Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Assessment of hyaluronic acid-conjugated pH-sensitive liposomes for prednisolone delivery to activated macrophages†
Andreia
Marinho
 ab,
Salette
Reis
ab,
Salette
Reis
 b and
Cláudia
Nunes
b and
Cláudia
Nunes
 *ac
*ac
aLAQV, REQUIMTE, Faculdade de Farmácia, Universidade do Porto, R. Jorge de Viterbo Ferreira 228, 4500-313 Porto, Portugal. E-mail: cdnunes@ff.up.pt
bLAQV, REQUIMTE, Faculdade de Ciências, Universidade do Porto, R. Campo Alegre s/n, 4169-007 Porto, Portugal
cLAQV, REQUIMTE, Instituto de Ciências Biomédicas Abel Salazar, Universidade do Porto, R. Jorge de Viterbo Ferreira 228, 4500-313 Porto, Portugal
First published on 29th April 2025
Abstract
Background: inflammatory diseases, including rheumatoid arthritis and osteoarthritis, are major health problems worldwide, often treated with glucocorticoids. These exert anti-inflammatory effects by modulating macrophages and other cells involved in the inflammatory response. While they can be highly effective in managing inflammation, long-term use of glucocorticoids is associated with significant side effects, highlighting the need for targeted strategies and controlled release in specific tissues and cells. We propose the use of hyaluronic acid-conjugated pH-sensitive liposomes to deliver prednisolone (LipoHA:PDP) to activated macrophages. Materials & methods: we evaluated the cytotoxicity and targeting potential of LipoHA:PDP using the THP-1 cell line. Results: LipoHA:PDP significantly inhibited the release of inflammatory mediators and reduced NF-κB translocation to the nucleus. The liposomes also decreased reactive oxygen species (ROS) production. Moreover, LipoHA:PDP prevented albumin denaturation, which impacts immune recognition, inflammation, and tissue damage. Conclusion: LipoHA:PDP was revealed to be a promising nanotherapy to enhance the therapeutic efficacy and efficiency of prednisolone on chronic inflammation long-term treatment.
1. Introduction
Inflammation is a complex immune defense process of the body1,2 that occurs in response to noxious stimuli.1 The inflammatory response that follows, in a controlled form (acute inflammation) has the function of restoring homeostasis,2,3 through the elimination of pathogens, cellular debris and inflammatory mediators.2 However, if this process is not properly regulated, inflammation can become chronic, favoring the development of autoimmune diseases, inflammatory diseases1,2 and even cancer.1The search for a detailed understanding of the inflammatory response has led to the development of a variety of anti-inflammatory agents, including glucocorticoids,2 such as prednisolone, considered an essential anti-inflammatory by the World Health Organization (WHO).4 Despite their potent anti-inflammatory effects,5,6 prolonged glucocorticoid therapy can lead to a multitude of serious side effects.1,3,7 For this reason, the European League Against Rheumatism (EULAR) recommends the use of glucocorticoids for short periods, functioning as a transition therapy to other classes of anti-rheumatic drugs.8 However, to overcome the limitations associated with glucocorticoids, nanoscale drug delivery systems have been developed, where liposomes have assumed a prominent role.
Due to the versatility that liposomes present, their applications are evident in various literature sources. Ferreira-Silva et al.9 describe the use of liposomes for rheumatoid arthritis (RA), highlighting the significant role of prednisolone. Metselaar et al. performed a comparative study between methylprednisolone (Depo-Medrol®, commercial drug) and PEGylated liposomes loaded with prednisolone sodium phosphate. In this clinical trial (phase 3), 150 patients were evaluated and the results showed that the liposomal formulation is more effective than Depo-Medrol® in the treatment of RA flares.10
Knowing that in inflammatory conditions local acidification occurs,2 pH-sensitive liposomes can be even more effective than conventional ones, delivering drugs at the inflamed site and releasing them in a controlled manner.11,12 In this context, HA-conjugated pH-sensitive liposomes were formulated to load prednisolone.13 Previously, it was demonstrated that the developed liposomes present favorable characteristics for an intravenous or intra-articular administration, the release of the drug under acidic conditions and in vitro (L929 and RAW 264.7 cell lines) citocompatibility.13
In the present study, the safety profile was established and the efficacy of the formulation was proved through anti-inflammatory and antioxidant activities assessment. Thus, the citocompatibility of liposomes in human macrophage cell line (THP-1) and red blood cells (RBCs) was evaluated. Using in vitro models of inflammation, the anti-inflammatory capacity of liposomes was also evaluated, including their ability to inhibit the release of inflammatory cytokines (interleukin (IL)-1β, IL-6, IL-8, TNF-α, and chemokine (C–C motif) ligand 3 (CCL-3)), as well as the ability of liposomes to inhibit the nuclear factor-κB (NF-κB) signalling pathway. As numerous pieces of evidence suggest, there is an intimate correlation between oxidative stress and inflammation,14 thus reactive oxygen species (ROS) induction and scavenging capacity of the formulations was assessed.
2. Experimental section
2.1 Materials
Hyaluronic acid (HA, 250 kDa) was a kind gift from Bloomage Biotechnology (Bloomage Biotechnology Corporation, Jinan, China). Prednisolone disodium phosphate (PDP) was purchased from Tokyo Chemical Industry Co. Ltd (Tokyo, Japan). 1,2-Dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE) and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl) (NBD-DSPE) were purchase from Avanti® Polar Lipids Inc. (Alabaster, ALA, USA). Chloroform and methanol were acquired from Thermo Fisher Scientific (Waltham, MA, USA) and Atom Scientific (Cheshire, UK). Cholesterol hemisuccinate (CHEMS), Triton™ X-100, resazurin sodium salt, ethyldimethyl-aminopropyl-carbodiimide (EDC), N-hydroxysuccinimide (NHS), 2,2′-azinobis-(3-ethybenzothiazoline-6-sulfonic acid) diammonium salt (ABTS), sodium chloride (NaCl), potassium persulphate, chlorpromazine, filipin, cytochalasin-D, sodium azide, HEPES hemisodium salt, trypan blue solution, propidium iodide, phorbol 12-myristate 13-acetate (PMA), 2′,7′-dichlorofluorescein diacetate (DCFH-DA) and bovine serum albumin (BSA) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Roswell Park Memorial Institute medium (RPMI), 0.25% Trypsin-EDTA (1×), Penicillin–Streptomycin (Pen-Strep) and Heat Inactivated Fetal Bovine Serum (FBS) (origin: South America) were purchased from Gibco® by Life TechnologiesTM (Loughborough, Leicestershire, UK). NF-κB p65 Antibody (F-6) Alexa Fluor® 647 was purchased from Santa Cruz Biotechnology (Heidelberg, Germany). Hoechst 33342® and CellMaskTM were purchased from Invitrogen by Thermo Fisher Scientific (Waltham, MA, USA). THP-1 macrophage cell line (ATCC® TIB-202™) was obtained from the American Type Culture Collection (ATCC). Ultrapure water (18.2 MΩ cm) was purified by an Ultra-pure water system (Healforce, Shanghai, China).2.2 Synthesis of DPPE–HA conjugate
HA–DPPE conjugate synthesis was achieved using carbodiimide chemistry previously described by Gouveia et al.13 Briefly, 100 mg of HA was dissolved in 50 mL of ultrapure water, followed by incubation with 0.5 g of EDC and 0.52 g of NHS at pH 4.0 for 2 h at 37 °C. Followed by the addition of 100 mg of DPPE to the activated HA resulting solution and further pH adjustment to 8.6 (borate buffer solution). This reaction was maintained for 24 h at 37 °C. At the end of the incubation, the excess reagents and by-products were separated by centrifugation (2870 × g, 4 °C and 30 min) and were repeatedly (at least 3 times) washed with PBS buffer, reducing the pH back to the physiological level. The solution was lyophilized using LyoQuest 85 plus v.407 Telstar freeze dryer (Telstar® Life Science Solutions, Terrassa, Spain), yielding a white dry powder.2.3 Preparation of pH-sensitive liposomes
pH-sensitive liposomes were prepared by the thin-film hydration methods previously described by Gouveia et al.13 Briefly, DPPE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CHEMs and DPPE
CHEMs and DPPE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CHEMS
CHEMS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPE–HA lipid solution of 10 mM, respectively, in a molar ratio of 6.5
DPPE–HA lipid solution of 10 mM, respectively, in a molar ratio of 6.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3.5 and 6.5
3.5 and 6.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3.5
3.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.03, were prepared in a round-bottom flask by dissolving the amounts of lipids in a chloroform
0.03, were prepared in a round-bottom flask by dissolving the amounts of lipids in a chloroform![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol mixture (3
methanol mixture (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2). For drug formulations, PDP was dissolved only in methanol and then added to the remaining organic phase. The organic phase solvents were evaporated using a rotary-evaporated under nitrogen flow, with the round-bottom flasks immersed in a bath at 42 °C. After this process, the thin film was hydrated with HEPES-buffered solution at pH 7.4, followed by vigorous vortex stirring. The formulations were extruded three- and- ten-times, respectively, through 600- and 100-nm polycarbonate filter membranes, under the pressure of nitrogen gas and above the main phase transition temperature of the lipids mixture (65 °C).
2). For drug formulations, PDP was dissolved only in methanol and then added to the remaining organic phase. The organic phase solvents were evaporated using a rotary-evaporated under nitrogen flow, with the round-bottom flasks immersed in a bath at 42 °C. After this process, the thin film was hydrated with HEPES-buffered solution at pH 7.4, followed by vigorous vortex stirring. The formulations were extruded three- and- ten-times, respectively, through 600- and 100-nm polycarbonate filter membranes, under the pressure of nitrogen gas and above the main phase transition temperature of the lipids mixture (65 °C).
2.4 Leakage of PDP from liposomes in simulated synovial fluid
In vitro leakage of PDP from liposomes was performed using a Slide-A-Lyzer dialysis device (Slide-A-Lyzer™ MINI Dialysis Device, MWCO 10 kDa, Thermo Scientific™, MA, USA). The leakage profile was assessed in simulated synovial fluid (SSF, composition is given in ESI,† Table S1).15 Lipo:PDP with and without HA modification (0.5 mL at 1.5 mM in SSF) was added into the upper compartment of the dialysis device and 14 mL of the SSF was added to the lower compartment. Devices were then placed in an incubator shaker (ES-60, Lan Technics, USA) at 125 rpm at 37 °C. At regular intervals, aliquots of 1 mL were collected from the lower compartment and replaced with 1 mL of fresh buffer heated at 37 °C. The PDP amount that passed through the dialysis membrane was quantified by measuring the absorbance (248 nm), using a UV-Vis spectrophotometer (V-660, Jasco Corporation, MD, USA). Free PDP was used as a control.Mathematical models for PDP-release kinetics (including zero-order and first-order equations, Higuchi, Hixcon–Crowell and Korsmeyer–Peppas) were applied to each release profile to evaluate the mechanism of PDP release. The fitting of each model was evaluated based on the correlation coefficient (R2) values obtained.
2.5 Cell culture conditions
Monocytes (THP-1 cell line) were cultured at 37 °C and 5% CO2 atmosphere in RPMI-1640 supplemented with 10% FBS (v/v) and 1% Pen-Strep (v/v).For in vitro cellular studies in macrophages, THP-1 cells differentiation into mature macrophages-like state (M0 macrophages) was induced through incubation with 20 ng mL−1 of PMA for 24 h at 37 °C and 5% CO2 atmosphere. After this incubation time cells were treated with PMA, the culture medium was replaced with fresh medium without PMA and incubated for 24 h at 37 °C and 5% CO2 atmosphere.
2.6 Safety profile
 | (1) |
 | (2) |
2.7 Efficacy studies
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events were recorded and the auto-fluorescence of cells in PBS was used as control.
000 events were recorded and the auto-fluorescence of cells in PBS was used as control.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 diluted in 1% BSA overnight in a humidified chamber at 4 °C. In the next day, cells were washed with PBS and the nucleus was stained with Hoescht 33342® (Thermo Fisher Scientific, Waltham, MA, USA) for 10 min at room temperature, before visualization under CLSM.
100 diluted in 1% BSA overnight in a humidified chamber at 4 °C. In the next day, cells were washed with PBS and the nucleus was stained with Hoescht 33342® (Thermo Fisher Scientific, Waltham, MA, USA) for 10 min at room temperature, before visualization under CLSM.
Images were acquired on a Leica Stellaris 8 confocal microscope (LeicaMicrosystems, Wetzlar, Germany) equipped with the Leica Application Suite X package (LAS X) using a λex/λem of 650/668 nm (NF-κB), and λex/λem of 405/461 nm (Hoechst 33342®), with a resolution of 1024 × 1024 using a 63X/1.4 oil immersion objective. Images were analyzed using Fiji ImageJ software (version 2.0, National Institutes of Health). CLSM studies were performed at the Imaging by Confocal and Fluorescence Lifetime Laboratory, CEMUP, Portugal.
 | (3) |
Additionally, cellular imaging was performed using CLSM. For this, the cells were prepared and treated as described for the ROS measurement by flow cytometry. After incubation with DCFH-DA, cells were incubated with CellMask™, for the membrane staining. After 10 min of incubation, cells were washed with PBS and fixed using Formalin solution. The nucleus was stained using Hoechst 33342® (10 min). Images were acquired on a Leica Stellaris 8 confocal microscope (LeicaMicrosystems, Wetzlar, Germany) equipped with the Leica Application Suite X package (LAS X) using a λex/λem of 485/530 nm (DCFH-DA), λex/λem of ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 649/666 nm (CellMask™) and λex/λem of 405/461 nm (Hoechst 33342®), with a resolution of 1024 × 1024 using a 63X/1.4 oil immersion objective. Images were analyzed using Fiji ImageJ software (version 2.0, National Institutes of Health). CLSM studies were performed at the Imaging by Confocal and Fluorescence Lifetime Laboratory, CEMUP, Portugal.
649/666 nm (CellMask™) and λex/λem of 405/461 nm (Hoechst 33342®), with a resolution of 1024 × 1024 using a 63X/1.4 oil immersion objective. Images were analyzed using Fiji ImageJ software (version 2.0, National Institutes of Health). CLSM studies were performed at the Imaging by Confocal and Fluorescence Lifetime Laboratory, CEMUP, Portugal.
 | (4) |
2.8 Statistical analysis
GraphPad Prism 10 Software (San Diego, CA, USA) was used to perform the statistical analysis. Statistical comparisons of the mean between groups were assessed by one-way or two-way ANOVA. Differences were considered significant with a p-value under 0.05. Data are represented as mean ± SD.3. Results and discussion
In the study by Gouveia et al.,13 LipoHA : PDP liposomes were comprehensively evaluated for their physicochemical properties, including size, zeta potential, encapsulation efficiency, load capacity, and morphology using transmission electron microscopy. Additionally, the biocompatibility of these liposomes was demonstrated through experiments on L929 and RAW 264.7 cell lines. The present study establishes the proof-of-concept efficacy of LipoHA : PDP liposomes (Fig. 1(A)) as an improved formulation for addressing arthritic conditions. Thus, the biocompatible concentrations of liposomes were established in THP-1 cell line and RBCs. Subsequently, the anti-inflammatory efficacy of liposomes, including their ability to inhibit the release of inflammatory mediators and NF-κB signalling pathway, was evaluated through in vitro models. And finally, their capacity to reduce oxidation was also assessed.3.1 Leakage of PDP from liposomes in simulated synovial fluid
Pioneers in the field of drug delivery systems, liposomes are the most successfully studied nanocarrier for local and systemic drug delivery.20 In the particular case of joint diseases, such as RA and osteoarthritis, intra-articular injections are still an attractive strategy for drug delivery.21 In these cases, the use of nanostructures such as liposomes the drug's efficacy and reduces off-target adverse effects.20 However, the joint environment may interfere with the performance of the liposomal formulations under study. In this sense, it becomes crucial to assess the release of PDP from liposomes in synovial fluid. Effectively, the composition of synovial fluid is more complex than the composition of solutions used in vitro for leakage studies.22 This is because the composition of synovial fluid is similar to the composition of plasma in terms of organic molecules and electrolytes.23 Contrary to what happens in plasma, synovial fluid still has a high content of HA, which is responsible for its viscosity.23In vivo, the amount of synovial fluid is very limited,22 which makes it difficult to use it in studies that would need to replicate the intrinsic environment of the joint. Therefore, to mimic this environment, solutions based on phosphate buffer and HA have been used, which allow recreating the viscosity of the synovial fluid,23 called simulated synovial fluid (SSF).Analyzing the data obtained (Fig. 1(B)), it is possible to observe that the amount of free PDP in the SSF is superior to the leakage of PDP in the liposomes, thus showing the controlled release of PDP from the liposomes, which constitutes an advantage for the intra-articular delivery of PDP. However, no clear differences are observed between Lipo:PDP and LipoHA:PDP. Despite this, the presence of HA in liposomes may be important for targeting purposes, since it can interact with the CD44 receptor expressed in most cells,24 including activated macrophages.13 The literature reports that the CD44-HA interaction plays an important defense role, being associated with the recruitment of leukocytes to inflamed sites.25
The release kinetics profile of PDP was studied by applying the mathematical models above mentioned (Section 2.4), and the main results are presented in Fig. 1(C). The values obtained for the correlations coefficient (R2) (ESI,† Table S3) show that the best fit, for both liposomal formulations, was obtained by the Higuchi model (Fig. 1(D)). This kinetics release model assumes that the drug's release occurs by a diffusion mechanism.26 In these situations, drug release is mediated by a concentration gradient.26,27 Mathematically, this diffusion is represented by Fick's law of diffusion,27,28 which states that the diffusion flux is proportional to the concentration gradient.28
3.2 Safety profile assessment
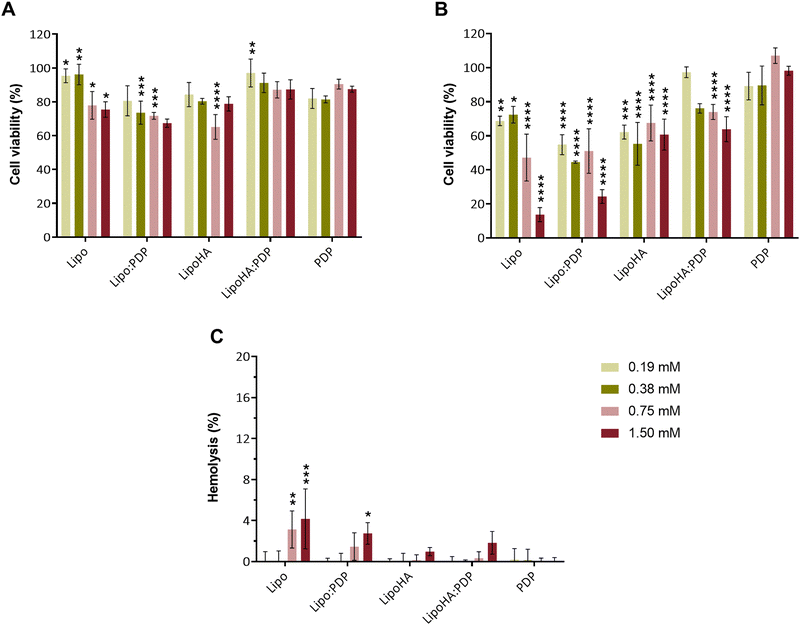
The cell viability of liposomal formulations with and without PDP was evaluated in monocytes and macrophages (THP-1 cells), after 24 h of incubation. The results (Fig. 2(A) and (B)) show that free PDP has no cytotoxic effects, with cell viability greater than 80% at all concentrations studied, both in macrophages and monocytes. The results obtained for differentiated cells (Fig. 2(A)) do not show cytotoxic effects associated with liposomal formulations, both in the absence and presence of PDP, at all concentrations used. On the other hand, in monocytes (Fig. 2(B)) it can be seen that except for LipoHA:PDP, all liposomal formulations have a viability percentage below 70%. For LipoHA:PDP there is an increase in the viability percentage, only at a concentration of 1.50 mM this value is slightly below 70%. In any case, it is important to mention that studies performed on monocytes are inherently associated with greater variability and errors since cells are not adhered. Thus, the loss of cells in an inhomogeneous way will influence the conversion of resazurin to resorufin, resulting in lower cell viability values.The literature reports the key role that macrophages play in the development of chronic inflammatory diseases, such as RA. In the particular case of this disease, there is an increase in the number of macrophages in the synovial tissue, which are responsible for the production of inflammatory cytokines and for contributing to the degradation of cartilage and bone.29 Therefore, the results obtained in the cell viability studies show the therapeutic potential of the formulations developed for the targeted delivery of PDP to activated macrophages.
As mentioned earlier, although prednisolone is referred to as an essential anti-inflammatory by the WHO, its prolonged use is associated with side effects. We also mentioned that these side effects could be overcomed using nanotechnology to develop drug delivery systems, such as liposomes. In the body, liposomes are exposed to a series of biological barriers that they need to overcome to exert the effect for which they were designed.30 One of these barriers is the bloodstream, since after administration the liposomes inevitably end up in contact with this fluid. Once in the blood, liposomes face a series of interactions with biological molecules that can lead to changes in surface chemistry, which in turn can lead to changes in biological response.16,31 As such, when drug delivery systems are designed to target a specific location after circulating in the bloodstream for long periods, their compatibility with the blood is of great importance.32
The hemocompatibility of the liposomal formulations was assessed through a hemolysis assay using human RBCs collected from 3 healthy donors. Hemolysis assay was carried out in a concentration range from 0.19 mM to 1.50 mM, and possible toxicity was analyzed after 1 h, through the percentage of hemolysis. The results presented in Fig. 2(C) show that the hemolysis values obtained were less than 5% in all cases, with a maximum hemolysis percentage of approximately 4%, obtained for Lipo at a concentration of 1.50 mM. According to the rate of hemolysis obtained, liposomes can be classified into (1) non-hemolytic – hemolysis rate below 2%, (2) slightly hemolytic – hemolysis rate between 2 and 5%, and (3) hemolytic – more than 5% of hemolysis.33,34 Although all hemolysis values obtained are less than 5%, the profile obtained indicates that the introduction of HA functionalization leads to lower hemolytic rates, so LipoHA:PDP is indicated for intravenous administration. Effectively, Mota et al., demonstrated that nanoparticles functionalized with HA (HA concentrations that vary between 4.7 × 10−5 to 8.34 μM) result in hemolytic rates lower than 1%.35 in another example, Janik-Hazuka et al., demonstrate that oil-core nanocapsules based on a derivative of HA, used for the administration of compounds derived from garlic oil, are capable of reducing the hemolytic rate to about half compared to the free compound.36 Similarly, Xiao et al., showed that the functionalization of cationic liposomes with HA also leads to a reduction (below 5%) in the hemolytic rate,37 which corroborates the results obtained.
3.3 Efficacy assessment
As described in the literature, macrophages play an essential role in the pathogenesis of inflammatory joint diseases, so the cellular studies in the following subsections will be carried out with differentiated cells.It is known that macrophages have the ability to internalize particles up to 300 nm by mechanisms of endocytosis mediated by clathrin, caveolae, phagocytosis and micropinocytosis.39 In the case of pH-sensitive liposomes, it is known that they are internalized, in their stable form, by endocytic pathways, undergoing subsequent destabilization in endosomes (acid pH).38 In this sense, to evaluate the mechanism of cellular uptake of liposomal formulations, several uptake inhibitors were used. Additionally, it was evaluated whether this cellular uptake mechanism is an active energy-dependent process.
To verify the uptake of Lipo and LipoHA was energy-dependent, a temperature of 4 °C and sodium azide were used. Fig. 3(B) shows a significant decrease in the internalization of both Lipo and LipoHA formulations at 4 °C. Reduced temperatures cause the plasma membrane to become rigid, making passive diffusion more challenging or even preventing it altogether.10 The uptake of Lipo and LipoHA in the presence of endocytosis inhibitors was also evaluated. For both formulations, the most evident effect is a decrease in cellular uptake in the presence of cytochalasin-D. This inhibitor is primarily associated with macropinocytosis and, to a lesser degree, with caveolae-mediated processes.39 It can also lead to the depolymerization of actin filaments present in the cell's cytoskeleton, which influences macropinocytosis-mediated internalization.40 Taken together, these data show that both Lipo and LipoHA formulations are internalized by energy-requiring mechanisms and that the main mechanism involved in the depolymerization of actin filaments. In LipoHA, the impact of cytochalasin-D is more pronounced, as it relates to the CD44 receptor to which HA can bind. Studies indicate the association of the cytoplasmic tail of the CD44 receptor with actin filaments,41,42 facilitating the binding of HA to CD44 via the actin cytoskeleton.43 Thus, inhibition of Lipo and LipoHA internalization by cytochalasin-D indicates that the interaction of the cytoplasmic domain of the CD44 receptor with the cytoskeleton is important for receptor localization and it can regulate CD44-HA binding,44 that is, blocking actin polymerization will inhibit HA binding.44,45
Chlorpromazine and filipin were also employed to assess the involvement of clathrin- and caveolae-mediated endocytosis, respectively. Chlorpromazine is a cationic compound that disrupts clathrin-mediated endocytosis by interfering with adaptor protein 2 (AP2), a protein complex essential for the formation of clathrin-coated vesicles.46 On the other hand filipin interacts with cholesterol in the cell membrane, disrupting its organization and thereby inhibiting caveolae-mediated endocytosis.47
Fig. 4(A)–(E) shows the values obtained for the secretion of interleukins (IL-1β, IL-6, IL-8, TNF-α and CCL-3) after exposure of activated macrophages to the developed liposomes. In general, liposomal formulations, and free PDP, are found to inhibit the release of inflammatory mediators, except for IL-8 (Fig. 4(C)) where there is no inhibition for Lipo and PDP. However Lipo:PDP and LipoHA:PDP can inhibit the release of IL-8. It is also important to note that for IL-6 (Fig. 4(B)) no results are presented for Lipo:PDP since it is below the detection limit, which is still indicative of a suppression of IL-6 release.
The findings align with existing literature, exemplified by Schweingruber et al.'s study, where liposomes (DPPC:DSPE-mPEG200:Chol) loaded with prednisolone demonstrated reduced expression of IL-1β and IL-6 in peritoneal macrophages isolated from C57BL/6 wild type mice.50 Additionally, Bartneck et al. demonstrated in another study that unloaded liposomes (DPPC:DSPE-PEG200:Chol) effectively inhibited the release of IL-1β, IL-6, and TNF-α.51
Taken together, the results obtained show that liposomal formulations can suppress the release of inflammatory mediators and thus regulate inflammation. Note further that the functionalization of liposomes with HA leads to greater suppression of the release of these inflammatory mediators, evident essentially in IL-1β, IL-8 and TNF-α. Except for IL-8, it is also verified that LipoHA:PDP has a superior suppression of inflammatory mediators. This shows LipoHA:PDP potential as anti-inflammatory agent.
Nuclear factor-κB (NF-κB) represents a family of transcription factors,52,53 that play an important role in responding to external stimuli,54 thus playing an important role in immune homeostasis.55 Despite this, inadequate regulation of NF-κB is associated with inflammatory diseases53,55–57 and has already been detected in human synovial tissue during the joint inflammation process.53,58
In non-stimulated cells, NF-κB is found in the cytoplasm in its inactive form, through interaction with the inhibitory protein IκB58–61 which masks NF-κB subunits.60 When cells are exposed to stimuli such as cytokines, LPS, UV radiation, and free radicals, NF-κB activation is induced.55,56 After the stimulus, phosphorylation of the IκB protein is triggered and its consequent degradation,55,56,58,59 which leads to the translocation of NF-κB to the nucleus where it can trigger the expression of several molecules’ flags.56,58,59 In all cell types involved in the inflammatory response, coordinated activation of this transcription factor occurs.58 It is also known to play a crucial role in the transcription of M1 macrophages53 and to be involved in the induction of pro-inflammatory cytokines such as IL-1β, IL-6, TNF-α,53,56,62 IL-856,62 and CCL-3.56
As depicted in Fig. 4(F) through confocal microscopy, NF-κB is primarily located in the cytosol of M0 macrophages. However, upon polarization to M1 macrophages (via LPS exposure), NF-κB translocates to the nucleus. Upon treatment of activated macrophages with either liposomal formulations or PDP, inhibition of NF-κB translocation to the nucleus is observed, indicating the anti-inflammatory efficacy of both liposomal formulations and PDP.
It is known that the glucocorticoid receptor (GR), after binding to glucocorticoids, is translocated to the nucleus where it regulates the expression of target genes acting as an anti-inflammatory and immunosuppressive agent.63 Indeed, GR induces the synthesis of the inhibitory protein IκB63,64 and represses NF-κB activation through protein–protein interactions that block access to the NF-κB binding site.63 Despite this, the literature already reposts other works that support the results obtained, as is the case of the work developed by Gouveia et al. where is reported that pH-responsive polymersomes are capable of reducing the translocation to the nucleus of NF-κB, both in the presence and absence of PDP, despite this, in the presence of PDP the effect is more pronounced.62
It is already established that protein denaturation is one of the causes of inflammatory and arthritic diseases.65,66 Pathologies such as rheumatoid arthritis and lupus erythematosus may originate from a type III hypersensitivity reaction.65–67 This reaction, during protein denaturation processes, leads to the production of antigens65,66 which, in turn, trigger the production of pro-inflammatory cytokines.67 Substances that prevent protein denaturation may act as a potential antiarthritic agent.65–67 In this sense, we evaluated the ability of PDP and liposomal formulations to prevent protein denaturation, where bovine serum albumin (BSA) was used as a model. Albumin represents about 60% of the total protein in the blood and undergoes denaturation when exposed to heat, expressing antigens that are associated with the type III hypersensitivity reaction.68
The relative percentage of inhibition of BSA denaturation (Fig. 4(G)) by the liposomal formulations, at 1.5 mM, ranged from 98.6% to 99.1%. In the case of PDP, the value obtained was 99.4%. These results show that the liposomes under study have potential as antiarthritic agents. However, similar to the results that have been obtained in previous assays, LipoHA and LipoHA:PDP show an improved inhibition over non-functionalizing liposomes.
We evaluated the intracellular generation of ROS in macrophages (Fig. 5(A) and (B)) in response to exposure to liposomal and PDP formulations. Fig. 5(A) shows the results obtained for the DCFH-DA assay using confocal microscopy. As evidenced by the images presented, it appears that the generation of ROS occurs both in the presence of liposomal formulations and PDP. Despite this, it appears that in all cases the generation of ROS is lower than in the presence of H2O2, used as a positive control. Similarly, the results obtained by flow cytometry (Fig. 5(B)) show that the developed formulations, as well as PDP, induce less ROS production than the control (H2O2). Notably, Lipo and PDP exhibit very similar inhibition, approximately 58%, while surface functionalization of liposomes with HA leads to reduced values, particularly noticeable in LipoHA:PDP, where values decrease to approximately 43%.
Although these results show that liposomal formulations slightly induce ROS production, this induction was not superior to PDP control, thus showing that liposomes can be useful, in this respect, as a vehicle for PDP. As mentioned, LipoHA:PDP was the formulation that presented the most promising results, which, once again, proves the potential application of this formulation for the administration of PDP.
While liposome formulations have been found to induce ROS production, the same is true of other examples in the literature. In the work developed by Wan et al., it is reported that liposomes (DSPC:DSPE-PEG) without drug induce ROS in the same percentage as the positive control. When liposomes are loaded with a drug, there is a decrease in ROS generation compared to unloaded liposomes, however, they have a higher percentage of ROS than free drug.74 In another study, Sanner et al., show that prednisolone is capable of inhibiting the generation of ROS, in platelets.75 Although they are a product of normal cellular metabolism,76,77 free radicals are highly reactive and capable of initiating cell-damaging chain reactions In this sense, the 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) scavenging assay was used to verify the antioxidant capacity of the developed liposomal formulations. The results present in Fig. 5(C) show an antioxidant effect for HEPES – the liposome production medium. Despite this, an antioxidant capacity of liposomal formulations and free PDP is also observed. These effects are visible on a large scale up to 1 h of testing, however, an antioxidant capacity superior to HEPES can be observed throughout the entire test time. It should be noted that LipoHA and LipoHA:PDP are more efficient than Lipo and Lipo:PDP.
Several studies have explored the use of liposome-based drug delivery systems and HA for the delivery of prednisolone. An example is the work reported by Tsai et al., which describes HA-containing prednisolone-encapsulated liposomes. Although this represents a distinct approach, it also emphasizes the benefits of using HA in combination with liposomal formulations to enhance anti-inflammatory effects.78 Despite the different design, our formulation shows a more controlled release profile (approximately 80% over 24 h), in contrast to the rapid release reported for the hydrogel system (nearly 100% in 150 min). Additionally, while Tsai et al. focused on nitric oxide reduction,78 our system demonstrated a broader modulation of inflammatory mediators. The liposomes also exhibited favorable safety features, including non-hemolytic behavior and no protein denaturation, further supporting their potential for therapeutic applications.
4. Conclusions
Inflammatory joint diseases often go undetected until they reach advanced stages due to the absence of specific biological markers. As there is currently no cure for these conditions, preventing joint degradation becomes paramount. While there are drugs available on the market that effectively manage inflammation, such as glucocorticoids, their prolonged use often results in undesirable side effects. To address these issues and maximize the potential of known molecules already used in clinical settings, nanotechnology has been developing drug delivery systems, with liposomes emerging as prominent players in this endeavor.Here we present pH-sensitive liposomes where prednisolone, a potent anti-inflammatory, is incorporated. The results demonstrate that liposomes can release PDP in a controlled way in an environment that mimics the joint environment. Furthermore, the developed liposomes do not show toxicity up to 1.5 mM concentration, in macrophages. The internalization of liposomes by macrophages and their ability to attenuate the release of inflammatory mediators was demonstrated. Moreover, similarly to what happens with prednisolone, liposomal formulations are capable of suppressing the activation of the NF-κB signalling pathway, which shows, once again, their anti-inflammatory capacity.
To increase targeting to activated macrophages, functionalization with HA was performed. This strategic step in the design of liposomal formulations proved to be of great importance. The results presented show that the internalization of liposomes may be dependent on the availability of the CD44 receptor expressed on the surface of macrophages. In addition, it is shown that functionalized liposomes, in general, have greater anti-inflammatory capacity, are non-hemolytic and do not promote protein denaturation. Given the formulation features, LipoHA:PDP has potential as an intravenous or intra-articular anti-inflammatory agent.
Thus, the pH-sensitive liposomes conjugated with HA hold promise as a nanotherapeutic approach for treating chronic inflammation, potentially mitigating the side effects linked to prolonged prednisolone therapy.
Author contributions
Andreia Marinho performed the whole experiments and wrote the first draft of the manuscript. Salette Reis provided helpful discussions. Cláudia Nunes designed, supervised the experiments, and prepared the final version of the manuscript. All authors have read and agree to the published version of the manuscript.Data availability
All data generated or analyzed during this study are included in this published article and its ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work received financial support from the PT national funds (FCT/MCTES, Fundação para a Ciência e Tecnologia and Ministério da Ciência, Tecnologia e Ensino Superior) through the project UID/50006 - Laboratório Associado para a Química Verde - Tecnologias e Processos Limpos. This research is supported by HEALTH-UNORTE I&D&I project (NORTE-01-0145-FEDER-000039) co-financed by European Regional Development Fund (ERDF), through the NORTE 2020 (Programa Operacional Regional do Norte 2014/2020). Andreia Marinho acknowledges funding from the project Norte-08-5369-FSE-000050. Cláudia Nunes thanks FCT (Fundação para a Ciência e Tecnologia) for funding through the Individual Call to Scientific Employment Stimulus [2022.05608.CEECIND/CP1724/CT0002]. The authors are grateful to Andreia Granja (LAQV/REQUIMTE, FFUP) for all technical assistance and in helping with manufacturing protocols for the flow cytometry and CLSM and to Ana Isabel Barbosa (LAQV/REQUIMTE, FFUP) for helping with the ABTS assay protocol. The authors are grateful for the technical support from the Imaging by Confocal and Fluorescence Lifetime Laboratory (CEMUP, UP) and from Manuela Barros and Vânia Dias from the Applied Chemistry Department from FFUP.References
- S. Ronchetti, G. Migliorati, S. Bruscoli and C. Riccardi, Clin. Sci., 2018, 132, 1529–1543 CrossRef CAS PubMed.
- Z. Deng and S. Liu, Drug Delivery Transl. Res., 2021, 11, 1475–1497 CrossRef CAS PubMed.
- A. E. Coutinho and K. E. Chapman, Mol. Cell. Endocrinol., 2011, 335, 2–13 CrossRef CAS PubMed.
- W. H. Organization, 22nd World Health Organization Model List of Essential Medicines, https://www.who.int/publications/i/item/WHO-MHP-HPS-EML-2021.02, (accessed March 9, 2022, 2022).
- E. Kauh, L. Mixson, M.-P. Malice, S. Mesens, S. Ramael, J. Burke, T. Reynders, K. Van Dyck, C. Beals, E. Rosenberg and M. Ruddy, Eur. J. Endocrinol., 2012, 166, 459–467 CAS.
- A. C. Liberman, M. L. Budziñski, C. Sokn, R. P. Gobbini, A. Steininger and E. Arzt, Front. Endocrinol., 2018, 9, 235 CrossRef PubMed.
- S. D. Reichardt, A. Amouret, C. Muzzi, S. Vettorazzi, J. P. Tuckermann, F. Lühder and H. M. Reichardt, Cells, 2021, 10, 2921 CrossRef CAS PubMed.
- S. S. Josef, B. M. L. Robert, B. Sytske Anne, K. Andreas, S. Alexandre, A. Daniel, C. Roberto, E. Christopher John, L. H. Kimme, E. P. Janet, S. Savia de, A. S. Tanja, T. Tsutomu, V. Patrick, L. W. Kevin, B. Alejandro, M. B. Joan, H. B. Maya, R. B. Gerd, B. Frank, C. Mario Humberto, C. Katerina, C. Catalin, C. Maurizio, A. d B. Alfons, A. Khadija El, F. Axel, F. João Eurico, G. Jacques-Eric, A. H. Espen, I. Annamaria, L. Kim, L. Zhanguo, B. M. Iain, F. M. Eduardo, N. Peter, P. Gyula, G. R. Gorica, R. Felice, R.-R. Andrea, S.-K. Hendrik, S. Nikolay, S. Anja, M. Annette van der Helm-van, D. Elsa van, P. M. V. V. Theodora, W. René and H. Désirée van der, Ann. Rheum. Dis., 2023, 82, 3 Search PubMed.
- M. Ferreira-Silva, C. Faria-Silva, P. Viana Baptista, E. Fernandes, A. Ramos Fernandes and M. L. Corvo, Pharmaceutics, 2021, 13, 454 CrossRef CAS PubMed.
- J. M. Metselaar, L. M. Middelink, C. H. Wortel, R. Bos, J. M. van Laar, H. E. Vonkeman, R. Westhovens, T. Lammers, S.-L. Yao, M. Kothekar, A. Raut and J. W. J. Bijlsma, J. Controlled Release, 2022, 341, 548–554 CrossRef CAS PubMed.
- S. R. Paliwal, R. Paliwal and S. P. Vyas, Drug Delivery, 2015, 22, 231–242 CrossRef CAS PubMed.
- M. Abri Aghdam, R. Bagheri, J. Mosafer, B. Baradaran, M. Hashemzaei, A. Baghbanzadeh, M. de la Guardia and A. Mokhtarzadeh, J. Controlled Release, 2019, 315, 1–22 CrossRef CAS PubMed.
- V. Gouveia, J. Lopes-de-Araújo, S. Costa Lima, C. Nunes and S. Reis, Nanomedicine, 2018, 13, 1037–1049 CrossRef CAS PubMed.
- T. Hussain, B. Tan, Y. Yin, F. Blachier, M. C. Tossou and N. Rahu, Oxid. Med. Cell. Longevity, 2016, 2016, 7432797 CrossRef PubMed.
- M. Marques, R. Löbenberg and M. Almukainzi, Dissolution Technol., 2011, 18, 15–28 CrossRef CAS.
- S. Moraes, A. Marinho, S. Lima, A. Granja, J. P. Araújo, S. Reis, C. T. Sousa and C. Nunes, Int. J. Pharm., 2021, 592, 120029 CrossRef CAS PubMed.
- P. Daram, S. R. Jitta, C. S. Shreedhara, C. S. Misra, K. Gourishetti and R. Lobo, S. Afr. J. Bot., 2021, 141, 313–321 CrossRef CAS.
- S. Chataut, S. Sharma, S. Sah, P. Shrestha, S. Nepali, S. Mallik, D. B. Poudel and R. Bhusal, Int. J. Prev. Med., 2015, 1, 201–206 Search PubMed.
- L. Esposito, A. I. Barbosa, T. Moniz, S. Costa Lima, P. Costa, C. Celia and S. Reis, Pharmaceutics, 2020, 12, 1149 CrossRef CAS PubMed.
- C. M. A. van Alem, J. M. Metselaar, C. van Kooten and J. I. Rotmans, Pharmaceutics, 2021, 13, 1004 CrossRef CAS PubMed.
- C. H. Evans, V. B. Kraus and L. A. Setton, Nat. Rev. Rheumatol., 2014, 10, 11–22 CrossRef CAS PubMed.
- E. L. Bortel, B. Charbonnier and R. Heuberger, Lubricants, 2015, 3, 664–686 CrossRef.
- M. Thing, N. Mertz, L. Ågårdh, S. W. Larsen, J. Østergaard and C. Larsen, J. Drug Delivery Sci. Technol., 2019, 49, 169–176 CrossRef CAS.
- A. Marinho, C. Nunes and S. Reis, Biomolecules, 2021, 11, 1518 CrossRef CAS PubMed.
- A. R. Jordan, R. R. Racine, M. J. P. Hennig and V. B. Lokeshwar, Front. Immunol., 2015, 6, 182 Search PubMed.
- G. Singhvi and M. Singh, Int. J. Pharm. Sci. Res., 2011, 2, 77–84 Search PubMed.
- N. A. Sawaftah, V. Paul, N. Awad and G. A. Husseini, IEEE Trans. NanoBiosci., 2021, 20, 565–576 Search PubMed.
- M. T. Thompson, in Intuitive Analog Circuit Design, ed. M. T. Thompson, Newnes, Boston, 2nd edn, 2014, pp. 53–86 DOI:10.1016/B978-0-12-405866-8.00003-6.
- C.-H. Lee and E. Y. Choi, J. Rheum. Dis., 2018, 25, 11–18 CrossRef.
- E. Blanco, H. Shen and M. Ferrari, Nat. Biotechnol., 2015, 33, 941–951 CrossRef CAS PubMed.
- D. Sobot, S. Mura and P. Couvreur, in Encyclopedia of Polymeric Nanomaterials, ed. S. Kobayashi and K. Müllen, Springer Berlin Heidelberg, Berlin, Heidelberg, 2015, pp. 1352–1360 DOI:10.1007/978-3-642-29648-2_227.
- S. Mourtas, G. P. Michanetzis, Y. F. Missirlis and S. G. Antimisiaris, J. Biomed. Nanotechnol., 2009, 5, 409–415 CrossRef CAS PubMed.
- M. Weber, H. Steinle, S. Golombek, L. Hann, C. Schlensak, H. P. Wendel and M. Avci-Adali, Front. Bioeng. Biotechnol., 2018, 6, 99 CrossRef PubMed.
- S. Aula, S. Lakkireddy, K. Jamil, A. Kapley, A. V. N. Swamy and H. R. Lakkireddy, RSC Adv., 2015, 5, 47830–47859 RSC.
- A. H. Mota, R. Direito, M. P. Carrasco, P. Rijo, L. Ascensão, A. S. Viana, J. Rocha, M. Eduardo-Figueira, M. J. Rodrigues, L. Custódio, N. Kuplennik, A. Sosnik, A. J. Almeida, M. M. Gaspar and C. P. Reis, Int. J. Pharm., 2019, 559, 13–22 CrossRef CAS PubMed.
- M. Janik-Hazuka, K. Kamiński, M. Kaczor-Kamińska, J. Szafraniec-Szczęsny, A. Kmak, H. Kassassir, C. Watała, M. Wróbel and S. Zapotoczny, Nanomaterials, 2021, 11, 1354 CrossRef CAS PubMed.
- S. Xiao, S. Huang, X. Yang, Y. Lei, M. Chang, J. Hu, Y. Meng, G. Zheng and X. Chen, Drug Delivery, 2023, 30, 2162156 CrossRef PubMed.
- X. Liu and G. Huang, Asian J. Pharm. Sci., 2013, 8, 319–328 Search PubMed.
- J. Lopes-de-Araújo, A. R. Neves, V. M. Gouveia, C. C. Moura, C. Nunes and S. Reis, Pharm. Res., 2016, 33, 301–314 CrossRef PubMed.
- D. A. Kuhn, D. Vanhecke, B. Michen, F. Blank, P. Gehr, A. Petri-Fink and B. Rothen-Rutishauser, Beilstein J. Nanotechnol., 2014, 5, 1625–1636 CrossRef PubMed.
- A. Ouhtit, M. E. Abdraboh, A. D. Hollenbach, H. Zayed and M. H. G. Raj, Cell Commun. Signaling, 2017, 15, 45 CrossRef CAS PubMed.
- A. Ouhtit, B. Rizeq, H. A. Saleh, M. D. M. Rahman and H. Zayed, Int. J. Biol. Sci., 2018, 14, 1782–1790 CrossRef CAS PubMed.
- C. R. Antonio and L. A. Trídico, Surg. Cosmet. Dermatol., 2021, 13, 20210006 Search PubMed.
- B. Ruffell and P. Johnson, Regulation and function of hyaluronan binding by CD44 in the immune system, https://www.glycoforum.gr.jp/article/13A1.html, (accessed 21 May 2022).
- M. Rochman, J. Moll, P. Herrlich, S. B. Wallach, S. Nedvetzki, R. V. Sionov, I. Golan, D. Ish-Shalom and D. Naor, Cell Adhes. Commun., 2000, 7, 331–347 CrossRef CAS PubMed.
- V. Francia, R.-S. Catharina, B. Guido and A. Salvati, Nanomedicine, 2019, 14, 1533–1549 CrossRef CAS PubMed.
- A. F. Amendoeira, A. Luz, R. Valente, C. Roma-Rodrigues, H. Ali, J. E. van Lier, F. Marques, P. V. Baptista and A. R. Fernandes, Int. J. Mol. Sci., 2023, 24, 3600 CrossRef CAS PubMed.
- V. Molnar, V. Matišić, I. Kodvanj, R. Bjelica, Ž. Jeleč, D. Hudetz, E. Rod, F. Čukelj, T. Vrdoljak, D. Vidović, M. Starešinić, S. Sabalić, B. Dobričić, T. Petrović, D. Antičević, I. Borić, R. Košir, U. P. Zmrzljak and D. Primorac, Int. J. Mol. Sci., 2021, 22, 9208 CrossRef CAS PubMed.
- P. Wojdasiewicz, Ł. A. Poniatowski and D. Szukiewicz, Mediators Inflammation, 2014, 2014, 561459 Search PubMed.
- N. Schweingruber, A. Haine, K. Tiede, A. Karabinskaya, J. van den Brandt, S. Wüst, J. M. Metselaar, R. Gold, J. P. Tuckermann, H. M. Reichardt and F. Lühder, J. Immunol., 2011, 187, 4310–4318 CrossRef CAS PubMed.
- M. Bartneck, F. M. Peters, K. T. Warzecha, M. Bienert, L. van Bloois, C. Trautwein, T. Lammers and F. Tacke, Nanomedicine, 2014, 10, 1209–1220 CrossRef CAS PubMed.
- V. Dixit and T. W. Mak, Cell, 2002, 111, 615–619 CrossRef CAS PubMed.
- T. Liu, L. Zhang, D. Joo and S.-C. Sun, Signal Transduction Targeted Ther., 2017, 2, 17023 CrossRef PubMed.
- P. Bąska and L. J. Norbury, Pathogens, 2022, 11, 310 CrossRef PubMed.
- K. De Bosscher, W. Vanden Berghe, L. Vermeulen, S. Plaisance, E. Boone and G. Haegeman, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 3919–3924 CrossRef CAS PubMed.
- J. A. Roman-Blas and S. A. Jimenez, Osteoarthr. Cartil., 2006, 14, 839–848 CrossRef CAS PubMed.
- J. Li, R.-S. Tang, Z. Shi and J.-Q. Li, Int. J. Rheum. Dis., 2020, 23, 1627–1635 CrossRef CAS PubMed.
- S. S. Makarov, Arthritis Res., 2001, 3, 200–206 CrossRef CAS PubMed.
- S. Rigoglou and A. G. Papavassiliou, Int. J. Biochem. Cell Biol., 2013, 45, 2580–2584 CrossRef CAS PubMed.
- J. Lugrin, N. Rosenblatt-Velin, R. Parapanov and L. Liaudet, Biol. Chem., 2014, 395, 203–230 CrossRef CAS PubMed.
- R. E. Simmonds and B. M. Foxwell, Rheumatology, 2008, 47, 584–590 CrossRef CAS PubMed.
- V. Gouveia, L. Rizzello, C. Nunes, A. Poma, L. Ruiz-Perez, A. Oliveira, S. Reis and G. Battaglia, Pharmaceutics, 2019, 11, 614 CrossRef CAS PubMed.
- G. Spinelli, G. Biddeci, A. Artale, F. Valentino, G. Tarantino, G. Gallo, F. Gianguzza, P. G. Conaldi, S. Corrao, F. Gervasi, T. S. Aronica, A. Di Leonardo, G. Duro and F. Di Blasi, Sci. Rep., 2021, 11, 22913 CrossRef CAS PubMed.
- R. Newton, Thorax, 2000, 55, 603–613 CrossRef CAS PubMed.
- E. d T. Bouhlali, A. Hmidani, B. Bourkhis, T. Khouya, M. Ramchoun, Y. Filali-Zegzouti and C. Alem, Heliyon, 2020, 6, e03436 CrossRef PubMed.
- D. B. Aidoo, D. Konja, I. T. Henneh and M. Ekor, Int. J. Inflammation, 2021, 2021, 1279359 Search PubMed.
- S. Qasim, Alamgeer, S. Kalsoom, M. Shahzad, I. A. Bukhari, F. Vohra and S. Afzal, ACS Omega, 2021, 6, 2074–2084 CrossRef CAS PubMed.
- H. Agarwal and V. K. Shanmugam, Inform. Med. Unlocked, 2019, 14, 6–14 CrossRef.
- X. Wang, D. Fan, X. Cao, Q. Ye, Q. Wang, M. Zhang and C. Xiao, Antioxidants, 2022, 11, 1153 CrossRef CAS PubMed.
- M. Mittal, M. R. Siddiqui, K. Tran, S. P. Reddy and A. B. Malik, Antioxid. Redox Signaling, 2014, 20, 1126–1167 CrossRef CAS PubMed.
- Y. Ranneh, F. Ali, A. M. Akim, H. A. Hamid, H. Khazaai and A. Fadel, Appl. Biol. Chem., 2017, 60, 327–338 CrossRef CAS.
- D.-F. Ding, Y. Xue, X.-C. Wu, Z.-H. Zhu, J.-Y. Ding, Y.-J. Song, X.-L. Xu and J.-G. Xu, J. Inflammation Res., 2022, 15, 5009–5026 CrossRef CAS PubMed.
- A. Haseeb, H. Mohammad Yusuf and A. Rizwan, in Reactive Oxygen Species, ed. A. Rizwan, IntechOpen, Rijeka, 2022, ch. 6 DOI:10.5772/intechopen.101333.
- J. Wan, J. Yang, W. Lei, Z. Xiao, P. Zhou, S. Zheng and P. Zhu, Int. J. Nanomed., 2023, 18, 579–594 CrossRef CAS PubMed.
- B. M. Sanner, U. Meder, W. Zidek and M. Tepel, Steroids, 2002, 67, 715–719 CrossRef CAS PubMed.
- B. Silpak, D. Rintu and B. Ena Ray, AIMS Biophys., 2017, 4, 596–614 Search PubMed.
- I. L. Elisha, J.-P. Dzoyem, L. J. McGaw, F. S. Botha and J. N. Eloff, BMC Complementary Altern. Med., 2016, 16, 307 CrossRef PubMed.
- W.-B. Tsai and C.-J. Chen, J. Biomed. Mater. Res., Part B, 2024, 112, e35453 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5ma00041f |
| This journal is © The Royal Society of Chemistry 2025 |