Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Enhanced biocompatibility of 3D printed resin parts via wet autoclave postprocessing: implications for stem cell organ-on-a-chip culture†
Alexander
Jönsson
 ,
Antonia
Iatrou
,
Louise
Wildfang
,
Dana J.
Neumann
,
Hakan
Gürbüz
,
Carina A. A.
Schoenmaker
,
Marlene Danner
Dalgaard
,
Pernille Rose
Jensen
,
Antonia
Iatrou
,
Louise
Wildfang
,
Dana J.
Neumann
,
Hakan
Gürbüz
,
Carina A. A.
Schoenmaker
,
Marlene Danner
Dalgaard
,
Pernille Rose
Jensen
 and
Martin
Dufva
and
Martin
Dufva
 *
*
Department of Health Technology, Technical University of Denmark, Oersteds plads 345C, DK-2800, Kgs Lyngby, Denmark. E-mail: dufva@dtu.dk
First published on 5th March 2025
Abstract
3D printed parts made from photocured resins are widely used in surgery, dentistry, medical devices, and organ-on-a-chip research due to their ease of fabrication and customization. However, extensive postprocessing is needed to reduce their cytotoxicity. In this study, we demonstrate that a 60-minute “wet” autoclave process significantly reduces leachates compared to many commonly used postprocessing methods. This reduction in leachates was observed across all four tested resins, indicating the general applicability of this method. Materials marketed as biocompatible did not affect stem cell growth and only had a minor effect on differentiation after a 60-minute wet autoclave treatment, unlike non-wet autoclaved parts, which showed marked effects. We assessed cellular function using morphology, viability assays, functional assays, and metabolomics. While no immediate effects were observed from the tested materials after wet autoclavation, transcriptomic analysis revealed that sub-cytotoxic levels of leachates downregulated extracellular matrix genes and upregulated genes related to cell adhesion and lipid and fatty acid metabolism. These changes could have long-term implications. In conclusion, the wet autoclave protocol described here is an easy-to-implement, standardized postprocessing step that reduces the exposure of organisms to resin leachates from 3D-printed parts.
Introduction
The introduction of 3D printing in the biomedical field has created a demand for biocompatible photosensitive resins and postprocessing methods that further reduce any adverse effects on cells and tissues. These resins are similar to UV curable dental cement and implants and thus share many of the same concerns. In their liquid state, before crosslinking, resins are cytotoxic1,2 since they contain harmful monomers and oligomers such as mixtures of acrylates, methacrylates, urethanes, epoxies, photoinitiators, photoabsorbers, and plasticizers.3In stereolithography (SLA), the technology that resin 3D-printers are based on, photopolymerization has an upper conversion limit of around 60%, leaving significant amounts of unreacted compounds in the printed parts.4 Another example is dental implants, which have been shown to leach traces of uncured 2-hydroxyethyl methacrylate (HEMA) and triethylene glycol dimethacrylate (TEGMA) that subsequently can be detected in the human circulation system,5 indicating systemic effects. Methacrylate leachates, such as HEMA, have been linked to oral mucosal irritation,6 and photoinitiator residues, like camphorquinone, can induce oxidative stress and cell death.3 Postprocessing generally reduces the toxic residues by leaching out from the 3D print and/or increases the material's cross-linking, resulting in a less harmful product.7 The efficiency of leaching out depends on the properties of the 3D printed part, such as geometry, composition and polymerization ratio, which typically occurs when the material is submerged in solvents. Studies have shown that uncured resin compounds can be extracted with both inorganic and organic solvents,8 and unbound compounds have been monitored using Raman spectroscopy,9,10 FTIR,10 and HPLC.11,12 A common approach to reducing the amount of leachable compounds is to add postprocessing leaching steps where 3D prints are placed in aqueous media for 1–10 days, allowing them to leach out uncured toxic substances before use.13–16 Other methods include washing with ethanol and other organic solvents9,17 and extended UV curing times18,19 or elevated temperature nitrogen processes.20 However, the results may vary depending on the resin and printer used.21 The success in making prints biocompatible, in terms of sustaining cell growth in vitro, varies. Some tests show decreased cell viability and adverse effects,22 while others demonstrate excellent biocompatibility for cell culture for weeks.14,16,23 Given the diversity of printers, resin formulation, postprocessing protocols, and cell types, it is difficult to collapse the current knowledge into one guideline. It can, however, be expected that reduction of unreacted compounds, contaminating the cell culture medium, is advantageous for reducing adverse effects on the cells.
In summary, while there is consensus on the biocompatibility issues of 3D printed resins, there is no consistent and quick protocol for improving biocompatibility in terms of cytotoxicity. Given that the biocompatibility of 3D prints increases when submerged in water for long periods,14,15,23 we here investigate if leaching could be quicker and more efficient through autoclavation in water. We assess leaching and biological effects, on adipose derived stem cells, of different postprocessing protocols using nuclear magnetic resonance (NMR), cell viability, transcriptomics and differentiation.
Methods
3D-printing
3D printed ring structures were designed to fit into 48-well titer plates (Nunc Thermo Scientific, Thermo Fisher, Denmark) using Fusion (Autodesk, California, USA). The ring structure had an outer diameter of 11 mm, an inner diameter of 6 mm, and a height of 3 mm. Three supporting legs of 0.5 mm each were included to elevate the ring from the bottom of the plate. Structures were printed with BM (Biomed Clear v1, Formlabs, Somerville, Massachusetts, USA), D90 (Biotough D90-MF, #P10161, 3DResyns, Barcelona, Spain), Dental LT v1 (Formlabs), and Clear v4 (Formlabs). Formlabs resins were printed using a low force stereolithography (LFS) 3D printer, Form 3B (Formlabs), with a layer thickness of 0.1 mm and predefined settings for each resin and support structures as dictated by the slicing software PreForm (v.3.28.0, Formlabs). D90 was printed using an LCD 3D printer, Sonic Mini 8K (Phrozen Technology, Hsinchu city, Taiwan), with a layer thickness of 0.05 mm, an exposure time of 5 seconds, and the necessary support structures designed using CHITUBOX (V1.9.4, Shenzhen, China) slicing software. The test structure was designed by Ameralabs (Kauno, Lithuania).Postprocessing
The protocols below are summarized in Table 1.| Short protocol | Long protocol | D90 specific protocols | ||||
|---|---|---|---|---|---|---|
| D90 | BM | D90 | BM | D90 protocol A | D90 protocol B | |
| a At 60 °C. b 20 min at 121 °C. c 20 min in 50 mL, 3 × 20 min in 50 mL, or 1 × 60 min in 500 mL. All at 121°. | ||||||
| Washing 1 | 20 min IPA, sonication | 20 min IPA in Form Wash | 20 min IPA, sonication | 20 min IPA in Form Wash | 20 min washing sol., sonication | 20 min washing sol., sonication |
| Washing 2 | — | — | 1 h IPA | 1 h IPA | 20 min washing sol., sonication | 20 min washing sol., sonication |
| Washing 3 | — | — | 1 h IPA | 1 h IPA | 20 min washing sol., sonication | 20 min washing sol., sonication |
| Postcuring | 30 mina | 30 mina | 120 mina | 120 mina | 30 mina | 30 mina |
| Add. step 1 | — | — | Overnight in MilliQ | Overnight in MilliQ | 10 min tester sol. | — |
| Add. step 2 | — | — | — | — | 10 min purification sol. | — |
| Autoclavation | Dryb or wetc | Dryb or wetc | Dryb or wetc | Dryb or wetc | Dryb or wetc | Dryb or wetc |
Short protocol: 3D printed structures were washed with isopropanol (IPA) for 20 min. For D90 this was done in a sonication bath and for Biomed Clear, Dental LT, and Clear it was done in a Formlabs FormWash station. Prints were then cured by UV light for 30 minutes at 60 °C in a UV light polymerization chamber, Formlabs FormCure.
Long protocol: 3D printed structures were first washed with IPA as described in the short protocol. The prints were then transferred to polyethylene bags containing IPA and incubated for 2 × 1 hour, with IPA changed between incubations. Prints were subsequently cured by UV light for 60 minutes at 60 °C in a Formlabs FormCure. Finally, the prints were incubated in Milli-Q water overnight.
D90 protocol A: manufacturer's recommended instructions. 3 × 20 minute sonication in 3DResyns cleaning solution (cleaning Fluid Bio, #P20397), with the cleaning solution changed between cycles, followed by 30-minute UV curing at 60 °C.
D90 protocol B: Following D90 protocol A above, the prints were treated with the Tester and Purification solutions (Cure Tester & Purification kit, #P11198) according to the manufacturer's specifications.
Dry autoclavation: 3D prints were dry autoclaved using a 60L Top-Loading & Vertical autoclave (Priorclave Ltd, UK) at 121 °C for 20 minutes in sterilization pouches.
1× wet autoclavation: 3D prints were placed in 50 mL Milli-Q water and autoclaved for 20 minutes at 121 °C using the same autoclave as above.
3× wet autoclavation: 3D structures were autoclaved three times according to the 1× wet autoclavation protocol, with Milli-Q water replaced after each round.
Long wet autoclavation: 3D structures were placed in 500 mL water and autoclaved for 60 minutes at 121 °C.
NMR analysis of leaching products
Dry or wet autoclaved 3D printed rings were placed in 48-well plates filled with 1 mL sterile Milli-Q water. The plates were incubated for 48 hours at 37 °C in a 5% CO2, 95% air atmosphere, and 100% relative humidity, simulating cell culture conditions. Milli-Q water without any ring was used as a negative control. After incubation, the water samples were collected and analyzed with nuclear magnetic resonance (NMR) spectroscopy. 1D 1H NMR spectra were acquired with a NOESY pre-saturation pulse sequence using a Bruker NMR spectrometer at 500 MHz, equipped with a sample changer (SampleJet). A total of 256 scans were collected using a spectral width of 12019 Hz, with a 2.7 s acquisition time resulting in 24 minutes acquisition time per sample (d1 = 4 s, aq = 2.7 s). NMR samples were prepared with 300 μL of the test sample and 250 μL phosphate buffer solution at pH 7.5 prepared in D2O with DSS (sodium trimethylsilylpropanesulfonate) as an internal reference. Spectra were manually corrected for phase and baseline distortions, and the chemical shift was normalized and referenced to DSS at 0.00 ppm using MestReNova software (Mestrelab Research, Santiago de Compostela, Spain). Ethanol (1.17–1.20 ppm and 3.8–3.6 ppm) contaminations were excluded from the analysis. Peaks observed in the test but not in the control samples were recorded as leachates. The corresponding peak integral from a control sample was subtracted from the respective peak to quantify the leachates.Cell culture
Adipose-derived stem cells (ADSCs) (StemPro, R7788115, Gibco, Thermo Fisher) were cultured in T-75 cell culture flasks (Nunc, Thermo Scientific, Thermo Fisher) in reduced serum culture medium (MSC Growth Medium 2 and Supplement mix, C-28009, PromoCell, Merck Life Science, Denmark) with 2 mM L-glutamine (25030081, Gibco, Thermo Fisher) at 37 °C in a 5% CO2, 95% air atmosphere, and 100% relative humidity. The medium was changed every 3–4 days. Cells were used at passages p2–p4. When cells reached 75–80% confluence, they were detached with TrypLE Express (without phenol red, 12604013, Gibco, Thermo Fisher) and seeded into 48-well plates, containing the 3D-printed rings, at densities of 2–2.5 × 104 cells per well. Wells without rings served as negative controls. For gene expression analysis and differentiation experiments, polystyrene rings, cut from 2 mm thick sheets, using a CO2 laser (FLUX Beambox, Flux Europe), were used as negative controls. Cells were allowed to grow for 72 hours at 37 °C in 5% CO2 before cell viability and staining assays. Cell growth was observed using a Zeiss Primovert microscope with 4× or 20× objectives (Carl Zeiss MicroImaging GmbH, Gottingen, Germany).Adipocyte differentiation
Cells grown as stated above were differentiated into adipocytes using the StemPro Adipogenesis Differentiation Kit (A1007001, Gibco, Thermo Fisher). After 72 hours, the growth medium was replaced with the differentiation medium included in the kit, supplemented with 10 mg mL−1 penicillin (100 U mL−1)–streptomycin (100 μg mL−1, 15140122, Gibco, Thermo Fisher), and maintained in culture.Cell viability
Cell growth and surface coverage in the presence of 3D printed structures were assessed using the live/dead assay (LIVE/DEAD Viability/Cytotoxicity Kit, L3224, Invitrogen, Thermo Fisher). Samples were stained according to the manufacturer's instructions and incubated for 60 minutes at 37 °C. Triplicates of each sample were used. After incubation, samples were imaged using a Zeiss AxioObserver Z1 epifluorescence microscope (Carl Zeiss MicroImaging GmbH). The 3D printed ring structures were removed from the wells before imaging due to their autofluorescence interfering with the signal from live cells. Images were processed using Zeiss Zen Blue 3.7 Lite Digital Imaging Software (Carl Zeiss Microscopy GmbH) and ImageJ v1.53.24To obtain quantitative results regarding cell growth, PrestoBlue HS Cell Viability Reagent (P50200, Invitrogen, Thermo Fisher) was used. Samples were stained with 10% (v/v) PrestoBlue for 15 minutes, following the manufacturer's protocol. After incubation, 90 μL of the sample were transferred to a 96-well plate to analyze fluorescence intensity at an excitation wavelength of 560 nm and an emission wavelength of 590 nm using a plate reader (Spark multimode microplate reader, TECAN, Mannedorf, Switzerland). Wells with media alone were used as background, which was subtracted from the measured values in the downstream analysis. After measurement, cells were washed twice with PBS, placed in fresh medium, and maintained in culture.
Red Oil O staining and quantification
Cultured cells were fixated in 10% formaldehyde and washed with 60% IPA. Cells were stained with 1 mM Red Oil O in IPA (Sigma Aldrich O-0625, Denmark) for 10 minutes and washed with water four times. After imaging, Red Oil O was extracted with 100 μL 100% IPA, and absorbance was measured at 500 nm.NMR analysis of metabolites during adipocyte differentiation
Four replicates of each medium sample were used, along with five replicates of MSC medium and adipocyte differentiation medium, respectively. 1D 1H NMR spectra were acquired as previously described. NMR samples were prepared with 400 μL of the medium sample, 100 μL Milli-Q water, and 100 μL phosphate buffer solution (600 mM, pH 7.4, 60% H2O, 40% D2O) including DSS and maleic acid as internal references. Spectra were manually corrected for phase and baseline distortions, and the chemical shift was calibrated to maleic acid at 5.995 ppm using MestReNova software. Peak integrals were measured for each metabolite, and the corresponding peak integral from the control media was used as a reference to determine the usage or production of metabolites.Transcriptomics
RNA was isolated using the RNA RNeasy Mini Kit (QIAGEN, 74106, Denmark) and RNase-Free DNase Set (QIAGEN, 79256) according to the manufacturer's instructions. RNA purity and quantity were determined using absorbance at 260/280 nm with a NanoDrop (Thermo Fisher). RNA quality was assessed using the Agilent RNA 6000 Nano Kit (5067-1511, Agilent, California, USA) and the 2100 Bioanalyzer Instrument (Agilent). Sequencing libraries were prepared using the Oxford Nanopore PCR-cDNA Barcoding Kit (SQK-PCB111.24, Oxford Nanopore, Oxford, UK) according to the manufacturer's instructions. cDNA quality and quantity were determined using the high sensitivity DNA Analysis Kit (5067-462, Agilent) and an Agilent bioanalyzer. Libraries were pooled and sequenced using an Oxford Nanopore Solo PromethION and PromethION flow cell according to the manufacturer's instructions. RNA-seq data were analyzed using the Galaxy server.25 The Minimap2 package26 with “PacBio/Oxford Nanopore read to reference mapping (-Hk19) (map-pb) to the human genome (hg38)” was used to map the reads. The aligned sequences per gene were counted with Featurecount27 and the count data were normalized with DESeq2.28 Genes were considered significantly differentially expressed with any fold change and an adjusted P-value <0.05 was used as cutoff. Biological processes were identified using the DAVID server.29Results and discussion
Wet autoclavation is fast and reduces leaching
The effect of wet autoclavation was investigated for two resins, BM and D90, that are advertised as biocompatible. We compared two previously published postprocessing protocols (“short” and “long”, Table 1)14,15,23 with two commercial protocols from 3Dresyn. The postprocessing steps of the protocols are described in detail in Table 1 and the overall experiment pipeline can be seen in Fig. 1A. For each postprocessing protocol, a standard “dry” autoclavation and an additional 20 min wet autoclavation, respectively, were investigated. NMR spectroscopy was used to quantify the leachates and the sums of all corresponding peak integrals, for the dominant peaks, are shown in Fig. 1B. For both materials, wet autoclavation resulted in reduction of leachates when compared to dry autoclavation, regardless of prior postprocessing. This effect was larger than the difference between the individual postprocessing steps. These results indicate that a 20 min wet autoclavation is an effective way to increase the leaching rate during postprocessing. We next investigated the effect of autoclavation duration. Rings were processed according to a short protocol (Table 1) and then subjected to either three rounds of 20-minute wet autoclavation, with water exchanged between each round, or a single 60-minute wet autoclavation. Leaching tests were performed as in Fig. 1A and evaluated by 1H NMR. For BM and D90, most peaks were observed in four ranges 1.2–2.5, 3.5–4.0, 5.5–6.5, and 6–8 ppm. Peaks in the 1–2 ppm range correspond to secondary (R2CH2) and tertiary aliphatic protons (R3CH); peaks around 3.5–4 ppm correspond to RyCHx motifs with an electronegative group attached, such as –OH; peaks around 5.5–6.5 ppm correspond to vinyl protons (–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2); and peaks in the 6.5–8 ppm range correspond to aromatic motifs.
