Open Access Article
Open Access ArticleRational design of indolyl acrylamides as antibacterial agents targeting multidrug-resistant Acinetobacter baumannii strains†
Velvett G.
Domínguez-Méndez
 a,
Rosa María
Chávez-Santos
a,
Rosa María
Chávez-Santos
 a,
Karol
Carrillo-Jaimes
a,
Karol
Carrillo-Jaimes
 b,
Alejandra
Hernández-Santoyo
b,
Alejandra
Hernández-Santoyo
 c,
Santos
Ramírez-Carreto
c,
Santos
Ramírez-Carreto
 c,
Armando
Hernandez-Garcia
c,
Armando
Hernandez-Garcia
 c,
Rodrigo
Aguayo-Ortiz
c,
Rodrigo
Aguayo-Ortiz
 d,
Corina-Diana
Ceapă
d,
Corina-Diana
Ceapă
 b,
José
Rivera-Chavéz
b,
José
Rivera-Chavéz
 b and
Roberto
Martínez
b and
Roberto
Martínez
 *a
*a
aDepartamento de Química Orgánica, Instituto de Química, Universidad Nacional Autónoma de México, Circuito Exterior, Ciudad Universitaria, Alcaldía Coyoacán, C.P. 04510, Cd de Mx, Mexico. E-mail: robmar@unam.mx
bLaboratorio of Microbiología, Departamento de Productos Naturales, Instituto de Química, Universidad Nacional Autónoma de México, Circuito Exterior, Ciudad Universitaria, Alcaldía Coyoacán, C.P. 04510, Cd de Mx, Mexico
cDepartamento de Química de Biomacromoléculas, Instituto de Química, Universidad Nacional Autónoma de México, Circuito Exterior, Ciudad Universitaria, Alcaldía Coyoacán, C.P. 04510, Cd de Mx, Mexico
dDepartamento de Farmacia, Facultad de Química, Universidad Nacional Autónoma de México, Circuito Exterior, Ciudad Universitaria, Alcaldía Coyoacán, C.P. 04510, Cd de Mx, Mexico
First published on 20th May 2025
Abstract
Antimicrobial resistance (AMR) has become a significant public health problem. This study investigated the structure–activity relationship of indole core molecules to uncover novel antimicrobials against resistant bacteria. Their antimicrobial evaluation against ESKAPEE bacteria showed superior efficacy compared to cefepime, meropenem, ciprofloxacin, and gentamicin against multidrug-resistant A. baumannii strain A-564, with minimum inhibitory concentration (MIC) values of 4.3 and 1.2 μg mL−1 for compounds 12e and 12j, respectively. Also, the same compounds showed better activity than cefepime for A. baumannii BAA ATCC 747 with MIC values of 1.2 and 4.4 μg mL−1. In addition, 12e and 12f showed activity against methicillin- and penicillin-resistant S. aureus with MIC values of 3.2 and 2.1 μg mL−1. Furthermore, the highly active compounds 12e and 12j exhibited low toxicity, with hemolysis values >40 μg mL−1. Preliminary examination of the mechanism of action revealed that 12e could exhibit dose-dependent inhibition of the AbFtsZ1–412 enzyme from strain XDR A-564, achieving 51% inhibition of GTPase activity at 32 μg mL−1, thus altering the binary fission process, which could be attributed to the fact that 12e binds to the GTP site and interferes with the function of the enzyme by inhibiting the formation of the Z-ring. Also, a cell viability assay indicates that cells treated with these compounds showed increased permeability, compromising the stability of the A. baumannii A-564 membrane. These results provided valuable information for further developing indolyl-acrylamides as new antimicrobial agents.
1. Introduction
Antimicrobial resistance (AMR) represents a significant global threat to public health.1 In 2019, AMR was associated with approximately 4.95 million deaths.2 The World Health Organization (WHO) projects that AMR could result in up to 10 million deaths per year by 2050.3 Clinically significant multidrug-resistant pathogens group under the ESKAPEE acronym, which includes: Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter spp., and Escherichia coli.4 These bacteria, prevalent in hospital settings, have developed multidrug resistance (MDR), extreme drug resistance (XDR) or pandrug resistance (PDR) making them particularly dangerous.5 The WHO recently updated its list of priority pathogens for antibiotic research and development, placing carbapenem-resistant A. baumannii among the top three bacteria in group one, classified as a critical priority.6 Clinical isolates of this bacterium, such as the extensively drug-resistant (XDR) strain A-564 from a Mexican patient, have shown resistance to various drug families, including quinolones, aminoglycosides, penicillins, cephalosporins, carbapenems, and macrolides.7 Hence, developing new and effective antibacterial drugs to address MDR and XDR strains is crucial.Designing bioactive molecules and conducting structure–function studies offer targeted and efficient approaches to drug discovery, reducing the time and resources required compared to the extensive testing of large chemical libraries. One of such alternative drug discovery approaches is searching for privileged structures – molecular frameworks that confer biological activities in diverse pharmacological targets.8 This approach has shown promising results in finding pharmacologically active compounds across a broad range of therapeutic areas. Several reviews on the topic of privileged structures8 support the idea that the indole ring is one of the best-known privileged structures and exerts various pharmacological activities depending on the functional groups joined to the moiety.9 Examples of bioactive compounds bearing the indole scaffold are panobinostat (1) for the treatment of multiple myeloma,10 ramosetron (2) as an antiemetic and as a treatment for irritable bowel syndrome,11 sumatriptan (3) for the treatment of migraine,12 and aterviridine (4) with activity against human immunodeficiency virus (HIV-1)13 (Fig. 1).
Moreover, the indole-containing scaffold is a key structural feature in many compounds that inhibit the growth of Gram-negative and Gram-positive bacteria.10 The synthesis of a series of 3-amino indoles was previously reported, and their antibacterial properties were evaluated against ESKAPEE group bacteria. Among these, compound 5 (Fig. 1) demonstrated significant inhibitory activity against ciprofloxacin-resistant A. baumannii and P. aeruginosa with a MIC of 8 μg mL−1 for both strains.14 Compound 6 (Fig. 1) demonstrated an even more potent activity against MDR and XDR strains, especially when combined with carbapenems. It also exhibited broad-spectrum efficacy against Enterobacteriaceae, with an MIC90 value of 1–2 μg mL−1. The study suggested that incorporating a primary amine into molecules that meet specific structural and rigidity requirements enhances accumulation in bacterial cells, thereby boosting antibacterial potency.15,16 Compound 7 (Fig. 1) showed promising activity with MIC values ranging from 1 μg mL−1 to 4.8 μg mL−1 against several drug-sensitive bacteria, including S. aureus ATCC 25923, S. aureus ATCC 26003, E. faecalis ATCC 29212, B. subtilis ATCC 63501, and E. coli ATCC 44568. Additionally, this antimicrobial agent exhibited activity against both Gram-positive and Gram-negative MDR bacteria, specifically E. coli BAA-196 and S. aureus ATCC 43300 (MRSA), with MIC values as low as 0.5 μg mL−1 for both strains.17 Interestingly, in the same year that compound 7 was reported, Yuan, W. et al.18 discovered a very similar indole derivative (compound 8), (Fig. 1) which presented excellent activity against S. aureus ATCC 29247 (ampicillin-resistant), E. faecalis 51575, and E. faecium 700221, both vancomycin-resistant, with MIC values between 2 and 4 μg mL−1. Upon investigating the antibacterial mechanism of compound 8, it was found that, at a concentration of 8 μg mL−1, this molecule inhibits 60% of the GTPase activity of the SaFtsZ enzyme. This closely aligns with previous observations of other indole derivatives, where bacterial growth inhibition was linked to the disruption of FtsZ activity.19 FtsZ is a temperature-sensitive filamentous protein involved in bacterial cell division that assembles into a circumferential structure known as the Z-ring.20 This process requires guanosine-5′-triphosphate (GTP) binding and hydrolysis (GTPase activity), along with stabilization and anchoring of FtsZ to cytoplasmic membrane-binding proteins. Once the Z-ring is formed, it recruits a series of additional proteins necessary for cytokinesis.21 To date, only a limited number of antimicrobial agents have been identified that bind to this protein and modulate its polymerization or GTPase activity.22 Nevertheless, as the FtsZ protein is highly conserved and essential across bacterial species, it represents a promising yet underexplored target for the development of new antimicrobial agents.22
This research centers on the design and synthesis of indole-based compounds, inspired by the remarkable antibacterial activity and mechanisms of action of compounds 7 and 8 (Fig. 1). By conducting a comprehensive structure–activity relationship (SAR) study, we aim to delve into their antimicrobial properties and elucidate potential modes of action, also showing that the strategic design of bioactive molecules and the application of structure–function studies provide a focused and efficient pathway for drug discovery, significantly cutting down on the time and resources needed compared to the broad testing of large chemical libraries. Our goal is to contribute to significant advancements in antimicrobial research, paving the way for the development of novel and effective antibacterial agents.
2. Results and discussion
2.1 Chemistry
Compounds 9a–c, 10a–c, 11a–h, 11a′, and 12a–j were synthesized through the efficient routes outlined in Scheme 1, demonstrating the effectiveness and productivity of our process. Initially, compounds 15a–c, 9a–c, and 18a–d were synthesized in two steps, with the first step involving the indole N–H substitution of compounds 13a–c with Boc, Ts, and p-Cl-benzyl groups. N-substitution with Boc was carried out using Boc2O and NaH in THF at rt for 24 h obtaining the N-Boc compounds 14a–c in 89–96% yields.23 The N-Ts intermediates 16a–c were obtained by stirring 13a–c with p-toluene sulfonyl chloride, NaOH, and catalytic amounts of benzyltriethylammonium chloride (TEBAC) in DCM at rt for 16 h, in 81–94% yields.24 Similarly, compounds 17a–d were prepared using 13a–d, p-chlorobenzyl chloride, NaOH and TEBAC. The second step was a Knoevenagel condensation reaction of 14c, 16a–c and 17a–d with malonic acid, via the Doebner modification to obtain 15a–c, 9a–c, and 18a–d in 84–93% yields.25Conversely, compounds 10a–c were obtained by acid hydrolysis of 15a–c with trifluoroacetic acid (TFA) in DCM, with yields between 68 and 78%.26 Likewise, the coupling of indolylacrylic acids 9a–c or 18a and the corresponding amine was achieved using HBTU and DIPEA in DCM/DMF (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 v/v), giving compounds 11a–h and 12a–j in 13–68% yields.27 Finally, compound 11a′ was prepared by stirring 19 with p-toluene sulfonyl chloride in DMC at rt for 24 h, followed by a coupling reaction with HBTU, DIPEA, and piperazine in DCM/DMF (1
3 v/v), giving compounds 11a–h and 12a–j in 13–68% yields.27 Finally, compound 11a′ was prepared by stirring 19 with p-toluene sulfonyl chloride in DMC at rt for 24 h, followed by a coupling reaction with HBTU, DIPEA, and piperazine in DCM/DMF (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 v/v) at rt for 5 h, obtaining compound 11a′ with a 43% yield.27 The structures of all compounds were confirmed by 1H and 13C NMR spectroscopy and high-resolution mass spectrometry (ESI-HRMS). For example, in the 1H NMR spectrum of compound 12e, the signals for the 2-N-amino-ethyl group are evident, including one triplet at approximately 2.70 ppm for C-2′ and one quartet at 3.22 ppm for C-1′. Two doublets were also observed at 6.58 and 7.51 ppm, corresponding to the unsaturated α–β system, with a J = 15.9 Hz indicating the trans conformation of the alkene. Moreover, the AA'BB′ system for the aromatic ring of the p-Cl-benzyl substituent was also observed.
3 v/v) at rt for 5 h, obtaining compound 11a′ with a 43% yield.27 The structures of all compounds were confirmed by 1H and 13C NMR spectroscopy and high-resolution mass spectrometry (ESI-HRMS). For example, in the 1H NMR spectrum of compound 12e, the signals for the 2-N-amino-ethyl group are evident, including one triplet at approximately 2.70 ppm for C-2′ and one quartet at 3.22 ppm for C-1′. Two doublets were also observed at 6.58 and 7.51 ppm, corresponding to the unsaturated α–β system, with a J = 15.9 Hz indicating the trans conformation of the alkene. Moreover, the AA'BB′ system for the aromatic ring of the p-Cl-benzyl substituent was also observed.
Additionally, we observed in the 13C NMR spectrum of 12e a peak at 166.18 ppm corresponding to the carbonyl group of the α–β unsaturated amide, as well as the carbons for the ethylenediamine motif at 41.09 and 41.42 ppm and the methylene at 48.65 ppm of the p-Cl-benzyl group. When the compound is 12j, the signals for the 2-N-amino-butyl group are a triplet at 2.99 ppm, with another at 3.36 ppm corresponding to C-1′ and C-4′, and a multiplet at 1.69–1.71 ppm corresponding to the two central methylene's CH2–2′ and CH2–3′. Also, the structure of the synthesized compound 12j, with a molecular formula C22H24BrClN3O, was supported by the HRMS spectrum, where the molecular ion peak ESI+m/z 460.07892 (M + H)+ is in concurrence with the calculated value of 460.07913. Similarly, the ESI-HRMS results were consistent with all the synthesized structures.
2.2 Antibacterial studies
Compounds 9a–c, 10a–c, 11a–j, 11a′, and 12a–j were screened against a panel of bacterial strains, including Acinetobacter baumannii ATCC BAA 747 and ATCC 17978, as well as the extensively drug-resistant (XDR) clinical strain of Mexican origin A-564, and against other bacteria of the ESKAPEE group: S. aureus (MRSA), S. aureus ATCC 25923, K. pneumoniae K2 (MDR), and P. aeruginosa P-48M (MDR). We used cefepime, gentamicin or colistin as controls (Table 1). MIC values were determined using a plate microdilution assay following the guidelines of the Clinical and Laboratory Standards Institute (CLSI).28 Percentages were afterwards calculated using GraphPad Prism 9.5.1.| Compounds | A. b 17978a | A. b BAA 747b | A. b A-564 (XDR)c | P. a 48-M (MDR)d | K. p K2 (MDR)e | S. a clinic (MRSA)f | S. a 25923g | Hemolysis HC50 |
|---|---|---|---|---|---|---|---|---|
| a A.b.17978, Acinetobacter baumannii ATCC 17978. b A.b. BAA 747, Acinetobacter baumannii ATCC BAA 747. c A.b. A-564 (XDR), Acinetobacter baumannii A-564 (XDR). d P.a. 48-M (MDR), Pseudomonas aeruginosa 48-M (MDR). e K.p. K2 (MDR), Klebsiella pneumoniae K2 (MDR). f S.a. MRSA, penicillin–methicillin-resistant Staphylococcus aureus. g S.a. ATCC 25923, Staphylococcus aureus ATCC 25923.1Hemolytic concentration 50 (HC50) (n = 3). *MIC values were calculated with the Gompertz equation using GraphPad Prism 9.5.1, (n = 3). NT = no test. | ||||||||
| 11a | 19.0 ± 1.1* | 7.8 ± 1.2* | 31.7 | >122.2 | NT | 48.8 | >73 | ≤47 |
| 11b | 19.7 ± 1.0* | 7.5 ± 1.2* | 44.3 | >110 | >66 | NT | >66 | >40 |
| 11c | >61.3 | 20.4 | 61.3 | >102 | NT | >61 | >61 | >40 |
| 11d | >63.5 | >21.1 | >105.8 | >105 | >63 | >63 | >63 | NT |
| 11e | >61.5 | >61.5 | >102.6 | >102 | >61 | >61 | >61 | NT |
| 11f | >63.9 | >63.9 | >106.6 | >106 | >63 | >106 | >63 | >40 |
| 11g | 8.3 ± 1.0* | 3.2 ± 2.3* | 10.2 ± 1.1* | 46.2 | >23 | 8.9 ± 3.4* | >69 | >40 |
| 11h | 13.7 ± 1.1* | 6.3 ± 1.1* | 9.6 ± 1.5* | 41.7 | >20 | 8.1 ± 1.1* | >62 | >40 |
| 11a′ | >61.6 | >41.1 | 51.3 | >102 | NT | NT | >61 | >40 |
| 12a | 20.9 ± 1.1* | 4.5 ± 1.2* | 13.7 ± 1.1* | >114 | >22 | >22 | >68 | ≤40 |
| 12b | 8.7 ± 1.1* | 8.7 ± 1.0* | 31 | >103 | >20 | >20 | >62 | >40 |
| 12c | >56.9 | 14.2 ± 1.1* | 29.6 | >94 | >56 | >18 | >56 | ≤40 |
| 12d | >59.0 | >59.0 | >59.0 | >98 | >19 | >19 | >59 | NT |
| 12e | 7.1 ± 1.1* | 1.2 ± 2.6* | 4.3 ± 1.1* | 23.2 ± 1.1* | 12.1 ± 1.2* | 3.2 ± 1.1* | 14.2 ± 1.0* | >40 |
| 12f | 7.4 ± 1.0* | 5.9 ± 3.5* | 8.8 ± 1.1* | 16.3 ± 1.0* | 9.7 ± 1.9* | 2.1 ± 2.1* | 11.6 ± 1.0* | >40 |
| 12g | 23.3 ± 1.0* | NT | 26.5 | >88 | >17 | >17 | >53 | >40 |
| 12h | 15.9 ± 1.1* | 9.6 ± 1.1* | 37.1 | NT | 18.5 | 6.6 ± 1.1* | >55 | NT |
| 12i | 10.23 ± 1.0* | 9.5 ± 1.0* | 15.4 ± 1.1* | 44.6 | >22 | 6.2 ± 1.2* | 21.8 ± 1.0* | <21 |
| 12j | 8.3 ± 1.1* | 4.4 ± 1.1* | 1.2 ± 1.3* | NT | 18.4 | 8.2 ± 1.9* | 16.5 ± 1.9* | >40 |
| Cefepime | 16.3 | 15.3 | >64 | NT | NT | NT | NT | NT |
| Meropenem | NT | NT | >32 | NT | NT | NT | NT | NT |
| Gentamicin | 0.95 | 0.95 | >64 | 0.95 | 0.13 | NT | NT | NT |
| Ciprofloxacin | NT | NT | >50 | NT | NT | NT | 8 | NT |
| Penicillin G | NT | NT | NT | NT | NT | >84 | NT | NT |
2.3 Structure–activity relationship discussion
We started with structural simplification to evaluate the antimicrobial structure–function relationship. The exchange of a basic group (guanidinium) for an acidic group was explored, resulting in compounds 9a–c (Fig. 2), evaluated at an initial concentration of 50 μg mL−1. Compounds 9a (C5–Br) and 9b (C5–Cl) showed 72% to 100% inhibition against A. baumannii BAA ATCC 747. Compound 9a stood out by achieving 39% inhibition against A. baumannii XDR strain A-564. Attempting to further simplify the structure, the tosyl indole group was replaced by a hydrogen, which resulted in a considerable decrease in potency of the resulting 10a–c molecules against all tested A. baumannii isolates (see ESI† Table S1, page S16), indicating the importance of the indole N-sulfonamide group for the antibacterial activity.29 These findings prompted the hypothesis that systematic exploration of structural modifications to the compounds could enhance their bioactivity. Compounds 9a–c were selected as hit molecules for further optimization (Fig. 2). Since piperazine-derived compounds have been previously reported as antibacterial agents, we replaced the carboxylic acid in the core structure with a piperazinyl group, thus obtaining 11a–c (Table 1).30 Compounds 11a–c, as well as the compounds resulting from the subsequent structural modifications, were evaluated against A. baumannii BAA ATCC 747, ATCC 17978, and XDR A-564 strains. The antibacterial activity of compound 11c against A. baumannii BAA ATCC 747 with a MIC of 20.4 μg mL−1 is comparable to that of the commercial antibiotic cefepime (MIC = 15.3 μg mL−1) (Table 1). Furthermore, it was observed that compound 11c exhibited a MIC value of 61.3 μg mL−1 against the XDR A-564 strain, which is better than the MIC value of the reference drug cefepime (MIC > 64 μg mL−1). Nevertheless, it did not show relevant activity against A. baumannii ATCC 17978 (MIC > 61.3 μg mL−1).Compounds 11a and 11b, with a (C5–Br) and (C5–Cl) substituent, respectively, showed better antibacterial activity than cefepime with values of 31.7 and 44.3 μg mL−1 respectively against A. baumannii XDR A-564 (Table 1). Compound 11a also showed an MIC of 7.8 μg mL−1 against A. baumannii BAA ATCC 747, indicating that the bromo substituent at C-5 effectively enhances the antibacterial activity against both strains.31 The MIC values were 2 times better than those of cefepime for both strains. Likewise, compound 11b presented a MIC = 7.5 μg mL−1 value lower than that of cefepime for strain BAA ATTC 747, but for strain XDR A-564, the potency decreased compared to 11a.
Based on these results, and aiming to improve the MIC value in the simpler compound 11c against A. baumannii strains, the NH group of the piperazinyl was replaced with O and S, resulting in 11e and 11f, respectively.32 These compounds lost their potency considerably against A. baumannii BAA ATCC 747, A. baumannii ATCC 17978, and A. baumannii XDR A-564, presenting MIC values above 60 μg mL−1.This finding suggested that the NH of the piperazinyl group is crucial for antibacterial potency. This hypothesis was confirmed experimentally by testing compound 11d, which showed a potency decrease (MIC > 20 μg mL−1) for all strains of A. baumannii. Similarly, the double bond influence of the α,β-unsaturated system was evaluated using compound 11a′. However, the potency was not significantly improved against any strain with a MIC > 40 μg mL−1.
The N-2-aminoethyl group was incorporated in compounds with higher activity 11a and 11b instead of a piperazinyl moiety to evaluate the influence of the rigidity of the piperazinyl group on bioactivity, generating compounds 11g (C5–Br) and 11h (C5–Cl). Compound 11g showed a significant bioactivity improvement against two ATCC strains of A. baumannii, with MIC values of 8.3 μg mL−1 for strain 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 978 and 3.2 μg mL−1 for strain BAA 747, thus increasing the potency of 11g two times compared to 11a against the BAA ATCC 747 strain. Evaluation of compound 11g against the XDR A-564 strain gave a MIC of 10.2 μg mL−1, indicating that its potency is six times higher than cefepime and three times higher than 11a. Previous studies have also reported that substituting the indole N-linked H with the p-Cl-benzyl group enhances the antimicrobial activity.33 Based on this, replacing the tosyl group with the p-Cl-benzyl group and keeping the piperazinyl group and the bromine at C-5 to generate compound 12a was proposed. Compound 12a showed a considerable increase in potency against strains BAA 747 and XDR A-564, exhibiting values of 4.5 μg mL−1 and 13.7 μg mL−1, respectively, indicating that 12a presents a significant increase in potency compared to cefepime and compound 11a. Both structure–activity relationship studies indicate that the N-2-aminoethyl group and the p-Cl-benzyl group significantly increase the activity against A. baumannii strains.
978 and 3.2 μg mL−1 for strain BAA 747, thus increasing the potency of 11g two times compared to 11a against the BAA ATCC 747 strain. Evaluation of compound 11g against the XDR A-564 strain gave a MIC of 10.2 μg mL−1, indicating that its potency is six times higher than cefepime and three times higher than 11a. Previous studies have also reported that substituting the indole N-linked H with the p-Cl-benzyl group enhances the antimicrobial activity.33 Based on this, replacing the tosyl group with the p-Cl-benzyl group and keeping the piperazinyl group and the bromine at C-5 to generate compound 12a was proposed. Compound 12a showed a considerable increase in potency against strains BAA 747 and XDR A-564, exhibiting values of 4.5 μg mL−1 and 13.7 μg mL−1, respectively, indicating that 12a presents a significant increase in potency compared to cefepime and compound 11a. Both structure–activity relationship studies indicate that the N-2-aminoethyl group and the p-Cl-benzyl group significantly increase the activity against A. baumannii strains.
As a consequence, molecular hybridization of both groups in the base structure of the indole ring was proposed, thus obtaining the 12e–h compounds.34 Evaluation of compound 12e (C5–Br) against ATCC strain BAA 747 revealed a MIC value of 1.2 μg mL−1, which is ten times more potent than cefepime. Moreover, this compound showed a remarkable increase in potency against the XDR A-564 with a MIC value of 4.3 μg mL−1, making it thirteen times more potent than cefepime and two times more potent than 11g, distinguishing 12e as a promising compound. Subsequently, to improve potency, bromine was retained at carbon 5, and a modification of the chain length (homologation) was proposed, replacing the N-2-aminoethyl group with aliphatic chains 12in = 3 and 12jn = 4.35 For the A. baumannii BAA ATCC 747 strain, compound 12i showed decreased antibacterial activity with a MIC value of 9.5 μg mL−1. However, for compound 12j, the MIC was 4.4 μg mL−1, indicating a homologous effect for this strain, with a value three times better than cefepime. The homology effect against strain XDR A-564 showed the next tendency. Compound 12i displayed a decrease in potency, with a MIC of 15.4 μg mL−1. At the same time, 12j showed a homologous effect, with a MIC of 1.2 μg mL−1, representing a significant increase in power compared to 12e, but a more than 50-fold increase compared to cefepime. In addition, all compounds underwent rigorous testing against other bacteria from the ESKAPEE group, including clinical isolates of penicillin–methicillin-resistant S. aureus resulting in very low MIC values of 3.2 μg mL−1 with 12e and of 2.1 μg mL−1 with 12f.
The chemical structure optimization steps and antimicrobial activity results presented in this study demonstrate a procedure that successfully enhanced the biological efficacy of a family of compounds by over tenfold through the derivatization of a privileged core structure. These findings underscore the potential of rational design and targeted structural modifications in drug development, highlighting a promising pathway for the creation of more potent antimicrobial agents. The final lead compounds 12e and 12j exhibit significant promise due to their superior bioactivity and potential to address various multidrug-resistant pathogens, marking a crucial step forward in limiting antimicrobial resistance.
2.4 Morphological effects of the indole family compounds on A. baumannii A-564 treated cells
Microscopic observations of the bacterial cell morphology have uncovered a significant finding. At a concentration of 2.1 μg mL−1, 12e has shown a remarkable effect on A. baumannii A-564 cells, increasing their length (B) compared to untreated cells (A) (Fig. 3). This increase in length indicates a potential mechanism causing abnormal bacterial cell division and cell death.36 The results strongly suggest that the synthesized compound has an inhibitory effect on GTPase activity, leading to instability and abnormal bacterial cell division, ultimately resulting in cell death. These findings could potentially lead to the development of new strategies for combating bacterial infections.2.5 Effects of indolyl-acrylamides derivatives on the GTPase activity of AbFtsZ1–412
The polymerization of the FtsZ protein, a pivotal process for cell division, is intricately linked to the rate of GTP hydrolysis.37 Previous studies by Carrillo-Jaimes, K. et al.,5 where they expressed and purified the AbFtsZ1–412 protein from the XDR A-564 strain, demonstrated that berberine can inhibit the GTPase activity of the AbFtsZ1–412 enzymes by no more than 50% at a concentration of 186 μg mL−1. Additionally, other compounds, such as sanguinarine and curcumin,5 previously reported as FtsZ inhibitors of E. coli and B. subtilis, were tested but no inhibition was observed for the GTPase activity of AbFtsZ1–412. Given the high conservation of this protein in bacterial species, we turned our attention to the potential of 12e and 12j to affect the GTPase activity of AbFtsZ1–412. This exploration could have far-reaching implications for our understanding of the antibacterial mechanism of indolyl-acrylamides analogs. The compounds were tested in the 8–46 μg mL−1 range. In the GTPase assay, 12e and 12j demonstrated concentration-dependent inhibition, reaching a maximum inhibition of 51 ± 5.4% and 30.5 ± 6.3% at 32 μg mL−1 (Fig. 3).2.6 Docking and molecular dynamics simulations of compounds 12e (A) and 12j (B) in the GTP binding site of FtsZ
The blind docking study revealed that both compounds strongly prefer the GTP binding site. Additionally, both compounds showed a highly similar binding mode at this site, depicted in gray (Fig. 4). Three independent 100 ns MD simulations showed that the protein backbone remained stable with an RMSD of less than 0.25 nm. However, both ligands exhibited a significant change in orientation at the site compared to the predicted docking pose. By analyzing the fraction of structures derived from clustering analysis (CF), the most representative pose for each ligand was identified. One of the most intriguing findings of our study is the unexpected similarity in the poses of both ligands. Despite this, the pose of compound 12e was only represented in 31.0% of the last 50 ns across the three simulations, while compound 12j's binding pose, despite its longer aliphatic chain, was found in 70.4%. Both compounds also exhibited highly similar profiles of hydrogen bond (HB) and hydrophobic (HI) interaction fractions (IFs). They formed hydrogen bonds with D213 in over 80.0% of the replicas, as well as a high percentage of hydrophobic contacts with N52 and Y209. Compound 12j also formed a hydrogen bond with D214 and hydrophobic interactions with K210 during the simulations. Despite these similarities, compound 12j may exhibit lower FtsZ inhibitory activity due to the potential increase in conformational entropy caused by its longer chain, which could affect its binding affinity.2.7 Effect of indolyl-acrylamides on membrane permeabilization
Previous studies have shown that specific FtsZ inhibitors can induce secondary effects on the bacterial membrane, leading to cell lysis.38 In A. baumannii, membrane disruption is a particularly relevant mechanism, given that this bacterium possesses a highly impermeable outer membrane that confers resistance to multiple antibiotics.39 The cell viability assay (Fig. 5) revealed that bacteria treated with 12e and 12j exhibited more dead cells (stained red) than the untreated control, in which no dead cells were observed. Moreover, in bacteria exposed to 12e, virtually no live cells (stained green) were detected. These results indicate that cells treated with these compounds showed increased permeability to propidium iodide (PI) (Fig. 5), suggesting that both compounds compromise the stability of the A. baumannii A-564 membrane, as PI is a marker that only penetrates damaged membranes.2.8 Hemolysis
