Liu, X. Chen, H. K. Bisoyi, X. Chen, X. Chen, Y. Xu, J. Li and Q. Li, Small, 2023, 19, 2205440 Search PubMed.
- X. Wang, Y. Li, K. Hasrat, L. Yang and Z. Qi, Small, 2023, 19, 2305101 CrossRef CAS PubMed.
- X. Wang, Y. Li and Z. Qi, Chem. – Asian J., 2024, 19, e202400311 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |




