Open Access Article
Open Access ArticleGreen synthesis and anti-inflammatory properties of zinc oxide nanoparticles from Fe’i and Cavendish banana extracts
Nabilla Ghina
Zavitri
ad,
Alia Putri
Syahbaniati
ad,
Rindia M.
Putri
 c,
Fenny Martha
Dwivany
ab,
Indra
Wibowo
c,
Fenny Martha
Dwivany
ab,
Indra
Wibowo
 *ad,
Daniel
Pramudita
*ad,
Daniel
Pramudita
 *ef and
Antonius
Indarto
*ef and
Antonius
Indarto
 *eg
*eg
aBiotechnology Study Program, School of Life Sciences and Technology, Institut Teknologi Bandung, Bandung, Indonesia. E-mail: indra.wibowo@itb.ac.id
bGenetics and Biotechnology Molecular Research Group, School of Life Sciences and Technology, Institut Teknologi Bandung, Bandung, Indonesia
cBiochemistry Division, Faculty of Mathematics and Natural Sciences, Institut Teknologi Bandung, Indonesia
dAnimal Development, Physiology, and Biomedical Science Research Group, School of Life Sciences and Technology, Institut Teknologi Bandung, Bandung, Indonesia
eDepartment of Chemical Engineering, Institut Teknologi Bandung, Bandung, Indonesia. E-mail: daniel.pramudita@itb.ac.id; antonius.indarto@itb.ac.id
fDepartment of Food Engineering, Institut Teknologi Bandung, Bandung, Indonesia
gDepartment of Bioenergy Engineering and Chemurgy, Institut Teknologi Bandung, Bandung, Indonesia
First published on 10th October 2025
Abstract
This study reports plant-assisted green synthesis of zinc oxide nanoparticles (ZnO NPs) using aqueous and methanolic extracts of Fe’i (Musa troglodytarum L.) and Cavendish (Musa acuminata) bananas, and evaluates their in vivo anti-inflammatory activity in zebrafish larvae. The extracts were qualitatively profiled and used for the synthesis of ZnO NPs, which were then characterized by UV-vis diffuse reflectance spectroscopy, X-ray diffraction (XRD), Fourier-transform infrared spectroscopy, scanning electron microscopy, and particle-size and zeta-potential measurements. Band-gap energies and structural parameters (crystallite size, lattice constants, and microstrain) were derived from optical and XRD data. All syntheses yielded wurtzite-phase ZnO with absorption maxima at 364–372 nm, band gaps of ∼3.28–3.41 eV, crystallite sizes of ∼14–15 nm, hydrodynamic sizes of 134–689 nm, and negative surface charges of −29.6 to −42.4 mV; SEM revealed flower-like, spherical-like, and snowflake morphologies. Acute toxicity testing in zebrafish gave LC50 values (mg L−1) of 5.65 (PTLA), 16.28 (PTLM), 9.47 (PCVA), and 8.17 (PCVM); LC25 was applied in efficacy assays. In a caudal-fin amputation model, treatment with all ZnO NPs significantly reduced neutrophil counts relative to the negative control (P ≤ 0.05), similarly reduced macrophage recruitment (comparable to dexamethasone), and diminished 48-h fin regrowth (P ≤ 0.05). In conclusion, banana-derived extracts afforded reproducible ZnO NPs with distinct physicochemical signatures and promising anti-inflammatory effects. Although the processing parameters were not fully optimized, these results justify further mechanistic studies and process intensification toward scalable biomedical applications.
Introduction
Zinc oxide nanoparticles (ZnO NPs) are widely studied because they combine useful optical, catalytic, and biomedical properties with relatively straightforward synthesis routes.1 Plant-assisted “green” synthesis provides a less-hazardous alternative using phytochemicals as reducing and stabilizing agents while allowing control over size, crystallinity, and surface chemistry through processing variables such as precursor concentration, pH, temperature, and time.1,2 Recent reviews emphasize that reporting band-gap energy, crystal structure, and particle statistics alongside synthesis conditions is essential for meaningful comparison across studies.1,3Banana-derived extracts have emerged as practical media for ZnO NP preparation because their polyphenols, carbohydrates, and pectins can act as reductants and capping ligands.4,5 Contemporary studies show that banana-extract-mediated ZnO NPs display expected wurtzite structures and application-relevant performance, supporting banana matrices as a robust green platform.4,6 Process-oriented reports indicate that extract composition and basic parameters can tune particle size and stability in these systems, underscoring the value of systematic comparisons.5,6
Fe’i banana (Musa troglodytarum) is notable for a distinct metabolite profile and documented differences from Cavendish that may influence reduction and capping behavior during synthesis.7–10 Genomic and biochemical evidence highlights non-climacteric behavior and carotenoid enrichment relative to Cavendish, suggesting a different phytochemical environment for nanoparticle formation.7 Yet Fe’i remains underexplored as a template for ZnO synthesis compared with common cultivars, creating an opportunity to test species-specific effects on material properties.1,7–10
Few comparative studies evaluate Fe’i versus Cavendish across aqueous and methanolic extracts while linking extract chemistry to ZnO properties and bioactivity, leaving a practical design gap.1,7–10 Meanwhile, studies show that ZnO NPs can modulate inflammatory pathways, including NF-κB, which motivates in vivo anti-inflammatory evaluation using rapid vertebrate models. However, direct side-by-side in vivo comparisons of green-synthesized ZnO from different banana sources remain limited, especially when standardized zebrafish assays are required for benchmarking.11–13
This study investigates the green synthesis of ZnO nanoparticles using the fruit and peel extracts of Fe’i and Cavendish under both aqueous and methanolic conditions. Fe’i banana extracts, which are underexplored in nanomaterials research, are systematically evaluated alongside Cavendish extracts within a single, matched design.7–10 This study characterizes nanoparticles’ structural, morphological, and optical properties, and relates these features to the phytochemical compositions of the extracts. Standardized methods and harmonized reporting are applied to ensure robust and meaningful comparison across species and solvents.1,3 To assess biological relevance, the study integrates physicochemical characterization with in vivo zebrafish assays. Specifically, caudal fin amputation is employed to quantify neutrophil and macrophage recruitment at the injury site, thereby linking extract composition and nanoparticle properties to acute inflammatory responses.11–16 This controlled, matched, side-by-side framework provides species- and solvent-dependent insights into the synthesis–property–bioactivity relationship of green-synthesized ZnO nanoparticles.1,4
Experimental
Preparation of plant extracts
Two solvents, namely distilled water and methanol (ACS grade, cat. no. 6501-04, CAS no. 67-56-1; Anhui Fulltime Specialized Solvent & Reagent Co., Ltd. China), were used to obtain Fe’i and Cavendish banana extracts, following Zavitri et al.17 with some modifications. Fe’i bananas (Musa troglodytarum L.) were sourced from Tasikmalaya Regency, West Java, Indonesia, whereas Cavendish bananas (Musa acuminata, Cavendish subgroup) were purchased from a local commercial supermarket prior to extraction. In aqueous extraction, banana peels were separated from the fruit, washed with distilled water, and then dried at room temperature. We sourced bananas with uniform ripeness (ripe), judged by external colour and pulp firmness for each cultivar, and processed the peel and pulp as specified. About 150 mL distilled water was poured into a 75 g mixture of banana peels and fruit (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The diluted mixture was heated at 80 °C for 120 minutes and then filtered with a Buchner funnel using Whatman filter paper no. 1 three times to separate the solid residue. The crude extract was stored at 4 °C for further experiments. In methanol extraction, the dried banana peels and fruits, about 10 g each (20 g in total), were ground into a soft powder and mixed with 200 mL methanol. The mixture was placed on an orbital shaker for 72 hours at room temperature and similarly filtered to remove the solid residue. The liquid was evaporated using a vacuum rotary evaporator at 40 °C. The crude extract was kept at 4 °C for further processing. From this point onward, the aqueous and methanolic extracts of Fe’i (Tongka Langit) banana are referred to as PTLA and PTLM, respectively. Similarly, those of Cavendish banana are referred to as PCVA and PCVM, respectively. Bioactive compounds from all extracts were identified using qualitative tests to detect phenolics, flavonoids, tannins, saponins, triterpenoids, steroids, and alkaloids. Gas chromatography–mass spectrometry (GC–MS) analysis was conducted for more detailed identification.
1). The diluted mixture was heated at 80 °C for 120 minutes and then filtered with a Buchner funnel using Whatman filter paper no. 1 three times to separate the solid residue. The crude extract was stored at 4 °C for further experiments. In methanol extraction, the dried banana peels and fruits, about 10 g each (20 g in total), were ground into a soft powder and mixed with 200 mL methanol. The mixture was placed on an orbital shaker for 72 hours at room temperature and similarly filtered to remove the solid residue. The liquid was evaporated using a vacuum rotary evaporator at 40 °C. The crude extract was kept at 4 °C for further processing. From this point onward, the aqueous and methanolic extracts of Fe’i (Tongka Langit) banana are referred to as PTLA and PTLM, respectively. Similarly, those of Cavendish banana are referred to as PCVA and PCVM, respectively. Bioactive compounds from all extracts were identified using qualitative tests to detect phenolics, flavonoids, tannins, saponins, triterpenoids, steroids, and alkaloids. Gas chromatography–mass spectrometry (GC–MS) analysis was conducted for more detailed identification.
Green synthesis of ZnO NPs
The green synthesis of ZnO NPs from the prepared banana extracts was conducted following Zavitri et al.17 with some modifications. Approximately 20 mL of the banana crude extract was diluted in 80 mL of distilled water. Subsequently, 6.42 mg of zinc nitrate in hydrate form (Zn(NO3)2·6H2O, Sigma-Aldrich; Merck KGaA, USA; catalog no. 228737-100G) was added and agitated using a magnetic stirrer for 10 minutes. The pH was adjusted to 12 by adding a 5 M solution of sodium hydroxide (NaOH). The suspension underwent oven drying at 60 °C for 1 hour or until a discernible white precipitate formed. After precipitation, the suspension was washed with a mixture of distilled water and ethanol (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) subsequent to decanting of the supernatant. The precipitate was then centrifuged at 4025 × g for 20 minutes at room temperature and incubated for 24 hours in an oven at 60 °C. Thereafter, the solid was subjected to furnace treatment at 400 °C for 2 hours in a crucible cup, yielding white powder comprising NPs. The synthesized product was safely stored in a hermetic tube for subsequent testing and characterization.
1) subsequent to decanting of the supernatant. The precipitate was then centrifuged at 4025 × g for 20 minutes at room temperature and incubated for 24 hours in an oven at 60 °C. Thereafter, the solid was subjected to furnace treatment at 400 °C for 2 hours in a crucible cup, yielding white powder comprising NPs. The synthesized product was safely stored in a hermetic tube for subsequent testing and characterization.
Characterization of ZnO NPs
The structure and size of the synthesized ZnO NPs were measured using SEM analysis (JSM 6510 LA, JEOL Ltd). Gold (99.9%) coating with 90-second sputtering at room temperature was performed to prepare the samples. Particle size distributions were determined from the SEM images using ImageJ software version 1.53t (ImageJ.org, USA). Meanwhile, the size distributions of the dissolved ZnO NPs were measured using a particle size analyzer (PSA). Zeta potential values, which indicate the surface charge of the synthesized ZnO NPs, were determined using a zeta potential analyzer (Horiba SZ-100). The optical absorption spectra of the ZnO NPs were acquired from a UV-visible DRS (UV-vis Thermo Scientific Evolution 2) within the wavelength range of 200–800 nm. Reflectance R was converted to the Kubelka–Munk function: F(R) = (1 − R)2/(2R). The direct band-gap of ZnO was estimated from the Tauc relation (F(R)hv)2vs. hv with the linear region extrapolated to the energy axis to obtain Eg, which is related to the edge wavelength λedge as Eg [eV] ≈ 1240/λedge [nm].18 XRD patterns were recorded on a D8 Advance diffractometer (Bruker AXS GmbH, Germany) using Cu-Kα radiation (λ = 1.5406 Å, 40 kV, 40 mA) at a scan rate of 1° min−1 over the 2θ range of 20–80°. The diffraction data were analyzed using OriginLab 2023b (OriginLab, USA) to determine the crystal structure and average crystallite size. The crystallite size was first estimated using the Scherrer equation: D = 0.94λ/β![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) cos
cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ, where D is the crystallite size, λ the X-ray wavelength, β the full width at half maximum, and θ is the Bragg angle. Interplanar spacings (d) were calculated using Bragg's law, while lattice constants were determined from the (100) and (002) reflections (a = 2d100/√3; c = 2d002), and the unit-cell volume was calculated as V = (√3/2)a2c. Microstrain (ε) and the crystallite size were further refined using Williamson–Hall (W–H) analysis in the uniform deformation model (UDM), where β
θ, where D is the crystallite size, λ the X-ray wavelength, β the full width at half maximum, and θ is the Bragg angle. Interplanar spacings (d) were calculated using Bragg's law, while lattice constants were determined from the (100) and (002) reflections (a = 2d100/√3; c = 2d002), and the unit-cell volume was calculated as V = (√3/2)a2c. Microstrain (ε) and the crystallite size were further refined using Williamson–Hall (W–H) analysis in the uniform deformation model (UDM), where β![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) cos
cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ was plotted against 4sin
θ was plotted against 4sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ; ε was obtained from the slope and D from the intercept. Dislocation density (δ) was estimated as δ = 1/D2.19,20 Additionally, FTIR analysis (Shimadzu Corporation; Prestige 21) at room temperature from 400 to 4000 cm−1 was conducted to identify functional groups and compound classes within both the banana extracts and the synthesized ZnO NPs.
θ; ε was obtained from the slope and D from the intercept. Dislocation density (δ) was estimated as δ = 1/D2.19,20 Additionally, FTIR analysis (Shimadzu Corporation; Prestige 21) at room temperature from 400 to 4000 cm−1 was conducted to identify functional groups and compound classes within both the banana extracts and the synthesized ZnO NPs.
Anti-inflammatory assays on zebrafish
The synthesized ZnO NPs with lethal concentrations LC25 and LC50 underwent toxicity tests in zebrafish larvae, according to the OECD 2013 protocol.21 Inflammation activity was assessed using the method reported by Sharif et al.22 with some modifications, which involved a caudal fin amputation procedure on zebrafish larvae. Adult wild-type zebrafish (Danio rerio) used for breeding and obtaining embryos/larvae were procured from Sifa Aquarium, Bogor, West Java, Indonesia. Viable embryos obtained through breeding were allowed to mature until one day post-fertilization (dpf) in E3 1× medium containing 0.003% PTU. Sixty zebrafish embryos (n = 30 per group, duplicate) were exposed to the synthesized ZnO NP suspension in a six-well plate (6 mL per well) for 6 h. The suspension concentration was set at the measured LC25, as previously applied in zebrafish embryo studies,23 to induce measurable biological responses while maintaining sufficient viability for analysis. Subsequently, the embryos were rinsed with E3 1× medium + 0.003% PTU and transferred to a fresh six-well plate for 48 hours. Dexamethasone (200 μM; Sigma-Aldrich, USA), a widely recognised anti-inflammatory drug, was utilized as the positive control and served as the reference compound to evaluate the anti-inflammatory properties of the ZnO NPs. E3 1× medium + 0.003% PTU without any additional compounds served as the negative control. Caudal fin amputation of each zebrafish larva (3–4 dpf) was conducted using a sterile surgical blade no. 13 (Onemed Bisturi, Indonesia) under a stereo microscope (Carl Zeiss, Germany) after being anaesthetised with 0.04% tricaine solution. Neutrophils and macrophages were evaluated at two time points (4 and 48 hours post-amputation, hpa) using Sudan black and neutral red staining, respectively, under a Zeiss stereo microscope and quantified using ImageJ. All experimental procedures received ethical approval from the Research Ethics Commission of Padjadjaran University (approval no. 1026/UN6.KEP/EC/2022).Statistical analysis
All quantitative results are reported as mean ± SD, with n stated in figure captions. Group differences were evaluated by one-way ANOVA (followed, where applicable, by Tukey's post hoc test). Unpaired Student's t-tests were used for pre-specified two-group contrasts. Statistical significance was set at α = 0.05 (two-tailed). Analyses were performed using GraphPad Prism v9.5.1.Results and discussion
Bioactive compound analysis in plant extracts
The results of phytochemical compound identification are tabulated in Table 1. It is evident that all extracts yield flavonoid and triterpenoid compounds. Additionally, phenolic compounds and tannins are detected in all extracts, except PTLM. Quantification using GC–MS (Fig. 1) showed that methanol extraction in general resulted in more bioactive compounds in the extracts compared to aqueous extraction. This may be due to the differences in polarity, such that different bioactive compounds were successfully dissolved during the extraction process. Additionally, Cavendish banana may contain higher quantities of bioactive compounds that are more soluble in methanol than in water.| Phyto-chemicals | Reactants (methods) | PTLA | PTLM | PCVA | PCVM |
|---|---|---|---|---|---|
| Note: PTLA: aqueous extract of Fe’i (Tongka Langit) banana, PTLM: methanolic extract of Fe’i banana, PCVA: aqueous extract of Cavendish banana, PCVM: methanolic extract of Cavendish banana. −: Not detected/not found. +: Detected in very small amounts (few). ++: Detected in moderate amounts (several). +++: Detected in high amounts (many/abundant). *Main secondary metabolites in the synthesis of ZnO NPs. | |||||
| Phenols* | Ferric chloride 5% | + | − | + | + |
| Tannins | Ferric chloride 1% | + | − | + | + |
| Saponins | Thermal | ++ | − | ++ | − |
| Flavonoids* | HCl + Mg | − | − | − | − |
| 2N H2SO4 | − | − | − | − | |
| 10% NaOH | + | + | + | + | |
| Triterpenoids | H2SO4 + CH3COOH anhydrate | +++ | + | ++ | ++ |
| Steroids | − | − | − | − | |
| Alkaloids* | Dragendorff's reagent | − | − | − | − |
Fe’i and Cavendish bananas show promise in nanoparticle synthesis due to their phytochemical composition. Extracts from both varieties contain compounds like polyphenols, terpenoids, flavonoids, sugars, and proteins, which can act as agents for converting metal salts into metal NPs. Studies have shown that both Fe’i and Cavendish banana extracts, when used in water or methanol solvents, contribute to the synthesis of NPs, with specific compounds such as flavonoids, tannins, alkaloids, glycosides, phenols, and pectins playing crucial roles in the process.24
ZnO NP characterization
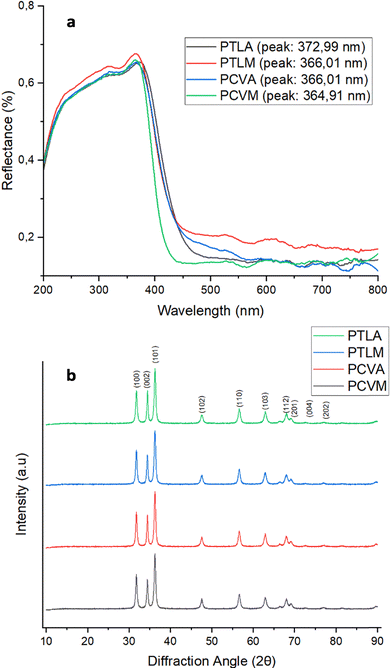
The characterization results of ZnO NPs using UV-vis DRS and XRD are presented in Fig. 2. The UV-vis DRS measurements revealed peaks in the wavelength range of 364–372 nm (Fig. 2a). This range is within the typical range for ZnO NPs (310–380 nm), which validates the successful synthesis of these NPs.25 Further analyzing the reflectance data in Fig. 2 using the Kubelka–Munk function and Tauc plot, the ZnO NPs display absorption edges at 386–414 nm, resulting in optical band-gap energies (Eg, in eV) of 3.179 (PVCA), 3.212 (PTLM), 3.139 (PVCM), and 2.995 (PTLA), respectively. These band-gap values fall within the typical range reported for plant-mediated ZnO nanoparticles and are close to the canonical bulk ZnO band gap (∼3.37 eV).1,25–30 Reports on banana-derived ZnO further show that extract chemistry and morphology can shift the apparent gap, with values spanning ∼3.0–3.6 eV depending on the synthesis method and nanostructure.1,26–28The results of XRD analysis (Fig. 2b) show that the crystals produced are 14–15 nm in size and have a hexagonal wurtzite shape. XRD analysis confirmed a hexagonal wurtzite crystal structure with an average diameter of 14–15 nm and identified crystal lattice planes including (100), (002), (101), (102), (110), (103), (200), and (202). Sharp peaks on (100), (002), and (101) lattice planes indicated optimal synthesis, with good peak separation suggesting the absence of intermediate complexes. The refined structural parameters of the biosynthesized ZnO are internally consistent across extracts. The lattice constants were a = 3.247–3.249 Å and c = 5.201–5.205 Å (V ≈ 47.55 Å3), matching standard wurtzite ZnO. The Debye–Scherrer crystallite sizes from (100)/(002)/(101) ranged from 17.2 to 18.6 nm; W–H analysis yielded D = 18.9–26.7 nm and ε = (1.1–2.6) × 10−3. The slightly larger D from W–H reflects the removal of strain broadening, as expected for green-synthesized ZnO. The corresponding dislocation density δ was (2.88–3.38) × 1015 m−2.19,20 The crystallite size obtained from XRD data (14–15 nm) represents the dimension of a single crystalline domain. In contrast, the larger sizes obtained from SEM and PSA correspond to secondary particles formed by the aggregation of several crystallites.31–34
Table 2 shows the measured properties of the dissolved NPs. It is shown that the particle sizes are in the range of 134–689 nm. The NPs obtained from methanolic extracts are smaller than those obtained from aqueous extracts. Additionally, the Fe’i banana extracts resulted in bigger NPs. The polydispersity index is the ratio between the weight average molecular weight and the number average molecular weight. It is thus positively correlated to the broadness of molecular weight distribution. The values indicate that the NPs obtained from the methanolic extract are more homogeneous, although still considerably polydisperse. From the zeta potentials, all the synthesized ZnO NPs showed moderate to good stability with the absolute values ranging from about −30 to −60 mV.32 Particle size analysis indicated average dissolved diameters of ∼100–700 nm with PI values of 0.35–0.75, consistent with prior banana extract syntheses, yet indicative of polydispersity likely driven by aggregation.
| No. | ZnO NPs | Mean (nm) | PI | Zeta potential (mV) |
|---|---|---|---|---|
| 1 | PTLA | 689.30 | 0.633 | −42.47 |
| 2 | PTLM | 249.80 | 0.356 | −29.53 |
| 3 | PCVA | 370.67 | 0.742 | −41.67 |
| 4 | PCVM | 134.07 | 0.524 | −32.63 |
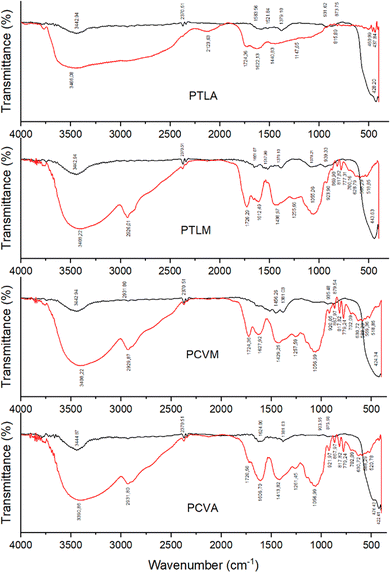
The results of FTIR analysis are shown in Fig. 3. The compounds predominantly found in the synthesized ZnO NPs are pectin, phenolic compounds, proteins, and carbohydrates. The broad peaks at 3442.94 cm−1 and 3444.87 cm−1 indicate the presence of O–H stretching and hydrogen bonded groups in alcohol. The presence of carboxylic groups from phenols or pectins in the extract is shown by the peak at 2931.80 cm−1. The peaks at 1651.07 cm−1 and 1624.06 cm−1 are associated with the –CQC stretching vibration of alkynes, while the peaks at 1521.84 and 1517.98 cm−1 indicate aromatic hydrocarbon nitro groups associated with protein. The observed peak from 873 up to 939 cm−1 indicates C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bending and C–H stretching vibrations from alkene and alkyne groups in carbohydrates. FTIR functional group identification results indicate the compounds present in each banana extract. The main compounds identified in the four synthesized ZnO NPs are pectin, phenolic compounds, protein, and carbohydrates, consistent with the results of phytochemical and GC–MS tests conducted on the extracts prior to synthesis. These biomolecules served as reducing, stabilizing, and capping agents in the synthesis of ZnO NPs. In the formation of ZnO NPs using zinc nitrate hexahydrate as the precursor, Zn2+ ions react with OH− groups provided by the phytochemical constituents to form zinc hydroxide [Zn(OH)2]. Subsequent transformation of these ions into ZnO leads to the nucleation and growth of ZnO NP crystals.18,33 All the characterization results mentioned above for the ZnO NPs synthesized in this study were similar to previous studies on the synthesis of ZnO NPs using banana peel extracts.24,27,31
C bending and C–H stretching vibrations from alkene and alkyne groups in carbohydrates. FTIR functional group identification results indicate the compounds present in each banana extract. The main compounds identified in the four synthesized ZnO NPs are pectin, phenolic compounds, protein, and carbohydrates, consistent with the results of phytochemical and GC–MS tests conducted on the extracts prior to synthesis. These biomolecules served as reducing, stabilizing, and capping agents in the synthesis of ZnO NPs. In the formation of ZnO NPs using zinc nitrate hexahydrate as the precursor, Zn2+ ions react with OH− groups provided by the phytochemical constituents to form zinc hydroxide [Zn(OH)2]. Subsequent transformation of these ions into ZnO leads to the nucleation and growth of ZnO NP crystals.18,33 All the characterization results mentioned above for the ZnO NPs synthesized in this study were similar to previous studies on the synthesis of ZnO NPs using banana peel extracts.24,27,31
The particle diameters were measured from representative SEM images using ImageJ, and the average values were calculated to obtain the mean particle size. SEM images showing the NP morphologies are presented in Fig. 4. ZnO NPs synthesized from aqueous extracts (PTLA and PVCA) exhibit flower/raspberry shapes, with average particle sizes of 301.23 nm and 192.10 nm, respectively. Methanolic extracts, on the other hand, yielded different morphologies: PTLM gave spherical particles, whereas PCVM resulted in a snowflake shape. PTLM NPs can be further classified into small particles (with an average diameter of 61.13 nm) and large-sized particles (587.77 nm). The PCVM flakes have an average length of 272.27 nm and a width of 43.55 nm.
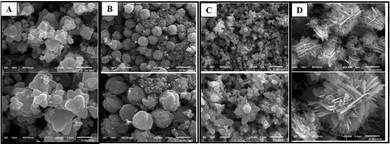 | ||
| Fig. 4 SEM images of the synthesized ZnO NPs: PTLA (A), PTLM (B), PCVA (C), and PCVM (D) at 1000 nm (top) and 500 nm (bottom) scales. | ||
In plant-mediated routes, pH, reaction temperature/duration, precursor concentration, and extract concentration are the primary determinants of size and morphology.1,3,24,25 Here, pH 12 was used to deprotonate polyphenols/carboxylates in the extract and shift Zn2+ speciation to zincate [Zn(OH)4]2−, thereby accelerating hydrolysis to Zn(OH)2/ZnO and increasing nucleation density.26,35,36 A sub-boiling temperature of 60 °C was chosen to promote reaction kinetics while preserving phytochemical reducing/capping functions; excessive heating risks ligand degradation and uncontrolled growth.26,36,37 A 1-h hold provided sufficient time for conversion under these kinetics without encouraging secondary growth or aggregation, which becomes more likely at longer durations.36,37 Finally, a conservative precursor concentration was selected to avoid supersaturation spikes that drive rapid growth and polydispersity; higher loadings typically yield larger, more aggregated particles.31,38 These literature-based settings enabled reproducible formation in our system; formal optimization of each factor was beyond the scope of the present study.35–37
Among the four systems, methanolic extracts yielded (i) slightly smaller hydrodynamic sizes, (ii) narrower PDIs, and (iii) more negative ζ-potentials than aqueous extracts, indicating more effective nucleation control and colloidal stabilisation. These trends align with the higher abundance of capping-competent metabolites (e.g., polyphenols/pectins) in methanolic extracts and with solvent-polarity effects on ligand adsorption.2 The microstructural metrics from XRD (ε, δ) remained comparable among batches, suggesting that the major differences arise from surface rather than bulk lattice effects.24,31
Toxicity testing of ZnO NPs in zebrafish
The toxicity testing of the green synthesized ZnO NPs involved a range finding test and a definitive test. In the range finding test, zebrafish embryos showed 100% mortality at a 100 mg L−1 concentration of all tested ZnO NP suspensions, while no mortality was observed at 0.01 mg L−1. The concentration ranges used in the definitive test are based on the results of the range finding test, following the OECD guidelines.21 A wider range of concentrations will be employed in the definitive test if mortality is detected at higher concentrations in the range finding test. PCVA showed 15% mortality at 10 mg L−1, leading to a concentration range for definitive test of 80 mg L−1 to 5 mg L−1. The other ZnO NPs showed various mortality percentages at different concentrations, resulting in concentration ranges for the definitive test between 20 mg L−1 and 1.25 mg L−1.The lethal concentration values obtained by probit analysis are given in Table 3. PTLM NPs fall into the hazardous category (LC50: 10–100 mg L−1), whereas PTLA, PCVA, and PCVM fall into the toxic category (LC50: 1–10 mg L−1) according to OECD guidelines. ZnO NPs, characterized by higher solubility, release Zn2+ ions, affecting zebrafish cells.1 Exposure affects reactive oxygen species (ROS) production and antioxidant capacity, causing oxidative damage.39 Dysfunctional gene transcription related to growth hormones may induce zebrafish abnormalities.40 Agglomeration of commercial ZnO NPs (20–30 nm) with an LC25 of 2.64 mg L−1 into larger particles in deionized water has been reported previously.23
| No. | ZnO NPs | LC25 (mg L−1) | LC50 (mg L−1) |
|---|---|---|---|
| 1 | PTLA | 3.64 | 5.66 |
| 2 | PTLM | 2.14 | 16.29 |
| 3 | PCVA | 4.09 | 9.47 |
| 4 | PCVM | 5.17 | 8.18 |
Dispersion factors, affected by size and surface charge, influence toxicity.41 The observed particle size of the synthesized ZnO NPs (134–689 nm) reflects polydispersity, which is known to influence biological responses. It is well documented that smaller ZnO NPs generally exhibit higher bioavailability and ROS induction, while larger particles tend to show reduced toxicity. Such heterogeneity may therefore provide a spectrum of biological effects relevant for biomedical applications.42 Embryo chorion pore sizes allow nanoparticle penetration, affecting embryos' oxygen exchange.43 Polydisperse ZnO NPs with large sizes may thus contribute to increased toxicity. Despite this, the LC25 values still indicate the low toxicity effect while allowing for the exploration of potential anti-inflammatory effects.23 As reported in a previous study,44 ZnO NPs at sublethal levels (LC10) exhibited minimal developmental toxicity while affecting gene expression in zebrafish. It is therefore recommended that future research be conducted at sub-toxic concentrations (e.g., LC10) in order to verify the anti-inflammatory effects independently of general toxicity.
In vivo study of the anti-inflammatory potential of the synthesized ZnO NPs in a zebrafish larvae model
Neutrophil and macrophage observations were carried out at 4 and 48 hours post-caudal fin amputation (hpa) to study the inflammatory response and fin regeneration (see Fig. 5). At 4 hours, neutrophil counts were higher than macrophage counts. Treatment with ZnO NPs significantly reduced neutrophil counts (PTLA ZnO NPs: 19.93 ± 4.68; PTLM ZnO NPs: 24.26 ± 5.8; PCVA ZnO NPs: 20.06 ± 5; and PCVM ZnO NPs: 23.46 ± 4.86) compared to the negative control (30.667 ± 5.41). The obtained values were close to that of the positive control, dexamethasone, which was 23.86 ± 5.59. This trend was also observed in macrophage counts for all ZnO NPs. At 48 hpa, further decreases were observed, although for macrophage counts the magnitudes were comparably lower than those of neutrophil counts. Compared to the results at 4 hpa, the overall trend was the same: the neutrophil and macrophage counts were close to the positive controls and lower than the negative controls.The regenerated caudal fin area was measured at 48 hpa due to the early innate immune response in zebrafish larvae. As shown in Fig. 6, treatment with ZnO NPs resulted in smaller areas (PTLA: 0.037 ± 0.01 mm2; PTLM: 0.038 ± 0.011 mm2; PCVM: 0.038 ± 0.008 mm2; and PCVA: 0.038 ± 0.011 mm2) than that of the negative control (0.051 ± 0.009 mm2). As observed for the neutrophil and macrophage counts, the regenerated areas were also similar to that of the positive control (0.038 ± 0.011 mm2).
The synthesized ZnO NPs exhibit anti-inflammatory effects akin to dexamethasone, indicated by the lower counts of immune cells (neutrophils and macrophages) and improved regeneration area. Similar findings have been reported for other green-synthesized ZnO nanomaterials, such as β-chitosan–ZnO and ZnO–cinnamic acid nanoparticles, which accelerated wound healing and down-regulated pro-inflammatory cytokines (IL-6, TNF-α, and IL-1β), further supporting the anti-inflammatory potential of ZnO NPs in vivo.45 ZnO NPs may attenuate neutrophil migration by reducing nitric oxide (NO) production and suppressing NF-κB activation, a transcription factor that regulates multiple pro-inflammatory cytokines. Consistent with this, ZnO NPs were shown to suppress NO production and inhibit NF-κB signaling in LPS-induced RAW 264.7 macrophages.46 Moreover, Zn2+ ions released from ZnO NPs can downregulate NF-κB signaling via induction of the zinc transporter ZIP8.47 Together, these mechanisms support our observation that the synthesized ZnO NPs reduce neutrophil recruitment, potentially through NF-κB-mediated inflammatory pathways. Additionally, ZnO NPs inhibit pro-inflammatory cytokines in macrophages, crucial for wound healing.48 Macrophages, a source of important inflammatory factors, play a critical role in tissue regeneration. Dexamethasone affects macrophage composition and delays tissue regeneration.22 ZnO NPs may similarly affect fin regeneration by inhibiting macrophage recruitment.49
Study limitations and future directions
This study has defined scope limitations, non-standardized extracts, fixed synthesis conditions, limited molecular readouts, and no formal scale-up trial that are routine to address and do not undermine our core findings (successful green synthesis of crystalline ZnO NPs with consistent XRD signatures and measurable bioactivity). Each limitation translates into a concrete, tractable improvement: (i) extract standardization (e.g., total phenolics/HPLC profiling) to narrow polydispersity; (ii) factorial optimization of precursor concentration, pH, temperature, and time to target PDI ≤ 0.2 and reconcile crystallite particle size gaps (with BET/aggregation controls); (iii) mechanistic assays (NO, cytokines, NF-κB) and Zn2+ partitioning (ICP-OES) to decouple ion- versus particle-driven effects; and (iv) a pilot scale-up using stirred-tank or continuous flow precipitation with in-line QA specifications. These steps constitute a clear roadmap for follow-up work and technology maturation.The aqueous, low-temperature, ambient-pressure route is intrinsically amenable to scale-up; however, successful translation to large-scale production requires proactive management of several predictable challenges. The most important is extract variability: natural fluctuations in phytochemical composition can shift nucleation and growth kinetics and widen particle-size distributions. This can be mitigated by standardizing extracts (for example, total phenolics or high-performance liquid chromatography fingerprints) and implementing closed-loop control of pH, temperature, and residence time. At higher volumes, micromixing and supersaturation control become critical; maintaining comparable power input per unit volume and residence-time distribution via stirred-tank cascades or continuous-flow mixers, supported by process analytical technology (for example, in-line turbidity or ultraviolet–visible spectroscopy and at-line dynamic light scattering and X-ray diffraction), will help maintain phase purity, average size/polydispersity index, and zeta potential within specifications. Bioorganic load and fouling may complicate reactor hygiene and downstream filtration/drying; practical mitigation strategies include extract clarification, low-temperature drying, and optional post-calcination. Environmental and cost considerations, notably zinc recovery from effluents and sodium hydroxide neutralization, should be addressed through routine wastewater treatment with zinc capture and reuse to improve material efficiency. For applications that require tighter regulatory control, Good Manufacturing Practice expectations, covering batch records, predefined acceptance criteria, change control, and stability protocols, must be planned from the outset. Taken together, these risks are tractable with standard chemical-engineering measures (extract standardization, design of experiments-guided operating windows, process analytical technology, and staged pilots ranging from litre to tens-of-litres scale), rendering the method scalable in principle and providing a clear agenda for optimization and industrial translation.
Conclusions
The green synthesis of ZnO NPs from aqueous and methanol extracts of Fe’i and Cavendish bananas was successfully achieved, with each exhibiting distinct characteristics corresponding to the banana species and solvent used. Anti-inflammatory tests conducted on zebrafish larvae showed that all synthesized ZnO NPs exhibited inhibitory effects on macrophage and neutrophil migration, as well as on the regeneration area at the zebrafish larval fin wound site, similar to the positive control (dexamethasone). This study highlights the potential of Fe’i and Cavendish banana extracts as agents for green synthesis of ZnO NPs with potential anti-inflammatory properties.Author contributions
N. G. Z.: investigation, data curation, formal analysis, validation, writing – original draft, writing – review & editing, visualization; A. P. S.: investigation, data curation, formal analysis, validation, writing – review & editing; R. M. P.: formal analysis, validation, resources, supervision; F. M. D.: investigation, formal analysis, validation; I. W.: conceptualization, supervision, validation, resources, funding acquisition, project administration; D. P.: visualization, formal analysis, writing – original draft, writing – review & editing; A. I.: conceptualization, supervision, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Data availability
Data for this article, including figures, are available from the Science Data Bank at https://doi.org/10.57760/sciencedb.28143.Acknowledgements
We thank the Research, Community Service, and Innovation Program supporting the Physiology, Animal Development, and Biomedical Science Research Group, School of Life Sciences and Technology, Institut Teknologi Bandung, for funding this study (Grant No. 52.A/IT1.C11.SK-PL/2022).References
- M. Y. Al-darwesh, S. S. Ibrahim and M. A. Mohammed, Results Chem., 2024, 7, 101368 CrossRef CAS.
- A. Alnehia, A. Al-Sharabi, A.-B. Al-Odayni, A. H. Al-Hammadi, F. H. AL-Ostoot, W. S. Saeed, N. A. Y. Abduh and A. Alrahlah, Bioinorg. Chem. Appl., 2023, 2023, 1–11 CrossRef PubMed.
- S. A. Hassanzadeh-Tabrizi, J. Alloys Compd., 2023, 968, 171914 CrossRef CAS.
- M. Q. Al-Khaial, S. Y. Chan, R. A. Abu-Zurayk and N. Alnairat, Inorganics, 2024, 12, 121 CrossRef CAS.
- K. S. Islam, Md. H. Mustak, Md. K. H. Shishir, Md. M. Karim and G. Md. A. Khan, South Afr. J. Chem. Eng., 2025, 52, 127–140 CrossRef.
- D. Jovanović, S. Bognár, V. Despotović, N. Finčur, S. Jakšić, P. Putnik, C. Deák, G. Kozma, B. Kordić and D. Šojić Merkulov, Foods, 2024, 13, 2643 CrossRef PubMed.
- Z. Li, J. Wang, Y. Fu, Y. Jing, B. Huang, Y. Chen, Q. Wang, X. B. Wang, C. Meng, Q. Yang and L. Xu, BMC Biol., 2022, 20, 186 CrossRef CAS PubMed.
- F. M. Dwivany, G. Stefani, A. Sutanto, H. Nugrahapraja, K. Wikantika, A. Hiariej, T. Hidayat, I. N. Rai and N. Sukriandi, HAYATI J. Biosci., 2020, 27, 258 CrossRef.
- L. Englberger, I. Darnton-Hill, T. Coyne, M. H. Fitzgerald and G. C. Marks, Food Nutr. Bull., 2003, 24, 303–318 CrossRef PubMed.
- I. Y. Pratiwi and O. Krisbianto, agriTECH, 2019, 39, 48 CrossRef.
- V. Aliko, L. Vasjari, E. S. Istifli, G. Gjonaj, F. Impellitteri, C. Faggio, E. Benedetti, S. Zugaro, A. Iannetta and M. Perugini, Aquat. Toxicol., 2024, 276, 107112 CrossRef CAS PubMed.
- J. Yang, D. Xiong and M. Long, Nanomaterials, 2025, 15, 1030 CrossRef CAS PubMed.
- A. A. Goma, A. R. Salama, H. G. Tohamy, R. R. Rashed, M. Shukry and S. E. El-Kazaz, Biol. Trace Elem. Res., 2024, 202, 4654–4673 CrossRef CAS PubMed.
- S. A. Renshaw, C. A. Loynes, D. M. I. Trushell, S. Elworthy, P. W. Ingham and M. K. B. Whyte, Blood, 2006, 108, 3976–3978 CrossRef CAS PubMed.
- L. Li, B. Yan, Y.-Q. Shi, W.-Q. Zhang and Z.-L. Wen, J. Biol. Chem., 2012, 287, 25353–25360 CrossRef CAS PubMed.
- T. A. Petrie, N. S. Strand, C. Tsung-Yang, J. S. Rabinowitz and R. T. Moon, Development, 2014, 141, 2581–2591 CrossRef CAS PubMed.
- N. G. Zavitri, A. P. Syahbaniati, R. K. Primastuti, R. M. Putri, S. Damayanti and I. Wibowo, Biomed. Rep., 2023, 19, 1–11 Search PubMed.
- M. H. Heng, Y. F. Win, E. S. G. Cheah, Y. B. Chan, Md. K. Rahman, S. Sultana, L.-H. Tey, L. S. Wong, S. Djearamane, Md Akhtaruzzaman and M. Aminuzzaman, Green Process Synth., 2024, 13, 20240071 CrossRef CAS.
- Y. Chan, V. Selvanathan, L.-H. Tey, Md Akhtaruzzaman, F. Anur, S. Djearamane, A. Watanabe and M. Aminuzzaman, Nanomaterials, 2022, 12, 3589 CrossRef CAS PubMed.
- Y. B. Chan, M. Aminuzzaman, L.-H. Tey, Y. F. Win, A. Watanabe, S. Djearamame and Md Akhtaruzzaman, Materials, 2023, 16, 5421 CrossRef CAS PubMed.
- Organisation for Economic Co-operation and Development (OECD), Test No. 236: Fish Embryo Acute Toxicity (FET) Test, OECD Guidelines for the Testing of Chemicals, Section 2, OECD Publishing, Paris, 2013 DOI:10.1787/9789264203709-en.
- F. Sharif, P. J. Steenbergen, J. R. Metz and D. L. Champagne, Wound Repair Regen., 2015, 23, 855–865 CrossRef PubMed.
- J. S. Choi, R.-O. Kim, S. Yoon and W.-K. Kim, PLoS One, 2016, 11, e0160763 CrossRef PubMed.
- A. Dmochowska, J. Czajkowska, R. Jedrzejewski, W. Stawiński, P. Migdał and M. Fiedot-Toboła, Int. J. Biol. Macromol., 2020, 165, 1581–1592 CrossRef CAS PubMed.
- R. S. Dangana, R. C. George and F. K. Agboola, Green Chem. Lett. Rev., 2023, 16 Search PubMed.
- V. N. Kalpana and V. Devi Rajeswari, Bioinorg. Chem. Appl., 2018, 2018, 1–12 CrossRef PubMed.
- H. Agarwal and V. Shanmugam, Bioorg. Chem., 2020, 94, 103423 CrossRef CAS PubMed.
- J. Singh, T. Dutta, K. H. Kim, M. Rawat, P. Samddar and P. Kumar, J. Nanobiotechnol., 2018, 16, 1–24 CrossRef PubMed.
- M. Thatyana, N. P. Dube, D. Kemboi, A.-L. E. Manicum, N. S. Mokgalaka-Fleischmann and J. V. Tembu, Nanomaterials, 2023, 13, 2616 CrossRef CAS PubMed.
- M. S. Jameel, A. A. Aziz and M. A. Dheyab, Green Process Synth., 2020, 9, 386–398 Search PubMed.
- J. Ruangtong, J. T-Thienprasert and N. P. T-Thienprasert, Mater. Today Commun., 2020, 24, 101224 CrossRef CAS.
- A. A. Barzinjy and H. H. Azeez, SN Appl. Sci., 2020, 2, 1–14 Search PubMed.
- V. Selvanathan, M. Aminuzzaman, L. X. Tan, Y. F. Win, E. S. Guan Cheah, M. H. Heng, L.-H. Tey, S. Arullappan, N. Algethami, S. S. Alharthi, S. Sultana, M. Shahiduzzaman, H. Abdullah and M. Aktharuzzaman, J. Mater. Res. Technol., 2022, 20, 2931–2941 CrossRef CAS.
- O. R. Vasile, I. Serdaru, E. Andronescu, R. Truşcă, V. A. Surdu, O. Oprea, A. Ilie and B. Ş. Vasile, Comptes Rendus Chim., 2015, 18, 1335–1343 CrossRef CAS.
- A. Bayrami, S. Parvinroo, A. Habibi-Yangjeh and S. Rahim Pouran, Artif. Cells Nanomed. Biotechnol., 2018, 46, 730–739 CrossRef CAS PubMed.
- J. K. Patra and K.-H. Baek, J. Nanomater., 2014, 2014 Search PubMed.
- J. Y. Song, E.-Y. Kwon and B. S. Kim, Bioprocess Biosyst. Eng., 2010, 33, 159–164 CrossRef CAS PubMed.
- M. Danaei, M. Dehghankhold, S. Ataei, F. Hasanzadeh Davarani, R. Javanmard, A. Dokhani, S. Khorasani and M. R. Mozafari, Pharmaceutics, 2018, 10, 57 CrossRef PubMed.
- X. Zhao, S. Wang, Y. Wu, H. You and L. Lv, Aquat. Toxicol., 2013, 136–137, 49–59 CrossRef CAS PubMed.
- Y. Dang, F. Wang and C. Liu, Chemosphere, 2018, 207, 365–376 CrossRef CAS PubMed.
- R. C. Murdock, L. Braydich-Stolle, A. M. Schrand, J. J. Schlager and S. M. Hussain, Toxicol. Sci, 2008, 101, 239–253 CrossRef CAS PubMed.
- D. Batir-Marin, M. Boev, O. Cioanca, I.-I. Lungu, G.-A. Marin, A. F. Burlec, A.-M. Mitran, C. Mircea and M. Hancianu, Antioxidants, 2025, 14, 489 CrossRef CAS PubMed.
- W. Bai, Z. Zhang, W. Tian, X. He, Y. Ma, Y. Zhao and Z. Chai, J. Nanoparticle Res., 2010, 12, 1645–1654 CrossRef CAS.
- S. Wang, H. Alenius, H. El-Nezami and P. Karisola, Nanomaterials, 2022, 12, 1247 CrossRef CAS PubMed.
- T. Ramachandran, K. G. Mohanraj, T. Mary Martin and M. S. K, Cureus, 2024, 16, e69861 Search PubMed.
- M.-H. Kim and H.-J. Jeong, J. Nanosci. Nanotechnol., 2015, 15, 6509–6515 CrossRef CAS PubMed.
- M.-J. Liu, S. Bao, M. Gálvez-Peralta, C. J. Pyle, A. C. Rudawsky, R. E. Pavlovicz, D. W. Killilea, C. Li, D. W. Nebert, M. D. Wewers and D. L. Knoell, Cell Rep., 2013, 3, 386–400 CrossRef CAS PubMed.
- M.-H. Kim, J.-H. Seo, H.-M. Kim and H.-J. Jeong, Eur. J. Pharmacol., 2014, 738, 31–39 CrossRef CAS PubMed.
- M. Nguyen-Chi, B. Laplace-Builhé, J. Travnickova, P. Luz-Crawford, G. Tejedor, G. Lutfalla, K. Kissa, C. Jorgensen and F. Djouad, Cell Death Dis., 2017, 8, e2979 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |