Open Access Article
Open Access ArticleDevelopment of p300-targeting degraders with enhanced selectivity and onset of degradation†
Graham P.
Marsh
 a,
Mark S.
Cooper
a,
Sean
Goggins
a,
Mark S.
Cooper
a,
Sean
Goggins
 a,
Stephen J.
Reynolds
a,
Dean F.
Wheeler
a,
Joel O.
Cresser-Brown
a,
Stephen J.
Reynolds
a,
Dean F.
Wheeler
a,
Joel O.
Cresser-Brown
 a,
Robert E.
Arnold
a,
Emily G.
Babcock
a,
Gareth
Hughes
a,
Darko
Bosnakovski
a,
Robert E.
Arnold
a,
Emily G.
Babcock
a,
Gareth
Hughes
a,
Darko
Bosnakovski
 bc,
Michael
Kyba
bc,
Samuel
Ojeda
d,
Drew A.
Harrison
d,
Christopher J.
Ott
de and
Hannah J.
Maple
bc,
Michael
Kyba
bc,
Samuel
Ojeda
d,
Drew A.
Harrison
d,
Christopher J.
Ott
de and
Hannah J.
Maple
 *a
*a
aBio-Techne (Tocris), The Watkins Building, Atlantic Road, Avonmouth, Bristol, BS11 9QD, UK. E-mail: hannah.maple@bio-techne.com
bLillehei Heart Institute, Minneapolis, USA
cDepartment of Pediatrics, University of Minnesota, Minneapolis, MN 55455, USA
dKrantz Family Center for Cancer Research, Massachusetts General Hospital, Charlestown, MA 02129, USA
eDepartment of Medicine, Harvard Medical School, Boston, MA 02115, USA
First published on 3rd March 2025
Abstract
p300 and CBP are paralogous epigenetic regulators that are considered promising therapeutic targets for cancer treatment. Small molecule p300/CBP inhibitors have so far been unable to differentiate between these closely related proteins, yet selectivity is desirable in order to probe their distinct cellular functions. Additionally, in multiple cancers, loss-of-function CREBBP mutations set up a paralog dependent synthetic lethality with p300, that could be exploited with a selective therapeutic agent. To address this, we developed p300-targeting heterobifunctional degraders that recruit p300 through its HAT domain using the potent spiro-hydantoin-based inhibitor, iP300w. Lead degrader, BT-O2C, demonstrates improved selectivity and a faster onset of action compared to a recently disclosed A 485-based degrader in HAP1 cells and is cytotoxic in CIC::DUX4 sarcoma (CDS) cell lines (IC50 = 152–221 nM), significantly reducing expression of CDS target genes (ETV1, ETV4, ETV5). Taken together, our results demonstrate that BT-O2C represents a useful tool degrader for further exploration of p300 degradation as a therapeutic strategy.
Introduction
CREB-binding protein (CBP, CREBBP, KAT3A) and E1A-binding protein (EP300, p300, KAT3B) are paralogous, multi-domain proteins that act as chromatin regulators and transcriptional co-activators. They contain an acetyltransferase (KAT) domain that catalyzes lysine acetylation at thousands of sites, often associated with transcriptional machinery, including the histone H3, lysine 27 acetylation (H3K27ac) mark at regulatory elements such as enhancers and promoters.1–3 Transcription factors associate with stretches of H3K27ac marked chromatin (known as ‘super-enhancer’ elements) and result in gene transcription that ultimately establishes cell identity and fate.4 p300/CBP are implicated in multiple diseases including cancer5 and small molecule inhibition of their KAT domains is considered a promising therapeutic strategy for a number of cancer types.6,7 On-target toxicity and altered acetyl-CoA metabolism as an acquired resistance mechanism, however, represent potential liabilities of p300/CBP KAT inhibitors.6,8,9CBP and p300 are multidomain proteins sharing 61% overall sequence identity but retain much higher sequence identity in their KAT domains (86%)5 (Fig. 1A). Small molecule inhibitors exhibiting selectivity for p300/CBP KAT domains have recently been discovered, offering useful chemical tools for probing acetyltransferase inhibition as a therapeutic strategy.1,9–14 Given the sequence similarity between p300 and CBP KAT domains, it is unsurprising that these small molecule inhibitors do not differentiate between the paralogs. Tools to aid in understanding differential roles for p300 and CBP are desirable, however, since it is well established that they present distinct genomic binding patterns.5,15–19 Furthermore, it has been shown that loss-of-function aberrations in the CREBBP gene, commonly present in multiple human cancer types (including lymphoma, leukemia, lung and bladder), set up a paralog dependent synthetic lethality with p300.20 A small molecule capable of selective ablation of p300 function could exploit this synthetic lethal relationship in cancers with loss-of-function CREBBP mutations.
 | ||
| Fig. 1 (A) p300 and CBP domain schematic (upper) with KAT domain sequence alignment for p300 and CBP (lower). Conserved regions are highlighted black21 and key residues mediating iP300w binding through H-bonds and Van der Waals interactions are underlined orange.10 Sequence alignment generated with using EMBL-EBI EMBOSS Needle pairwise sequence alignment tool (UniProt and NCBI accession numbers: Q09472 and NP_004371). (B) Chemical structures of iP300w and A 485, with the solvent facing exit vector region highlighted blue. (C) Co-crystal structure of p300 in complex with iP300w (PDBID 7LJE) with the acetamide exit vector indicated (black dashed circle).10 | ||
An alternative small molecule approach to achieve enhanced selectivity between closely related proteins is targeted protein degradation (TPD). TPD utilizes small molecules termed proteolysis targeting chimeras (PROTACs) or degraders, to degrade a target protein. Degraders are heterobifunctional small molecules that bind to a target protein and an E3 ligase, forming a ternary complex and facilitating ubiquitin transfer and polyubiquitination of the target protein, directing it to the proteasome for degradation. The degrader itself can engage in multiple rounds of target protein degradation, providing a catalytic mode of action and the potential for sub-stoichiometric dosing.22
Degraders present opportunities to achieve enhanced selectivity through formation of distinct E3 ligase-target interfaces in the ternary complex, influenced through subtle differences in the degrader chemical structure. For example, degraders developed from the pan-selective kinase inhibitor foretinib were able to achieve p38-MAPK family isoform selectivity. Degraders selective for both p38α and p38δ were developed through alterations in the exit vector of the E3 ligase ligand used, ultimately driving selectivity through altering the ternary complex structure.23,24
A bromodomain-recruiting dual p300/CBP degrader ‘dCBP-1’ was recently developed to provide a chemical tool to explore the phenotypic consequences of p300/CBP chemical knockdown.25 dCBP-1 treatment in multiple myeloma cellular models potently downregulated levels of oncogenic MYC and caused near complete loss of H3K27 acetylation, which could not be recapitulated with equivalent doses of p300/CBP inhibitors. A further study demonstrated that it is possible to degrade p300 with some selectivity by converting the p300/CBP HAT-domain inhibitor A 485 (Fig. 1B) into a degrader, called ‘JQAD1’. JQAD1 treatment caused loss of H3K27ac at core regulatory circuitry enhancers and was shown to selectively degrade p300 in neuroblastoma cell lines, however after 48 hour administration, degradation of CBP was also observed.19
A p300/CBP inhibitor structurally related to A 485 was recently disclosed and demonstrated improved activity in a number of different models (iP300w, Fig. 1B).9–12 A co-crystal structure of p300 in complex with iP300w reveals that the solvent facing acetamide moiety offers a potential exit vector for degrader design (Fig. 1C). Here we investigate iP300w as a warhead ligand for degrader development with the aim of developing a more selective chemical tool for p300 degradation.
Results & discussion
Having identified a viable solvent-facing exit vector for linker attachment, the iP300w warhead ligand was synthesized with a modified chemical handle allowing for degrader assembly. A carboxylic acid was installed in place of the terminal N-methylacetamido functionality appending the pyrazole group. Chemical synthesis proceeded according to the published route11 whereby a Bucherer–Bergs reaction and oxone-mediated benzylic oxidation sequence produced spiro-hydantoin 2, and subsequent alkylation deploying homochiral bromide 4 gave rise to key intermediate 5. Suzuki coupling enabled facile installation of the required pyrazole unit bearing a pendant tert-butyl ester which, after a two-stage reduction and fluorination progression, could be deprotected to unveil carboxylic acid 9 ready for degrader construction (Scheme 1).A series of degraders was then designed and prepared by coupling pre-assembled degrader building blocks (E3 ligase ligand and linker constructs with amino-functionalized termini) to the modified iP300w warhead ligand by means of conventional amide coupling chemistries (Table 1). The degrader panel was designed to explore recruitment of two E3 ligases most commonly harnessed for degrader development: cereblon (CRBN) and von Hippel–Lindau (VHL) using different ligands and exit vectors. A range of linkers were explored using PEG and alkyl chains together a rigidified linker to develop a preliminary understanding of the effect of linker length and type on the degradation profile. The list of putative structures was filtered to select a panel of compounds that fall within physicochemical guidelines for degraders including HBD ≤5 and TPSA ≤250 Å2.23,26,27
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, hydrogen bond donor count (HBD), hydrogen bond acceptor count (HBA), number of rotatable bonds (RB), topological surface area (TPSA). Physicochemical properties were calculated using ChemDraw® version 15.0.0 with the following exceptions: clog
P, hydrogen bond donor count (HBD), hydrogen bond acceptor count (HBA), number of rotatable bonds (RB), topological surface area (TPSA). Physicochemical properties were calculated using ChemDraw® version 15.0.0 with the following exceptions: clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, calculated using the consensus value from SwissADME (http://www.swissadme.ch)28
P, calculated using the consensus value from SwissADME (http://www.swissadme.ch)28
The published A 485-based p300 degrader, JQAD1 (Table 2) contains a relatively long (C11) alkyl linker and utilizes 5′ pomalidomide as the E3 ligase ligand. We therefore designed an iP300w-based degrader (BT-O2C) as a ‘matched pair’ compound, mimicking the linker length and E3 ligase ligand of JQAD1, while taking account for the overall longer warhead ligand exit vector provided by iP300w. A matched-pair negative control compound for BT-O2C was also synthesized (BT-O2C-N), incorporating the well-established N-methyl ‘bump’ on the glutarimide moiety to remove E3 ligase binding capability.
| Compound | % CBP degradation (24 hours, 100 nM) | % p300 degradation (24 hours, 100 nM) | ||
|---|---|---|---|---|
| HiBiT assay | Immunoassay | HiBiT assay | Immunoassay | |
| dCBP-1 | 89% | 86% | 91% | 83% |
| JQAD1 | 23% | −10% | 37% | −13% |
| BT-O2F | −6% | −17% | 60% | 55% |
| BT-O2C | −1% | −12% | 57% | 43% |
We assessed p300/CBP degradation in the human haploid cell line HAP1 using a HiBiT cellular degradation assay.29 Briefly, p300 or CBP were endogenously N-terminal tagged with a HiBiT tag using CRISPR/Cas9 genome editing in a HAP1 cell line stably expressing LgBiT. A luminescence read-out was used to assess protein levels across a period of 48 hours following treatment with degraders over a concentration range (100 nM–10 μM). dCBP-1 was used as a positive control for dual degradation of both targets. Degradation at the 48 hour timepoint (Fig. 2) revealed that several iP300w-based degraders induce degradation of p300, with limited degradation of CBP. The CRBN-recruiting compounds (BT-O2C-H) showed a preference toward longer linker lengths, with negligible degradation for the shortest linker length (BT-O2D). The VHL-recruiting compounds based on the prototypical VHL ligand, VH 032 (BT-O2I, BT-O2J), did not degrade either target, while compounds based on the VH 101 scaffold using the phenolic exit vector (BT-O2K, BT-O2L) exhibited potent p300 degradation. The degraders incorporating a rigidified linker and diverse CRBN-recruiting ligands (BT-O2M-P) exhibited overall lower activity. Interestingly the phenyl dihydrouracil- and phenyl amino glutarimide-based degraders (BT-O2N and BT-O2O, respectively) show an apparent reversal in activity, with some limited degradation of CBP observed with negligible effect on p300. The A 485-based JQAD1 clearly degraded p300 as expected, but also induced marked degradation of CBP at the 48 hour timepoint, to a greater degree than the iP300w-based series. The control degrader, dCBP-1 potently degraded both targets, as expected. To assess potential confounding results due to compound-induced toxicity, we additionally assessed viability of HAP1 cells following 24 hour treatment with selected degraders. No significant loss of viability was observed up to the maximum treatment concentration (10 μM) (Fig. S1†).
Full HiBiT assay degradation profiles of lead compounds BT-O2C and BT-O2F, together with dCBP-1 and JQAD1 (Fig. 3, degradation profiles for all compounds tested provided in Fig. S2†) revealed differences in onset of degradation across the different series. dCBP-1 potently degraded both target proteins within 1 hour, while JQAD1 exhibited a much slower onset of action; selective p300 degradation was observed only after 24 hours with maximal degradation at 48 hours and concomitant degradation of CBP at this timepoint. In contrast, the iP300w-based degraders displayed potent degradation after 6 hours incubation and reached Dmax at 24 hours. The negative control for BT-O2C, BT-O2C-N showed no evidence of target degradation as expected (Fig. 3).
Lead degraders were next assessed by immunoassay using capillary-based electrophoresis (Simple Western™ technology by Bio-Techne) and validated selective antibodies for CBP and p300 (Fig. 4). HAP1 cells treated with compounds for 24 hours revealed that BT-O2C potently degrades p300 while sparing CBP, while JQAD1 exhibits limited activity against either protein (Fig. 4A). BT-O2F also (but to a slightly lesser extent) degraded p300 with selectivity over CBP, while BT-O2H and BT-O2K were less potent compounds in this assay (Fig. 4B). It is interesting to note that the Simple Western platform and HiBiT assay data, while directionally consistent display some clear differences. For example, higher Dmax values were observed for BT-O2C, BT-O2H, BT-O2F and dCBP-1 by immunoassay compared to the HiBiT assay. Additionally, JQAD1 appeared almost inactive by immunoassay, compared to clear p300 degradation in the HiBiT assay at the same concentrations and timepoint. Quantifying the percentage degradation of lead compounds and controls at a single dose and timepoint highlights these differences clearly (Table 2).
The reason for these differences is not altogether clear, but does highlight the importance of employing multiple, orthogonal assays where possible to assess degradation. It is possible that proteasomal degradation of HiBiT-tagged p300/CBP could in some instances leave the HiBiT 11mer peptide intact and able to bind LgBiT, providing a degree of false negative readout.30 It has also been noted that the HiBiT tag, while minimal in size, can cause stabilization of proteins when introduced at the N-terminus.31
To assess mechanism of action for BT-02C, HAP1 cells were co-treated with degrader and either the neddylation inhibitor MLN 4924, or the proteasome inhibitor carfilzomib (Fig. 5). Degradation of p300 was rescued by co-treatment with either inhibitor, demonstrating that BT-O2C is mechanistically dependent on both neddylation to activate the CUL4CRBN and on proteasome activity.
The effect of p300-targeting degraders in CIC-DUX4 sarcoma (CDS) cell lines was then assessed. CDS is a rare and aggressive cancer occurring predominantly in children and young adults. The CIC::DUX4 fusion resulting from aberrant translocation results in the expression of the oncoprotein CIC::DUX4 and transcriptional activation. A direct interaction between CIC::DUX4 and p300 has recently been proven,32 adding to the body of evidence suggesting that pharmacological perturbation of p300 may represent an attractive therapeutic intervention for CDS.11,12,32
Cell viability was measured in two CDS cell lines, NCC-CDS1-X1 and KITRA, following treatment with lead degrader BT-O2C, JQAD1 and the parent inhibitor compounds iP300w and A 485 for 48 or 72 hours (Fig. S3†). BT-O2C and JQAD1 displayed broadly similar potencies in both cell lines. IC50 values calculated from this data at the 72 h timepoint in NCC-CDS-X1 cells were 152 ± 39 nM (BT-O2C) and 173 ± 53 nM (JQAD1), and at the same timepoint in KITRA cells, IC50 values were 221 ± 46 nM (BT-O2C) and 104 ± 19 nM (JQAD1). Both degrader compounds were overall less potent than parent inhibitors iP300w and A 485, which could plausibly be attributed to reduced cell permeability for the degrader molecules versus the inhibitors.
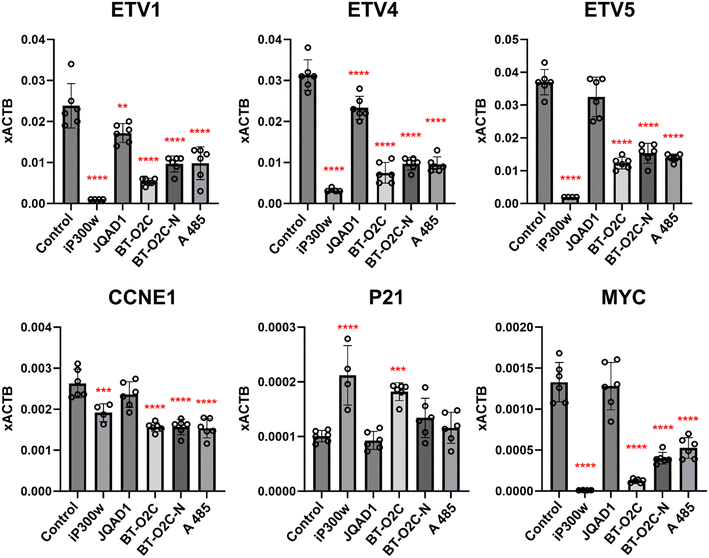
To further evaluate the effect of degradation versus inhibition in the NCC-CDS-X1 cell line, expression of CDS target genes ETV1, ETV4 and ETV5 were monitored by RT-qPCR following 6 hour compound treatment (Fig. 6). Genes involved in cell cycle regulation (P21 and CCNE1), and known p300 target, c-MYC, were also monitored. BT-O2C reduced CDS target gene expression, displaying significantly more activity than JQAD1 in this experiment. BT-O2C and the negative control, BT-O2C-N lacked statistically significant differences in response in this experiment and so degradation was assessed in this cell line by capillary electrophoresis (Fig. S4†). BT-O2C was unable to induce degradation of p300 under these conditions. The effect of compound treatment on histone acetylation was next assessed in NCC-CDS-X1 cells. H3K18 and H3K27 are well characterized substrates of p300/CBP; inhibition or degradation of p300/CBP will result in a loss of the H3K18ac and H3K27ac marks. Cells were treated with the inhibitors iP300w and A 485 (both at 0.25 μM) or degraders JQAD1 and BT-O2C (both at 4 μM) for 24 hours (Fig. S5†). As expected, iP300w and A 485 treatment significantly reduces the acetylation marks. In contrast no clear effect was observed following treatment with JQAD1, while a slight decrease in H3K27 acetylation was observed following BT-O2C treatment.
During preparation of this manuscript, a p300-selective degrader (MC-1) was reported, utilizing a HAT-domain inhibitor and recruiting VHL as the E3 ligase.33 In addition to the growing number of dual p300/CBP degraders (HAT and BRD-domain targeting) and inhibitors, we anticipate that the paralog-targeting degraders (JQAD1, MC-1 and BT-O2C) will be helpful tools to further understand p300 and CBP biology and their potential as therapeutic targets.
Conclusion
The exquisite selectivity that heterobifunctional degraders (or PROTACs) can confer offers an attractive approach to targeting closely related proteins. Here we pursued the selective degradation of the chromatin regulator, p300, over its paralog CBP. Targeting p300 selectively offers the potential to exploit the paralog dependent synthetic lethality resulting from cancers with loss-of-function CREBBP mutations. Further there is growing evidence that p300 inhibition or degradation offers a potential therapeutic approach to target the aggressive pediatric soft tissue tumor, CDC-DUX4 sarcoma. We developed iP300w-based degraders that elicit proteasome-mediated degradation of p300 with improved selectivity over CBP and faster onset of degradation. Lead degrader BT-O2C was profiled for activity in CIC::DUX4 sarcoma cell lines and demonstrated a dose-responsive potent cytotoxic effect. We further demonstrate that BT-O2C treatment significantly reduces expression of CIC::DUX4 target genes ETV1, ETV4 and ETV5 but its activity was not statistically distinguishable from a matched pair negative control (BT-O2C-N) and degradation of p300 was not observed in this cell line. We anticipate that BT-O2C will be a useful tool compound for further exploration of p300 degradation as a therapeutic strategy but note that differences in response between cell lines should be taken into careful account.Methods
Chemical synthesis
See ESI† for synthetic methods.Cell lines
HAP1 cells [male] were obtained from Horizon Discovery and grown in IMDM media supplemented with 10% fetal bovine serum (FBS). NCC-CDS-X1 (CIC-DUX4 sarcoma cell line, a generous gift from Tadashi Kondo) cells were cultured in RPMI with 10% FBS, Glu and P/S, Kitra-SRS (CIC-DUX4 sarcoma cell line, a generous gift from Hidetatsu Otani) cells were cultured in DMEM/10% FBS/Glu/P/S.11 Their identity as CDS cells was confirmed by PCR for CIC-DUX4. The immortalized human myoblast LHCN-M2 cell line was cultured in proliferation medium: F10 supplemented with 20% FBS, 2-mercaptoethanol 1× (GIBCO), 10−9 M dexamethasone (Sigma), 10 ng mL−1 bFGF (Peprotech), and Glu/P/S.34 All cells were cultured at 37 °C in a 5% CO2 atmosphere.HiBiT assays
HiBiT assays were performed as previously described.29 In brief, HAP1-HiBiT cells were seeded into 384 well plates in IMDM-0 (GIBCO 12440-053) + 2% FBS (GIBCO A52567-01) + 1% PenStrep (GIBCO 15140-122). Cells are incubated at 37 °C, 5% CO2 overnight. The following day 20 μL of a 1% endurazine solution in IMDM was added and incubated for 3 hours at 37 °C, 5% CO2. Compounds were delivered to the assay plate with a JANUS workstation pintool, plates were sealed and loaded onto a Perkin Elmer Envision plate reader. Luminescence readings were taken at indicated time points.Capillary electrophoresis (Simple Western technology)
Simple Western assays were performed as previously described.25 In brief, cells were plated into 6-well plates followed by overnight incubation before treatment. Cells were harvested and lysed in RIPA buffer containing HALT protease inhibitor cocktail (Pierce). Protein extraction was facilitated by passing all samples through a gauge 28 Micro-Fine IV insulin syringe (BD). Capillary-based immunoassays were performed using a standard Wes platform, powered by Simple Western technology. Lysates were loaded onto Wes plates at 0.8 μg μL−1 total protein. Wes staining was conducted using the following antibodies: vinculin (Bethyl, A302-535A), CBP (Cell Signaling, D6C5), p300 (Cell Signaling, D2X6N).Western blot
Cell lysates were prepared using RIPA buffer supplemented with a protease inhibitor cocktail (Complete, Roche). Proteins were separated on 10% SDS-PAGE gels and transferred to PVDF membranes. The membranes were incubated with primary antibodies diluted in 5% skim milk in TBST overnight at 4 °C. Following primary antibody incubation, the membranes were incubated with HRP-conjugated secondary antibody for 1 hour at RT. After washing with TBST, protein bands were visualized using Pierce ECL Western blotting substrate (Thermo Scientific). The primary antibodies used for Western blot analysis were GAPDH-HRP (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000, Proteintech 60004), rabbit anti-histone H3K18Ac (1
5000, Proteintech 60004), rabbit anti-histone H3K18Ac (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, Abcam ab1191), and rabbit anti-histone H3K27Ac (1
500, Abcam ab1191), and rabbit anti-histone H3K27Ac (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, Abcam ab1791, lot: GR3297878-1). The secondary antibody was HRP-conjugated anti-rabbit (1
500, Abcam ab1791, lot: GR3297878-1). The secondary antibody was HRP-conjugated anti-rabbit (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000, Jackson ImmunoResearch 111-035-003, lot: 149393).
5000, Jackson ImmunoResearch 111-035-003, lot: 149393).
Cell viability (ATP) assay
Cell lines were plated in a 96-well dish (1 × 105 cells per well), and the following day were treated with iP300w or its stereoisomers. ATP assays were performed using CellTiter-Glo® Luminescent Cell Viability Assay (Promega) according to the manufacturer's instructions. Luminescence was analyzed on POLARstar Optima Microplate Reader (BMG Labtech, Offenburg, Germany).RNA isolation and quantitative real-time RT-PCR (RT-qPCR)
RNA was extracted using an RNA extraction kit (Zymo) and cDNA was made using 500 ng of total RNA with oligo-dT primer and Verso cDNA Synthesis Kit (Thermo Scientific) following the manufacturer's instructions. qPCRs were performed by using Premix Ex Taq or SYBR-Green Master Mixes (Takara). The following probes: B2M (Hs00187842_m1) and MYC (Hs00153408_m1); and primer sets ETV1 (F: 5′ GGC TGT ATC AGA GCG TAT TGT C and R: 5′ CAC TGG GTC GTG GTA CTC CT), ETV4 (F: 5′ GTC ACT TCC AGG AGA CGT GG and R: 5′ ATA GGC ACT GGA GTA AAG GCA C) and ETV5 (F: 5′ TCT GAG CTG TCG TCT TGT AGC C and R: 5′ GTT ATT GGC TTG AAC CCA GAG G), P21 (F: 5′ GTC AGG CTG GTC TGC CTC CG and R: 5′ CGG TCC CGT GGA CAG GAG CAG), CCNE1 (F: 5′-TTT TTG CAG GAT CCA GAT GA and R: 5′-TGC ACG TTG AGT TTG GGT AA) and ACTIN (F: 5′-TGG CCG TCA GGC AGC TCG TA and R: 5′-GCG ACG AGG CCC AGA GCA AG) were used. Gene expression levels were normalized to that of B2M or ACTIN and analyzed with 7500 System Software using the ΔCT method (Applied Biosystems).Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There is no conflict of interest to declare.Acknowledgements
D. B. is supported by the NIAMS grant R01AR081228, Department of Defense grant HT9425-23-1-0456 and the Children's Cancer Research Fund, Minneapolis, MN. C. J. O. acknowledges support of this work from the Paula and Rodger Riney Foundation. M. K. was supported in part by the National Institute for Arthritis and Musculoskeletal and Skin Diseases (R01 AR055685).References
- L. M. Lasko, C. G. Jakob, R. P. Edalji, W. Qiu, D. Montgomery, E. L. Digiammarino, T. M. Hansen, R. M. Risi, R. Frey, V. Manaves, B. Shaw, M. Algire, P. Hessler, L. T. Lam, T. Uziel, E. Faivre, D. Ferguson, F. G. Buchanan, R. L. Martin, M. Torrent, G. G. Chiang, K. Karukurichi, J. W. Langston, B. T. Weinert, C. Choudhary, P. de Vries, A. F. Kluge, M. A. Patane, J. H. V. Drie, C. Wang, D. McElligott, E. Kesicki, R. Marmorstein, C. Sun, P. A. Cole, S. H. Rosenberg, M. R. Michaelides, A. Lai and K. D. Bromberg, Discovery of a Selective Catalytic P300/CBP Inhibitor That Targets Lineage-Specific Tumours, Nature, 2017, 550(7674), 128–132, DOI:10.1038/nature24028.
- V. V. Ogryzko, R. L. Schiltz, V. Russanova, B. H. Howard and Y. Nakatani, The Transcriptional Coactivators P300 and CBP Are Histone Acetyltransferases, Cell, 1996, 87(5), 953–959, DOI:10.1016/S0092-8674(00)82001-2.
- A. J. Bannister and T. Kouzarides, The CBP Co-Activator Is a Histone Acetyltransferase, Nature, 1996, 384(6610), 641–643, DOI:10.1038/384641a0.
- D. Hnisz, B. J. Abraham, T. I. Lee, A. Lau, V. Saint-André, A. A. Sigova, H. A. Hoke and R. A. Young, Super-Enhancers in the Control of Cell Identity and Disease, Cell, 2013, 155(4), 934–947, DOI:10.1016/j.cell.2013.09.053.
- B. M. Dancy and P. A. Cole, Protein Lysine Acetylation by P300/CBP, Chem. Rev., 2015, 115(6), 2419–2452, DOI:10.1021/cr500452k.
- S. D. Whedon and P. A. Cole, KATs off: Biomedical Insights from Lysine Acetyltransferase Inhibitors, Curr. Opin. Chem. Biol., 2023, 72, 102255, DOI:10.1016/j.cbpa.2022.102255.
- S. J. Hogg, O. Motorna, L. A. Cluse, T. M. Johanson, H. D. Coughlan, R. Raviram, R. M. Myers, M. Costacurta, I. Todorovski, L. Pijpers, S. Bjelosevic, T. Williams, S. N. Huskins, C. J. Kearney, J. R. Devlin, Z. Fan, J. S. Jabbari, B. P. Martin, M. Fareh, M. J. Kelly, D. Dupéré-Richer, J. J. Sandow, B. Feran, D. Knight, T. Khong, A. Spencer, S. J. Harrison, G. Gregory, V. O. Wickramasinghe, A. I. Webb, P. C. Taberlay, K. D. Bromberg, A. Lai, A. T. Papenfuss, G. K. Smyth, R. S. Allan, J. D. Licht, D. A. Landau, O. Abdel-Wahab, J. Shortt, S. J. Vervoort and R. W. Johnstone, Targeting Histone Acetylation Dynamics and Oncogenic Transcription by Catalytic P300/CBP Inhibition, Mol. Cell, 2021, 81(10), 2183–2200.e13, DOI:10.1016/j.molcel.2021.04.015.
- T. R. Bishop, C. Subramanian, E. M. Bilotta, L. Garnar-Wortzel, A. R. Ramos, Y. Zhang, J. N. Asiaban, C. J. Ott, C. O. Rock and M. A. Erb, Acetyl-CoA Biosynthesis Drives Resistance to Histone Acetyltransferase Inhibition, Nat. Chem. Biol., 2023, 19(10), 1215–1222, DOI:10.1038/s41589-023-01320-7.
- M. C. Crawford, D. R. Tripu, S. A. Barritt, Y. Jing, D. Gallimore, S. C. Kales, N. V. Bhanu, Y. Xiong, Y. Fang, K. A. T. Butler, A. LeClair, N. P. Coussens, A. Simeonov, B. A. Garcia, C. C. Dibble and L. Meier, Comparative Analysis of Drug-like EP300/CREBBP Acetyltransferase Inhibitors, ACS Chem. Biol., 2023, 18(10), 2249–2258, DOI:10.1021/acschembio.3c00293.
