Open Access Article
Open Access ArticleLight enhanced cytotoxicity and antitumoral effect of a ruthenium-based photosensitizer inspired from natural alkaloids†
Gennaro
Sanità‡
 a,
Maria Laura
Alfieri‡
b,
Barbara
Carrese
c,
Serena
Damian
b,
Vincenza
Mele
c,
Gaetano
Calì
d,
Brigida
Silvestri
a,
Maria Laura
Alfieri‡
b,
Barbara
Carrese
c,
Serena
Damian
b,
Vincenza
Mele
c,
Gaetano
Calì
d,
Brigida
Silvestri
 e,
Sebastiano
Marra
e,
Susan
Mohammadi
f,
Giuseppina
Luciani
g,
Paola
Manini
e,
Sebastiano
Marra
e,
Susan
Mohammadi
f,
Giuseppina
Luciani
g,
Paola
Manini
 *b and
Annalisa
Lamberti
*b and
Annalisa
Lamberti
 *c
*c
aInstitute of Applied Sciences and Intelligent Systems Unit of Naples, National Research Council, Naples, Italy
bDepartment of Chemical Sciences, University of Naples Federico II, Naples, Italy. E-mail: pmanini@unina.it
cDepartment of Molecular Medicine and Medical Biotechnology, University of Naples Federico II, Naples, Italy. E-mail: annalisa.lamberti@unina.it
dInstitute of Endocrinology and Experimental Oncology, National Research Council, Naples, Italy
eDepartment of Civil, Architectural and Environmental Engineering, University of Naples Federico II, Naples, Italy
fScuola Superiore Meridionale, Naples, Italy
gDepartment of Chemical, Materials and Industrial Production Engineering, University of Naples Federico II, Naples, Italy
First published on 25th October 2024
Abstract
In this work, we report on the synthesis and properties of a new sensitizer for photodynamic therapy applications, constituted by a ruthenium(II) complex (1) featuring a ligand inspired from natural isoquinoline alkaloids. The spectroscopic analysis revealed that 1 is characterized by an intense red emission (λem = 620 nm, Φ = 0.17) when excited at 550 nm, a low energy radiation warranting for a safe therapeutic approach. The phototoxicity of 1 on human breast cancer (Hs578T) and melanoma (A375) cell lines was assessed after irradiation using a LED lamp (525 nm, total fluence 10 J cm−2). In vitro biological assays indicated that the cytotoxicity of 1 was significantly enhanced by light reaching IC50 values below the micromolar threshold. The cell damage induced by 1 proved to be strictly connected with the overproduction of reactive oxygen species (ROS) responsible for mitochondrial dysfunction leading to the activation of caspases and then to apoptosis, and for DNA photocleavage leading to cell cycle arrest.
Introduction
The severe impact of tumoral pathologies, representing one of the major causes of mortality worldwide, has stimulated, in the last century, the research for more efficient therapeutic treatments.1–5 Although cisplatin and its analogues have certainly been proven to be highly effective as metal-based drugs against several types of epithelial cancer (e.g., ovarian, bladder and testicular cancer),6–9 significant side effects (including nephrotoxicity, peripheral neurotoxicity and emetogenesis) and frequent induction of drug resistance together with restricted therapeutic activity against the most common tumors such as colon and breast cancer limited their clinical applications.10–12 Hence, there is an urgent need for the development and clinical introduction of alternative therapies and antineoplastic agents. In this regard, photodynamic therapy (PDT) has recently emerged as a valuable choice with respect to surgical procedures, radiotherapy and chemotherapy thanks to its minimal invasiveness, lack of adverse side effects, safety and targeted therapeutic approach.13–17Known photosensitizers currently used for clinical applications are based on tetrapyrrolic moieties, including for example porphyrins, chlorins and phthalocyanines.18–21 However, these compounds share similar drawbacks including complications in the synthesis and purification, low water solubility, weak photostability, and lack of tumor tissue selectivity aside from poor tissue penetration of shorter-wavelength visible light, which limit their potential use.22–25 Consequently, different attempts have been made over the years to improve the effectiveness of the PDT treatment. Among others, the use of transition metal complexes26–30 and more specifically of ruthenium(II) complexes featuring polypyridyl ligands31–38 was found to be a valuable alternative for PDT owing to the outstanding photophysical properties of ruthenium(II) complexes.31,34,39,40
The great potential of ruthenium(II) complexes relies also on: 1) the multiple mechanisms through which they can enter cells, such as passive diffusion, active transport and endocytosis; 2) the different cellular targets they can address, such as cell nuclei, mitochondria or lysosomes; 3) the way they can damage and kill cancer cells, i.e. via DNA intercalation, protein interaction and ROS production; 4) the low systemic toxicity and selective antimetastatic properties.41–44 Overall, this evidence prompted the design of different kinds of ruthenium(II) complexes to look for the best performing PS for PDT application. This kind of investigation is possible considering that the photophysical and chemical properties of ruthenium(II)-based complexes (e.g., charge, solubility, ligand conformations and metal- and ligand-based redox potentials) can be finely tuned through the proper selection of the ligand–metal combination and designing appropriate geometries for specific interactions with biological targets, which thus makes them attractive for photobiological applications.45–48
In this connection, we recently explored the possibility of taking advantage of biologically relevant ligands to build bio-inspired transition metal complexes.49 By pursuing this strategy, the PS may be easily recognized by the cellular environment and can accumulate in the tissues. Previous studies have demonstrated, for example, the high and selective cytotoxicity of ruthenium(II) complexes experiencing the presence of β-carboline ligands. By taking advantage of the presence of a heterocyclic platform common to many natural and synthetic alkaloids,50,51 the β-carboline ligand can camouflage the metal and foster the diffusion of the ruthenium(II) complex into the cellular target via specific receptors. By this way, high and selective cytotoxicity and antitumor activity against various cancer cells through multiple mechanisms (i.e. interfering with DNA synthesis, inhibiting DNA topoisomerases I and II) have been reached.52–54
By pursuing a similar approach, herein we report on the synthesis and characterization of the photo-physical properties of a ruthenium(II) complex (1) featuring a ligand inspired from natural isoquinoline alkaloids (Fig. 1). On the basis of literature data reporting on the use of PDT as an excellent alternative in the treatment and diagnosis of breast cancer and melanoma compared to the conventional surgery, chemotherapy and radiotherapy,55,56 we have assayed for the first time the light-promoted enhanced cytotoxicity of 1 on the human breast carcinoma cell line (Hs578T) and human melanoma cell line (A375) to assess the potential use of the complex as an efficient PS.
Results and discussion
Synthesis and characterization of the ruthenium complex 1
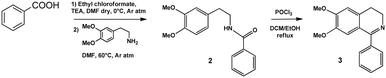
In the present paper, we report on the synthesis and characterization of a new PS based on a ruthenium(II) complex (1) featuring a C^N ligand (3) inspired from natural isoquinoline alkaloids. The ligand 3 was prepared via a two-step synthetic procedure starting from a commercially available derivative of the neurotransmitter dopamine, namely O,O-dimethyldopamine (Scheme 1). The latter was first used to form the amide 2 by reaction with ethyl chloroformate and benzoic acid, then Bischler–Napieralski intramolecular cyclization was carried out to afford the desired ligand 3.The identity of both the amide 2 and the ligand 3 was confirmed by NMR spectroscopy and MALDI mass spectrometry (Fig. S1–S3†).
The complex 1 was synthesized according to a procedure reported in the literature.54 A suspension of cis-Ru(bpy)2Cl2 in a mixture of ethanol and water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v)) was treated under an argon atmosphere with the ligand 3 and triethylamine under reflux conditions (Scheme 2). After 3 h, the reaction mixture was cooled down and treated with a water solution of NH4PF6. The formation of a dark-red solid was observed; this latter was collected by centrifugation to give the complex 1 in good yield (74%).
1 (v/v)) was treated under an argon atmosphere with the ligand 3 and triethylamine under reflux conditions (Scheme 2). After 3 h, the reaction mixture was cooled down and treated with a water solution of NH4PF6. The formation of a dark-red solid was observed; this latter was collected by centrifugation to give the complex 1 in good yield (74%).
Mono- and bidimensional NMR spectroscopy (Fig. S5–S10†) was carried out to confirm the structure of the complex 1.
In particular, the 1H and 1H,1H COSY spectra displayed a series of signals both in the aromatic and aliphatic proton regions. Four multiplets at 3.46, 2.82, 2.73 and 2.41 ppm were ascribable to the methylene protons of the isoquinoline unit along with the four singlets at 3.93 and 3.91 ppm, relative to the proton of the methoxyl group, and at 7.56 and 6.85 ppm due to the aromatic protons. The spectrum was completed by the signals of two 2,2′-bipyridine units along with the signals of the protons of the phenyl residue. The 13C, 1H,13C HSQC and 1H,13C HMBC spectra allowed for the complete assignment of all the resonances, definitely supporting the structural identification of the complex 1. In good agreement were the data obtained from the MALDI mass spectrometry analysis, revealing the presence of the peaks of the two ionic portions in the positive (m/z 680, [Ru(bpy)23]+) and negative (m/z 145, [PF6]−) ion modes (Fig. S12 and S13†). The complex 1 exhibited good solubility in PBS at the micromolar concentration, unlike the parent compound Ru(bpy)3(PF6)2, suggesting its potential use for biological assays and therapeutic purposes.
Photophysical properties of the ruthenium complex 1
To check for the performances of the complex 1 as a PS, we investigated its photophysical properties by UV-visible and emission spectroscopy. The spectra were registered on dilute solutions (1 × 10−5 M) in organic solvent (CH2Cl2) and in phosphate buffer saline solution (PBS) at pH 7.4 and compared with those from the ligand 3 and the complex Ru(bpy)3(PF6)2 taken as a reference (Table 1).UV-vis λmax, nm (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε, M−1 cm−1) ε, M−1 cm−1) |
UV-vis λmax,d nm (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε, M−1 cm−1) ε, M−1 cm−1) |
PL λem, nm (λecc, nm) | Φ%e (λem, nm) | PL λem,d,e nm (λecc, nm) | |
|---|---|---|---|---|---|
| a Determined in diluted solutions (1 × 10−5 M). b Determined in CH2Cl2. c Determined in ACN. d Determined in PBS solution (pH 7.4). e Determined relatively to fluorescein (Φ = 0.9 in a 0.1 M solution of NaOH). | |||||
| 1 | 294 (4.87), 352 (4.05), 430 (3.96), 504 (4.07), 558 (3.97), 606 (sh) | 290, 341, 439, 482, 550 (sh) | 620 (550) | 0.17 | 665 (550) |
| Ru(bpy)3(PF6)2c | 288 (4.98), 390 (sh), 426 (sh), 451 (4.17) | Low solubility | 615 (451) | 0.09 | Low solubility |
The UV-vis spectrum of 1 (Fig. 2) displayed the typical profile of ruthenium(II) complexes with polypyridyl ligands (see data from Ru(bpy)3(PF6)2). In detail, it has been possible to note the presence of: a) an intense maximum (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε > 4 M−1 cm−1) below 300 nm assigned to the spin-allowed π–π* ligand centred (LC) transitions (see the UV-vis spectrum of 3 for analogies); b) a series of intense maxima (log
ε > 4 M−1 cm−1) below 300 nm assigned to the spin-allowed π–π* ligand centred (LC) transitions (see the UV-vis spectrum of 3 for analogies); b) a series of intense maxima (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε ≈ 4 M−1 cm−1) in the range of 300–500 nm assigned to metal-to-ligand charge transfer (1MLCT) transitions; c) weak and broad absorption maxima at longer wavelengths (>500 nm) ascribable to spin forbidden metal-to-ligand charge transfer (3MLCT) transitions. The absorption maxima of 1 proved to be red shifted with respect to those from Ru(bpy)3(PF6)2 as a consequence of the reduction of the HOMO–LUMO gap due to the higher electron-releasing properties of the C^N ligand via σ-donation from the carbanion with respect to 2,2′-bipyridine (N^N).57
ε ≈ 4 M−1 cm−1) in the range of 300–500 nm assigned to metal-to-ligand charge transfer (1MLCT) transitions; c) weak and broad absorption maxima at longer wavelengths (>500 nm) ascribable to spin forbidden metal-to-ligand charge transfer (3MLCT) transitions. The absorption maxima of 1 proved to be red shifted with respect to those from Ru(bpy)3(PF6)2 as a consequence of the reduction of the HOMO–LUMO gap due to the higher electron-releasing properties of the C^N ligand via σ-donation from the carbanion with respect to 2,2′-bipyridine (N^N).57
 | ||
| Fig. 2 UV-vis (blue trace) and emission (red trace) spectra of 1 in CH2Cl2 (solid line) and in PBS at pH 7.4 (dotted line). | ||
The complex 1 exhibited a red emission with a maximum set at 620 nm and an emission quantum yield of Φ = 0.17 (relative to fluorescein) comparable to those reported in the literature for similar complexes and higher than the parent compound Ru(bpy)3(PF6)2.
The photophysical properties of 1 have been investigated also in phosphate buffer saline (PBS) at pH 7.4; the aim was to assess if any change in the optical properties of 1 may occur when incubated in the medium for cell culture biological assays and to evaluate the stability of the complex with time.
As shown in Fig. 2, the absorption profile of 1 showed a slight reduction of the intensity of some maxima, such as the one at 558 nm, and a significant red shift of the emission maximum (from 620 to 665 nm). Under these conditions, the complex 1 also proved to be stable over 24 h incubation time. These data have been taken into account for the design of the biological assays. A spectrophotometric approach was also pursued to evaluate the hydrophobic/hydrophilic character of the ruthenium complex. The log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value measured for 1 proved to be −2.06 suggesting a moderate hydrophilic character.
P value measured for 1 proved to be −2.06 suggesting a moderate hydrophilic character.
Phototoxicity of the ruthenium complex 1: in vitro biological assays
The phototoxic activity of 1 was evaluated against two human cancer cell lines: the breast carcinoma cells Hs578T and the melanoma cells A375. In each experiment, the cells were treated with the ruthenium complex at a selected concentration and then incubated under dark conditions or after irradiation for 30 minutes at 525 nm, corresponding to the wavelength that activates the ruthenium complex 1 (Fig. 3).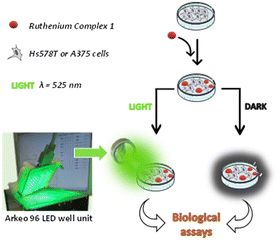 | ||
| Fig. 3 Schematic representation of the experimental procedure followed to assess the phototoxicity of the ruthenium complex 1 on Hs578T or A375 cell lines. | ||
The irradiation was performed by using an Arkeo 96 led well unit provided by Cicci Research s.r.l., a portable device constituted by an array of 96 LEDs with 12 independent lines, fitting with the 96 well plates used for the biological tests. All the irradiation sessions were carried out by using a total fluence of 10 J cm−2.
After the incubation, the metabolic functionalities of the cells were evaluated through a combined multiparametric approach based on the following assays/analyses: 1) the CellTiter-GLO® and Live Cell Explorer® assays to evaluate the vitality of cells; 2) the flow cytometry and confocal microscopy analyses to assess the cellular uptake of the complex; 3) the propidium iodide assay to evaluate the onset of apoptosis; 4) the Caspase-Glo® assay to estimate the activity of caspases; 5) the western blot analysis to evaluate the expression of the anti-apoptotic protein Bcl-xL and the pro-apoptotic protein Bak; 6) the ROS-Glo® H2O2 assay to measure the amount of intracellular ROS; 7) the agarose gel electrophoresis analysis to check for the ability of 1 to induce the DNA photocleavage.