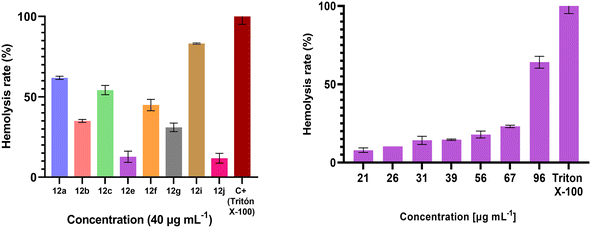
Hemolysis, the breakdown of red blood cells, can be caused by exogenous factors and lead to health complications. The hemolysis assay, a tool that can be used to evaluate the safety and usefulness of new compounds with promising biological activity, has shown promising results.40,41 All compounds were tested at 40 μg mL−1. Interestingly, the hemolytic properties of the most active compounds against A. baumannii XDR A-564 and A. baumannii BAA ATCC 747, 12e and 12j, were >40 μg mL−1 (Fig. 6), indicating that the compounds present more selectivity against microorganisms with a lower effect on red blood cells.3. Conclusions
In summary, a novel library of indolyl-acrylamides analogs was synthesized by an in vitro MIC activity-guided structure–activity relationship (SAR) study, and their in vitro antibacterial activity was evaluated. The results show that these compounds exhibit up to ten fold increased potency against resistant and extremely resistant bacteria when they present the following structural features: 1) a bromo bonded at the C-5 position of the indole ring, 2) a p-chlorobenzyl group joined to indole nitrogen, 3) an increase in the rotational degrees of freedom of the N-amino group bonded to the C-1 carbonyl moiety, and 4) a homology effect due to chain length. Significant antibacterial activity against various strains, including those multidrug-resistant, was obtained with these modifications (Fig. 7). Importantly, the antibacterial activities observed against A. baumannii A-564 XDR ranged from 4.3 μg mL−1 to 1.2 μg mL−1, significantly lower compared to that of cefepime (MIC > 64 μg mL−1), among others. Furthermore, our research has uncovered compounds demonstrating potent antibacterial activity against methicillin-resistant S. aureus (MRSA), with compounds 12e and 12f emerging as standout performers with MIC values of 3.2 μg mL−1 and 2.1 μg mL−1, respectively. The MIC values of several of these compounds are equally promising against Gram-negative strains, such as drug-resistant K. pneumoniae and P. aeruginosa, suggesting a common cellular target or multiple mechanisms of action. These findings, coupled with additional biochemical evaluations that reveal at least one mode of action of these compounds, suggest a potential breakthrough. Furthermore, compounds 12e and 12j have demonstrated the potential to inhibit 50% of the enzymatic activity of AbFtsZ1–412. Notably, both compounds were able to inhibit A. baumannii growth at concentrations lower than those required to inhibit AbFtsZ1–412, suggesting the involvement of additional mechanisms in their antibacterial effect and, consequently, in their antimicrobial activity. The ability of 12e and 12j to disrupt membrane integrity could represent an additional mechanism enhancing their efficacy against A. baumannii. Importantly, 12e and 12j exhibited negligible hemolytic activity compared to their MIC values against A. baumannii strains, suggesting a possible selectivity for prokaryotic membranes. Antimicrobials with multiple mechanisms of action offer significant advantages in combating antimicrobial resistance (AMR). They reduce the likelihood of resistance development by targeting multiple pathways, enhance efficacy and provide broad-spectrum activity against a range of pathogens, including MDR and XDR strains. By effectively targeting resistant strains, these antimicrobials also help mitigate the spread of resistance genes, contributing to better long-term control of AMR. This multifaceted approach represents a promising strategy for developing more effective and sustainable treatments. However, further studies are needed to fully understand the factors driving this selective interaction with bacterial membranes. The implications of these findings are profound, suggesting that novel indolyl-acrylamides could be a beacon of hope in the fight against drug-resistant bacteria.4. Biological assay
4.1 Antibacterial assay
The antimicrobial activities of compounds 9a–c, 10a–c, 11a–h, 11a′, and 12a–j were evaluated according to the method described in CLSI guidelines M07-A10 (microdilution assay) (CLSI 2015).42 The bacterial strain was suspended in Müller–Hinton broth at 37 °C for 12 h, and turbidity was adjusted to 0.5 McFarland standard units (108 CFU mL−1) equivalent to 0.08–0.13 optical density units (600 nm). Molecules were evaluated at concentrations between 0.1 and 250 μM dissolved in DMSO. The microdilution setup consisted of 10 μL of bacterial suspension, 2 μL of sample (compounds), and 88 μL of Müller–Hinton broth. The optical density of the plate was then measured at 600 nm (OD600) using a BioTek Cytation 7 cell imaging multimode reader plate reader. The plate was then incubated at 37 °C for 24 h, and the measurement was repeated to calculate the % inhibition and growth rate. The % inhibition was calculated using eqn (1), and the minimum inhibitory concentrations of the compounds were calculated by nonlinear regression using a modified Gompertz function with GraphPad Prism 9.5.1.5,42 | (1) |
4.2 Microscopic visualization
Cells of strain A. baumannii A-564 were grown in Müller–Hinton medium and incubated for 24 h, then the bacterial culture was re-suspended in Müller–Hinton broth, and the turbidity was adjusted to 0.5 McFarland standard units (108 CFU mL−1) equivalent to 0.08–0.13 optical density units (600 nm). The microdilution setup consisted of 10 μL of bacterial suspension, 2 μL of 12e (in DMSO), and 88 μL of Müller–Hinton broth; in 4 wells of the plate, there was no compound and in 4 other wells, the 0.5 MIC of the test compound was cultured at 37 °C for 24 h. Cells for morphology studies were collected and stained with Gram stain. Images were captured using a Zeiss Imager.M2 microscope with an Axiocam 503.2 camera.364.3 Hemolysis assay
Rabbit erythrocytes were resuspended in 0.9% w/v NaCl and centrifuged at 3000 × g for 10 minutes. This procedure was repeated until a clear supernatant was obtained. The erythrocytes were then resuspended in a 50 mM phosphate buffer, pH 7.4, containing 150 mM NaCl to prepare a 20% v/v solution. For the assay, 96-well flat-bottom microplates were used. Each well was loaded with 50 μL of buffer, 50 μL of erythrocytes, and 2 μL of each compound to be tested (diluted in DMSO), to achieve a final volume of 102 μL per well. The plates were incubated for 4 h at 37 °C. After incubation, 25 μL of the supernatant was transferred to a new plate, and absorbance was measured at 405 nm using a BioTek Cytation 7 cell imaging multimode reader.43 All experiments were performed in triplicate, using Triton X-100 as a positive control and DMSO as a negative control. Compounds were tested at a concentration of 40 μg mL−1, and the percentage of hemolysis was calculated using the following formula: | (2) |
4.4 Enzymatic assay of AbFtsZ1–412
The evaluation of enzyme inhibition was performed following a protocol reported.5 The assay was performed in a 96-well plate, employing a buffer solution of 50 mM Tris, 200 mM KCl, and 2.5 mM MgCl2 at pH 7.0, GTP as a substrate (50 μM), AbFtsZ1–412 enzyme (4 μM) and 12e and 12j to be evaluated (20–100 μM in DMSO). Then, the plate was incubated for 30 min at 37 °C; after this time, a malachite green solution (malachite green 79%, molybdate 20%, Tween 1%) was added, and then the absorbance was read at 630 nm using a BioTek Cytation 7 cell imaging multimode plate reader. All experiments were performed in triplicate, using berberine (500 μM) as a positive control and DMSO as a negative control. The activity results were calculated with eqn (3) and are expressed as a percentage (%). | (3) |
4.5 Cell viability assay LIVE/DEAD
The cell viability of the A. baumannii XDR strain A-564 was assessed using a LIVE/DEAD BacLight Bacterial Viability Kit, following the manufacturer's instructions (Thermo Fisher Scientific, MA, USA). Bacterial cultures were grown to the late log phase, then centrifuged at 5000 × g and resuspended in 2 mL of culture medium to achieve an OD600 of 5. The cells were transferred to a 96-well plate and exposed to twice the MIC of compounds 12e and 12j for 6 hours at 37 °C in 100 μL reaction volume. Following treatment, cultures—including the untreated negative control—were harvested and washed with 0.85% NaCl by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 minutes at 4 °C. The cell pellet was resuspended in 0.85% NaCl, and a fluorescent nucleic acid stain mixture containing SYTO 9 and propidium iodide (PI) was added. SYTO 9 penetrates all cells, regardless of membrane integrity, binds to nucleic acids, and fluoresces green. In contrast, PI only crosses damaged bacterial membranes, binding to nucleic acids and fluorescing red, thus serving as a marker of membrane integrity. Stained cells were incubated at room temperature for 15 minutes in the dark with constant shaking. Fluorescence analysis was performed using a BioTek Cytation 5 multimode cell imaging reader (Agilent) equipped with a 40× objective and filters for fluorescein and Texas Red. Image processing was carried out using BioTek Gen5 Software (Agilent) for imaging and microscopy.
000 × g for 10 minutes at 4 °C. The cell pellet was resuspended in 0.85% NaCl, and a fluorescent nucleic acid stain mixture containing SYTO 9 and propidium iodide (PI) was added. SYTO 9 penetrates all cells, regardless of membrane integrity, binds to nucleic acids, and fluoresces green. In contrast, PI only crosses damaged bacterial membranes, binding to nucleic acids and fluorescing red, thus serving as a marker of membrane integrity. Stained cells were incubated at room temperature for 15 minutes in the dark with constant shaking. Fluorescence analysis was performed using a BioTek Cytation 5 multimode cell imaging reader (Agilent) equipped with a 40× objective and filters for fluorescein and Texas Red. Image processing was carried out using BioTek Gen5 Software (Agilent) for imaging and microscopy.
5. Docking and molecular dynamics simulations of compounds 12e and 12j in the GTP site of FtsZ
5.1 Ligand preparation
The tridimensional structures of compounds 12e and 12j were generated from the SMILES representation using the OpenBabel toolbox.44 Both compounds underwent energy minimization using the Universal Force Field (UFF) implemented in the same software. Subsequently, a second round of geometry optimization and energy minimization was conducted with Gaussian 16,45 using the B3LYP46,47 density functional and the 6-31G(d) basis set.5.2 Protein preparation
A homology model of the FtsZ protein from A. baumannii was constructed using the SwissModel web server.48 We employed the FtsZ crystal structure from Pseudomonas aeruginosa (PDB ID: 1OFU, resolution: 2.10 Å) as the template for our model. This structure was chosen as the template since it had the highest sequence identity with the FtsZ sequence from A. baumannii, based on the BLAST analysis conducted in UniProt49 with the UniProtKB with the 3D structure target database. The assessment of the generated structure showed that the model has acceptable global model quality estimate (GMQE) and QMEANDisCo global scores,50 with values of 0.64 and 0.78, respectively. Additionally, the Ramachandran plot indicated that 96.38% of the backbone dihedral angles are in the favored region. MolProbity51 revealed no Cβ deviations or bad bonds and only 10 bad angles, and only 3.77% of residues were identified as rotamer outliers.5.3 Molecular docking
To identify the most reliable binding site and binding mode of both 12e and 12j in AbFtsZ, we carried out a blind docking study using QuickVina-W.52 We set a search box covering the entire surface of the structure and an exhaustiveness value of 50 for the docking to ensure thorough exploration. From the 20 docking runs, we selected the binding pose with the best score for the molecular dynamics simulations.5.4 Molecular dynamics
The selected binding poses of compounds 12e and 12j within the GTP site of AbFtsZ were subjected to three independent 100 ns molecular dynamics (MD) simulations using the AMBER14SB53 force field in GROMACS 2021.54 The ACPYPE package55 was used to parametrize the molecules within the AMBER force field framework and generate the necessary GROMACS input files. To carry out the MD simulations, the protein–ligand complexes were initially solvated in a cubic box using the TIP3P water model. Sodium and chloride ions were added randomly to neutralize the system and achieve a concentration of 0.15 M. Subsequently, an energy minimization step was applied, followed by a 1 ns equilibration period in both the canonical (NVT) and isothermal–isobaric (NPT) ensembles. Once equilibrated, the system underwent a 100 ns non-restrained NPT production run. The temperature was maintained at 300 K using the V-rescale algorithm,56 while the pressure was controlled at 1.0 bar using the Parrinello-Rhaman barostat.57 All hydrogen-containing bonds were constrained using the LINCS algorithm.58 Additionally, a Lennard–Jones potential with a shift function was employed within a range of 1.0 to 1.2 nm, while electrostatic interactions were computed within a 1.2 nm cutoff radius. Long-range electrostatic interactions were calculated using the PME method.59Here, the root-mean-square deviation (RMSD) of the AbFtsZ backbone and ligand heavy atoms after least squares fit to the protein backbone were computed for each MD simulation replica. The last 50 ns of the three MD replicas were collectively analyzed, determining the most representative pose of the compounds using a ligand RMSD-based clustering analysis with a cutoff value of 0.2 nm and employing the Gromos method. The number of hydrogen bonds and hydrophobic interactions formed during the simulations was also computed using the GROMACS-built in tools and the stand-alone application of PLIP v2.3.0.60 Figures and plots were generated using PyMOL61 and Gnuplot,62 respectively.
Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
Velvett G. Domínguez-Méndez: investigation, methodology, writing; Rosa Ma. Chávez Santos: investigation, methodology; Karol Carrillo-Jaimes: data curation, investigation, methodology, writing – original draft; Alejandra Hernández-Santoyo: methodology, investigation; Santos Ramírez-Carreto: methodology, writing; Armando Hernández-Garcia: methodology, visualization, writing; Rodrigo Aguayo-Ortiz: investigation, methodology, software, supervision, writing – review & editing; Corina-Diana Ceapă: conceptualization, formal analysis, investigation, methodology, visualization, writing review and editing, Corina-Diana Ceapă: conceptualization, formal analysis, investigation, methodology, visualization, writing – review and editing; José Rivera-Chávez: data curation, formal analysis, methodology, supervision and writing – review and editing; Roberto Martínez: project administration, funding acquisition, conceptualization, supervision, and writing – review and editing.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
We acknowledge financial support from DGAPA-UNAM (grant IN204619). The work of AHG and SRC has been supported by UNAM-PAPIIT IN226624. VGDM thanks SECIHTI for a PhD fellowship (CVU 963609), SRC thanks UNAM-DGAPA for the postdoctoral fellowship awarded (period: October 2024–September 2025). RMCS thanks SECIHTI for the postdoctoral fellowship awarded (period: September 2023–August 2025), (CVU: 269079). We are also grateful for the technical support provided by Carmen Garcia, Angeles Peña, Javier Perez, Elizabeth Huerta, Isabel Chavez, Teresa Ramirez Apan and Antonio Nieto Camacho (IQ-UNAM).References
- C. J. Iwu-Jaja, A. Jaca and I. F. Jaja, et al., Preventing and managing antimicrobial resistance in the African region: A scoping review protocol, PLoS One, 2021, 16, e0254737 Search PubMed.
- C. J. L. Murray, K. S. Ikuta and F. Sharara, et al., Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis, Lancet, 2022, 399, 629–655 CrossRef CAS PubMed.
- T. R. Walsh, A. C. Gales, R. Laxminarayan and P. C. Dodd, Antimicrobial Resistance: Addressing a Global Threat to Humanity, PLoS Med., 2023, 20, e1004264 CrossRef PubMed.
- S. Ruekit, A. Srijan and O. Serichantalergs, et al., Molecular characterization of multidrug-resistant ESKAPEE pathogens from clinical samples in Chonburi, Thailand (2017-2018), BMC Infect. Dis., 2022, 22, 695 Search PubMed.
- K. Carrillo-Jaimes, C. A. Fajardo-Hernández and F. Hernández-Sedano, et al., Antibacterial Activity and AbFtsZ Binding Properties of Fungal Metabolites Isolated from Mexican Mangroves, Rev. Bras. Farmacogn., 2024, 34, 564–576 CrossRef CAS.
- WHO publishes list of bacteria for which new antibiotics are urgently needed https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed Oct 16, 2024).
- M. Y. Cruz-Muñiz, L. E. López-Jacome and M. Hernández-Durán, et al., Repurposing the anticancer drug mitomycin C for the treatment of persistent Acinetobacter baumannii infections, Int. J. Antimicrob. Agents, 2017, 49, 88–92 CrossRef PubMed.
- R. W. DeSimone, K. S. Currie, S. A. Mitchell, J. W. Darrow and D. A. Pippin, Privileged structures: applications in drug discovery, Comb. Chem. High Throughput Screening, 2004, 7(5), 473–494 CrossRef CAS PubMed.
- F. R. de Sá Alves, E. J. Barreiro and C. A. M. Fraga, From nature to drug discovery: the indole scaffold as a “privileged structure”, Mini-Rev. Med. Chem., 2009, 9, 782–793 CrossRef PubMed.
- N. Chadha and O. Silakari, Indoles as therapeutics of interest in medicinal chemistry: Bird's eye view, Eur. J. Med. Chem., 2017, 134, 159–184 Search PubMed.
- X. Rabasseda, Ramosetron, a 5-HT3 receptor antagonist for the control of nausea and vomiting, Drugs Today, 2002, 38, 75–89 CrossRef CAS PubMed.
- Subcutaneous Sumatriptan International Study Group, Treatment of Migraine Attacks with Sumatriptan, N. Engl. J. Med., 1991, 325, 316–321 CrossRef PubMed.
- G. D. Morse, R. C. Reichman and M. A. Fischl, et al., Concentration-targeted phase I trials of atevirdine mesylate in patients with HIV infection: dosage requirements and pharmacokinetic studies, Antiviral Res., 2000, 45, 47–58 CrossRef CAS PubMed.
- T. Vedekhina, A. Lukin, E. Rogacheva, L. Kraeva and M. Krasavin, Zn(OTf)2-catalyzed arenehydrazination of protected propargylamines leading to 3-amidoindoles, Tetrahedron Lett., 2020, 61, 151430 CrossRef CAS.
- J. Brem, T. Panduwawala and J. U. Hansen, et al., Imitation of β-lactam binding enables broad-spectrum metallo-β-lactamase inhibitors, Nat. Chem., 2022, 14, 15–24 CrossRef CAS PubMed.
- M. F. Richter, B. S. Drown and A. P. Riley, et al., Predictive compound accumulation rules yield a broad-spectrum antibiotic, Nature, 2017, 545, 299–304 CrossRef CAS PubMed.
- M. Song, S. Wang and Z. Wang, et al., Synthesis, antimicrobial and cytotoxic activities, and molecular docking studies of N-arylsulfonylindoles containing an aminoguanidine, a semicarbazide, and a thiosemicarbazide moiety, Eur. J. Med. Chem., 2019, 166, 108–118 CrossRef CAS PubMed.
- W. Yuan, Z. Yu and W. Song, et al., Indole-core-based novel antibacterial agent targeting FtsZ, Infect. Drug Resist., 2019, 12, 2283–2296 CrossRef CAS PubMed.
- N. Sun, Y.-Y. Zheng and R.-L. Du, et al., New application of tiplaxtinin as an effective FtsZ-targeting chemotype for an antimicrobial study, MedChemComm, 1909, 2017, 8 Search PubMed.
- X. Yang, Z. Lyu and A. Miguel, et al., GTPase activity–coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis, Science, 2017, 355, 744–747 Search PubMed.
- A. W. Bisson-Filho, Y.-P. Hsu and G. R. Squyres, et al., Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division, Science, 2017, 355, 739–743 CrossRef CAS.
- R. R. Battaje, R. Piyush, V. Pratap and D. Panda, Models versus pathogens: how conserved is the FtsZ in bacteria?, Biosci. Rep., 2023, 43, BSR20221664 CrossRef CAS PubMed.
- Y. Qiao, T. Si, M.-H. Yang and R. A. Altman, Metal-Free Trifluoromethylation of Aromatic and Heteroaromatic Aldehydes and Ketones, J. Org. Chem., 2014, 79, 7122–7131 CrossRef CAS PubMed.
- P. Guo, W. Sun and Y. Liu, et al., Stereoselective Synthesis of Vinylcyclopropa[b]indolines via a Rh-Migration Strategy, Org. Lett., 2020, 22, 5978–5983 CrossRef CAS PubMed.
- Q. Xu, L. Huang and J. Liu, et al., Design, synthesis and biological evaluation of thiazole- and indole-based derivatives for the treatment of type II diabetes, Eur. J. Med. Chem., 2012, 52, 70–81 CrossRef CAS PubMed.
- S. J. Taylor, A. K. Padyana and A. Abeywardane, et al., Discovery of potent, selective chymase inhibitors via fragment linking strategies, J. Med. Chem., 2013, 56, 4465–4481 Search PubMed.
- G. Jin, S. Lee and M. Choi, et al., Chemical genetics-based discovery of indole derivatives as HCV NS5B polymerase inhibitors, Eur. J. Med. Chem., 2014, 75, 413–425 Search PubMed.
- CLSI, Methods for Antimicrobial Susceptibility Test for Bacteria That Grow Aerobically Approved Standard-Eight Edition, CLSI document M07-A8, Clinical and Laboratory Standards Institute, Wayne, PA, 2009.
- S. Kumar Verma, R. Verma and F. Xue, et al., Antibacterial activities of sulfonyl or sulfonamide containing heterocyclic derivatives and its structure-activity relationships (SAR) studies: A critical review, Bioorg. Chem., 2020, 105, 104400 CrossRef CAS PubMed.