CH2); and peaks in the 6.5–8 ppm range correspond to aromatic motifs.
 | ||
| Fig. 1 Quantification of leachates from 3D-printed rings consisting of BM and D90. (A) Overview of the experimental process. (B) NMR analysis of leached samples following the different postprocessing protocols (n = 4, mean ± SD, statistical analysis with Welch's t-test, P < 0.05). Examples of spectra can be found in Fig. S1 (ESI†). | ||
For BM, we observed aliphatic and aromatic protons, while vinyl protons were not detected. For D90, aliphatic protons, vinyl protons and aromatic protons were detected. One peak (2.05 ppm) in BM did not react to wet autoclavation but was consistently close to background levels (Fig. 2A). All other peaks in D90 and BM were reduced to near the detection limit after wet autoclavation. All peaks were reduced more with longer wet autoclavation (3 × 20 min or 1 × 60 min) compared to a single 20 min wet autoclavation, consistent with a diffusion mechanism. A single 60 min wet autoclavation was either superior, or similar, to three 20 min autoclavations (Fig. 2A and B).
 | ||
| Fig. 2 Effect of different autoclavation protocols (mean ± SD, n = 3). (A) Leachates from BM after wet autoclavation with varying duration. (B) Equivalent for D90 resin. | ||
Acrylates and methyl acrylates exhibit peaks associated with aliphatic and vinyl protons, while the photoinitiator used in BM, diphenyl(2,4,6-trimethylbenzoyl)phosphine oxide (TPO-L), exhibits peaks around 7–8 ppm due to its aromatic rings. Specifically, the composition of BM is listed to be: proprietary acrylate monomer(s) (50–70%), urethane dimethacrylate (25–45%), proprietary methacrylate monomer(s) (7–10%), and TPO-L (1–2%) on the manufacturer's home page.
Therefore, we speculate that the leachates are mainly uncured methacrylate and acrylic monomers/oligomers, as well as photoinitiator residues.
Two additional materials, Formlabs Clear and Dental LT, were also tested in the same way to further validate the method. While Dental LT and Formlabs Clear show more peaks than BM and D90, these resins displayed a similar, time dependent, inverse relation between wet autoclavation time and the resulting concentrations of leachates in the test (Fig. S2, ESI†). The effects of short and long wet autoclavations were similar for the four resins, suggesting that wet autoclavation is a generic method to remove leachates from resin 3D prints. Presumably, even longer autoclavations, with periodic water change, may decrease the leached products even further. This would, however, require higher sensitivity than in our current study to quantify. It should be noted that not all resins can survive autoclavation, as is the case with Formlabs Clear (Fig. S3, ESI†). In contrast, BM and D90 maintain their structure after autoclavation, as seen in the test print while Formlabs Clear delaminated. This is consistent with reports from the manufacturer, claiming negative effects on the mechanical properties of Formlabs Clear, after steam autoclavation, but no noticeable effects on BM.
Growth of stem cells in proximity to the postprocessed materials
Adipocyte derived stem cells (ADSCs) were seeded in wells with rings of the 3D-printed materials, as seen in Fig. 3A. Wells without any ring were used as negative controls (Fig. 3B). The rings were either dry autoclaved or wet autoclaved for 1 × 60 min, after processing with the short protocol (Table 1). Incubation with dry autoclaved D90 resulted in almost complete loss of cells due to either poor cell adhesion or cell death (Fig. 3C). Dry autoclaved BM, in contrast, resulted in cell growth in the center but not under the rings. As seen in Fig. 1B, BM and D90 show comparable levels of leached products after short postprocessing and dry autoclavation. This suggests that there is no general correlation between the amount of leaching product and the cytotoxicity of the material. The compositions of the two materials differ, as shown in Fig. 2, and it is likely that D90 contains one or more compounds, not found in BM, that contribute to its cytotoxicity after dry autoclavation.In the case of wet autoclaved rings, cells could grow under the rings, where the concentration of leached chemicals would be the highest, for both materials (Fig. 3E and F). Using the 3 × 20 min protocol instead did not result in different growth patterns (Fig. S4, ESI†). The morphology of the ADSCs was also similar in cultures with processed rings and cultures without rings (Fig. S5, ESI†). Presto blue metabolic activity correlated with the growth patterns of the cells, where cells under dry autoclaved D90 showed very low activity while under all other conditions they showed comparable activity to the control (Fig. 3G). Overall, the wet autoclaved rings seemed to not have a noticeable effect on the ADSCs.
Effect of materials on ADSC transcriptome
To understand the molecular changes induced by the 3D-printed materials, transcriptomics was used. Cells from cultures in the presence of 3D-printed rings, as shown in Fig. 3, were analyzed using RNA Seq. A laser-cut polystyrene (PS) ring of similar dimensions was used as a material control for the 3D printed rings, instead of a no ring culture, ensuring that any changes in the cells caused by growth under the rings were accounted for. Examples of under ring effects could be enrichment of secreted differentiation factors30 and lower oxygen and nutrient levels due to diffusion limitations. The full lists of regulated genes are shown in Table S1A (ESI†) for BM and Table S1B (ESI†) for D90. Compared to the PS control, 0.2% and 0.5% of the detected genes were differentially expressed for coculture with BM and D90, respectively. The fewer regulated genes in BM may be due to fewer samples (n = 2 for BM compared to n = 4 for D90) and not because it has less effects on the cells compared to D90. Detailed analysis of differentially expressed genes showed that many were associated with the extracellular matrix and cell adhesion, lipid and fatty acid metabolism, and cholesterol synthesis and transport for both materials (Fig. 4A and Table S2 for BM and Table S3 for D90, ESI†). D90 also affected carbon metabolism, with multiple genes associated with the pentose phosphate pathway (Table S4, ESI†). Specifically, 6-phosphogluconolactonase (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2(FC) = 1.41) and 6-phosphogluconate dehydrogenase (log
2(FC) = 1.41) and 6-phosphogluconate dehydrogenase (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2(FC) = 1.3) were upregulated in the oxidative phase of the pentose phosphate pathway, likely leading to an increased production of NADPH. This could be further facilitated by the upregulation of phosphoglucose isomerase (log
2(FC) = 1.3) were upregulated in the oxidative phase of the pentose phosphate pathway, likely leading to an increased production of NADPH. This could be further facilitated by the upregulation of phosphoglucose isomerase (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2(FC) = 0.78), balancing glucose-6-phosphate to supply the pentose phosphate pathway. It is likely that the upregulation of transketolase (log
2(FC) = 0.78), balancing glucose-6-phosphate to supply the pentose phosphate pathway. It is likely that the upregulation of transketolase (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2(FC) = 0.78) thus aids in returning glyceraldehyde 3-phosphate to glycolysis. NADPH is, among other things, needed for the synthesis of fatty acids and cholesterol, both seemingly upregulated here. Many extracellular matrix associated genes were down regulated (Table 2). Examples are collagens (1A1, 3A1 and 6A3), decorin, periostin and metalloprotease 3. Some of these proteins are either involved in TGFβ signaling or are regulated by TGFβ signaling. Of note, seemingly all differentially expressed genes directly related to the ECM are downregulated for both materials, whereas genes related to cell adhesion and migration are upregulated. These include ITGA4 (log2(FC) = 7.62) for BM, or ABI3BP (log2(FC) = 2.01) and ANKRD28 (log2(FC) = 4.93) for D90. Others have shown with gene expression profiling that dental cement similarly affects cell adhesion31 as well as oxidative processes.31,32 Analyzing the base mean, i.e. the level of expression in the paired BM-PS and D90-PS samples, respectively, for the regulated genes with significant differential expression for both materials, showed high concordance in expression levels between the D90 and BM groups (Fig. 4B). Note that genes like POSTN and SPARC belong to the top 200 highest expressed genes in the cells. The degree of regulation (log2(FC)) also showed high concordance between the D90 and BM samples, with nine genes expressed and regulated similarly by both materials across different biological functions, a majority of which are related to the ECM and fatty acid metabolism, indicating similar effects of the two materials on the cells (Fig. 4C).
2(FC) = 0.78) thus aids in returning glyceraldehyde 3-phosphate to glycolysis. NADPH is, among other things, needed for the synthesis of fatty acids and cholesterol, both seemingly upregulated here. Many extracellular matrix associated genes were down regulated (Table 2). Examples are collagens (1A1, 3A1 and 6A3), decorin, periostin and metalloprotease 3. Some of these proteins are either involved in TGFβ signaling or are regulated by TGFβ signaling. Of note, seemingly all differentially expressed genes directly related to the ECM are downregulated for both materials, whereas genes related to cell adhesion and migration are upregulated. These include ITGA4 (log2(FC) = 7.62) for BM, or ABI3BP (log2(FC) = 2.01) and ANKRD28 (log2(FC) = 4.93) for D90. Others have shown with gene expression profiling that dental cement similarly affects cell adhesion31 as well as oxidative processes.31,32 Analyzing the base mean, i.e. the level of expression in the paired BM-PS and D90-PS samples, respectively, for the regulated genes with significant differential expression for both materials, showed high concordance in expression levels between the D90 and BM groups (Fig. 4B). Note that genes like POSTN and SPARC belong to the top 200 highest expressed genes in the cells. The degree of regulation (log2(FC)) also showed high concordance between the D90 and BM samples, with nine genes expressed and regulated similarly by both materials across different biological functions, a majority of which are related to the ECM and fatty acid metabolism, indicating similar effects of the two materials on the cells (Fig. 4C).
| Gene | Function | BMa | D90a |
|---|---|---|---|
| a Mean log2(FC) for the respective material compared to PS. Adjusted P < 0.05. For values in bold 0.1 > adjusted P > 0.05. | |||
| DPT (dermatopontin) | Binds to decorin; increases TGFβ activity.33 Increases collagen fibrillogenesis.34 Suppresses WNT signalling.35 Suppressor of cancer.36 | −2.03 | −2.31 |
| SPARC (secreted protein acidic and cysteine rich) | Promotes metastasis in highly malignant cancers.37 Tumour suppressor in less malignant cancers.37 Regulates TGFβ induced epithelial-to-mesenchymal transition.37 Increases COL1 expression via activation of TGFβ.38 Marker for gastric cancer.39 | −0.75 | −1.00 |
| POSTN (periostin) | Upregulation gives tumour progression, inflammation and fibrosis.40 Associated with cardiac diseases.40 | −0.95 | −0.93 |
| MFAP4 (microfibril associated protein 4) | Deletion is linked to pathogenesis of Smith–Magenis syndrome.41 Decreased secretion is linked to leukoencephalopathy with vanishing white matter.42 Decreased expression associated with poor prognosis in oral squamous cell carcinoma.43 | −0.91 | −1.20 |
| MMP3 (matrix metallopeptidase 3) | Increased expression in nerve cells by cell stress and leads to neurodegeneration.44 Upregulation is associated with metastasis in breast cancer.45 | −3.13 | −3.74 |
| DCN (decorin) | Suppresses pro-tumour cues:46 suppresses TGFβ signalling and TLR2/4 signalling leading to suppression of inflammation. Inhibitor of numerous receptor tyrosine kinases including EGRF. | n.s. | −1.39 |
| ANXA2 (annexin A2) | Vascular haemostasis; pro-fibro lysis. Overexpressed in haemorrhaging cancers.47 Expression correlates with poor survival of cancer patients.48 | n.s. | −0.51 |
| FBLN5 (fibulin 5) | Mutated forms linked to pathogenesis of cutis laxa49 and age-related macular degeneration.50 Decreased expression associated with unfavorable prognosis of high-grade serous ovarian carcinoma.51 | n.s. | −1.24 |
| LUM (lumican) | Plays a role in maintaining corneal transparency and scleral integrity.52 Involved in TGFβ signalling.53 Polymorphism associated with high myopia.54 Increased expression associated with poor prognosis in gastric cancer.55 | n.s. | −0.64 |
| TNC (tenascin C) | Mutated form linked to pathogenesis of nonsyndromic hearing loss.56 Highly expressed in cancer tumors.57 Modulates resistance to apoptosis and drug resistance in pancreatic cancer.58 | n.s. | −3.29 |
| COL3A1 (collagen type III alpha 1 chain) | Mutated forms give Ehlers–Danlos syndrome.59 Marker for gastric cancers.39 Marker for cancer associated fibroblast together with COL1A1 and SPARC.60 | n.s. | −1.63 |
| COL6A3 (collagen type VI alpha 3 chain) | Mutated forms are associated with muscle diseases.61 Increased expression associated with poor prognosis in colon cancer.62 | n.s. | −0.89 |
| COL1A1 (collagen type I alpha 1 chain) (COL1A1) | Regulated by TGFβ.63 Mutated in osteogenesis imperfecta.64 Highly expressed in cancers and implicated in cell proliferation, migration and EMT.65 Marker for gastric cancer.39 | −1.08 | −0.96 |
| BGN (biglycan) | Gene defects cause severe thoracic aortic aneurysm and dissection.66 Increased expression associated with enhanced invasion and migration ability in endometrial cancer.67 | −1.22 | n.s. |
| TIMP3 (TIMP metallopeptidase inhibitor 3) | Decreased expression associated with pathogenesis of diabetic kidney disease.68 Antitumour effect, downregulated in many cancers.69 | −2.12 | n.s. |
| TGFBI (transforming growth factor beta induced) | Mutated forms associated with pathogenesis of corneal dystrophies.70 Increased expression associated with poorer clinical outcomes in many cancers.71 | −1.28 | n.s. |
It would be reasonable to observe morphological changes in the adherent cells if the extracellular microenvironment was changed, but we could not observe any differences in ADSC morphology, in the presence of 3D-printed materials, compared to normal polystyrene well cultures (Fig. S4, ESI†). However, non-wet autoclaved rings resulted in no or few cells under the rings (Fig. 3C and D). An explanation could be that cells detach due to decreased ECM production. It has previously been observed that long term culture of mouse intestinal organoids, suspended in Transwell-like 3D printed inserts, grow bigger compared to organoids at the bottom of a cell culture well,14 suggesting that 3D prints do not have a large impact on the cell physiology or organoid organization. Earlier studies on PC12 cells exposed to polydimethylsiloxane (PDMS), during differentiation, showed that nearly 700 genes were differentially expressed.72 As with 3D-printed materials, it is known that PDMS also leaches monomers and crosslinkers. Despite this, no morphological effects could be observed.33 This indicates that effects of 3D-prints (or other materials) may be cell dependent, and these effects are not large enough to elicit a clear morphological or physiological response, or that cells are compensating for the external cues.
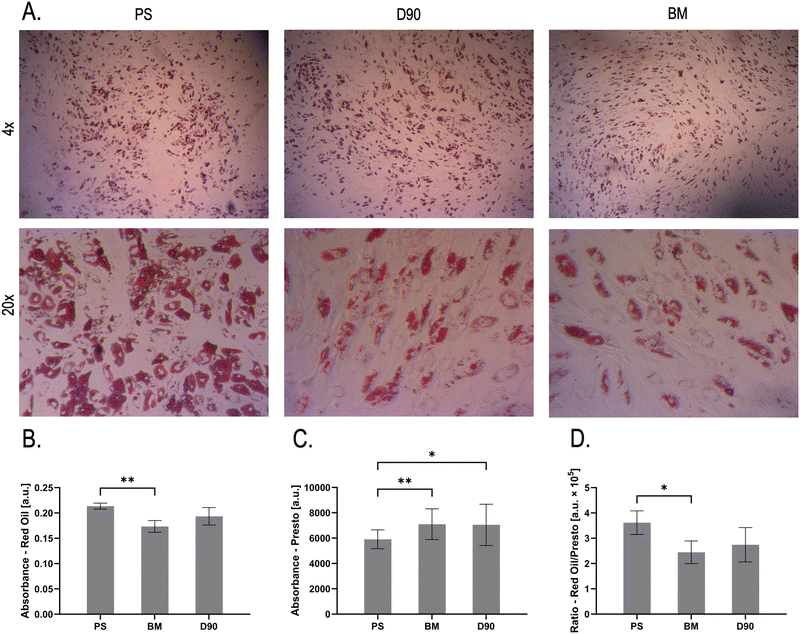
Effects of materials on the differentiation of ADSCs into adipocytes
Next, we tested the effects of the 60 min wet autoclavation on cell differentiation. We again used a laser-cut polystyrene ring as the negative control.ADSCs were grown and differentiated in cultures with rings present. Cells differentiated into adipocytes in the aperture as well as under the ring for all materials (Fig. S6–S8, ESI†), showing characteristic lipid vesicles and expected morphology. Red Oil O stain was used to measure differentiation (Fig. 5A and B), and a Presto Blue assay was used to compare metabolic activity and, by extension, the cell number (Fig. 5C). The PS rings showed lower metabolic activity than in the cultures of either 3D-printed material. This is likely due to the lack of “feet” on the laser cut ring, since only the laser-cut burr raises the ring off the bottom of the well. This could result in a reduced supply of nutrients, and subsequently slower growth, under the ring. In contrast, the degree of differentiation, when corrected for metabolic activity, seems to be slightly higher in the PS control than in either 3D-printed material. However, the difference is only significant for BM (Fig. 5D). This suggests that while the 3D-printed materials have no detrimental effect on the growth of ADSCs (Fig. 3G), differentiation might be slightly reduced.
Effects of materials on metabolism during differentiation
Lastly, medium composition from differentiating ADSCs with the three different ring materials was analyzed using 1H NMR spectroscopy (Fig. 6). The consumption and production of metabolites in the medium were calculated by subtracting the start level in pure medium from the respective timepoints. The results show similar levels of measured metabolites before (D0) and during differentiation (D7 and D14) for all tested materials. An increased usage of amino acids is observed at D7 of differentiation compared to D0 and D14, which suggested increased protein synthesis during early adipose differentiation. Amino acid usage then diminished again during the last week of differentiation. A similar pattern was seen for glucose, correlating with an overall increase in metabolism, specifically anabolism, during the first week of differentiation towards an adipocyte phenotype. No clear metabolic difference was found between the three ring materials, indicating that both BM and D90 rings are comparable to PS in terms of ADSC differentiation into adipocytes. Of note, the glucose consumption of ADSCs is predicted not to be significantly changed in the transcription data above, i.e., no change in the expression of hexokinase, for either of the two 3D-printed materials (Tables S1A and B, ESI†). This is also supported by the metabolic data.Conclusions
Wet autoclavation is here shown to be a promising method to reduce the cytotoxicity of resin 3D-printed parts. NMR analysis shows that this method reduces the amount of unreacted compounds that can be leached from four commercial resins (BM, D90, Formlabs Clear, and Formlabs Dental LT), showing broad applicability. Compared to many currently used alternatives, this method is fast, uses no hazardous chemicals, and has the added benefit of sterilizing the 3D-printed parts. Improved cell viability is shown in cultures of ADSCs, after incubation with wet autoclaved parts compared to dry autoclaved parts, for both advertised biocompatible resins (BM and D90) to the point where metabolic activity is comparable to a control.For undifferentiated ADSCs transcriptomic analysis shows strong indications of altered expression profiles. Specifically, for both materials, genes related to the composition of the ECM are upregulated, while genes related to cell adhesion are generally downregulated. For D90, clear indications of an increased production of NADPH, through the pentose phosphate pathway, can also be seen. This could be linked to the upregulation of cholesterol and fatty acid synthesis that is also seen. Differentiation of ADSCs into adipocytes, likewise, seems to be slightly reduced, but no adverse effects on morphology can be seen.
It stands to reason that further reduction of the leachable compounds will also reduce the effects on cells and tissues. This could potentially be achieved by longer wet autoclavation, and this will be investigated in a future study.
In summary, wet autoclavation has the potential to reduce leaching of unreacted compounds from 3D-printed parts, improving their compatibility with cell cultures, when compared to alternative commonly used methods. The method is fast, uses no hazardous chemicals, and additionally sterilizes the parts for direct use in in vitro cultures.
Author contributions
AI, LW, HG, AJ, and MDD collected data; DJN, CAA, MDD, MD, and AJ analysed transcriptomics data; LW, AJ, AI, and PRJ analysed NMR data; all participated in the design of experiments; MD, AJ, LW, and AI wrote the manuscript and MD, MDD and PRJ supervised the research.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
The study was funded by the H2020 project GUTVIBRATION—953201 and the Danish research council, DFF grant number 3105-00152B. NMR data were acquired using infrastructure supported by the Novo Nordisk Foundation (NNF 19OC0055825). usegalaxy.eu is supported by the German Federal Ministry of Education and Research grant 031L0101C and de.NBI-epi. RNA Seq experiments were performed at the DTU Multi Assay Core (DMAC) core facility at the Technical University of Denmark.Notes and references
- H. B. Rogers, L. T. Zhou, A. Kusuhara, E. Zaniker, S. Shafaie, B. C. Owen, F. E. Duncan and T. K. Woodruff, Chemosphere, 2021, 270, 129003 CAS.
- M. Walpitagama, M. Carve, A. M. Douek, C. Trestrail, Y. Bai, J. Kaslin and D. Wlodkowic, Aquat. Toxicol., 2019, 213, 105227 CAS.
- M. Carve and D. Wlodkowic, Micromachines, 2018, 9, 91 Search PubMed.
- J. L. Ferracane and J. R. Condon, Dent. Mater., 1992, 8, 290–295 CAS.
- C. A. De Souza Costa, A. B. L. Do Nascimento and H. M. Teixeira, Dent. Mater., 2002, 18, 543–551 Search PubMed.
- H. Schweikl, C. Petzel, C. Bolay, K. A. Hiller, W. Buchalla and S. Krifka, Biomaterials, 2014, 35, 2890–2904 CAS.
- J. R. C. Dizon, C. C. L. Gache, H. M. S. Cascolan, L. T. Cancino and R. C. Advincula, Technologies, 2021, 9, 61 CrossRef.
- M. A. Cebe, F. Cebe, M. F. Cengiz, A. R. Cetin, O. F. Arpag and B. Ozturk, Dent. Mater., 2015, 31, e141–e149 CrossRef CAS PubMed.
- J. Jeršovaitė, U. Šarachovaitė, I. Matulaitienė, G. Niaura, D. Baltriukienė and M. Malinauskas, Front. Bioeng. Biotechnol., 2023, 11, 1167753 Search PubMed.
- M. A. Gauthier, I. Stangel, T. H. Ellis and X. X. Zhu, Biomaterials, 2005, 26, 6440–6448 CrossRef CAS PubMed.
- T. Kawahara, Y. Nomura, N. Tanaka, W. Teshima, M. Okazaki and H. Shintani, J. Dent., 2004, 32, 277–283 CrossRef CAS PubMed.
- E. Berghaus, T. Klocke, R. Maletz and S. Petersen, J. Mater. Sci.: Mater. Med., 2023, 34, 23 CrossRef CAS PubMed.
- R. P. Rimington, A. J. Capel, D. J. Player, R. J. Bibb, S. D. R. Christie and M. P. Lewis, Macromol. Biosci., 2018, 18, e1800113 CrossRef PubMed.
- A. A. Dogan and M. Dufva, Sci. Rep., 2022, 12, 3694 CrossRef CAS PubMed.
- A. A. Dogan and M. Dufva, Biotechnol. Bioeng., 2023, 120, 1667–1677 CrossRef CAS PubMed.
- K. Piironen, M. Haapala, V. Talman, P. Järvinen and T. Sikanen, Lab Chip, 2020, 20, 2372–2382 RSC.
- Y. Xu, A. B. Xepapadeas, B. Koos, J. Geis-Gerstorfer, P. Li and S. Spintzyk, Dent. Mater., 2021, 37, e314–e327 CrossRef CAS PubMed.
- N. K. Hwangbo, N. E. Nam, J. H. Choi and J. E. Kim, Polymers, 2021, 13, 4410 CrossRef CAS PubMed.
- C. Hart, C. M. Didier, F. Sommerhage and S. Rajaraman, Biosensors, 2020, 10, 152 CrossRef CAS PubMed.
- E. Bayarsaikhan, J. H. Lim, S. H. Shin, K. H. Park, Y. B. Park, J. H. Lee and J. E. Kim, Polymers, 2021, 13, 1180 CrossRef CAS PubMed.
- N. P. MacDonald, F. Zhu, C. J. Hall, J. Reboud, P. S. Crosier, E. E. Patton, D. Wlodkowic and J. M. Cooper, Lab Chip, 2016, 16, 291 RSC.
- S. Kreß, R. Schaller-Ammann, J. Feiel, J. Priedl, C. Kasper and D. Egger, Materials, 2020, 13, 3011 CrossRef PubMed.
- M. Leth Jepsen, A. Willumsen, C. Mazzoni, A. Boisen, L. Hagner Nielsen and M. Dufva, Adv. Biosyst., 2020, 4, e1900289 CrossRef PubMed.
- C. A. Schneider, W. S. Rasband and K. W. Eliceiri, Nat. Methods, 2012, 9, 671–675 CrossRef CAS PubMed.
- T. G. Community, L. A. L. Abueg, E. Afgan, O. Allart, A. H. Awan, W. A. Bacon, D. Baker, M. Bassetti, B. Batut, M. Bernt, D. Blankenberg, A. Bombarely, A. Bretaudeau, C. J. Bromhead, M. L. Burke, P. K. Capon, M. Čech, M. Chavero-Díez, J. M. Chilton, T. J. Collins, F. Coppens, N. Coraor, G. Cuccuru, F. Cumbo, J. Davis, P. F. De Geest, W. de Koning, M. Demko, A. DeSanto, J. M. D. Begines, M. A. Doyle, B. Droesbeke, A. Erxleben-Eggenhofer, M. C. Föll, G. Formenti, A. Fouilloux, R. Gangazhe, T. Genthon, J. Goecks, A. N. G. Beltran, N. A. Goonasekera, N. Goué, T. J. Griffin, B. A. Grüning, A. Guerler, S. Gundersen, O. J. R. Gustafsson, C. Hall, T. W. Harrop, H. Hecht, A. Heidari, T. Heisner, F. Heyl, S. Hiltemann, H.-R. Hotz, C. J. Hyde, P. D. Jagtap, J. Jakiela, J. E. Johnson, J. Joshi, M. Jossé, K. Jum’ah, M. Kalaš, K. Kamieniecka, T. Kayikcioglu, M. Konkol, L. Kostrykin, N. Kucher, A. Kumar, M. Kuntz, D. Lariviere, R. Lazarus, Y. Le Bras, G. Le Corguillé, J. Lee, S. Leo, L. Liborio, R. Libouban, D. L. Tabernero, L. Lopez-Delisle, L. S. Los, A. Mahmoud, I. Makunin, P. Marin, S. Mehta, W. Mok, P. A. Moreno, F. Morier-Genoud, S. Mosher, T. Müller, E. Nasr, A. Nekrutenko, T. M. Nelson, A. J. Oba, A. Ostrovsky, P. V. Polunina, K. Poterlowicz, E. J. Price, G. R. Price, H. Rasche, B. Raubenolt, C. Royaux, L. Sargent, M. T. Savage, V. Savchenko, D. Savchenko, M. C. Schatz, P. Seguineau, B. Serrano-Solano, N. Soranzo, S. K. Srikakulam, K. Suderman, A. E. Syme, M. A. Tangaro, J. A. Tedds, M. Tekman, W. Cheng (Mike) Thang, A. S. Thanki, M. Uhl, M. van den Beek, D. Varshney, J. Vessio, P. Videm, G. Von Kuster, G. R. Watson, N. Whitaker-Allen, U. Winter, M. Wolstencroft, F. Zambelli, P. Zierep and R. Zoabi, Nucleic Acids Res, 2024, 52, W83–W94 Search PubMed.
- H. Li, Bioinformatics, 2018, 34, 3094–3100 CrossRef CAS PubMed.
- Y. Liao, G. K. Smyth and W. Shi, Bioinformatics, 2014, 30, 923–930 CrossRef CAS PubMed.
- M. I. Love, W. Huber and S. Anders, Genome Biol., 2014, 15, 1–21 CrossRef PubMed.
- B. T. Sherman, M. Hao, J. Qiu, X. Jiao, M. W. Baseler, H. C. Lane, T. Imamichi and W. Chang, Nucleic Acids Res., 2022, 50, W216–W221 CrossRef CAS PubMed.
- M. Hemmingsen, S. Vedel, P. Skafte-Pedersen, D. Sabourin, P. Collas, H. Bruus and M. Dufva, PLoS One, 2013, 8, e63638 CrossRef CAS PubMed.
- S. G. Cho, J. W. Lee, J. S. Heo and S. