 Open Access Article
Open Access ArticleMXenes for exosome detection: a new frontier in biomolecular analysis
Siavash
Iravani
 *a,
Atefeh
Zarepour
b,
Arezoo
Khosravi
cd,
Ali
Zarrabi
*a,
Atefeh
Zarepour
b,
Arezoo
Khosravi
cd,
Ali
Zarrabi
 *e,
Ehsan
Nazarzadeh Zare
*e,
Ehsan
Nazarzadeh Zare
 f,
Rajender S.
Varma
f,
Rajender S.
Varma
 g and
Pooyan
Makvandi
g and
Pooyan
Makvandi
 *hi
*hi
aIndependent Researcher, W Nazar ST, Boostan Ave, Isfahan, Iran. E-mail: siavashira@gmail.com
bDepartment of Biology, Faculty of Arts and Sciences, Kocaeli University, 41001, İzmit, Kocaeli, Turkey
cDepartment of Genetics and Bioengineering, Faculty of Engineering and Natural Sciences, Istanbul Okan University, Istanbul 34959, Turkey
dGraduate School of Biotechnology and Bioengineering, Yuan Ze University, Taoyuan 320315, Taiwan
eDepartment of Biomedical Engineering, Faculty of Engineering and Natural Sciences, Istinye University, Istanbul 34396, Turkey. E-mail: alizarrabi@gmail.com
fSchool of Chemistry, Damghan University, Damghan 36716-45667, Iran
gCentre of Excellence for Research in Sustainable Chemistry, Department of Chemistry, Federal University of São Carlos, 13565-905 São Carlos – SP, Brazil
hThe Quzhou Affiliated Hospital of Wenzhou Medical University, Quzhou People's Hospital, 324000, Quzhou, Zhejiang, China. E-mail: pooyanmakvandi@gmail.com; pooyan.makvandi@wmu.edu.cn
iResearch and Innovation Cell, Rayat Bahra University, Mohali-140301, Punjab, India
First published on 27th June 2025
Abstract
Exosomes, the small extracellular vesicles secreted by cells, hold immense potential as biomarkers for disease diagnosis, monitoring, and therapeutic development. MXenes and their composites have emerged as promising materials for exosome detection, showcasing remarkable attributes such as remarkable electrical conductivity, mechanical flexibility, large surface area, and tunable surface chemistry. These characteristics position MXenes as optimal candidates for biosensing applications, enabling the effective capture and analysis of exosomes, which are vital in cell communication and disease progression. However, significant challenges persist in the practical use of MXenes for exosome detection, notably pertaining to the reproducibility and stability of these materials in diverse biological environments. Furthermore, optimizing MXene functionalization for selectivity towards specific exosomes remains an ongoing task. Recent innovations, including hybrid MXene-based sensors integrated with nanomaterials and machine learning algorithms for data analysis, promise significant improvements in detection accuracy and real-time monitoring capabilities, paving the way for accessible point-of-care diagnostic devices. This review delves into the transformative applications of MXenes and their composites in exosome detection, emphasizing their unique properties that enhance biosensing capabilities. By showcasing recent advancements, current challenges, and future perspectives, it underscores how MXene-based (bio)sensors are poised to develop more accurate and early disease detection systems using exosomes.
1. Introduction
In recent years, MXenes have emerged as a fascinating class of two-dimensional (2D) materials, captivating researchers with their unique properties. These materials, composed of transition metal carbides, nitrides, or carbonitrides, exhibit excellent conductivity, high surface area, rich surface functionalities, and tunable physicochemical properties.1–4 Thus, they are finding innovative applications in various fields, including biosensing.5–7 Exosomes, nano-sized extracellular vesicles, play crucial roles in intercellular communication and disease progression (Fig. 1).8 Detecting exosomes efficiently can provide valuable insights into health diagnostics and therapeutic monitoring.9,10 Notably, exosomes have emerged as pivotal components in the realm of liquid biopsies. Their role in disease diagnosis, particularly for cancers, is gaining traction due to their unique properties.8 These nanoscale vesicles carry vital biomolecules, including proteins, lipids, and nucleic acids, which reflect the physiological status of their cells of origin. Consequently, they serve as a non-invasive source of biomarkers, enabling early detection and monitoring of various diseases.11,12 The ability to detect cancer early through exosomes presents a promising approach for effective treatment. Commonly used methods for detecting exosomes, such as optical and electrochemical assays, immunoreaction assays, and aptamer-based detection, face several challenges. These include low purity of isolated exosomes, tedious and time-consuming procedures, and the requirement for expensive and sophisticated instruments, which limit their widespread application. Immunoaffinity techniques, while highly selective, are hindered by the high cost and instability of antibodies. Aptamer-based methods offer advantages in stability, easy chemical modification, and cost but are limited by the availability of suitable aptamers. Additionally, techniques like fluorescence, surface plasmon resonance (SPR), and surface-enhanced Raman scattering (SERS) provide sensitive detection but often require complex setups. Chromatography and microfluidic methods improve isolation and characterization but still face issues such as low reproducibility, large sample demands, and challenges in achieving high sensitivity and purity.13–15 Overall, despite advances in nanomaterial-based biosensors and lab-on-chip devices that enhance sensitivity and multifunctionality, the detection and isolation of exosomes remain technically challenging due to these limitations, including the need for extensive sample purification, elevated false-positive rates, and difficulties in labeling due to the small size of exosomes.11,13,16,17Several studies have focused on utilizing electrochemical, optical, and electrochemiluminescence (ECL) biosensors for detecting exosomes.18 These technologies have been applied to detect exosomes released from various cancer cell types, including breast, ovarian, pancreatic, lung, and cervical cancers. While exosomes can identify a range of external and internal biomarkers when conjugated with specific recognition elements, most current biosensor designs predominantly focus on CD9 and CD63. Consequently, developing novel biosensors that offer selective and sensitive recognition of exosomes remains a pressing challenge in the field.11,13,16,17 In the context of exosome detection, MXenes offer several unique properties that enhance their potential as advanced biosensors.19,20 Their exceptional electrical conductivity enables rapid electron transfer, crucial for sensitive detection of low-abundance biomarkers.21 This property enhances the sensitivity of electrochemical biosensors, allowing for detecting low concentrations of exosomes. With a large surface area, MXenes allow for extensive functionalization, improving specificity by facilitating the attachment of biomolecules like antibodies or aptamers.22 This enhances the sensitivity of the detection method, enabling the identification of exosomes at low concentrations. Their tunable surface chemistry enables customization for selective interaction with exosomes, while lightweight and flexible characteristics promote the development of portable sensing devices for point-of-care applications.23,24 Additionally, their biocompatibility makes them suitable for use in biological environments, positioning MXenes as promising candidates for advancing exosome detection technologies and contributing to personalized medicine solutions.25–27 Moreover, the combination of MXenes with other nanomaterials, such as gold nanoparticles or graphene, can lead to synergistic effects.4,28 These hybrid materials can enhance signal amplification, thereby increasing detection limits. Additionally, the integration of MXenes into microfluidic devices allows for real-time monitoring, which is essential for clinical applications.7,29
The applications of MXenes and their composites in exosome detection are diverse (Table 1). The ultrasensitive colorimetric aptasensor constructed using MXenes and the biotin–streptavidin system allows for the early diagnosis of cancer.30,31 It can also be used to evaluate the effectiveness of cancer treatments and assess the prognosis of cancer patients. The detection method exhibits excellent sensitivity, with a low limit of detection (LOD) and a wide linear range, making it suitable for clinical applications. Furthermore, MXenes-based aptasensors demonstrate satisfactory reproducibility, stability, and selectivity, ensuring reliable and accurate detection results. These properties make MXenes a promising class of nanomaterials for the detection of exosomes and pave the way for their potential applications in clinical cancer detection and other biomedical fields.30–33
| Biosensor type | Structures and materials | Limit of detection (LOD) | Linear range | Ref. |
|---|---|---|---|---|
| An electrochemical biosensor | Fe4[Fe(CN)6]3 (Prussian blue) on the surface of MXene (Ti3C2) as hybrid nanoprobes; a CD63 aptamer-modified poly(amidoamine) (PAMAM)-Au NP electrode interface | 229 particles μL−1 | 5 × 102 particles μL−1 to 5 × 105 particles μL−1 | 34 |
| An extended-gate field-effect transistor (EGFET)-type biosensor | CD9 aptamer and MXene (Ti3C2Tx) | 10.64 pM for CD9 proteins; in the clinical test: 6.41 × 102 exosomes per mL | 10 pM to 1 μM in the buffer; in the clinical test: 1 × 103 to 1 × 107 exosomes per mL | 35 |
| A portable electrochemical aptasensor | Cubic AuPt dendritic nanocrystals/Ti3C2; an aptamer CD63 modified graphene oxide was immobilized on a screen-printed carbon electrode (SPCE) as the substrate materials for the direct capture and detection of colorectal carcinoma exosomes | Down to 20 exosomes μL−1 | 100 exosomes μL−1 to 5.0 × 105 exosomes μL−1 | 36 |
| A sandwich-type biosensor | Amino-functionalized Fe3O4 nanoparticles; CD63 aptamer attached Fe3O4 nanoprobes; MXene (Ti3C2) nanosheets modified with epithelial cell adhesion molecule (EpCAM) aptamer | 43 particles μL−1 | 102 particles μL−1 to 107 particles μL−1 | 37 |
| An electrochemiluminescence (ECL) sensor | MoS2 quantum dots-MXene heterostructure; Au nanoparticles@biomimetic lipid layer | 10 fM | From 30 fM to 20 nM | 38 |
| An enzyme-assisted photoelectrochemical (PEC) biosensor | MXene (Ti3C2)/CdS composites | 7.875 × 104 particles per mL | 7.3 × 105 particles per mL to 3.285 × 108 particles per mL | 39 |
| A metasurface-regulated ECL-based biosensor | A hybrid plasmonic-dielectric metasurface consisting of Au nanorings and TiO2 nanoparticles derived from MXene | 21 particles per mL | 102 to 106 particles per mL | 40 |
| Aptamer-functionalized magnetic MXene-based nanoplatform | An aptamer-functionalized magnetic MXene composite, including Fe3O4, Ti3C2, poly(ethylenimine), 3,3′-dithiodipropionic acid di (N-hydroxysuccinimide ester) (DSP), CD63 aptamer, FAM-ssDNA | 4.21 × 104 particles per mL | 105–1010 particles per mL | 41 |
Herein, we explore the applications and advancements of MXenes in the detection of exosomes. We delve into the unique properties and synthesis methods of MXenes, highlighting their capabilities for selective exosome capture. Moreover, different MXene-based sensor platforms are discussed, showcasing the versatility of MXenes in achieving sensitive and real-time detection. The article also addresses the challenges and future perspectives in MXene-based exosome detection, thereby emphasizing the need for continued research and development. Overall, MXenes offer promising avenues for enhancing our understanding of exosomes and their role in health and disease, paving the way for novel diagnostic approaches and therapeutic interventions.
2. Exosome extraction for detection: a critical step in clinical sample analysis
Isolating exosomes effectively is paramount for understanding their biological functions and potential applications in diagnostics and therapeutics. Various isolation techniques have emerged, each with its advantages and limitations. One of the most commonly used methods is ultracentrifugation.42 This technique involves subjecting cell culture supernatants to high-speed centrifugation, which separates exosomes based on their size and density. While ultracentrifugation is effective, it can be time-consuming and may lead to the co-isolation of other contaminants, such as protein aggregates. Moreover, this method often requires specialized equipment that may not be readily available in all laboratories.42 Another prominent technique is precipitation-based isolation. This method utilizes commercially available kits that employ polymers to precipitate exosomes from biological fluids. Precipitation is relatively quick and straightforward, making it accessible to many researchers. However, it may not achieve the same purity levels as ultracentrifugation and can also lead to the co-isolation of non-exosomal proteins and lipids.43 Size exclusion chromatography (SEC) offers an alternative approach that relies on the size of exosomes for separation.44 In this method, samples are passed through a column filled with porous beads, allowing smaller molecules to pass through while retaining larger exosomes. SEC is advantageous due to its ability to purify exosomes without harsh conditions, preserving their integrity. However, the process can be less efficient in terms of yield compared to ultracentrifugation.44 Additionally, immunoaffinity capture techniques utilize antibodies that specifically recognize surface markers on exosomes.45 This protocol provides a high level of specificity and purity. However, it requires prior knowledge of the exosomal markers of interest and may not be suitable for all exosome types.45Ultrafiltration is a widely utilized technique that employs membrane filters with defined pore sizes to separate exosomes from larger particles and contaminants.46 By applying pressure, liquid samples pass through the membrane, allowing smaller molecules to diffuse while retaining exosomes. This method is relatively quick and can handle large sample volumes. However, the efficiency of ultrafiltration can depend on the membrane's pore size and the viscosity of the sample, which may affect the overall yield.46,47 Additionally, microfluidic separation represents an exciting advancement in exosome isolation technology.48 This approach uses micro-scale devices to manipulate fluids, allowing for precise control over the separation process. By integrating various mechanisms, such as size-based filtering and affinity capture, microfluidic devices can effectively isolate exosomes with high purity and yield. Moreover, these systems often require smaller sample volumes, making them suitable for clinical applications where sample availability is limited. However, the complexity of microfluidic systems may pose challenges in terms of design and implementation.48 Charge-based isolation techniques capitalize on the surface charge of exosomes. This method involves using charged membranes or materials that attract or repel exosomes based on their electrostatic properties. By altering the ionic strength or pH of the solution, researchers can selectively isolate exosomes with varying surface charges. Charge-based isolation can enhance purity while minimizing the co-isolation of contaminants. However, it may not be universally applicable to all exosome types, as variations in surface charge can influence the efficiency of isolation.49 By understanding and optimizing these isolation techniques, researchers can enhance the yield and purity of exosomes, unlocking their potential in medical research and clinical applications.
3. Advancements in MXene-based exosome detection
The quest for superior interface materials is paramount in the realm of optoelectronic devices. Selecting the right materials hinges on their performance, functionality, and stability. Among these, MXene has emerged as a frontrunner.50 The integration of MXenes into exosome detection platforms holds great potential for various biomedical applications, including early diagnosis of diseases, monitoring disease progression, and studying exosome-mediated cell communication. The use of MXenes and their composites for exosome detection offers several advantages. Firstly, their unique structure allows for the efficient immobilization of capture molecules, such as antibodies or aptamers, onto their surface. This enables selective recognition and binding of exosomes, facilitating their detection. Furthermore, MXenes can be integrated into various sensing platforms, including field-effect transistors (FETs), optical, ECL, and electrochemical sensors, to enable label-free and sensitive detection of exosomes. The high conductivity of MXenes ensures efficient signal transduction, enhancing the sensitivity and reliability of exosome detection. Notably, MXenes can be functionalized with different molecules or nanoparticles to further enhance their sensing performance. For instance, the surface of MXenes can be altered with fluorescent dyes or plasmonic nanoparticles, thereby enabling signal amplification and multiplexed detection of exosomes.5,51–53 However, its susceptibility to oxidation, especially in hot and humid conditions, poses significant challenges. This vulnerability can detrimentally impact photovoltaic efficiency, urging the need for innovative solutions. In response to these challenges, Wang et al.54 developed a 2D-semiconductor/0D-plasma heterojunction, dubbed MXene-TA-Au-PEG (MTAP). This novel composite not only shielded MXene from the dreaded effects of stacking and oxidation but also showcased remarkable optoelectronic properties. The integration of these features positioned MTAP as an exceptional candidate for high-performance photovoltaic applications. Furthermore, MTAP's unique characteristics extended to the enhancement of surface plasmon resonance (SPR) spectroscopy. This improvement facilitated the direct, real-time detection of tumor cell exosomes. Astonishingly, it achieved LOD as low as 0.28 particles mL−1. Notably, the as-prepared MTAP addressed the conventional “three S” requirements—sensitivity, specificity, and stability. It merged photoelectric enhancement with interfacial antifouling and oxidative stabilization, creating a multifaceted tool for sensors. The analysis of serum samples further highlighted MTAP's promising application in clinical diagnostics.54 This study not only sheds light on the fabrication of 2D/0D heterojunctions but also inspires future innovations in sensing chip development. The possibilities are vast, paving the way for enhanced optoelectronic performance and groundbreaking clinical solutions.The development of MXene-based biosensors for exosome detection represents a significant advancement in the field of exosome research and paves the way for the development of innovative diagnostic tools. Researchers developed an ultrasensitive ECL biosensor tailored for the detection of exosomes and their surface proteins.55 The innovative design employed an in situ formation of gold nanoparticles (Au NPs) decorated on Ti3C2 MXenes, enhanced further by aptamer modification, creating a hybrid known as AuNPs-MXenes-Apt (Fig. 2(A)). To begin with, the strategy focused on efficiently capturing exosomes using an electrode interface modified with a CD63 aptamer. This specific recognition was crucial, as CD63 is a well-known exosomal marker. Simultaneously, the in situ formation of Au NPs on the single-layer Ti3C2 MXenes, modified with aptamers, showcased the versatility of MXenes. Remarkably, MXenes served a dual purpose: they acted as both reductants and stabilizers. This eliminated the need for additional reductants or stabilizers, streamlining the process. The resulting AuNPs–MXenes–Apt hybrid exhibited exceptional recognition capabilities for exosomes. Additionally, it provided a catalytic surface characterized by the electrocatalytic activity of Au NPs, particularly those with predominant (111) facets. This feature significantly enhanced the ECL signal of luminol, leading to improved sensitivity. Consequently, this ECL biosensor demonstrated an impressive detection limit of 30 particles per μL for exosomes derived from the HeLa cell line. This limit was over 1000 times lower than the conventional ELISA method. Furthermore, the biosensor operated within a linear range from 102 to 105 particles per μL. Moreover, the platform showed remarkable selectivity towards exosomes and their surface proteins derived from various tumor cell lines, including HeLa, OVCAR, and HepG2 cells. Notably, it enabled sensitive and accurate detection of exosomes from human serum.55 This advancement implies that the ECL biosensor stands as a feasible, sensitive, and reliable tool for exosomes detection, particularly in exosome-related clinical diagnostics. Zhang et al.56 developed a sensitive ECL biosensor specifically designed for exosome detection (Fig. 2(B)). The biosensor employed aptamer-modified MXenes (Ti3C2) nanosheets as the ECL nanoprobe, capitalizing on their large surface area, excellent conductivity, and remarkable catalytic properties. To efficiently capture exosomes, the electrode surface was functionalized with an aptamer that recognized the EpCAM protein. This strategic modification enabled high-affinity binding of exosomes onto the electrode surface. Additionally, the ECL nanoprobe itself could recognize the exosomes, significantly amplifying the ECL signals of luminol during detection. As a result of this approach, a highly sensitive ECL biosensor for detecting MCF-7 exosomes was successfully developed. The detection limit achieved was an impressive 125 particles per μL, which is over 100 times lower than that of traditional ELISA methods. The biosensor's effectiveness was further validated through successful detection of MCF-7 exosomes in serum samples.56
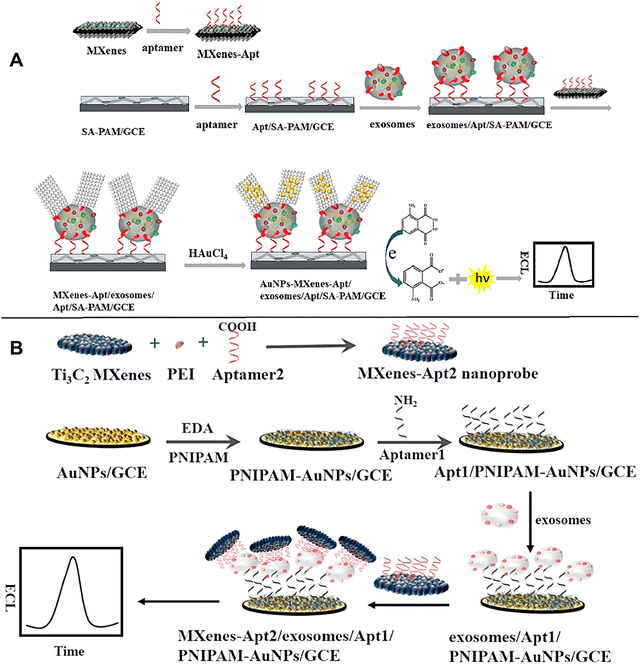 | ||
| Fig. 2 (A) The ECL biosensor operates on the principle of utilizing in situ formed gold nanoparticles, which are decorated on MXenes (Ti3C2) nanoprobes, to facilitate the detection of exosomes. Reproduced with permission from ref. 55 Copyright 2020 American Chemical Society. (B) The ECL biosensor operates on a sophisticated principle designed to amplify the detection of exosome activity effectively; the biosensor utilizes AuNPs integrated with MXenes (Ti3C2). Reproduced with permission from ref. 56 Copyright 2018 Elsevier. Polyethyleneimine (PEI), glassy carbon electrode (GCE), ethylenediamine (EDA), poly(N-isopropylacrylamide), carboxylic acid terminated (PNIPAM), aptamer (Apt), sodium alginate (SA), poly(acrylamide) (PAM). | ||
Exosomes have emerged as noninvasive biomarkers, offering exciting potential for disease prediction and diagnosis, particularly in the context of cancer-related public health concerns.10 In one study, researchers developed special Cy3 labeled CD63 aptamer (Cy3-CD63 aptamer)/MXene (Ti3C2) nanocomplexes.57 This configuration served as a self-standard ratiometric fluorescence resonance energy transfer (FRET) nanoprobe, enabling the quantitative detection of exosomes. Accordingly, the mechanism initiated with the selective adsorption of the Cy3-CD63 aptamer onto the Ti3C2 nanosheets. This interaction was facilitated by hydrogen bonding and metal chelation between the aptamer and MXenes. Initially, the fluorescence signal from the Cy3-CD63 aptamer was quenched due to FRET occurring between the Cy3 dye and the MXenes. Upon introducing exosomes, which specifically bind to the aptamer, the fluorescence signal of Cy3 significantly was recovered. This recovery occurred because the aptamer, now attached to the exosome's CD63 protein, was released from the MXenes surface, effectively restoring the fluorescence. Importantly, the self-fluorescence signal from the MXenes remained relatively unchanged throughout the process, serving as a reliable standard reference. Utilizing this self-standard turn-on FRET biosensing platform, the detection limit for exosomes was determined to be an impressive 1.4 × 103 particles per mL, which was over 1000 times lower than that of traditional ELISA methods. This fluorescence sensor also demonstrated versatility in identifying multiple biomarkers on the exosome surface and distinguishing between different types of exosomes through fluorescent confocal scanning microscopy imaging.57 In another study, Fang et al.58 developed a dual-mode biosensor integrating ECL and photothermal detection methods for exosome analysis. This biosensor utilized cutting-edge materials, specifically black phosphorous quantum dots (BPQDs) and MXenes, to enhance signal amplification. At the heart of the biosensor, BPQDs played a crucial role by catalyzing the oxidation of Ru(dcbpy)32+. This study marks the first time BPQDs have been employed as a coreactant in an ECL system. The self-enhanced Ru(dcbpy)32+@BPQDs ECL system generated a robust ECL signal by minimizing energy loss and shortening the distance required for electron transfer, thereby improving the overall efficiency. In addition to BPQDs, MXenes were incorporated due to their large specific surface area and exceptional conductivity. These properties contributed to increased immobilization of Ru(dcbpy)32+ and BPQDs on the biosensor surface, further amplifying the ECL signal. Moreover, both BPQDs and MXenes exhibited remarkable photothermal effects, which were cleverly utilized to develop a photothermal biosensor for exosome analysis. This dual-modality approach not only enriched the applications of MXenes and BPQDs in biodetection but also offered a highly effective and reliable method for detecting exosomes.58 This work presents a pioneering dual-mode probe combining MXenes and BPQDs, paving the way for advanced biosensing technologies. The resulting biosensor holds significant promise for enhancing exosome detection and advancing cancer diagnostics, thereby contributing to the growing field of personalized medicine.
Cancer often progresses silently, reaching terminal stages where treatment becomes unfeasible. Although treatment options are limited, early diagnosis can significantly enhance survival rates and reduce recurrence. Exosomes, biomolecules released by cancer cells, present promising opportunities for clinical diagnosis.59 In this context, a sensitive electrochemical biosensor was developed that leverages the in situ generation of Prussian Blue on the surface of MXene (Ti3C2), creating hybrid nanoprobes known as PB-MXene, specifically for the detection of exosomes and their surface proteins.34 To achieve specificity, a CD63 aptamer-modified poly(amidoamine) (PAMAM)–Au NP electrode interface was fabricated. This interface was designed to bind selectively with the CD63 protein present on exosomes derived from OVCAR cells. Notably, the CD63-modified MXene served as an effective nanocarrier, accommodating numerous aptamers that adsorb onto the exosomes. The MXene enabled the in situ generation and high-efficiency loading of Prussian Blue, which significantly amplified the electrochemical signal at a low potential. This feature was particularly advantageous, as it minimized interference from electrochemically active species that could compromise detection accuracy. The dual amplification effect achieved through this approach allowed for highly selective and sensitive electrochemical detection of exosomes. Remarkably, the electrochemical biosensor exhibited high specificity even in complex serum samples, underscoring its potential for clinical diagnostic applications.34
Su et al.19 developed a label-free aptasensor, ingeniously designed with CuNi bimetallic metal–organic frameworks (MOFs), enhanced by MXene quantum dots. This innovative sensor displayed a promising approach for exosome sensing. Accordingly, the CuNi–MOFs were functionalized with dual specific aptamers, namely CD63 and MUC1. These aptamers served as optimal nanocarriers and catalysts, facilitating the recognition and capture of target exosomes. Simultaneously, the MXene quantum dots., known for their excellent electron transfer properties, could significantly boost the electrochemical catalytic performance of the aptasensor. Furthermore, the 2D bimetallic CuNi–MOF nanosheets had a large specific surface area, providing abundant exposed active sites for aptamer connection. Consequently, this could lead to successful specific capture and detection of exosomes. The electrochemical aptasensor exhibited a remarkable linear detection range, spanning from 1 × 10−1 to 1 × 10−6 particles per μL, with an impressively LOD of 5 particles per μL. Notably, the aptasensor exhibited commendable electrochemical stability during exosome detection. This stability opens new avenues for clinical applications, particularly in tumor diagnosis.19 MXene has gained recognition in the construction of optoelectronic interfaces due to its remarkable properties. But, its hydrophilicity and metastable surface make it prone to oxidation, leading to degradation of its characteristics, which hinders practical applications.60 One study addressed these limitations by growing MOF in situ on the MXene surface through heterojunction engineering.60 This approach could effectively suppress direct contact between reactive molecules and the inner layer of MXene, preserving its inherent advantages while enhancing stability and optoelectronic performance. The resulting MXene@MOF heterojunction exhibited dual photoelectric gain, confirming its suitability as an interface sensitization layer material for surface plasmon resonance (SPR) applications. The study delved into the performance of the MXene@MOF material and its potential mechanisms for enhancing SPR, utilizing a combination of experimental data and simulation calculations (FDTD/DFT). To explore practical applications, a MXene@MOF/peptides-SPR sensor was constructed for the rapid and sensitive detection of cancer marker exosomes. The MXene@MOF heterojunction exhibited outstanding performance as an interfacial material, significantly enhancing SPR while effectively binding bioprobes. This capability permitted the SPR sensor to achieve fast and sensitive detection of Exo-PD-L1, with an LOD of 5.24 particles mL−1. Furthermore, when tested with actual serum samples, the sensor displayed excellent discrimination between positive and negative cancer groups, achieving an impressive area under the curve (AUC) value of 0.9421. These results underscored the potential of the MXene@MOF SPR sensor in providing reliable and efficient cancer diagnostics.60 This work presents a forward-looking strategy for designing interface materials with superior photoelectric performance, paving the way for advanced detection technologies in biomedical applications.
Among exosomes, the cluster of differentiation 9 (CD9) protein stands out as a crucial exosomal biomarker for detecting exosomes. In one study, a CD9 aptamer was synthesized and integrated it into an extended-gate field-effect transistor (EGFET)-type biosensor, featuring a disposable sensing membrane (Fig. 3).35 This innovative design aimed to demonstrate the feasibility of detecting exosomes within a clinical setting. The selection of nucleic acid sequences that specifically target the CD9 protein was systematically evaluated using the exponential enrichment (SELEX) technique, which is vital for optimizing ligand specificity. The detection of exosomes was achieved by monitoring electrical signal changes on the extended gate, which utilized an Au microelectrode. Thus, the integration of MXene and EGFET technologies presents several advantages, such as a low signal-to-noise ratio, ease of functionalization, and high electrical activity. These features empower the biosensor to perform effectively in environments where target concentrations are low and off-target molecules are present. Given the diagnostic challenges often associated with early cancer detection, the CD9 aptamer/MXene-modified biosensor exhibited significant potential for detecting tumor-derived exosomes (TEXs) on the EGFET platform. The ability of MXenes to amplify the signal, coupled with the specific binding capabilities of the CD9 aptamer, enhances the biosensor's sensitivity and specificity. This innovative approach offers a promising solution for overcoming complex diagnostic limitations, paving the way for more accurate and reliable early cancer diagnoses. By efficiently identifying TEXs, this biosensor may contribute to improved patient outcomes through timely interventions and personalized treatment strategies.35 These results highlight the biosensor's reliability and low error rates, suggesting its effectiveness in real-world applications.
 | ||
| Fig. 3 The process of exosome detection utilizing the EGFET aptasensor: in the first step, MXenes are incorporated into the biosensor design to serve as signal amplifiers. Their unique properties enhance the sensitivity and efficiency of the detection process. Next, the CD9-26 aptamer is conjugated onto the surface of the MXene material. This aptamer specifically targets and binds to the CD9 protein present on the exosome's surface. Once the CD9 protein on the exosome binds with the CD9-26 aptamer, changes in the electrical signals are detected through the extended-gate field-effect transistor. This interaction facilitates the accurate detection of exosomes, demonstrating the aptasensor's effectiveness in identifying exosomal biomarkers in a clinical environment. Reproduced with permission from ref. 35 Copyright 2023 American Chemical Society. | ||
A novel electrochemical biosensor was designed for the sensitive detection of exosomes, utilizing hierarchical Au nanoarray-modified MXene (Ti2CTx) membranes.61 The MXene nanosheets were prepared as foundational building blocks for preparing 2D membranes, employing a vacuum filtration technique to create an effective sensing platform. To enhance the conductivity of the MXene membrane, hierarchical Au nanoarrays were deposited in situ onto the surface for the first time. This combination of the MXene membrane, characterized by its large specific surface area, and the Au nanoarrays, known for their excellent conductivity, results in a higher electrocatalytic activity and an increase in active sites for aptamer immobilization. In this innovative strategy, the composite membrane was modified with an EpCAM-recognizing aptamer that specifically captured target exosomes. Furthermore, these target exosomes were designed to anchor another aptamer targeting CD63, which significantly enhanced the sensing sensitivity and accuracy of the biosensor. As a result, this biosensor displayed remarkable performance for exosome detection, achieving a low detection limit of 58 particles per μL, with a linear detection range spanning from 1 × 102 to 1 × 107 particles per μL. Additionally, the biosensor exhibited satisfactory electrochemical stability and robust anti-interference capabilities when detecting exosomes in real serum samples.61 Wang et al.62 developed an ultrasensitive colorimetric aptasensor for exosome detection, utilizing a dual-effect amplification method based on the biotin–streptavidin system and MXenes nanomaterial (Fig. 4). The high specific surface area of MXenes facilitated enhanced loading of both the aptamer and biotin, significantly improving the sensor's performance. By leveraging the biotin–streptavidin system, they could increase the quantity of horseradish peroxidase-linked (HRP-linked) streptavidin, which in turn considerably amplifies the color signal generated by the aptasensor. The proposed colorimetric aptasensor demonstrated exceptional sensitivity, achieving a detection limit of 42 particles per μL, with a linear detection range extending from 102 to 107 particles per μL. Additionally, the constructed aptasensor exhibited satisfactory reproducibility, stability, and selectivity in its performance. These results confirmed the promising application of exosomes in clinical cancer detection, underscoring the potential of this aptasensor to enhance diagnostic capabilities and improve patient outcomes.62
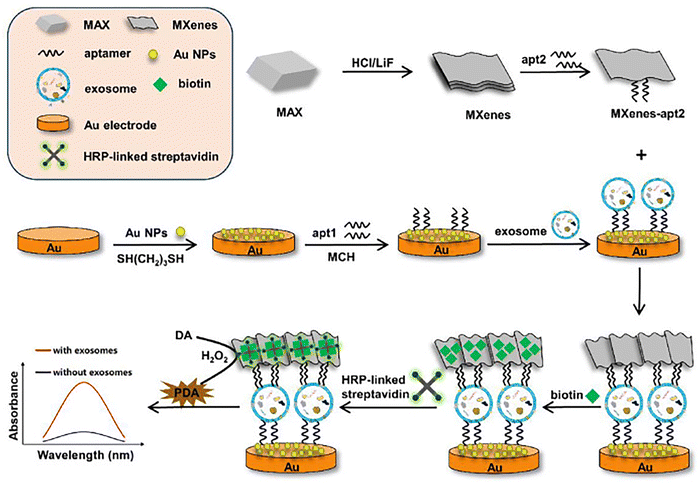 | ||
| Fig. 4 The construction of the colorimetric sensor with several systematic steps to ensure optimal performance for exosome detection. 6-Mercapto-1-hexanol (MCH), horseradish peroxidase (HRP), dopamine (DA), brown polydopamine (PDA). Reproduced with permission from ref. 62 Copyright 2023 Elsevier. | ||
Biosensors have been designed using a sandwich approach with promising detection capabilities for exosomal programmed cell death 1 ligand 1 (ExoPD-L1). However, conventional PD-L1 antibodies, peptides, and aptamers commonly bind to the extracellular region, often competing for overlapping binding sites. This limitation hinders the effective fabrication of biosensors. Zhou et al.63 introduced a novel strategy to specifically identify and analyze ExoPD-L1 by leveraging the non-selective trapping effect of MXene (Ti3C2Tx, where X = –O, –F, –OH) on exosomes. This approach involved the formation of Ti–O–P complexation, which facilitated the selective capture of peptide-functionalized Au@MPBA (4-mercaptophenylboronic acid)@SiO2 surface-enhanced Raman scattering (SERS) tags on ExoPD-L1. The resulting biosensor exhibited both hypersensitive and reliable performance in exosome detection, achieving a LOD of 20.74 particles per mL, with a linear detection range from 102 to 5 × 106 particles per mL. Furthermore, the biosensor exhibited outstanding stability and resistance to interference when detecting ExoPD-L1 in clinical serum samples. This capability enabled efficient differentiation between breast cancer patients and healthy controls.63 This work provides valuable insights into the design of biosensors for exosome detection, showcasing a replicable template for sandwich immunoassay detection applicable to various sensor types, including SERS. The innovative use of MXenes and SERS tags paves the way for enhanced biosensing strategies, ultimately contributing to improved cancer diagnostics and monitoring.
Sun et al.64 combined SERS and deep learning to profile exosomes. Accordingly, the use of MXene-coated gold@silver core@shell nanoparticles could significantly enhance the SERS signal through both electromagnetic enhancement (EM) and chemical enhancement (CM) (Fig. 5). This dual enhancement mechanism allowed the proposed sensing platform to achieve a dynamic range of 0.5 × 1010 to 2.0 × 1011 EVs mL−1, with an LOD of 1.7 × 109 EVs mL−1. This sensitivity is crucial for accurately detecting low concentrations of exosomes present in biological samples like plasma, where early-stage disease markers may be scarce. Following the SERS detection, a sophisticated deep-learning classification algorithm was employed. By utilizing residual neural networks, the algorithm efficiently extracted features from complex Raman spectra, enabling robust data interpretation. This integration of advanced machine learning techniques with spectroscopic methods enhanced diagnostic accuracy. The preliminary validation demonstrated the method's effectiveness in distinguishing thyroid cancer patients from healthy controls, achieving a remarkable diagnostic accuracy of 96.0%. Additionally, the algorithm exhibited an accuracy (∼86.6%) in staging cancer patients, further underscoring its potential in clinical applications.64 This innovative approach of utilizing SERS combined with deep learning for label-free profiling of exosomes represents a significant advancement in liquid biopsy technology. It not only enhances the sensitivity and specificity of cancer diagnostics but also paves the way for broader applications in personalized medicine. As the field continues to evolve, the integration of advanced nanosensing platforms and artificial intelligence will likely play a crucial role in transforming disease detection and monitoring strategies.
 | ||
| Fig. 5 This illustration involves several key steps: (a) collection of clinical samples from patients, (b) isolation of exosomes, (c) fabrication of a MXene-coated gold@silver core@shell nanoparticle substrate for SERS measurement, (d) deep learning-assisted analysis of the acquired data, and (e) structures of the basic blocks involved in each step. Reproduced with permission from ref. 64 Copyright 2024 Elsevier. | ||
Effectively capturing and accurately identifying cancer exosomes within complex biomatrices remains a challenging endeavor. The large size and non-conductivity of exosomes pose significant obstacles to achieving highly sensitive electrochemical or ECL detection.65 To address these challenges, Nie et al.65 developed a novel nanoarchitecture comprising a Ti3C2Tx–Bi2S3–x heterostructure with an engineered lipid layer (Fig. 6). This innovative design could effectively overcome the limitations associated with traditional detection methods. The engineered lipid layer played a dual role: it specifically captured and efficiently fused with CD63 positive exosomes while also exhibiting excellent antifouling properties in biological matrices. This feature is crucial for maintaining sensor performance in the presence of complex biological components. Furthermore, the incorporation of a MUC1 aptamer-modified Ti3C2Tx–Bi2S3–x heterostructure enhanced the specificity of the detection system. This aptamer was tailored to identify and bind to gastric cancer exosomes trapped within the engineered lipid layer, ensuring precise targeting during analysis. In this self-luminous Faraday cage-type sensing system, the as-prepared heterostructure, enriched with sulfur vacancies, effectively extended the outer Helmholtz plane. This modification amplified the ECL signal, leading to improved sensitivity in detection. As a result, this sensor could detect tumor exosomes present in the ascitic fluid of cancer patients without the need for additional purification steps.65 This work provides a new pathway for the sensitive detection of exosomes and other large-sized vesicles, offering significant implications for cancer diagnostics and monitoring. The integration of engineered lipid layers with advanced nanomaterials represents a promising approach to overcoming existing challenges in exosome research, fostering enhanced clinical applications in the future.
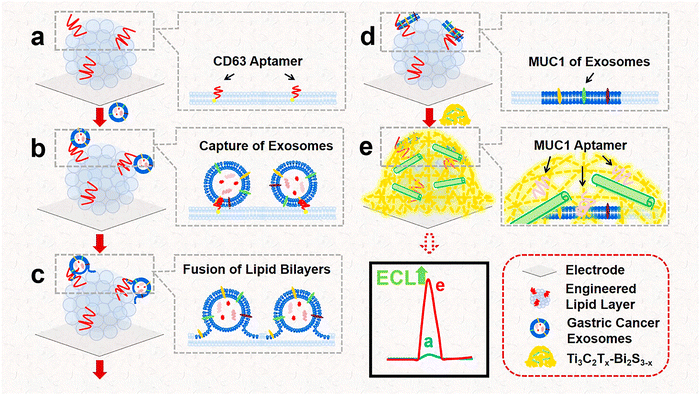 | ||
| Fig. 6 The fabrication of the Ti3C2Tx–Bi2S3–x heterostructure and engineered lipid layer for detecting gastric cancer exosomes involves a series of meticulous steps: (a) the first step involved creating the engineered lipid layer directly on the electrode surface. (b) Next, the engineered lipid layer facilitated the capture of gastric cancer exosomes through specific interactions. (c) Following this, the lipid bilayers of the exosomes merged with the engineered lipid layer. (d) This merging resulted in the development of fusion bodies, which encapsulated the gastric cancer exosomes within the lipid environment. (e) Finally, the Ti3C2Tx–Bi2S3–x heterostructure was incorporated to form a Faraday cage-type sensor, enhancing detection capabilities. Reproduced with permission from ref. 65 Copyright 2023 American Chemical Society. | ||
Qiu et al.39 developed an enzyme-assisted photoelectrochemical (PEC) biosensor for the quantification of exosomes. This approach utilized the in situ fabrication of MXene (Ti3C2)/CdS composites, combined with magnetic separation technology and hybridization chain reaction (HCR). Initially, exosomes were selectively captured between aptamer-labeled magnetic beads (CD63-MBs) and a cholesterol-labeled DNA anchor. The specially designed anchor ends served as triggers, enriching alkaline phosphatase (ALP) through HCR. The ALP catalyzed the conversion of sodium thiophosphate, leading to the generation of sulfide ions (S2−). These ions then reacted with Cd2+ ions to facilitate the in situ formation of CdS on the MXene, resulting in an increased photocurrent. The MXene-anchored PEC method enabled the quantitative detection of exosomes, showing notable stability, enhanced sensitivity/selectivity, and rapid response times.39 Such this PEC biosensor holds significant promise as a routine bioassay technique for the accurate quantification of exosomes, particularly in breast cancer diagnostics.
By focusing on saliva exosomes, this integrated system offers a non-invasive approach to identifying asthma-related biomarkers. Asthma, a chronic respiratory condition affecting millions of children globally, often remains undiagnosed or misdiagnosed.66 Traditional methods, while effective, can be uncomfortable and stressful for young patients. Herein lies the beauty of the ECL sensor. It enables the detection of specific exosomal content, which correlates with asthma development and exacerbation. Moreover, the technology harnesses the power of electrochemiluminescence, providing enhanced sensitivity and specificity. As a result, healthcare professionals can achieve quicker results. This accelerated diagnosis can lead to timely interventions, ultimately improving patient outcomes. Additionally, the use of saliva as a diagnostic medium is a game-changer. Saliva collection is simple, painless, and can be conducted in various settings, including schools and clinics. Thus, it reduces the barriers to testing, ensuring more children receive the necessary evaluations.67 Li et al.67 have pioneered a remarkable surface plasmon coupling ECL technique. This cutting-edge approach targeted salivary exosomes, aiming to enhance the diagnosis of childhood asthma (Fig. 7). At the core of this innovation lied the integration of SnS2 quantum dots with MXene. By forming Ohmic junctions, these components functioned as efficient ECL emitters. Remarkably, the creation of these junctions could lead to a significant reduction in contact resistance. Consequently, this improvement boosted charge injection efficiency, resulting in a stunning 2.76-fold enhancement of the ECL signal. Furthermore, a self-assembled surface plasmonic Bi@SiO2 array was meticulously constructed. In this system, when the ECL emitted from SnS2 quantum dots@MXene interacts with the electronic oscillations of the Bi@SiO2 nanoparticle array, a striking effect occurs. The luminescence intensity experienced a further amplification. This dynamic interaction not only intensified the signal but also facilitated the regulation into a directionally polarized output. This phenomenon, known as the SPC-ECL effect, showcased the sophistication of this advanced sensing mechanism. Utilizing this innovative ECL technique, the successful detection of CD9-exosomes in saliva could be achieved. This capability holds immense promise, especially in analyzing both acute exacerbations and chronic persistence of childhood asthma.67 This integrated system represents a significant advancement in respiratory health diagnostics, allowing for precise detection of asthma-related biomarkers. In another study, Li et al.68 developed a nano-sensing system, showcasing a luminescent Faraday cage mode tailored for detecting miRNA-221-5p in clinical saliva exosomes to assess asthma (Fig. 8). The process began with the in situ synthesis of copper nanoclusters (Cu NCs) on the defects of Ti3CN nanosheets, leveraging the nano-confinement effect. This innovative approach paved the way for a highly effective detection system. The remarkable Electronic metal-support interactions between the MXene and the anchored Cu NCs resulted in Cu NCs@MXene demonstrating strong luminescence performance. Alongside this, it exhibited high electrochemical activity and commendable stability properties. This synergy significantly enhances the overall functionality of the sensing system. Moreover, the MXene nanosheets, known for their excellent conductivity and flexibility, played a crucial role. They expanded the outer Helmholtz plane, which in turn improved the sensing ability of the system. In a further advancement, Cu nanocone/Bi nanoparticles arrays were meticulously constructed through step-by-step electrodeposition. Thanks to the surface plasmon coupling effect, these Cu nanocone/Bi NP arrays converted the ECL signal of Cu NCs@MXene-Faraday cage into directional emission, achieving an impressive 13.4-fold enhancement. To finalize the design, a thin phospholipid film and capture DNA were meticulously modified onto the surface of the Cu nanocone/Bi NPs array, establishing a sensing interface for miRNA-221-5p detection in saliva exosomes. This advanced nano-sensing system demonstrated a detection range of 1.0 × 10−16 to 1.0 × 10−8 mol L−1, with a remarkably low detection limit of just 34 aM.68 The application of such MXene-based biosensors in clinical analysis marks a significant step forward in asthma evaluation.
 | ||
| Fig. 7 This illustration showcases the ECL sensing system that incorporates SnS2 quantum dots@MXene Ohmic junctions and a Bi@SiO2 nanoparticle array, crucial for the detection of CD9-exosomes. The first step (A) involves creating the SnS2 QDs@MXene heterojunction, where the quantum dots are synthesized to enhance electrochemiluminescence. This combination forms a robust junction that reduces contact resistance, thereby improving charge injection efficiency. Following this, the Bi@SiO2 nanoparticle array is assembled (B), designed to optimize surface plasmon resonance. The arrangement of Bi nanoparticles within the SiO2 matrix is essential for maximizing the interaction with the emitted ECL from the SnS2 QDs@MXene junction. Finally, the sensing process for CD9-exosome detection occurs when the ECL interacts with the Bi@SiO2 array (C), leading to amplified luminescence and enabling sensitive identification of these exosomes in saliva. Carboxymethyl chitosan (CMCS), catalyzing hairpin self-assembly (CHA), (3-aminopropyl)triethoxysilane (APTES), carboxymethyl chitosan (CMCS). Reproduced with permission from ref. 67 Copyright 2024 American Chemical Society. | ||
 | ||
| Fig. 8 The nano-sensing system, integrating Cu NCs@MXene-Faraday cage and Cu nanocone/Bi nanoparticles, exemplifies an innovative approach to detect miRNA-221-5p in saliva exosomes for asthma evaluation. (A) The process commences with the synthesis of copper nanoclusters on Ti3CN nanosheets, which enhances luminescence and electrochemical properties. (B) During the ECL sensing phase, the modified sensing interface captures the target miRNA, producing a measurable ECL signal. (C) Additionally, the surface plasmon coupling effect within the Cu nanocone/Bi NPs arrays amplifies the ECL signal, achieving a 13.4-fold enhancement in detection sensitivity. Tetramethylammonium hydroxide (TMAOH), glutathione (GSH). Reproduced with permission from ref. 68 Copyright 2024 Elsevier. | ||
4. Challenges
While MXenes offer exciting opportunities for exosome detection, overcoming these challenges will be essential for their successful implementation in clinical diagnostics and research applications:4.1. Surface functionalization
Effective functionalization of MXenes is crucial for selective binding to exosomes.24,69 For instance, the synergistic amplification of amino-functionalized Fe3O4 nanoparticles and MXene nanosheets exhibited promising potential in exosome detection.37 Achieving the right balance of stability, biocompatibility, and specificity in the functionalization process can be complex. One significant challenge is achieving uniform and stable functionalization across the MXene surface. Variability in the attachment of biomolecules—such as antibodies, peptides, or nucleic acids—can lead to inconsistent binding properties, resulting in poor sensitivity and specificity for target exosomes. Moreover, non-specific binding to other components in biological samples can further complicate the detection process, leading to false positives or negatives. Another challenge involves selecting appropriate functionalization strategies that preserve the intrinsic properties of MXenes. Some functionalization techniques may alter the electrical and optical characteristics of MXenes, potentially diminishing their sensing capabilities. Balancing the need for effective functionalization with the preservation of MXene performance is crucial, yet difficult to achieve. Notably, the stability of the functionalized surface over time is a concern. Environmental factors, such as humidity, temperature, and exposure to biological fluids, can affect the integrity of the functionalized layers. This degradation may lead to reduced sensor performance and reliability. Furthermore, developing scalable and cost-effective functionalization methods is necessary for practical applications.4.2. Stability and sensitivity
MXenes are known for their chemical stability, but long-term stability and durability of the biosensors need to be evaluated.21,70–72 Factors such as biofouling, degradation of capture molecules, or mechanical stress could affect the performance of the sensors over time. Developing protective coatings or incorporating self-cleaning mechanisms may help address these challenges. Additionally, MXenes can be sensitive to environmental conditions, such as humidity and pH, which may affect their stability and performance in biosensing applications. Ensuring that MXenes maintain their properties over time and under varying conditions is essential for reliable exosome detection. While MXenes offer excellent conductivity and surface area for exosome detection, further improvements are needed to enhance the sensitivity and selectivity of the biosensors. Developing advanced surface functionalization strategies and optimizing the capture molecules could address these challenges.Working with MXenes presents a myriad of challenges, particularly when it comes to their stability in humid conditions. Oxidation can significantly impair their desirable characteristics, as moisture in the air can lead to degradation. This process not only diminishes their electrical conductivity but also affects their mechanical properties. Researchers often find themselves grappling with this issue, which necessitates the development of protective coatings or alternative storage methods. Such solutions are essential to maintain the integrity of MXenes over time and enhance their applicability in various fields. Moreover, inconsistent synthesis methods pose another significant hurdle in the utilization of MXenes. Variability in the fabrication process can result in variations in material quality, making it difficult to reproduce reliable results. This inconsistency undermines the potential of MXenes for widespread use in applications like energy storage and biomedical sensing. To overcome this obstacle, the establishment of standardized protocols for synthesis is crucial. By ensuring uniformity, researchers can pave the way for more reproducible outcomes, ultimately enhancing the reliability of MXenes. Functionalizing MXenes to target specific exosomes without compromising their unique properties adds yet another layer of complexity. The challenge lies in striking the right balance between effective targeting and retaining the inherent characteristics that make MXenes attractive. The intricacies of biological samples, such as blood, exacerbate this issue. These samples contain a complex mix of proteins and other molecules that can interfere with the performance of MXenes. Thus, developing effective functionalization strategies that maintain their functionality while navigating this biological complexity is a pressing need. To tackle these multifaceted challenges, innovative solutions emerge. Implementing greener synthesis methods enhances the scalability of MXenes significantly. Additionally, utilizing protective coatings can shield these materials from oxidative degradation, thus extending their lifespan. Establishing standardized protocols provides a framework for consistent synthesis, while focusing on targeted functionalization can lead to more effective applications in diagnostics and therapeutics.
4.3. Reproducibility
The synthesis of MXenes can lead to variations in their properties,73–75 which may affect the reproducibility of results in exosome detection assays. Standardizing the synthesis and functionalization processes is necessary to ensure consistent performance across different experiments.76–78 One of the primary factors contributing to reproducibility issues is the variability in MXene synthesis and functionalization processes. Differences in preparation methods, such as the choice of precursor materials or etching techniques, can lead to batch-to-batch inconsistencies in the properties of MXenes, affecting their performance as biosensors. Even slight deviations in experimental conditions can yield different results, complicating efforts to replicate findings across studies. Notably, the complexity of biological samples further complicates reproducibility. Variations in sample collection, processing, and storage can introduce inconsistencies that affect the isolation and characterization of exosomes. These discrepancies can lead to differences in the concentration and composition of exosomes analyzed, resulting in varied detection outcomes. To address this, researchers must establish standardized protocols for sample handling and analysis, ensuring that results are consistent and reliable across different laboratories. Another challenge lies in the interpretation of results. The data obtained from MXene-based sensors can be influenced by numerous factors, including environmental conditions and the presence of interfering substances. This variability can complicate the analysis and lead to difficulties in drawing definitive conclusions. To enhance reproducibility, it is essential to develop robust analytical frameworks and quality control measures that account for these variables.4.4. Complex biological samples
Biological fluids, such as blood or urine, contain a complex mixture of proteins, lipids, and other biomolecules that can interfere with exosome detection.79,80 Developing MXene-based sensors that can effectively differentiate exosomes from other components in these samples is a significant challenge. The analysis of complex biological samples poses significant challenges for MXene-based technologies aimed at exosome detection. One major issue is the presence of various biological components, such as proteins, lipids, and other extracellular vesicles, which can interfere with the detection process. These substances may produce background noise or compete for binding sites, leading to decreased sensitivity and specificity in the detection of target exosomes. Consequently, researchers must develop robust pre-treatment protocols to isolate exosomes effectively from these complex matrices, which can be both time-consuming and labor-intensive. Additionally, the heterogeneity of exosomes themselves complicates detection efforts. Exosomes vary widely in size, composition, and surface markers, depending on their cellular origin and physiological state.81 This diversity can result in variations in how different exosome populations interact with MXene sensors, making it difficult to achieve consistent results across different sample types. Moreover, the dynamic nature of exosomes—affected by factors such as disease state or environmental conditions—can further complicate their characterization and detection. Notably, reproducibility can be a significant concern when dealing with complex biological samples. Variations in sample collection, storage, and processing can introduce inconsistencies that affect the performance of MXene-based biosensors. To overcome these challenges, it is essential to establish standardized protocols for sample preparation and analysis, ensuring that results are reliable and comparable across different studies and clinical settings. By addressing these challenges, MXene technologies can enhance their applicability in the analysis of complex biological samples, ultimately contributing to more effective diagnostics in personalized medicine.4.5. Detection limitations
While MXenes can enhance sensitivity, achieving the detection limits required for early disease diagnosis remains a challenge.7,72,82–84 Further optimization of the sensing platforms and techniques is needed to improve the detection capabilities of MXenes for low-abundance exosomes. Detection limitations of MXene-based technologies for exosome analysis present significant hurdles in realizing their full potential in clinical applications. One of the primary challenges is achieving sufficient sensitivity to detect low-abundance exosomal biomarkers, which are often crucial for early diagnosis of diseases such as cancer. While MXene-based composites exhibit excellent electrical and optical properties, factors like background noise and interference from other biomolecules in complex biological samples can obscure signal detection. This can lead to false negatives or inaccurate results, undermining the reliability of the diagnostic tool. Another challenge lies in the specificity of MXene sensors. As the functionalization of MXenes is crucial for targeting specific exosomes, the development of effective bioconjugation strategies remains complex. Non-specific binding to other components in the sample can result in cross-reactivity, complicating the interpretation of results. Moreover, the dynamic nature of exosomes, which can vary significantly in size, composition, and charge, adds another layer of complexity to their detection. Notably, the stability of the MXene materials themselves can impact their performance over time. Degradation or oxidation of MXenes may alter their sensing capabilities, leading to reduced accuracy and reliability.85,86 Addressing these detection limitations will require ongoing research and innovation focused on enhancing the sensitivity, specificity, and stability of MXene-based biosensors.4.6. Regulatory and clinical validation
For MXene-based exosome detection technologies to be adopted in clinical settings, they must undergo rigorous testing and validation to meet regulatory standards.87,88 This process can be time-consuming and requires substantial investment. For MXene-based exosome sensors to be adopted in clinical settings, rigorous validation and clinical studies are required. Understanding the correlation between exosome biomarkers and disease states, as well as establishing reliable detection thresholds, will be crucial for successful clinical translation. Challenges in clinical translation studies of MXene-based technologies for exosome detection present significant barriers to their successful implementation in real-world healthcare settings. One primary obstacle is the lack of standardized protocols for the isolation and characterization of exosomes, which can lead to inconsistencies in study outcomes. Variability in exosome preparation methods affects the reproducibility and comparability of results across different research groups, complicating the validation process needed for clinical application. Moreover, the transition from laboratory settings to clinical environments requires rigorous testing to ensure that MXene sensors maintain their performance under diverse biological conditions, such as varying pH levels or the presence of interfering substances. Regulatory hurdles also pose challenges, as obtaining approval from health authorities can be a lengthy and complex process. MXene-based technologies ought to demonstrate safety, efficacy, and specificity in detecting exosomes before they can be marketed for clinical use. This requires extensive preclinical and clinical trials, which can be resource-intensive and time-consuming. Additionally, gaining acceptance from clinicians and healthcare providers is crucial, as they must be convinced of the benefits and reliability of integrating these new diagnostic tools into their practice.4.7. Scalability and cost
The production of MXenes and their integration into sensing devices must be scalable and cost-effective for widespread use. Addressing these economic factors is crucial for translating research findings into practical applications. Standardization of fabrication protocols and quality control measures will be essential to enable large-scale production of MXene-based exosome sensors.5,89,90 Additionally, the scalability and cost of MXene-based technologies for exosome detection pose significant challenges that must be addressed to facilitate widespread adoption. Notably, the synthesis of MXenes often involves complex and time-consuming processes, such as selective etching of precursor materials, which can be difficult to scale up for mass production.75,91,92 Achieving consistent quality and performance across large batches is crucial, as any variability can impact the reliability of diagnostic results. Furthermore, the need for specialized equipment and controlled environments during the synthesis process adds to the production costs, making it less accessible for smaller laboratories or resource-limited settings. Notably, the functionalization of MXenes to enhance their specificity and sensitivity introduces further complexities, often requiring expensive reagents and meticulous procedures.24,69 These factors contribute to the overall cost of developing MXene-based biosensors, which may hinder their commercial viability compared to established diagnostic technologies. Furthermore, integrating MXenes with other platforms can increase costs, as the combined systems may require additional materials, components, and expertise for implementation. To overcome these challenges, researchers are exploring more efficient synthesis methods, such as scalable liquid-phase exfoliation and green chemistry approaches that reduce costs and environmental impacts.4.8. Integration with other technologies
MXene-based biosensors can benefit from integration with other emerging technologies such as microfluidics, artificial intelligence, and data analysis algorithms.77 The combination of these technologies could enhance the overall performance and diagnostic capabilities of MXene-based exosome sensors. One major hurdle is ensuring compatibility between MXenes and the diverse range of materials and platforms they are being combined with, such as microfluidics or electrochemical sensors. Differences in surface chemistry and physical properties can lead to suboptimal interactions, compromising the performance of the overall system. Furthermore, the stability of MXenes in various environments poses another challenge; their susceptibility to oxidation and degradation can affect their functionality when integrated with other technologies. Additionally, achieving uniform distribution and reproducibility during the fabrication process can be difficult, impacting the reliability of the final product. Another challenge lies in the complexity of signal transduction mechanisms when combining MXenes with advanced sensing technologies. Researchers must develop robust protocols to accurately interpret signals generated from these integrated systems, ensuring that they yield meaningful and reproducible results. Moreover, scaling up these integrated technologies for widespread use while maintaining performance consistency remains a significant obstacle.4.9. The extraction of exosomes from clinical samples
The extraction of exosomes from clinical samples presents several significant challenges that impact the accuracy and reliability of downstream analyses. One of the main difficulties lies in isolating exosomes efficiently from complex biological fluids, which contain a mixture of proteins, lipids, and other extracellular vesicles of similar size and density. Achieving high purity without compromising yield is particularly challenging, as contaminants can interfere with quantification and biomarker detection. Additionally, clinical samples often have limited volume, making it essential to use extraction methods that maximize recovery while preserving exosome integrity. Another limitation is balancing the trade-offs between different isolation techniques. Ultracentrifugation, the traditional gold standard, is time-consuming and requires specialized equipment, which limits its practicality in clinical settings. On the other hand, precipitation kits and combination methods offer faster processing and higher yields but may introduce variability or incomplete removal of contaminants. Standardizing protocols to ensure reproducibility and consistency across laboratories remains a critical hurdle. Overcoming these challenges is essential to fully harness the potential of exosomes as reliable diagnostic and prognostic biomarkers in clinical practice.5. Future perspectives
The future perspectives of MXenes and their composites for exosome detection are promising, driven by additional explorations in materials science and biosensing technologies. Several key areas are presented in this section where MXenes could significantly impact exosome detection in the future:5.1. Enhanced functionalization techniques
Future research may focus on developing more efficient and versatile functionalization methods for MXenes. This could involve the use of novel bioconjugation strategies or the incorporation of multiple recognition elements to improve specificity and sensitivity for various exosome subtypes. By modifying the surface chemistry of MXenes, researchers can tailor their properties to selectively capture specific exosomes, thereby improving sensitivity and specificity. Recent advancements in functionalization strategies include the use of bioconjugation methods, which allow for the precise attachment of antibodies, aptamers, or other biomolecules to the MXene surface.23 This targeted approach not only facilitates the recognition of exosomal markers but also enhances the binding affinity, leading to more reliable detection outcomes. Additionally, the incorporation of functional groups, such as carboxyl or amine groups, can significantly improve the interaction between MXenes and exosomes, further boosting their effectiveness in biosensing applications. Moreover, employing hybrid materials—combining MXenes with nanomaterials—can amplify signal responses, resulting in enhanced detection limits.5.2. Integration with advanced sensing platforms
MXenes can be integrated into a variety of sensing platforms, such as electrochemical sensors, optical sensors, and field-effect transistors.72,93–95 Future developments may lead to hybrid systems that combine the strengths of different sensing modalities, enhancing detection capabilities and enabling real-time monitoring of exosome levels in complex biological samples. By combining MXenes with cutting-edge technologies such as microfluidics, surface plasmon resonance, and electrochemical detection systems, researchers can enhance the sensitivity and specificity of exosome analysis. This synergy allows for the development of multifunctional sensors capable of detecting low-abundance biomarkers, thus improving early diagnosis of diseases like cancer and neurodegenerative disorders. Furthermore, the incorporation of nanotechnology enables the miniaturization of sensing devices, facilitating their use in portable applications. These integrated platforms not only streamline the detection process but also allow for multiplexing capabilities, enabling simultaneous analysis of multiple exosomal biomarkers. Such advancements can lead to a more comprehensive understanding of disease mechanisms and patient status. As the integration of MXenes and their derivatives with advanced sensing platforms continues to progress, it holds immense potential for creating next-generation diagnostic tools that are faster, more accurate, and capable of delivering real-time insights into health conditions.5.3. Point-of-care applications
Point-of-care (POC) devices represent a significant advancement in exosome detection, allowing for rapid and on-site analysis. These portable devices are designed for use in clinical settings or remote locations, providing timely results without the need for complex laboratory equipment. By employing simple sample processing techniques and user-friendly interfaces, POC devices can facilitate the detection of exosomes in bodily fluids, enhancing diagnostic capabilities and enabling personalized medicine. The portability and ease of use of MXene-based sensors could facilitate their application in POC diagnostics.32,96 Future advancements may focus on miniaturizing these sensors and developing user-friendly interfaces, making it possible to detect exosomes in remote or resource-limited settings. By leveraging the high sensitivity and tunable properties of MXenes, researchers are creating compact devices capable of delivering real-time results, which is crucial for timely medical interventions. For instance, MXene-based sensors can quickly analyze patient samples, providing immediate insights into disease markers without the need for extensive laboratory infrastructure. This capability is particularly beneficial in resource-limited environments, where access to advanced diagnostic tools may be restricted. Moreover, the integration of user-friendly interfaces and smartphone connectivity enhances the practicality of these devices, allowing healthcare professionals to interpret results swiftly and make informed decisions. Additionally, lab-on-a-chip (LOC) devices have emerged as powerful tools for exosome detection. These microfluidic systems integrate multiple laboratory functions on a single chip, enabling the isolation, characterization, and analysis of exosomes in a compact format. LOC devices can streamline workflows, reduce sample volumes, and provide rapid results, making them particularly advantageous for high-throughput applications.97 By incorporating various detection methods within the chip, researchers can achieve efficient and sensitive exosome analysis.5.4. Personalized medicine
As the understanding of exosome roles in various diseases deepens, MXenes could play a crucial role in personalized medicine.98 By enabling the detection of specific exosome biomarkers associated with individual patients, MXene-based technologies could help tailor treatment strategies and monitor therapeutic responses more effectively. By harnessing the unique properties of MXenes, researchers can develop highly sensitive biosensors that identify specific exosomal biomarkers linked to various diseases. This enables clinicians to obtain detailed insights into a patient's condition, facilitating early diagnosis and targeted therapies. For instance, the ability to detect cancer-related exosomes can lead to more effective treatment plans that are customized to the molecular characteristics of a patient's tumor. Additionally, the non-invasive nature of exosome collection from bodily fluids, coupled with the rapid and accurate detection capabilities of MXene-based sensors, enhances the feasibility of implementing personalized medicine strategies in routine clinical practice.5.5. Multifunctional platforms
Future research may explore the development of multifunctional MXene-based platforms that not only detect exosomes but also provide additional functionalities, such as drug delivery or therapeutic applications.99,100 These innovative systems combine the unique properties of MXenes with other materials, enabling simultaneous detection and analysis of multiple biomarkers. For instance, researchers are integrating MXenes with fluorescence and electrochemical sensors, creating a hybrid platform that not only detects exosomes but also provides quantitative data on their composition.96,101 This multifaceted approach enhances diagnostic capabilities, allowing for a more comprehensive understanding of disease states. Furthermore, the adaptability of MXene materials facilitates the development of sensors that can be altered for various applications, from cancer diagnostics to monitoring therapeutic responses. As these multifunctional platforms evolve, they hold the potential to develop personalized medicine, offering rapid, reliable, and simultaneous detection of multiple exosome types, ultimately leading to improved patient outcomes and tailored treatment strategies.5.6. Sustainability and scalability
Future developments may also focus on sustainable production methods for MXenes and their composites, ensuring that their synthesis is environmentally friendly and economically viable.76,91 This will be important for the widespread adoption of MXene-based technologies in healthcare. Innovations in synthesis methods, such as using green chemistry approaches, have shown promise in reducing the environmental impact associated with MXene fabrication. Moreover, researchers are exploring the utilization of abundant and non-toxic precursors, which can make the production process more sustainable. On the scalability front, achieving mass production of high-quality MXenes remains a challenge. However, advancements in manufacturing techniques, including mechanochemical synthesis and scalable liquid-phase exfoliation, are paving the way for large-scale production.102,103 By addressing these sustainability and scalability issues, MXenes can transition from laboratory-scale applications to widespread use in clinical settings, ultimately contributing to a greener and more accessible future in exosome detection technologies.Greener techniques for the synthesis of MXenes focus on developing environmentally friendly, safer, and more sustainable methods compared to conventional approaches that often involve hazardous chemicals like hydrofluoric acid (HF). Some of these greener methods include electrochemical etching, molten salt etching, alkali etching, and fluoride-free chemical etching.104 The key advantage herein lies in avoiding the use of highly corrosive and environmentally harmful fluorinated acids like HF, which are commonly used in conventional etching techniques. Electrochemical etching is considered one of the safest and greenest techniques as it operates at room temperature, uses mild aqueous electrolytes, and avoids toxic reagents, thereby reducing chemical waste and energy consumption. Alkali etching with solutions such as sodium hydroxide (NaOH) offers a fluorine-free alternative with relatively straightforward processing and lower environmental impact, although high temperature and concentrated alkali conditions pose scalability challenges. Molten salt etching uses high-temperature molten salts (e.g., LiCl, KCl) to selectively remove the aluminum layers from MAX phases, providing improved safety and reduced environmental hazards compared to acid-based methods, but requires further optimization for large-scale production. Fluoride-free etching using acids like HCl or HNO3 also reduces toxic fluoride use but may face issues with etching control and repeatability.76,91,104 Additionally, mechanochemical synthesis of MXenes stands out as environmentally friendly method. This approach significantly reduces the need for solvents and harsh chemicals, enhancing sustainability compared to traditional methods. Conventional synthesis often involves high temperatures and complex chemical reactions, which can be detrimental to the environment. In contrast, mechanochemistry simplifies the process, minimizing ecological impact while maintaining efficiency and effectiveness in producing high-quality MXenes.102 These greener synthesis routes not only minimize the use of toxic chemicals but also enhance the scalability and cost-effectiveness of MXene production. They enable the fabrication of MXenes with desirable surface terminations and properties suitable for diverse applications such as energy storage, environmental remediation, and biomedicine. The green-synthesized MXenes exhibit good quality, hydrophilicity, and biocompatibility, making them promising for sustainable technologies. However, challenges remain in optimizing these strategies for industrial-scale production, including controlling product quality, reducing energy input, and managing byproducts responsibly. Additional explorations are directed at refining these greener approaches to make MXene synthesis more sustainable and commercially viable while maintaining or enhancing their unique material properties.76,91
6. Conclusion
MXenes and their composites have recently garnered attention for their potential in exosome detection. These materials exhibit unique properties, including high electrical conductivity, excellent mechanical flexibility, and tunable surface chemistry. Consequently, they are ideal candidates for biosensing applications. Researchers have developed MXene-based sensors that can effectively capture and analyze exosomes, which play a crucial role in cell communication and disease progression. This advancement allows for the non-invasive detection of biomarkers, leading to early diagnosis of various conditions, including cancer. Despite the significant progress, challenges remain in the practical application of MXenes for exosome detection. One major hurdle is the reproducibility and stability of MXene materials in different biological environments. Additionally, the functionalization of MXenes to enhance selectivity towards specific exosomes requires further optimization. Notably, the integration of these materials into existing detection platforms also poses a challenge, as compatibility with current technologies remains a concern. These obstacles hinder the widespread adoption of MXene-based sensors in clinical settings, limiting their potential impact on personalized medicine.Additional explorations aim to overcome current challenges by developing more stable and reproducible MXene materials. Moreover, advancements in nanotechnology and surface modification techniques will likely enhance the selectivity and sensitivity of these sensors. Recent breakthroughs include the development of hybrid MXene-based sensors that integrate nanostructures like gold nanoparticles, significantly enhancing sensitivity and specificity. Researchers are now employing advanced surface functionalization techniques to create selective binding sites tailored for specific exosome types, which improves detection accuracy. Additionally, the incorporation of machine learning algorithms into data analysis is developing the interpretation of sensor signals, enabling real-time monitoring of exosome profiles. These cutting-edge innovations not only promise enhanced performance but also pave the way for portable and user-friendly diagnostic devices, making them accessible for point-of-care applications.
Author contributions
Siavash Iravani: supervision, conceptualization, writing – review & editing, drafted Sections 1, 3, and 5; Atefeh Zarepour: writing – review & editing, drafted Section 2; Arezoo Khosravi: visualization, graphical abstract, writing – review & editing, drafted Section 2; Ali Zarrabi: supervision, writing – review & editing, drafted Sections 3, and 5; Ehsan Nazarzadeh Zare: writing – review & editing, drafted Sections 3 and 4; Rajender S. Varma: writing – review & editing, drafted Sections 3 and 4; Pooyan Makvandi: supervision, writing – review & editing, drafted Sections 3 and 4.Conflicts of interest
Author(s) declare no conflict of interest.Data availability
No data was used for the research described in the article.References
- R. Akhter and S. S. Maktedar, J. Materiomics, 2023, 9, 1196–1241 CrossRef.
- B. Anasori and Y. Gogotsi, Graphene 2D Mater., 2022, 7, 75–79 CrossRef.
- S. Iravani and R. S. Varma, Chem. Commun., 2022, 58, 7336–7350 RSC.
- E. Mostafavi and S. Iravani, Nano-Micro Lett., 2022, 14, 130 CrossRef CAS.
- D. H. Ho, Y. Y. Choi, S. B. Jo, J. M. Myoung and J. H. Cho, Adv. Mater., 2021, 33, 2005846 CrossRef CAS.
- V. Chaudhary, V. Khanna, H. T. Ahmed Awan, K. Singh, M. Khalid, Y. K. Mishra, S. Bhansali, C. Z. Li and A. Kaushik, Biosens. Bioelectron., 2023, 220, 114847 CrossRef CAS.
- J. Yoon, M. Shin, J. Lim, J.-Y. Lee and J.-W. Choi, Biosensors, 2020, 10, 185 CrossRef CAS.
- D. T. Nurrohman, N.-F. Chiu, Y.-S. Hsiao, Y.-J. Lai and H. S. Nanda, Biosensors, 2024, 14, 307 CrossRef CAS.
- S. Singh, A. Numan and S. Cinti, Biosens. Bioelectron., 2022, 216, 114635 CrossRef CAS.
- H. Xu and B. C. Ye, TrAC, Trends Anal. Chem., 2020, 123, 115773 CrossRef CAS.
- H. K. Kordasht and M. Hasanzadeh, Anal. Methods, 2020, 12, 2795–2811 RSC.
- M. I. Mosquera-Heredia, L. C. Morales, O. M. Vidal, E. Barceló, C. Silvera-Redondo, J. I. Vélez and P. Garavito-Galofre, Biomedicines, 2021, 9, 1061 CrossRef.
- S. Sonbhadra, Mehak and L. M. Pandey, Biosensors, 2023, 13, 802 CrossRef CAS.
- X. Ma, Y. Hao and L. Liu, Int. J. Nanomed., 2021, 16, 7575–7608 CrossRef CAS.
- S. M. I. Bari, F. B. Hossain and G. G. Nestorova, Sensors, 2021, 21, 7645 CrossRef.
- X. Zhu, Y. Zhang, M. Liu and Y. Liu, Biosens. Bioelectron., 2021, 171, 112730 CrossRef CAS.
- V. P. Chavda, A. Pandya, L. Kumar, N. Raval, L. K. Vora, S. Pulakkat, V. Patravale, Salwa, Y. Duo and B. Z. Tang, Nanotoday, 2023, 49, 101771 CrossRef CAS.
- X. Zhu, H. Chen, Y. Zhou, J. Wu, S. Ramakrishna, X. Peng, H. S. Nanda and Y. Zhou, Curr. Opin. Biomed. Eng., 2021, 18, 100280 CrossRef CAS.
- H. Su, X. You, Q. P. Wang, L. Li, M. Ge, L. Yang, W. F. Dong and Z. Chang, J. Mater. Chem. C, 2024, 12, 8561–8568 RSC.
- N. Arab, M. Hosseini and G. Xu, Biosens. Bioelectron., 2024, 265, 116623 CrossRef CAS.
- M. P. Bilibana, Adv. Sen. Energy Mater., 2023, 2, 100080 CrossRef.
- D. L. Pawara, R. S. Tade, S. N. Nangare, P. O. Patil, P. K. Deshmukh, B. A. Vyas, S. B. Bari and M. P. More, J. Ind. Eng. Chem., 2024, 145, 1–19 Search PubMed.
- R. Ibragimova, P. Erhart, P. Rinke and H. P. Komsa, J. Phys. Chem. Lett., 2021, 12, 2377–2384 CrossRef CAS.
- M. Mozafari and M. Soroush, Mater. Adv., 2021, 2, 7277–7307 RSC.
- L. Chen, X. Dai, W. Feng and Y. Chen, Acc. Mater. Res., 2022, 3, 785–798 CrossRef CAS.
- R. Garg and F. Vitale, MRS Bull., 2023, 48, 283–290 CrossRef CAS.
- H. Huang, C. Dong, W. Feng, Y. Wang, B. Huang and Y. Chen, Adv. Drug Delivery Rev., 2022, 184, 114178 CrossRef CAS.
- S. Iravani, A. Zarepour, E. Nazarzadeh Zare, P. Makvandi, A. Khosravi and A. Zarrabi, FlatChem, 2024, 48, 100759 CrossRef CAS.
- Y. Li, S. Huang, S. Peng, H. Jia, J. Pang, B. Ibarlucea, C. Hou, Y. Cao, W. Zhou, H. Liu and G. Cuniberti, Small, 2023, 19, 2206126 CrossRef CAS.
- C. Zhu, L. Li, Z. Wang, M. Irfan and F. Qu, Biosens. Bioelectron., 2020, 160, 112213 CrossRef CAS.
- P. K. Kalambate, N. S. Gadhari, X. Li, Z. Rao, S. T. Navale, Y. Shen, V. R. Patil and Y. Huang, TrAC, Trends Anal. Chem., 2019, 120, 115643 CrossRef CAS.
- D. Khorsandi, J.-W. Yang, Z. Ülker, K. Bayraktaroğlu, A. Zarepour, S. Iravani and A. Khosravi, Microchem. J., 2024, 197, 109874 CrossRef CAS.
- M. Huang, Z. Gu, J. Zhang, D. Zhang, H. Zhang, Z. Yang and J. Qu, J. Mater. Chem. B, 2021, 9, 5195–5220 RSC.
- H. Zhang, Z. Wang, F. Wang, Y. Zhang, H. Wang and Y. Liu, Talanta, 2021, 224, 121879 CrossRef CAS.
- J. An, H. Park, J. Kim, H. Park, T. H. Kim, C. Park, J. Kim, M. H. Lee and T. Lee, ACS Sens., 2023, 8, 3174–3186 CrossRef CAS.
- W. Feng, P. Xu, M. Wang, G. Wang, G. Li and A. Jing, Micromachines, 2023, 14, 138 CrossRef.
- L. Zhuang, Q. You, X. Su, Z. Chang, M. Ge, Q. Mei, L. Yang, W. Dong and L. Li, Sensors, 2023, 23, 3508 CrossRef CAS.
- Y. Guo, Y. Nie, P. Wang, Z. Li and Q. Ma, Talanta, 2023, 259, 124559 CrossRef CAS.
- Z. Qiu, D. Fan, X. H. Xue, J. Zhang, J. Xu, H. Lyu and Y. Chen, RSC Adv., 2022, 12, 14260–14267 RSC.
- Z. Liang, P. Wang, Z. Li, W. Li and Q. Ma, Anal. Chem., 2024, 96, 16443–16452 CrossRef CAS.
- H. Cui, T. Zheng, N. Qian, X. Fu, A. Li, S. Xing and X. F. Wang, Small, 2024, 20, 2402434 CrossRef CAS.
- D. Yang, W. Zhang, H. Zhang, F. Zhang, L. Chen, L. Ma, L. M. Larcher, S. Chen, N. Liu and Q. Zhao, Theranostics, 2020, 10, 3684 CrossRef CAS.
- S. Cho, H. C. Yang and W. J. Rhee, Process Biochem., 2020, 88, 197–203 Search PubMed.
- Y. Q. Koh, F. B. Almughlliq, K. Vaswani, H. N. Peiris and M. D. Mitchell, Front. Biosci., 2018, 23, 865–874 CrossRef CAS.
- P. Sharma, S. Ludwig, L. Muller, C. S. Hong, J. M. Kirkwood, S. Ferrone and T. L. Whiteside, J. Extracell. Vesicles, 2018, 7, 1435138 CrossRef.
- S. Busatto, G. Vilanilam, T. Ticer, W.-L. Lin, D. W. Dickson, S. Shapiro, P. Bergese and J. Wolfram, Cells, 2018, 7, 273 CrossRef CAS.
- J. Y. Kim, W.-K. Rhim, Y.-I. Yoo, D.-S. Kim, K.-W. Ko, Y. Heo, C. G. Park and D. K. Han, J. Tissue Eng., 2021, 12, 20417314211008626 CrossRef.
- Y. T. Kang, E. Purcell, C. Palacios-Rolston, T. W. Lo, N. Ramnath, S. Jolly and S. Nagrath, Small, 2019, 15, 1903600 CrossRef CAS.
- T. S. Martins, M. Vaz and A. G. Henriques, Anal. Bioanal. Chem., 2023, 415, 1239–1263 CrossRef CAS.
- R. K. Mishra, J. Sarkar, K. Verma, I. Chianella, S. Goel and H. Y. Nezhad, Open Ceram., 2024, 18, 100596 CrossRef CAS.
- N. Gao, J. Zhao, X. Zhu, J. Xu, G. Ling and P. Zhang, Acta Biomater., 2022, 154, 1–22 CrossRef CAS.
- S. Iravani and R. S. Varma, ACS Biomater. Sci. Eng., 2021, 7, 1900–1913 CrossRef CAS.
- S. Iravani and R. S. Varma, Nanomaterials, 2022, 12, 3360 CrossRef CAS.
- Y. Wang, S. Zhong, J. Lee and H. Chen, Adv. Funct. Mater., 2024, 34, 2405272 Search PubMed.
- H. Zhang, Z. Wang, F. Wang, Y. Zhang, H. Wang and Y. Liu, Anal. Chem., 2020, 92, 5546–5553 Search PubMed.
- H. Zhang, Z. Wang, Q. Zhang, F. Wang and Y. Liu, Biosens. Bioelectron., 2019, 124–125, 184–190 Search PubMed.
- Q. Zhang, F. Wang, H. Zhang, Y. Zhang, M. Liu and Y. Liu, Anal. Chem., 2018, 90, 12737–12744 CrossRef CAS.
- D. Fang, D. Zhao, S. Zhang, Y. Huang, H. Dai and Y. Lin, Sens. Actuators, B, 2020, 305, 127544 Search PubMed.
- Z. Wang, Z. Zhang, Y. Zhang, X. Xu, T. Shen, H. Pan and D. Chang, Talanta, 2023, 265, 124848 Search PubMed.
- Y. Wang, C. Xu, Y. Zhou, J. Lee, Q. Chen and H. Chen, Small, 2024, 20, 2308897 Search PubMed.
- Q. You, L. Zhuang, Z. Chang, M. Ge, Q. Mei, L. Yang and W. F. Dong, Biosens. Bioelectron., 2022, 216, 114647 CrossRef CAS.
- Z. Wang, Z. Zhang, H. Pan and D. Chang, Anal. Biochem., 2023, 676, 115233 CrossRef CAS.
- Y. Zhou, Q. Chen, S. Zhong, H. Liu, K. Koh and H. Chen, Biosens. Bioelectron., 2023, 237, 115493 CrossRef CAS.
- X. Sun, B. Chen, Z. Li, Y. Shan, M. Jian, X. Meng and Z. Wang, Chem. Eng. J., 2024, 488, 150835 CrossRef CAS.
- Y. Nie, P. Wang, S. Wang, Q. Ma and X. Su, ACS Sens., 2023, 8, 1850–1857 Search PubMed.
- P. L. Au-Doung, J. C. Chan, O. Y. H. Kui, M. K. Y. Ho, Y. T. Cheung, J. K. W. Lam, H. K. Chan, J. Brannan, K. C. C. Chan, A. M. Li and S. S. Y. Leung, Respir. Res., 2024, 25, 194 CrossRef.
- Z. Li, Z. Liang, P. Wang, W. Li, Y. Li, N. Liu and Q. Ma, Nano Lett., 2024, 24, 15878–15885 CrossRef CAS.
- Z. Li, P. Wang, Z. Liang, D. Wang and Q. Ma, Chem. Eng. J., 2024, 493, 152650 CrossRef CAS.
- S. Jung, U. Zafar, L. S. Kumar Achary and C. M. Koo, EcoMat, 2023, 5, e12395 Search PubMed.
- S. K. Bhardwaj, H. Singh, M. Khatri, K.-H. Kim and N. Bhardwaj, Biosens. Bioelectron., 2022, 202, 113995 Search PubMed.
- S. Hajian, D. Maddipatla, B. B. Narakathu and M. Z. Atashbar, Front. Sens., 2022, 3, 1006749 Search PubMed.
- D. Lu, H. Zhao, X. Zhang, Y. Chen and L. Feng, Biosensors, 2022, 12, 820 Search PubMed.
- B. Fu, J. Sun, C. Wang, C. Shang, L. Xu, J. Li and H. Zhang, Small, 2021, 17, 2006054 CrossRef CAS.
- X. Guan, Z. Yang, M. Zhou, L. Yang, R. Peymanfar, B. Aslibeiki and G. Ji, Small Stuct., 2022, 3, 2200102 CrossRef CAS.
- K. R. G. Lim, M. Shekhirev, B. C. Wyatt, B. Anasori, Y. Gogotsi and Z. W. Seh, Nat. Synth., 2022, 1, 601–614 CrossRef CAS.
- S. Iravani, Ceram. Int., 2022, 48, 24144–24156 Search PubMed.
- S. Iravani, A. Khosravi, E. Nazarzadeh Zare, R. S. Varma, A. Zarrabi and P. Makvandi, RSC Adv., 2024, 14, 36835–36851 Search PubMed.
- S. Iravani and R. S. Varma, Matter, 2022, 5, 3574–3576 Search PubMed.
- E. I. Yakubovich, A. G. Polischouk and V. I. Evtushenko, Biochem. (Moscow), Suppl. Ser., 2022, 16, 115–126 Search PubMed.
- N. Dilsiz, Transl. Oncol., 2024, 50, 102121 CrossRef CAS.
- R. Kalluri and V. S. LeBleu, Science, 2020, 367, eaau6977 CrossRef CAS.
- M. R. Ali, M. S. Bacchu, M. R. Al-Mamun, M. I. Hossain, A. Khaleque, A. Khatun, D. D. Ridoy, M. Aly Saad Aly and M. Z. Hossain Khan, Crit. Rev. Anal. Chem., 2022, 54(6), 1381–1398 CrossRef.
- S. Alwarappan, N. Nesakumar, D. Sun, T. Y. Hu and C.-Z. Li, Biosens. Bioelectron., 2022, 205, 113943 CrossRef CAS.
- A. Koyappayil, S. G. Chavan, Y.-G. Roh and M.-H. Lee, Biosensors, 2022, 12, 454 Search PubMed.
- J. Jiang, S. Bai, J. Zou, S. Liu, J.-P. Hsu, N. Li, G. Zhu, Z. Zhuang, Q. Kang and Y. Zhang, Nano Res., 2022, 15, 6551–6567 Search PubMed.
- R. A. Soomro, P. Zhang, B. Fan, Y. Wei and B. Xu, Nano-Micro Lett., 2023, 15, 108 CrossRef CAS.
- B. Farasati Far, N. Rabiee and S. Iravani, RSC Adv., 2023, 13, 34562–34575 RSC.
- N. Rabiee and S. Iravani, Mater. Chem. Horiz., 2023, 2, 171–184 Search PubMed.
- Z. U. D. Babar, B. D. Ventura, R. Velotta and V. Iannotti, RSC Adv., 2022, 12, 19590–19610 RSC.
- A. Khunger, N. Kaur, Y. K. Mishra, G. R. Chaudhary and A. Kaushik, Mater. Lett., 2021, 304, 130656 CrossRef CAS.
- T. Amrillah, C. A. Che Abdullah, A. Hermawan, F. N. Indah Sari and V. N. Alviani, Nanomaterials, 2022, 12, 4280 Search PubMed.
- F. Bu, M. M. Zagho, Y. Ibrahim, B. Ma, A. Elzatahry and D. Zhao, Nanotoday, 2020, 30, 100803 Search PubMed.
- A. Maleki, M. Ghomi, N. Nikfarjam, M. Akbari, E. Sharifi, M.-A. Shahbazi, M. Kermanian, M. Seyedhamzeh, E. Nazarzadeh Zare, M. Mehrali, O. Moradi, F. Sefat, V. Mattoli, P. Makvandi and Y. Chen, Adv. Funct. Mater., 2022, 32, 2203430 Search PubMed.
- A. Sinha, Dhanjai, H. Zhao, Y. Huang, X. Lu, J. Chen and R. Jain, TrAC, Trends Anal. Chem., 2018, 105, 424–435 Search PubMed.
- G. Yang, F. Liu, J. Zhao, L. Fu, Y. Gu, L. Qu, C. Zhu, J. J. Zhu and Y. Lin, Coord. Chem. Rev., 2023, 479, 215002 Search PubMed.
- L. Karadurmus, S. I. Kaya, A. Cetinkaya and S. A. Ozkan, TrAC, Trends Anal. Chem., 2023, 165, 117145 CrossRef CAS.
- M. S. Chiriacò, M. Bianco, A. Nigro, E. Primiceri, F. Ferrara, A. Romano, A. Quattrini, R. Furlan, V. Arima and G. Maruccio, Sensors, 2018, 18, 3175 CrossRef.
- D. J. Beetler, D. N. D. Florio, K. A. Bruno, T. Ikezu, K. L. March, L. T. Cooper Jr, J. Wolfram and D. Fairweather, Mol. Aspects Med., 2022, 91, 101155 CrossRef.
- S. Iravani and R. S. Varma, Matter, 2023, 6, 2628–2629 CrossRef CAS.
- A. A. P. R. Perera, K. A. U. Madhushani, B. T. Punchihewa, A. Kumar and R. K. Gupta, Materials, 2023, 16, 1138 Search PubMed.
- L. Lorencova, P. Kasak, N. Kosutova, M. Jerigova, E. Noskovicova, A. Vikartovska, M. Barath, P. Farkas and J. Tkac, Microchim. Acta, 2024, 191, 88 Search PubMed.
- S. Iravani, A. Zarepour, E. Nazarzadeh Zare, P. Makvandi, A. Khosravi, R. S. Varma and A. Zarrabi, Mater. Adv., 2024, 5, 8404–8418 Search PubMed.
- P. Chavalekvirat, W. Hirunpinyopas, K. Deshsorn, K. Jitapunkul and P. Iamprasertkun, Precis. Chem., 2024, 2, 300–329 Search PubMed.
- A. J. Y. Wong, K. R. G. Lim and Z. W. Seh, J. Mater. Res., 2022, 37, 3988–3997 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2025 |