Cytotoxicity of the ruthenium complex 1
The cytotoxic activity of 1 was assessed by the CellTiter-GLO® and Live Cell Explorer® assays. In the first one, the cell viability was evaluated by measuring the levels of ATP, which is a marker of the presence of metabolically active cells. In the Live Cell Explorer assay, the cell viability was determined through the activity of intracellular esterases that convert calcein-AM, a non-fluorescent cell-permeant compound, into calcein, an anionic fluorescent compound.Both assays were carried out on Hs578T and A375 cancer cell lines (Fig. 4) and on a mammary breast fibrocystic disease cell line (MCF10a), selected as the non-tumoral control cell line (Fig. S14†).
The assays were performed after incubation for 24 h with different amounts of a PBS solution of 1 under dark conditions or after irradiation at 525 nm.
The results reported in Fig. 4 and in Fig. S16† showed that the complex 1 exhibited a dose-dependent toxicity that proved to be significantly enhanced by irradiation. As a matter of fact, the CellTiter-GLO® assay revealed that the half-maximal inhibitory concentrations (IC50) of 1, set at 1.1 and 1.4 μM under dark conditions in the case of the Hs578T and A375 cell lines, drop down at 0.5 and 0.6 μM after irradiation (Fig. 4A and B). These data, also supported by the results from the Live Cell Explorer assay (Fig. 4C and D), indicated that the cytotoxic effect of the ruthenium complex 1 is enhanced by light reaching IC50 values below the micromolar threshold, quite lower with respect to cisplatin (IC50 = 30.3 and 4.9 μM in the case of Hs578T58 and A375 (ref. 59) cell lines, respectively) and other similar ruthenium complexes.
Similar results were also observed in the case of the MCF10a cell line with IC50 = 1.2 μM under dark conditions and IC50 = 0.4 μM after irradiation (Fig. S14 and S15†). These data indicated that the cytotoxic effect of 1 was not selective toward tumoral cells, suggesting that the application/accumulation of the photosensitizer on the damaged tissues was necessary before irradiation.
Finally, control experiments carried out in the absence of the photosensitizer revealed that the irradiation step did not cause loss in cell viability, in agreement with Lifshits.60 Overall, these preliminary data suggested the potential use of 1 as a photosensitizer for local therapeutic applications.
As detailed in the next sections, to delineate the mechanism of action of 1, other biological assays have been carried out.
Cellular uptake
The uptake of 1 by Hs578T and A375 cell lines was investigated by flow cytometry and confocal microscopy, exploiting the intrinsic fluorescence of the ruthenium complex. For the flow cytometry analysis, the cells were incubated with the complex 1 at 1 μM and 10 μM (Hs578T) and at 1.5 μM and 15 μM (A375), for 3 and 6 h.The results showed a concentration dependent uptake in both cell lines already after 3 h of incubation, while no increase was observed after 6 h of incubation. In particular, in Hs578T cells mean fluorescence intensity (MFI) increases of about 5 and 12 were observed by using 1 μM and 10 μM, respectively, whereas in A375 cells an MFI increase of about 10 and 30 was detected by using 1.5 μM and 15 μM, respectively (Table 2). The results suggest a quick cellular uptake and a very good cell membrane permeability of the complex 1.
| Time (h) | Hs578T | A375 | ||
|---|---|---|---|---|
| MFI (a.u.) | MFI (a.u.) | |||
| 3 | Control | 2.56 ± 0.12 | Control | 0.85 ± 0.04 |
| 3 | 1 (1 μM) | 14.16 ± 0.87 | 1 (1.5 μM) | 9.2 ± 0.01 |
| 3 | 1 (10 μM) | 35.38 ± 4.15 | 1 (15 μM) | 29.4 ± 1.37 |
| 6 | Control | 2.85 ± 0.33 | Control | 0.63 ± 0.07 |
| 6 | 1 (1 μM) | 14.82 ± 0.95 | 1 (1.5 μM) | 7.24 ± 0.34 |
| 6 | 1 (10 μM) | 35.33 ± 2.1 | 1 (15 μM) | 16.7 ± 1.56 |
For confocal microscopy analysis, both cell lines were incubated with the complex 1. After 6 h of incubation, the images revealed the presence of the complex in the perinuclear area confirming that the cellular uptake occurred (Fig. 5). Furthermore, in good agreement with the data obtained by flow cytometry analysis, the red fluorescence signal from 1 proved to be more intense in the case of Hs578T cells than in A375 cells.
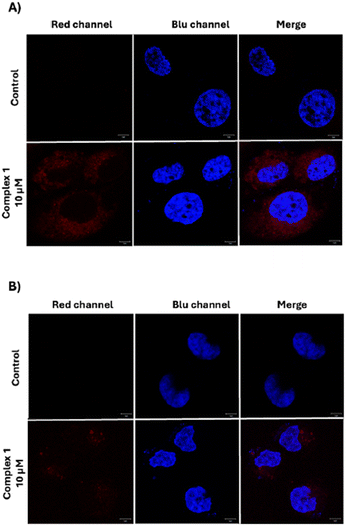 | ||
| Fig. 5 Representative confocal microscopy images of Hs578T cells (A) and A375 cells (B). Cell nuclei were stained with Hoechst 33342, complex 1 is visible as a red color. Scale bars: 5 μm. | ||
Apoptosis promoted by 1
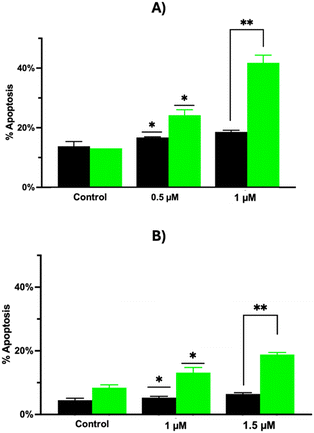
To evaluate the mechanisms by which 1 can promote cell death, the effect of the ruthenium complex on the induction of apoptosis was investigated by propidium iodide staining and flow cytometry. To this aim, Hs578T and A375 cell lines were incubated with 1 both under dark conditions and after irradiation. The data reported in Fig. 6 showed that: a) 1 can promote the apoptotic death on both cell lines; b) the effect is more pronounced after irradiation; c) the apoptosis measured in treated cells compared to the control was in a dose dependent manner. In particular, the percentage of apoptotic cells after irradiation was 24.2 and 41.8 in the case of Hs578T cells (concentration of 1 = 0.5 and 1 μM, respectively), and 13.2 and 18.8 in the case of A375 cells (concentration of 1 = 1 and 1.5 μM, respectively). These values proved to be quite high with respect to the same experiments carried out under dark conditions, with a percentage of apoptotic cells of 16.8 and 18.6 measured in the case of Hs578T cells (concentration of 1 = 0.5 and 1 μM, respectively), and of 5.3 and 6.4 measured in the case of A375 cells (concentration of 1 = 1 and 1.5 μM, respectively).To clarify the mechanism of cell apoptosis induced by 1, the activities of caspase-9 and caspase-3/7 were examined by using specific luminogenic substrates (Caspase-Glo® assay). Caspase-9 is the initiator caspase related to the intrinsic or mitochondrial pathway of apoptosis. When activated, caspase-9 cleaves and activates downstream effector caspase-3 and -7, thus resulting in apoptosis.61 To evaluate caspase activation, Hs578T and A375 cell lines were incubated with 1 and analyzed both under dark conditions and after 4.5 h post irradiation (this incubation time was chosen taking into account that the activation of caspases in the apoptotic pathway is an early event).62–64 Under all tested conditions, both cell lines showed an increase of the caspase activity in a dose-dependent manner after irradiation, compared to dark conditions. The caspase 3/7 activity increased after irradiation, with an enhancement of about 2 and 2.8 times in Hs578T cells (Fig. 7A) with the concentration of 1 set at 0.5 and 1 μM, respectively, and of about 2.2 and 3.2 times in A375 cells (Fig. 7B) with the concentration of 1 set at 1 and 1.5 μM, respectively.
 | ||
| Fig. 7 Caspases 3/7 and 9 activities. Caspase-GLO® 3/7 assay in Hs578T cells (A) and in A375 cells (B); Caspase-GLO® 9 assay in Hs578T cells (C) and in A375 cells (D). *** P ≤ 0.001. | ||
The caspase-9 activity proved to be significantly enhanced in Hs578T cells (Fig. 7C), with an increase of about 7.2 and 8.4 times after administration of 1 (0.5 and 1 μM, respectively), and to a lesser extent in A375 cells (Fig. 7D), with an increase of about 2.2 and 2.3 times after incubation of 1 (1 and 1.5 μM, respectively). In both cases, no increase of caspase activity was observed when the incubation was carried out under dark conditions. These data suggest that an intrinsic apoptosis pathway could be photoactivated in both tested cell lines after incubation with the ruthenium complex.
Mitochondrial dysfunction induced by 1
Based on the results obtained from the caspase assays, the possible role of 1 in promoting mitochondrial dysfunction was investigated by evaluating the effects of the ruthenium complex on the expression of the Bcl-2 protein family.The Bcl-2 proteins are involved in the regulation of apoptosis through the control of mitochondrial membrane permeability and the release of cytochrome c and/or Smac/Diablo. They include anti-apoptotic proteins, such as Mcl-1, Bcl-2, and Bcl-xL, and pro-apoptotic proteins, including Bax, Bad, Bak, Bid, and Bim.65
Therefore, the effects of 1 on the expression levels of the anti-apoptotic protein Bcl-xL and the pro-apoptotic protein Bak were evaluated by performing a western blot analysis in Hs578T and A375 cell lines treated with the ruthenium complex and irradiated at a wavelength of 525 nm. The results indicated that 1 is able to inhibit the expression of the Bcl-xL protein and stimulate the expression of the Bak protein, with an overall reduction of the Bcl-xL/Bak ratio (Fig. 8).
Accumulation of reactive oxygen species (ROS) induced by 1
One of the possible mechanisms by which ruthenium(II) complexes can induce apoptosis and autophagy in cancer cells is via the overproduction of ROS through a complex series of fast reactions triggered by light irradiation.66–69ROS are normally produced in normal metabolic living cells. At low concentrations, these species have an important role in cell signaling, while at high concentrations, they can become toxic due to the interaction with proteins, DNA, lipids, and other biological molecules affecting their functioning. To investigate the possible implication of 1 in the formation of ROS, the ruthenium complex has been administered to Hs578T and A375 cell lines and the amount of intracellular ROS was measured after light irradiation by the ROS-Glo® H2O2 assay.
The data reported in Fig. 9 show an increase of the ROS production after irradiation in both cell lines. The effect was more evident in the Hs578T cell line with a ROS production up to 1.8 times higher with respect to the control experiments, whereas in the case of the A375 cell line, the ROS production was 1.3 times higher compared to the control. In both cases, the results suggest that ROS-mediated pathways can trigger apoptosis.
 | ||
| Fig. 9 ROS levels determined in Hs578T (A) and in A375 (B) cell lines after incubation with 1 and light irradiation. *** P ≤ 0.001. | ||
DNA photocleavage activity
Metal-based antitumoral drugs such as cisplatin can perform their therapeutic action by inducing DNA damage.70 To investigate this possibility, the DNA photocleavage activity of 1 was assessed by agarose gel electrophoresis. In particular, the pEGFP-N3 plasmid DNA was incubated for 72 h with different amounts of the ruthenium complex both under dark or light conditions and then subjected to gel electrophoresis to evaluate the abundance of the supercoiled form with respect to the circular (cleaved) one.As shown in Fig. 10, the complex 1 does not induce significant DNA cleavage under dark conditions, whereas after irradiation a dose-dependent reduction of the supercoiled form is visible. This result indicates that the DNA cleavage is determined by photoinduced processes mediated by 1 and supports the possibility of using 1 as a photosensitizer to induce cellular toxicity by DNA damage.71
 | ||
| Fig. 10 Photocleavage of pEGFP-N3 plasmid DNA induced by 1 under dark and light irradiation conditions. | ||
Conclusions
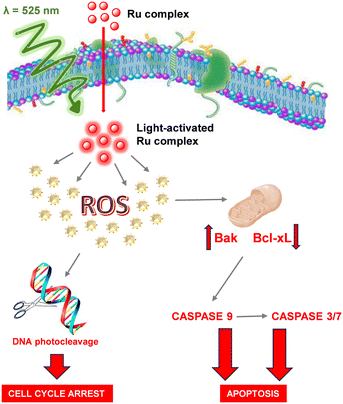
In the frame of a research activity aimed at testing the potential impact of luminescent ruthenium complexes as effective sensitizers for the photodynamic treatment of cancer, we have reported herein the synthesis of a new complex (1) featuring, besides 2,2′-bipyridine, a ligand inspired from natural isoquinoline alkaloids.The investigation of the photophysical properties of the complex allowed us to assess that, both in dichloromethane and in PBS, low energetic radiation (a green light, λ = 525 nm) was necessary to promote the sensitization of the complex.
This represents an important requisite for a safer approach to the photodynamic treatment. A set of biological tests was carried out on two selected human cell lines, breast cancer (Hs578T) and melanoma (A375), to assess the phototoxicity of the ruthenium complex 1 and to investigate the mechanisms of cellular damage. What emerged is that 1 is cytotoxic for both cell lines, in a more effective way toward Hs578T, and that the cytotoxicity is enhanced by light reaching IC50 values below the micromolar threshold, quite lower with respect to cisplatin and other similar ruthenium complexes. Light irradiation proved to be fundamental in causing the overproduction of ROS, a key event triggering a sequence of processes leading to cell death (Fig. 11). The increase in the levels of the proapoptotic protein Bak and the decrease in the levels of the antiapoptotic protein Bcl-xL were also evident after the photoactivation of 1, resulting in the activation of caspase-3/7/9 and the onset of apoptosis. Finally, experiments carried out on pEGFP-N3 plasmid DNA showed the ability of 1 to promote the DNA photocleavage.
Overall, these results pointed out that 1 can explicate its phototoxicity via a combined action that leads to the impairment of the main cellular activities and finally to death, thus opening a way to its possible use in the treatment of cancer.
Materials and methods
Materials and general methods
Benzoic acid, O,O-dimethyldopamine, cis-dichlorobis(2,2′-bipyridine)ruthenium(II), ethyl chloroformate and triethylamine were purchased from Sigma Aldrich. Phosphate-buffered saline (PBS) was purchased from Gibco (Grand Island, NY, USA). A Live Cell ExplorerTM live-cell labeling kit was purchased from AAT Bioquest® (Sunnyvale, CA, USA). Plasmid pEGFP-N3 (4700bp) was purchased from Addgene (Watertown, MA, USA). CellTiter-GLO® Luminescent Cell Viability Assay, Caspase-Glo® 3/7 Assay, Caspase-Glo® 9 Assay and ROS-Glo™ H2O2 Assay kits were purchased from Promega (Madison, WI, USA).UV-visible and emission spectra have been recorded on Jasco V-560 and Jasco FP-750 instruments. Quantum efficiencies (Φ) have been calculated using fluorescein (Φ = 0.9 in a 0.1 M solution of NaOH) as a reference. 1H and 13C NMR spectra have been registered on a Bruker DRX (400 MHz) instrument. 1H,1H COSY, 1H,13C HSQC, 1H,13C HMBC and NOESY experiments have been run at 400.1 MHz using standard pulse programs. MALDI mass spectra have been recorded on an AB Sciex TOF/TOF 5800 instrument using 2,5-dihydroxybenzoic acid as the matrix. The spectra represent the sum of 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 laser pulses from randomly chosen spots per sample position.
000 laser pulses from randomly chosen spots per sample position.
Elemental analysis has been performed on a Thermo Scientific Flash Smart V CHNS/CHNS instrument.
Synthesis of 1-phenyl-6,7-dimethoxy-3,4-dihydroisoquinoline (3)
A solution of benzoic acid (500 mg, 4 mmol) in 28 mL of dry dimethylformamide was cooled with an ice bath, and under stirring and an argon atmosphere, and then triethylamine (400 mg, 3.9 mmol) and ethyl chloroformate (394 mg, 3.6 mmol) were added. After 1 h, O,O-dimethyldopamine (634 mg, 3.5 mmol) was added dropwise. The mixture was stirred at room temperature for 1 h and then heated at 50–60 °C for 1 h. The mixture was evaporated under reduced pressure and the residue was extracted with an aqueous solution of NH4Cl (10% w/w) and chloroform (three times) and then with phosphate buffer (0.1 M, pH 7.4) and chloroform (three times). The organic layers were collected, dried over anhydrous sodium sulphate, filtered and evaporated under reduced pressure to afford the pure N-(3,4-dimethoxyphenethyl)benzamide 2 (969 mg, 98%).1H NMR (400 MHz, CDCl3) δ ppm: 7.83 (dd, J = 8.2, 2.3 Hz, 2H), 7.71 (t, J = 8.2 Hz, 1H), 7.59 (t, J = 8.2 Hz, 2H), 6.56 (bs, 1H), 4.31 (s, 3H, –OCH3), 4.28 (s, 3H, –OCH3), 4.12 (dt, J = 6.4, 6.0 Hz, 2H), 3.32 (t, J = 6.4 Hz, 2H).
A solution of the amide 2 (969 mg, 3.4 mmol) in 11.7 mL of a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 11 (v/v) mixture of ethanol/dichloromethane was treated under stirring with POCl3 (3.28 mL, 35 mmol) and kept under reflux. After 5 h, petroleum ether was added (8 mL), and the mixture was kept under reflux. After 12 h, the reaction mixture was filtered and the solid was rinsed with water and treated with an aqueous solution of K2CO3 until pH 10 was reached. Then, the mixture was extracted with chloroform and the organic layers were dried with anhydrous sodium sulphate, filtered, and evaporated under reduced pressure. The crude residue was purified by liquid chromatography on silica gel using dichloromethane/methanol 95
11 (v/v) mixture of ethanol/dichloromethane was treated under stirring with POCl3 (3.28 mL, 35 mmol) and kept under reflux. After 5 h, petroleum ether was added (8 mL), and the mixture was kept under reflux. After 12 h, the reaction mixture was filtered and the solid was rinsed with water and treated with an aqueous solution of K2CO3 until pH 10 was reached. Then, the mixture was extracted with chloroform and the organic layers were dried with anhydrous sodium sulphate, filtered, and evaporated under reduced pressure. The crude residue was purified by liquid chromatography on silica gel using dichloromethane/methanol 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 (v/v) as an eluent to give the pure 1-phenyl-6,7-dimethoxy-3,4-dihydroisoquinoline 3 (885 mg, 95%).
5 (v/v) as an eluent to give the pure 1-phenyl-6,7-dimethoxy-3,4-dihydroisoquinoline 3 (885 mg, 95%).
1H NMR (400 MHz, CDCl3) δ ppm: 7.61 (m, 2H), 7.40 (m, 3H), 6.78 (m, 2H), 3.82 (s, 3H, –OCH3), 3.80 (t, J = 7.28 Hz, 2H), 3.73 (s, 3H, –OCH3), 2.75 (t, J = 7.28 Hz, 2H). 13C NMR (100 MHz, CDCl3) δ ppm: 167.0, 151.3, 147.2, 138.5, 132.8, 129.6, 128.9, 128.2, 121.3, 111.9, 110.4, 56.2, 56.1, 47.2, 26.0. ESI+-MS: m/z 268.
Synthesis of [Ru(bpy)23][PF6] (1)
cis-Dichlorobis(2,2′-bipyridine)ruthenium(II) (100 mg, 0.21 mmol) was suspended in 20 mL of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) solution of water/ethanol under a positive pressure of argon and then the ligand 3 (85 mg, 0.32 mmol) and triethylamine (31.2 g, 0.31 mmol) were added. The resulting mixture was kept under reflux and an argon atmosphere for 3 h, and then it was cooled down to room temperature and concentrated under reduced pressure. Water was added until the starting volume was reached, and then the mixture was centrifuged to remove the solid. NH4PF6 (175 mg, 1.1 mmol) was added to the supernatant and the formation of a dark red solid consisting of 1 was observed; this latter was collected by centrifugation, washed with water, and dried under reduced pressure (126 mg, 74%).
1 (v/v) solution of water/ethanol under a positive pressure of argon and then the ligand 3 (85 mg, 0.32 mmol) and triethylamine (31.2 g, 0.31 mmol) were added. The resulting mixture was kept under reflux and an argon atmosphere for 3 h, and then it was cooled down to room temperature and concentrated under reduced pressure. Water was added until the starting volume was reached, and then the mixture was centrifuged to remove the solid. NH4PF6 (175 mg, 1.1 mmol) was added to the supernatant and the formation of a dark red solid consisting of 1 was observed; this latter was collected by centrifugation, washed with water, and dried under reduced pressure (126 mg, 74%).
1H NMR (400 MHz, CD2Cl2) δ ppm: 8.4–8.2 (m, 3H), 8.21 (d, J = 8.0 Hz, 2H), 8.02 (d J = 8.0 Hz, 2H), 7.94 (t, J = 8.0 Hz, 1H), 7.84–7.77 (m, 4H), 7.60 (d, J = 8.0 Hz, 1H), 7.55 (s, 1H), 7.4–7.3 (m, 2H), 7.3–7.2 (m, 2H), 6.9–6.8 (m, 2H), 6.81 (s, 1H), 6.51 (m, 1H), 3.92 (s, 3H, –OCH3), 3.90 (s, 3H, –OCH3), 3.46 (m, 1H), 2.8–2.7 (m, 2H), 2.37 (m, 1H). 13C NMR (100 MHz, CD2Cl2) δ ppm: 157.6, 156.6 (2C), 154.8 (2C), 151.3 (2C), 149.5, 148.7 (2C), 147.3, 136.1 (2C), 134.6, 133.6, 133.3, 131.6 (2C), 127.9, 126.8, 126.3, 126.2 (2C), 125.3, 122.8, 122.7 (4C), 121.5 (2C), 111.4, 110.5, 56.3, 55.9, 50.3, 28.1. MALDI-MS: m/z 680 ([M–PF6]+). Anal. calc. for C37H32F6N5O2PRu: C 53.88%, H 3.91%, N 8.49%, found: C 53.12%, H 3.88%, N 8.40%.