- S. Mazzotta, T. Cebrero-Cangueiro and L. Frattaruolo, et al., Exploration of piperazine-derived thioureas as antibacterial and anti-inflammatory agents. In vitro evaluation against clinical isolates of colistin-resistant Acinetobacter baumannii, Bioorg. Med. Chem. Lett., 2020, 30, 127411 CrossRef CAS PubMed.
- F. Song, Z. Li and Y. Bian, et al., Indole/isatin-containing hybrids as potential antibacterial agents, Arch. Pharm., 2020, 353, 2000143 CrossRef CAS PubMed.
- P. Liu, Y. Jiang and L. Jiao, et al., Strategies for the Discovery of Oxazolidinone Antibacterial Agents: Development and Future Perspectives, J. Med. Chem., 2023, 66, 13860–13873 Search PubMed.
- W. Hong, J. Li and Z. Chang, et al., Synthesis and biological evaluation of indole core-based derivatives with potent antibacterial activity against resistant bacterial pathogens, J. Antibiot., 2017, 70, 832–844 Search PubMed.
- C. Zeng, S. R. Avula, J. Meng and C. Zhou, Synthesis and Biological Evaluation of Piperazine Hybridized Coumarin Indolylcyanoenones with Antibacterial Potential, Molecules, 2023, 28, 2511 CrossRef CAS PubMed.
- G. Bahatheg, R. Kuppusamy and M. Yasir, et al., Short Tryptamine-Based Peptoids as Potential Therapeutics for Microbial Keratitis: Structure-Function Correlation Studies, Antibiotics, 2022, 11, 1074 CrossRef CAS PubMed.
- N. Sun, R.-L. Du and Y.-Y. Zheng, et al., Antibacterial activity of N-methylbenzofuro[3,2-b]quinoline and N-methylbenzoindolo[3,2-b]-quinoline derivatives and study of their mode of action, Eur. J. Med. Chem., 2017, 135, 1–11 CrossRef CAS PubMed.
- L. Carro, Recent Progress in the Development of Small-Molecule FtsZ Inhibitors as Chemical Tools for the Development of Novel Antibiotics, Antibiotics, 2019, 8, 217 Search PubMed.
- D. A. Ramirez-Diaz, A. Merino-Salomón and F. Meyer, et al., FtsZ induces membrane deformations via torsional stress upon GTP hydrolysis, Nat. Commun., 2021, 12, 3310 CrossRef CAS PubMed.
- F. Perez, A. M. Hujer and K. M. Hujer, et al., Global Challenge of Multidrug-Resistant Acinetobacter baumannii, Antimicrob. Agents Chemother., 2007, 51, 3471–3484 CrossRef CAS PubMed.
- I. P. Sæbø, M. Bjørås, H. Franzyk, E. Helgesen and J. A. Booth, Optimization of the Hemolysis Assay for the Assessment of Cytotoxicity, Int. J. Mol. Sci., 2023, 24, 2914 CrossRef PubMed.
- N. Liaqat, N. Jahan, K. ur Rehman, T. Anwar and H. Qureshi, Green synthesized silver nanoparticles: Optimization, characterization, antimicrobial activity, and cytotoxicity study by hemolysis assay, Front. Chem., 2022, 10, 952006 CrossRef CAS PubMed.
- R. J. W. Lambert and J. Pearson, Susceptibility testing: accurate and reproducible minimum inhibitory concentration (MIC) and non-inhibitory concentration (NIC) values, J. Appl. Microbiol., 2000, 88, 784–790 CrossRef CAS PubMed.
- S. A. Li, R. J. Zheng and K. Sue, et al., Discovery and Preliminary Structure-Activity Investigation of 3-Substituted-1H-imidazol-5-yl-1H-indoles with In Vitro Activity towards Methicillin-Resistant Staphylococcus aureus, Antibiotics, 2022, 11, 1450 CrossRef CAS PubMed.
- N. M. O'Boyle, M. Banck and C. A. James, et al., Open Babel: An open chemical toolbox, Aust. J. Chem., 2011, 3, 33 Search PubMed.
- M. J. Frisch, G. W. Trucks and H. B. Schlegel, et al., Gaussian 16, Revision A.03, Gaussian, Inc., Wallingford, CT, 2016 Search PubMed.
- C. Lee, W. Yang and R. G. Parr, Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density, Phys. Rev. B: Condens. Matter Mater. Phys., 1988, 37, 785–789 CrossRef CAS PubMed.
- A. D. Becke, Density-functional thermochemistry. III. The role of exact exchange, J. Chem. Phys., 1993, 98, 5648–5652 CrossRef CAS.
- A. Waterhouse, M. Bertoni and S. Bienert, et al., SWISS-MODEL: Homology modelling of protein structures and complexes, Nucleic Acids Res., 2018, 46, W296–W303 CrossRef CAS PubMed.
- A. Bateman, M.-J. Martin and S. Orchard, et al., UniProt: the universal protein knowledgebase in 2021, Nucleic Acids Res., 2021, 49, D480–D489 CrossRef PubMed.
- G. Studer, C. Rempfer and A. M. Waterhouse, et al., QMEANDisCo—distance constraints applied on model quality estimation, Bioinformatics, 2020, 36, 1765–1771 CrossRef CAS PubMed.
- C. J. Williams, J. J. Headd and N. W. Moriarty, et al., MolProbity: More and better reference data for improved all-atom structure validation, Protein Sci., 2018, 27, 293–315 CrossRef CAS PubMed.
- N. M. Hassan, A. A. Alhossary, Y. Mu and C. K. Kwoh, Protein-Ligand Blind Docking Using QuickVina-W with Inter-Process Spatio-Temporal Integration, Sci. Rep., 2017, 7, 1–13 CrossRef CAS PubMed.
- J. A. Maier, C. Martinez and K. Kasavajhala, et al., ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB, J. Chem. Theory Comput., 2015, 11, 3696–3713 CrossRef CAS PubMed.
- M. J. Abraham, T. Murtola and R. Schulz, et al., GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers, SoftwareX, 2015, 19–25 CrossRef.
- A. W. Sousa da Silva and W. F. Vranken, ACPYPE - AnteChamber PYthon Parser interfacE, BMC Res. Notes, 2012, 5, 367 CrossRef PubMed.
- G. Bussi, D. Donadio and M. Parrinello, Canonical sampling through velocity rescaling, J. Chem. Phys., 2007, 126, 1–7 CrossRef PubMed.
- M. Parrinello and A. Rahman, Strain fluctuations and elastic constants, J. Chem. Phys., 1982, 76, 2662–2666 CrossRef CAS.
- B. Hess, H. Bekker, H. J. C. Berendsen and J. G. E. M. Fraaije, LINCS: A linear constraint solver for molecular simulations, J. Comput. Chem., 1997, 18, 1463–1472 CrossRef CAS.
- T. Darden, D. York and L. Pedersen, Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems, J. Chem. Phys., 1993, 98, 10089 CrossRef CAS.
- M. F. Adasme, K. L. Linnemann and S. N. Bolz, et al., PLIP 2021: expanding the scope of the protein–ligand interaction profiler to DNA and RNA, Nucleic Acids Res., 2021, 49, W530–W534 CrossRef CAS PubMed.
- L. Schrödinger, The PyMOL Molecular Graphics System, Version 2.5.4, Schrödinger, LLC, 2022 Search PubMed.
- T. Williams, C. Kelley and C. Bersch, et al.Gnuplot 5.0: An Interactive Plotting Program, 2017 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5md00145e |
| This journal is © The Royal Society of Chemistry 2025 |