- Z. Ji, R. F. Clark, V. Bhat, T. Matthew Hansen, L. M. Lasko, K. D. Bromberg, V. Manaves, M. Algire, R. Martin, W. Qiu, M. Torrent, C. G. Jakob, H. Liu, P. A. Cole, R. Marmorstein, E. A. Kesicki, A. Lai and M. R. Michaelides, Discovery of Spirohydantoins as Selective, Orally Bioavailable Inhibitors of P300/CBP Histone Acetyltransferases, Bioorg. Med. Chem. Lett., 2021, 39, 127854, DOI:10.1016/j.bmcl.2021.127854.
- D. Bosnakovski, E. T. Ener, M. S. Cooper, M. D. Gearhart, K. A. Knights, N. C. Xu, C. A. Palumbo, E. A. Toso, G. P. Marsh, H. J. Maple and M. Kyba, Inactivation of the CIC-DUX4 Oncogene through P300/CBP Inhibition, a Therapeutic Approach for CIC-DUX4 Sarcoma, Oncogenesis, 2021, 10(10), 68, DOI:10.1038/s41389-021-00357-4.
- D. Bosnakovski, M. T. da Silva, S. T. Sunny, E. T. Ener, E. A. Toso, C. Yuan, Z. Cui, M. A. Walters, A. Jadhav and M. Kyba, A Novel P300 Inhibitor Reverses DUX4-Mediated Global Histone H3 Hyperacetylation, Target Gene Expression, and Cell Death, Sci. Adv., 2019, 5(9), eaaw7781, DOI:10.1126/sciadv.aaw7781.
- J. E. Wilson, G. Patel, C. Patel, F. Brucelle, A. Huhn, A. S. Gardberg, F. Poy, N. Cantone, A. Bommi-Reddy, R. J. Sims, R. T. Cummings and J. R. Levell, Discovery of CPI-1612: A Potent, Selective, and Orally Bioavailable EP300/CBP Histone Acetyltransferase Inhibitor, ACS Med. Chem. Lett., 2020, 11(6), 1324–1329, DOI:10.1021/acsmedchemlett.0c00155.
- Y. Yang, R. Zhang, Z. Li, L. Mei, S. Wan, H. Ding, Z. Chen, J. Xing, H. Feng, J. Han, H. Jiang, M. Zheng, C. Luo and B. Zhou, Discovery of Highly Potent, Selective, and Orally Efficacious P300/CBP Histone Acetyltransferases Inhibitors, J. Med. Chem., 2020, 63(3), 1337–1360, DOI:10.1021/acs.jmedchem.9b01721.
- Y. F. M. Ramos, M. S. Hestand, M. Verlaan, E. Krabbendam, Y. Ariyurek, M. Van Galen, H. Van Dam, G.-J. B. Van Ommen, J. T. Den Dunnen, A. Zantema and P. A. C. 'T Hoen, Genome-Wide Assessment of Differential Roles for P300 and CBP in Transcription Regulation, Nucleic Acids Res., 2010, 38(16), 5396–5408, DOI:10.1093/nar/gkq184.
- R. A. Henry, Y.-M. Kuo and A. J. Andrews, Differences in Specificity and Selectivity Between CBP and P300 Acetylation of Histone H3 and H3/H4, Biochemistry, 2013, 52(34), 5746–5759, DOI:10.1021/bi400684q.
- S. Martire, J. Nguyen, A. Sundaresan and L. A. Banaszynski, Differential Contribution of P300 and CBP to Regulatory Element Acetylation in mESCs, BMC Mol. Cell Biol., 2020, 21(1), 55, DOI:10.1186/s12860-020-00296-9.
- V. I. Rebel, A. L. Kung, E. A. Tanner, H. Yang, R. T. Bronson and D. M. Livingston, Distinct Roles for CREB-Binding Protein and P300 in Hematopoietic Stem Cell Self-Renewal, Proc. Natl. Acad. Sci. U. S. A., 2002, 99(23), 14789–14794, DOI:10.1073/pnas.232568499.
- A. D. Durbin, T. Wang, V. K. Wimalasena, M. W. Zimmerman, D. Li, N. V. Dharia, L. Mariani, N. A. M. Shendy, S. Nance, A. G. Patel, Y. Shao, M. Mundada, L. Maxham, P. M. C. Park, L. H. Sigua, K. Morita, A. S. Conway, A. L. Robichaud, A. R. Perez-Atayde, M. J. Bikowitz, T. R. Quinn, O. Wiest, J. Easton, E. Schönbrunn, M. L. Bulyk, B. J. Abraham, K. Stegmaier, A. T. Look and J. Qi, EP300 Selectively Controls the Enhancer Landscape of MYCN -Amplified Neuroblastoma, Cancer Discovery, 2022, 12(3), 730–751, DOI:10.1158/2159-8290.CD-21-0385.
- H. Ogiwara, M. Sasaki, T. Mitachi, T. Oike, S. Higuchi, Y. Tominaga and T. Kohno, Targeting P300 Addiction in CBP -Deficient Cancers Causes Synthetic Lethality by Apoptotic Cell Death Due to Abrogation of MYC Expression, Cancer Discovery, 2016, 6(4), 430–445, DOI:10.1158/2159-8290.CD-15-0754.
- X. Liu, L. Wang, K. Zhao, P. R. Thompson, Y. Hwang, R. Marmorstein and P. A. Cole, The Structural Basis of Protein Acetylation by the P300/CBP Transcriptional Coactivator, Nature, 2008, 451(7180), 846–850, DOI:10.1038/nature06546.
- M. Békés, D. R. Langley and C. M. Crews, PROTAC Targeted Protein Degraders: The Past Is Prologue, Nat. Rev. Drug Discovery, 2022, 21(3), 181–200, DOI:10.1038/s41573-021-00371-6.
- B. E. Smith, S. L. Wang, S. Jaime-Figueroa, A. Harbin, J. Wang, B. D. Hamman and C. M. Crews, Differential PROTAC Substrate Specificity Dictated by Orientation of Recruited E3 Ligase, Nat. Commun., 2019, 10(1), 131, DOI:10.1038/s41467-018-08027-7.
- A. Gopalsamy, Selectivity through Targeted Protein Degradation (TPD), J. Med. Chem., 2022, 65(12), 8113–8126, DOI:10.1021/acs.jmedchem.2c00397.
- R. Vannam, J. Sayilgan, S. Ojeda, B. Karakyriakou, E. Hu, J. Kreuzer, R. Morris, X. I. H. Lopez, S. Rai, W. Haas, M. Lawrence and C. J. Ott, Targeted Degradation of the Enhancer Lysine Acetyltransferases CBP and P300, Cell Chem. Biol., 2021, 28(4), 503–514.e12, DOI:10.1016/j.chembiol.2020.12.004.
- H. J. Maple, N. Clayden, A. Baron, C. Stacey and R. Felix, Developing Degraders: Principles and Perspectives on Design and Chemical Space, MedChemComm, 2019, 10(10), 1755–1764, 10.1039/C9MD00272C.
- S. D. Edmondson, B. Yang and C. Fallan, Proteolysis Targeting Chimeras (PROTACs) in ‘beyond Rule-of-Five’ Chemical Space: Recent Progress and Future Challenges, Bioorg. Med. Chem. Lett., 2019, 29(13), 1555–1564, DOI:10.1016/J.BMCL.2019.04.030.
- A. Daina, O. Michielin and V. Zoete, SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules, Sci. Rep., 2017, 7, 42717 CrossRef PubMed.
- P. Kumar Tiwari, S. Reddy Doda, R. Vannam, M. Hudlikar, D. A. Harrison, S. Ojeda, S. Rai, A.-S. Koglin, A. Nguyen Gilbert and C. J. Ott, Exploration of Bromodomain Ligand-Linker Conjugation Sites for Efficient CBP/P300 Heterobifunctional Degrader Activity, Bioorg. Med. Chem. Lett., 2024, 102, 129676, DOI:10.1016/j.bmcl.2024.129676.
- A. F. Kisselev, T. N. Akopian, K. M. Woo and A. L. Goldberg, The Sizes of Peptides Generated from Protein by Mammalian 26 and 20 S Proteasomes, J. Biol. Chem., 1999, 274(6), 3363–3371, DOI:10.1074/jbc.274.6.3363.
- M. P. Schwalm, A. Krämer, A. Dölle, J. Weckesser, X. Yu, J. Jin, K. Saxena and S. Knapp, Tracking the PROTAC Degradation Pathway in Living Cells Highlights the Importance of Ternary Complex Measurement for PROTAC Optimization, Cell Chem. Biol., 2023, 30(7), 753–765.e8, DOI:10.1016/j.chembiol.2023.06.002.
- A. Bakaric, L. Cironi, V. Praz, R. Sanalkumar, L. C. Broye, K. Favre-Bulle, I. Letovanec, A. Digklia, R. Renella, I. Stamenkovic, C. J. Ott, T. Nakamura, C. R. Antonescu, M. N. Rivera and N. Riggi, CIC-DUX4 Chromatin Profiling Reveals New Epigenetic Dependencies and Actionable Therapeutic Targets in CIC-Rearranged Sarcomas, Cancers, 2024, 16(2), 457, DOI:10.3390/cancers16020457.
- X. Chen, M. C. Crawford, Y. Xiong, A. B. Shaik, K. F. Suazo, L. G. Bauer, M. S. Penikalapati, J. H. Williams, K. V. M. Huber, T. Andressen, R. E. Swenson and J. L. Meier, Paralogue-Selective Degradation of the Lysine Acetyltransferase EP300, JACS Au, 2024, 4(8), 3094–3103, DOI:10.1021/jacsau.4c00442.
- D. Bosnakovski, M. D. Gearhart, E. A. Toso, E. T. Ener, S. H. Choi and M. Kyba, Low Level DUX4 Expression Disrupts Myogenesis through Deregulation of Myogenic Gene Expression, Sci. Rep., 2018, 8(1), 16957, DOI:10.1038/s41598-018-35150-8.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1 – ATP viability assay in HAP1 cells. Fig. S2 – degradation profiles of p300 and CBP in HAP1 cells using a HiBiT assay. Fig. S3 – ATP viability assay in CIC::DUX4 sarcoma cell lines. Fig. S4 – degradation profiles in NCC-CDS-X1 cells using capillary electrophoresis. Fig. S5 – histone lysine acetylation in NCC-CDS-X1 cells. Chemical synthesis – full experimental. Fig. S6–S8 – analytical characterization data for BT-O2C. See DOI: https://doi.org/10.1039/d4md00969j |
| This journal is © The Royal Society of Chemistry 2025 |