Cell culture
The human breast carcinoma cell line (Hs578T), mammary breast fibrocystic disease cell line (MCF10a) and human melanoma cell line (A375) were obtained from the American Type Tissue Collection (Rockville, MD, USA). Hs578T and A375 cells were grown in DMEM supplemented with 10% heat inactivated fetal bovine serum (FBS) (GIBCO), 100 U mL−1 penicillin, 100 mg mL−1 streptomycin, and 1% L-glutamine. MCF10a cells were grown in DMEM supplemented with a mammary epithelial cell growth medium bullet kit (Lonza),100 nM cholera toxin (Sigma Aldrich) and 10% heat inactivated FHS (Lonza). All cell lines were grown at 37 °C in a 5% CO2 atmosphere.Cell viability assays
The CellTiter-GLO® assay and Live Cell Explorer® assay were performed according to the manufacturer's instructions. For the CellTiter-GLO® assay, cells were seeded into 96 opaque-walled plates at a density of 5 × 103 cells per well for Hs578T and 2.5 × 103 for A375 and, after 24 h, were incubated with 1 at different concentrations (0.25, 0.5, 1, 1.5 μM). After 12 h of incubation, the culture medium was replaced with a fresh medium, and the measurement was carried out after 12 additional hours under both dark conditions and light conditions (total fluence 10 J cm−2). Cell viability was assessed by recording the luminescence signal for 0.25 s per well using a multilabel reader (PerkinElmer, Waltham, MA, United States).For the Live Cell Explorer assay, Hs578T and A375 were seeded into 96-well microplates at a density of 5 × 103 cells per well for Hs578T and 2.5 × 103 for A375 and, after 24 h, were incubated with 1 at two selected concentrations: 0.5 and 1 μM in the case of Hs578T and at 1 and 1.5 μM in the case of A375. The analysis was carried out by using the same experimental conditions described for the CellTiter-GLO® assay. Cell viability was assessed by fluorescence microscopy.
Cellular uptake
To evaluate the concentration and time dependent intracellular internalization, the Hs578T and A375 cell lines were plated on six-well plates and incubated with complex 1 (1 μM and 10 μM for Hs578T and 1.5 μM and 15 μM for A375) for 3 h and 6 h at 37 °C. After the incubation time, the cells were collected and washed three times with PBS. The mean fluorescence intensity (MFI) of the cells containing complex 1 was measured by using a flow cytometer (Miltenyi).For confocal microscopy, cells (5 × 103 per coverslip) were plated on 10 mm glass coverslips placed on the bottom of a 24-well plate, allowed to attach for 24 h under normal cell culture conditions and then incubated with complex 1 at a concentration of 10 μM (Hs578T) and 15 μM (A375) for 6 h at 37 °C. The cells were washed with PBS, fixed in 2% formaldehyde for 10 min, and washed 3 times with PBS. The cell nuclei were then stained with Hoechst 33342 (Invitrogen, Carlsbad, CA, United States). The cells were then spotted on microscope slides and analyzed on an inverted and motorized microscope (Axio Observer Z.1) equipped with a 63×/1.4 Plan-Apochromat objective. The attached laser-scanning unit (LSM 700 4× pigtailed laser 405–488–555–639; Zeiss, Jena, Germany) enabled confocal imaging. For excitation, 405 and 555 nm lasers were used. Fluorescence emission was revealed using a main dichroic beam splitter and a variable secondary dichroic beam splitter. The signal in the red channel was acquired by means of a 640 long pass filter. Double staining fluorescence images were acquired separately using ZEN 2012 software in the red and blue channels at a sampling of 1024 × 1024 pixels, with the confocal pinhole set to one Airy unit and then saved in TIFF format.
Apoptosis assay
Cell apoptosis was assessed by propidium iodide (PI) staining and flow cytometry after incubating Hs578T and A375 cells (20 × 103) with 1 (0.5 and 1 μM in the first case and 1 and 1.5 μM in the second case). The assay was performed after 24 h incubation time under dark conditions, while under light conditions after 16 h incubation time, the culture medium was replaced with a fresh medium, and the cells were irradiated at 525 nm for 30′ (total fluence 10 J cm−2) and then were grown for supplementary 8 h before performing the assay. The latter was carried out by washing the cells in PBS and suspending them in 200 mL of a solution containing 0.1% sodium citrate, 0.1% Triton X-100 and 50 μg mL−1 propidium iodide (Sigma Chemical Co.). Following incubation at 4 °C for 30 min in the dark, the cell DNA content was analyzed using MACSQuant Analyzer 10 (Miltenyi Biotec) and the percentage of cells in the hypodiploid region was calculated.Caspase activity assay
Caspase-9 and caspase 3/7 activities were evaluated by using the Promega Caspase-GLO® assays, following the manufacturer's instructions. Hs578T cells were seeded into 96-well opaque microplates at a density of 5 × 103 cells per well for 24 h before incubation with 1 (0.5 and 1 μM), while A375 cells were seeded into 96-well opaque microplates at a density of 2.5 × 103 cells per well 24 h before incubation with 1 (1 and 1.5 μM). The assay was performed after 24 h incubation time under dark conditions, while under light conditions after 18 h incubation time, the cells were irradiated at 525 nm for 30′ (total fluence 10 J cm−2) and then were grown for supplementary 4.5 h before performing the assay. The caspase activity was assessed by recording the luminescence signal for 0.25 s per well using a multilabel reader (PerkinElmer, Waltham, MA, United States).Western blotting analysis
Hs578T and A375 cell lines were treated with 1 at two selected concentrations (0.5 and 1 μM in the first case and 1 and 1.5 μM in the second case) and after 18 h incubation time, were irradiated at 525 nm for 30′ (total fluence 10 J cm−2) and then were grown for supplementary 12 h before performing the assay. The latter was carried out by collecting the cells, washing them twice in PBS and resuspending them in 20–40 μL of radio immunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl pH 7.4, 1% NP40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 μg mL−1 aprotinin, leupeptin, pepstatin, 1 mM Na3VO4, 1 mM NaF) for 30 min on ice. The supernatant was then recovered by centrifugation (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 min at 4 °C) and the protein concentration was determined by a modified Bradford method using the Bio-Rad protein assay and compared with the bovine serum albumin standard curve.72 Cytosolic proteins (20 μg) were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis, electrotransferred to a PVDF (polyvinylidene difluoride) membrane (Merck Millipore, MA, USA) and reacted with Bcl-xL monoclonal antibodies (H5) (Santa Cruz, TX, USA), Bak rabbit monoclonal antibodies (Y164) (Abcam, Cambridge, UK) and b-actin (C4) (Santa Cruz). Blots were then developed using enhanced chemiluminescence detection reagents (western blotting luminol reagent, Santa Cruz) and exposed to X-ray films. All films were scanned for densitometric analysis using ImageJ 1.41 software.
000g for 30 min at 4 °C) and the protein concentration was determined by a modified Bradford method using the Bio-Rad protein assay and compared with the bovine serum albumin standard curve.72 Cytosolic proteins (20 μg) were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis, electrotransferred to a PVDF (polyvinylidene difluoride) membrane (Merck Millipore, MA, USA) and reacted with Bcl-xL monoclonal antibodies (H5) (Santa Cruz, TX, USA), Bak rabbit monoclonal antibodies (Y164) (Abcam, Cambridge, UK) and b-actin (C4) (Santa Cruz). Blots were then developed using enhanced chemiluminescence detection reagents (western blotting luminol reagent, Santa Cruz) and exposed to X-ray films. All films were scanned for densitometric analysis using ImageJ 1.41 software.
Reactive oxygen species (ROS) assay
A bioluminescence assay (ROS-Glo®H2O2 assay) was used to detect ROS production, according to the manufacturer's instructions. Hs578T and A375 cell lines were used under the same experimental conditions of the caspase assays and ROS levels were determined 30 min only after light irradiation by recording the luminescence signal for 0.25 s per well using a multilabel reader.DNA cleavage
The pEGFP-N3 plasmid DNA (200 ng) was mixed with different aliquots of a solution of 1 in 5 mM Tris-HCl and 50 mM NaCl pH 7.5 (final concentrations: 0, 1, 5, 10, 20, 50, 100 μM) at 37 °C for 72 h. The incubation was carried out under dark conditions or after irradiation at 525 nm for 30′ (total fluence: 10 J cm−2). The samples were then analyzed by gel electrophoresis on 0.8% agarose w/v containing ethidium bromide (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) in TAE (Tris-acetate–EDTA) buffer to evaluate the conversion of the supercoiled form of the plasmid into the circular form.
000) in TAE (Tris-acetate–EDTA) buffer to evaluate the conversion of the supercoiled form of the plasmid into the circular form.
Statistical analysis
The results of the biological assays are expressed as the mean ± SD of three independent experiments. The statistical significance of differences among groups was evaluated using analysis of variance, through the software GraphPad Prism 9.0. The significance was accepted at a confidence level of 95% (P < 0.05).Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
GS and MLA: investigation, methodology, writing – original draft; BC, SD, VM and GC: methodology; SMa and SMo: investigation; BS and GL: conceptualization and methodology; PM and AL: project administration, conceptualization, writing – original draft, writing – review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study has been carried out in the frame of the project POR Campania FESR 2014–2020 “SATIN”. The authors would like to thank the flow cytometry facility of the Department of Molecular Medicine and Medical Biotechnology of the University of Napoli Federico II.References
- B. Youden, R. Jiang, A. J. Carrier, M. R. Servos and X. Zhang, ACS Nano, 2022, 16, 17497–17551 Search PubMed.
- S. Uthaman, G. Cutshaw, S. Ghazvini and R. Bardhan, ACS Appl. Mater. Interfaces, 2023, 15, 50708–50720 CAS.
- P. Jing, Y. Luo, Y. Chen, J. Tan, C. Liao and S. Zhang, Bioconjugate Chem., 2023, 34, 366–376 Search PubMed.
- K. Li, D. Yang and D. Liu, Bioconjugate Chem., 2023, 34, 283–301 Search PubMed.
- F. Bray, J. Ferlay, I. Soerjomataram, R. L. Siegel, L. A. Torre and A. Jemal, Ca-Cancer J. Clin., 2018, 68, 394–424 Search PubMed.
- X. Huang, R. Huang, S. Gou, Z. Wang and H. Wang, Bioconjugate Chem., 2017, 28, 1305–1323 Search PubMed.
- R. Ranasinghe, M. L. Mathai and A. Zulli, Heliyon, 2022, 8, e10608 Search PubMed.
- S. A. Fahmy, F. Ponte, I. M. Fawzy, E. Sicilia, U. Bakowsky and H. M. E. S. Azzazy, Molecules, 2020, 25, 5926 CAS.
- C. Zhang, C. Xu, X. Gao and Q. Yao, Theranostics, 2022, 12, 2115–2132 CAS.
- B. Oronsky, C. M. Ray, A. I. Spira, J. B. Trepel, C. A. Carter and H. M. Cottrill, Med. Oncol., 2017, 34, 1–7 CAS.
- A. E. Freimund, J. A. Beach, E. L. Christie and D. D. L. Bowtell, Hematol./Oncol. Clin. North Am., 2018, 32, 983–996 CrossRef.
- Y. J. Chern and I. T. Tai, Cancer Biol. Med., 2020, 17, 842 CAS.
- S. Kwiatkowski, B. Knap, D. Przystupski, J. Saczko, E. Kędzierska, K. Knap-Czop, J. Kotlińska, O. Michel, K. Kotowski and J. Kulbacka, Biomed. Pharmacother., 2018, 106, 1098–1107 CrossRef.
- A. F. Dos Santos, D. R. Q. De Almeida, L. F. Terra, M. S. Baptista and L. Labriola, J. Cancer Metastasis Treat., 2019, 5, 25 Search PubMed.
- M. A. Saad and T. Hasan, Bioconjugate Chem., 2022, 33, 2041–2064 CrossRef CAS.
- P. De Silva, M. A. Saad, H. C. Thomsen, S. Bano, S. Ashraf and T. Hasan, J. Porphyrins Phthalocyanines, 2020, 24, 1321–1360 CrossRef.
- M. Yi, B. Xiong, Y. Li, W. Guo, Y. Huang and B. Lu, Eur. J. Med. Chem., 2023, 247, 115084 CrossRef CAS PubMed.
- M. Gallardo-Villagrán, D. Y. Leger, B. Liagre and B. Therrien, Int. J. Mol. Sci., 2019, 20, 3339 Search PubMed.
- S. Ghosh, K. A. Carter and J. F. Lovell, Biomaterials, 2019, 218, 119341 CrossRef CAS PubMed.
- M. Miretti, C. G. Prucca, T. C. Tempesti and M. T. Baumgartner, Curr. Med. Chem., 2021, 28, 5339–5367 Search PubMed.
- M. Qindeel, S. Sargazi, S. M. Hosseinikhah, A. Rahdar, M. Barani, V. K. Thakur, S. Pandey and R. Mirsafaei, ChemistrySelect, 2021, 6, 14082–14099 CrossRef CAS.
- J. Karges, U. Basu, O. Blacque, H. Chao and G. Gasser, Angew. Chem., Int. Ed., 2019, 58, 14334–14340 CrossRef CAS.
- P. S. Felder, S. Keller and G. Gasser, Adv. Ther., 2020, 3, 1900139 CrossRef.
- J. Zhang, C. Jiang, J. P. Figueiró Longo, R. B. Azevedo, H. Zhang and L. A. Muehlmann, Acta Pharm. Sin. B, 2018, 8, 137–146 CrossRef PubMed.
- F. Heinemann, J. Karges and G. Gasser, Acc. Chem. Res., 2017, 50, 2727–2736 CrossRef CAS PubMed.
- H. Huang, S. Banerjee, K. Qiu, P. Zhang, O. Blacque, T. Malcomson, M. J. Paterson, G. J. Clarkson, M. Staniforth, V. G. Stavros, G. Gasser, H. Chao and P. J. Sadler, Nat. Chem., 2019, 11, 1041–1048 CrossRef CAS PubMed.
- F. Javani Jouni, V. Abdollahi, S. Zadehmodarres, H. Abbasinia, M. Asnaashari and J. Zafari, Lasers Med. Sci., 2022, 37, 1175–1180 CrossRef.
- L. K. McKenzie, H. E. Bryant and J. A. Weinstein, Coord. Chem. Rev., 2019, 379, 2–29 CrossRef CAS.
- V. Novohradsky, A. Rovira, C. Hally, A. Galindo, G. Vigueras, A. Gandioso, M. Svitelova, R. Bresolí-Obach, H. Kostrhunova, L. Markova, J. Kasparkova, S. Nonell, J. Ruiz, V. Brabec and V. Marchán, Angew. Chem., Int. Ed., 2019, 58, 6311–6315 Search PubMed.
- J. Zhang, Y. Ma, K. Hu, Y. Feng, S. Chen, X. Yang, J. Fong-Chuen Loo, H. Zhang, F. Yin and Z. Li, Bioconjugate Chem., 2019, 30, 1658–1664 CAS.
- J. Liu, C. Zhang, T. W. Rees, L. Ke, L. Ji and H. Chao, Coord. Chem. Rev., 2018, 363, 17–28 CAS.
- M. Martínez-Alonso and G. Gasser, Ruthenium polypyridyl complex-containing bioconjugates, Coord. Chem. Rev., 2021, 434, 213736 Search PubMed.
- M. Jakubaszek, B. Goud, S. Ferrari and G. Gasser, Chem. Commun., 2018, 54, 13040–13059 RSC.
- J. Shum, P. K. K. Leung and K. K. W. Lo, Inorg. Chem., 2019, 58, 2231–2247 CAS.
- M. Lin, S. Zou, X. Liao, Y. Chen, D. Luo, L. Ji and H. Chao, Chem. Commun., 2021, 57, 4408–4411 RSC.
- J. Karges, S. Kuang, F. Maschietto, O. Blacque, I. Ciofini, H. Chao and G. Gasser, Nat. Commun., 2020, 11, 1–13 Search PubMed.
- S. Paul, P. Kundu, P. Kondaiah and A. R. Chakravarty, Inorg. Chem., 2021, 60, 16178–16193 CAS.
- Y. Chen, W. Li, Y. Yang, R. Zhong, H. Hu, C. Huang, J. Chen, L. Liang and Y. Liu, Eur. J. Med. Chem., 2023, 257, 115541 CrossRef CAS.
- S. Kuang, L. Sun, X. Zhang, X. Liao, T. W. Rees, L. Zeng, Y. Chen, X. Zhang, L. Ji and H. Chao, Angew. Chem., Int. Ed., 2020, 59, 20697–20703 CrossRef CAS.
- J. A. Roque, P. C. Barrett, H. D. Cole, L. M. Lifshits, G. Shi, S. Monro, D. Von Dohlen, S. Kim, N. Russo, G. Deep, C. G. Cameron, M. E. Alberto and S. A. McFarland, Chem. Sci., 2020, 11, 9784–9806 RSC.
- L. Zeng, P. Gupta, Y. Chen, E. Wang, L. Ji, H. Chao and Z. S. Chen, Chem. Soc. Rev., 2017, 46, 5771–5804 RSC.
- H. Huang, B. Yu, P. Zhang, J. Huang, Y. Chen, G. Gasser, L. Ji and H. Chao, Angew. Chem., Int. Ed., 2015, 54, 14049–14052 CrossRef CAS PubMed.
- J. Cervinka, A. Gobbo, L. Biancalana, L. Markova, V. Novohradsky, M. Guelfi, S. Zacchini, J. Kasparkova, V. Brabec and F. Marchetti, J. Med. Chem., 2022, 65, 10567–10587 CrossRef CAS PubMed.
- B. Demoro, A. Bento-Oliveira, F. Marques, J. C. Pessoa, L. Otero, D. Gambino, R. F. M. De Almeida and A. I. Tomaz, Molecules, 2019, 24, 2861 CrossRef CAS.
- C. B. Bo, Q. Bu, X. Li, G. Ma, D. Wei, C. Guo, B. Dai and N. Liu, J. Org. Chem., 2020, 85, 4324–4334 CrossRef CAS PubMed.
- S. K. Gupta and J. Choudhury, ChemCatChem, 2017, 9, 1979–1984 CrossRef CAS.
- R. Sole, M. Bortoluzzi, A. Spannenberg, S. Tin, V. Beghetto and J. G. De Vries, Dalton Trans., 2019, 48, 13580–13588 RSC.
- A. Gandioso, A. Vidal, P. Burckel, G. Gasser and E. Alessio, ChemBioChem, 2022, 23, e202200398 CAS.
- V. Criscuolo, C. T. Prontera, M. Pavone, O. Crescenzi, M. G. Maglione, P. Tassini, S. Lettieri, P. Maddalena, C. Borriello, C. Minarini and P. Manini, ACS Omega, 2019, 4, 2009–2018 CAS.
- Z. Adamski, L. L. Blythe, L. Milella and S. A. Bufo, Toxins, 2020, 12, 210 Search PubMed.
- A. Rajput, R. Sharma and R. Bharti, Mater. Today: Proc., 2022, 48, 1407–1415 CAS.
- Y. Chen, M. Y. Qin, J. H. Wu, L. Wang, H. Chao, L. N. Ji and A. L. Xu, Eur. J. Med. Chem., 2013, 70, 120–129 CrossRef CAS PubMed.
- Y. Chen, M. Y. Qin, L. Wang, H. Chao, L. N. Ji and A. L. Xu, Biochimie, 2013, 95, 2050–2059 CrossRef CAS PubMed.
- J. Chen, F. Peng, Y. Zhang, B. Li, J. She, X. Jie, Z. Zou, M. Chen and L. Chen, Eur. J. Med. Chem., 2017, 140, 104–117 CrossRef CAS PubMed.
- C. N. Honors, C. A. Kruger and H. Abrahamse, Technol. Cancer Res. Treat., 2018, 17, 1–15 Search PubMed.
- E. Ostanska, D. Aebisher and D. Bartusik-Aebisher, Biomed. Pharmacother., 2021, 137, 111302 CrossRef CAS.
- T. C. Motley, L. Troian-Gautier, M. K. Brennaman and G. J. Meyer, Inorg. Chem., 2017, 56, 13579–13592 CrossRef CAS.
- J. Lin, X. Chen, M. Sun, X. Qu, Y. Wang, C. Li, X. Li, L. Zhao, Z. Su and H. Ye, Oncol. Lett., 2021, 21, 247 CrossRef CAS PubMed.
- A. Mitra and R. Ghosh, Mol. Biol. Rep., 2021, 48, 5993–6005 CrossRef CAS PubMed.
- L. M. Lifshits, J. A. Roque III, P. Konda, S. Monro, H. D. Cole, D. von Dohlen, S. Kim, G. Deep, R. P. Thummel, C. G. Cameron, S. Gujar and S. A. McFarland, Chem. Sci., 2020, 11, 11740–11762 RSC.
- X. Wen, Z. Lin, B. Liu and Y. Wei, Cell Proliferation, 2012, 45, 217–224 CrossRef CAS PubMed.
- M. Fussenegger, J. E. Bailey and J. Varner, Nat. Biotechnol., 2000, 18, 768–774 CAS.
- C. P. Popolin, J. P. B. Reis, A. B. Becceneri, A. E. Graminha, M. A. P. Almeida, R. S. Corrêa, L. A. Colina-Vegas, J. Ellena, A. A. Batista and M. R. Cominetti, PLoS One, 2017, 12, e0183275 CrossRef PubMed.
- J. Caia, Q. Zheng, H. Huang and B. Li, Photodiagn. Photodyn. Ther., 2018, 21, 257–262 CrossRef.
- S.-H. Lai, W. Li, J.-H. Yao, B.-J. Han, G.-B. Jiang, C. Zhang, C.-C. Zeng and Y.-J. Liu, J. Photochem. Photobiol., B, 2016, 158, 39–48 CrossRef CAS PubMed.
- C. O. D'Sousa Costa, J. H. Araujo Neto, I. R. S. Baliza, R. B. Dias, L. d. F. Valverde, M. T. A. Vidal, C. B. S. Sales, C. A. G. Rocha, D. R. M. Moreira, M. B. P. Soares, A. A. Batista and D. P. Bezerra, Onco Targets Ther, 2017, 8, 104367–104392 Search PubMed.
- D.-Y. Zhang, Y. Zheng, C.-P. Tan, J.-H. Sun, W. Zhang, L.-N. Ji and Z.-W. Mao, ACS Appl. Mater. Interfaces, 2017, 9, 6761–6771 CrossRef CAS.
- G.-B. Jiang, W.-Y. Zhang, M. He, Y.-Y. Gu, L. Bai, Y.-J. Wang, Q.-Y. Yi and F. Du, Spectrochim. Acta, Part A, 2020, 227, 117534 CrossRef CAS PubMed.
- R. C. Gilson, K. C. L. Black, D. D. Lane and S. Achilefu, Angew. Chem., 2017, 129, 10857–10860 CrossRef.
- R. L. Lucaciu, A. C. Hangan, B. Sevastre and L. S. Oprean, Molecules, 2022, 27, 6485 Search PubMed.
- S.-Q. Zhang, T.-T. Meng, J. Li, F. Hong, J. Liu, Y. Wang, L.-H. Gao, H. Zhao and K.-Z. Wang, Inorg. Chem., 2019, 58, 14244–14259 Search PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4md00600c |
| ‡ G. S. and M. L. A. contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |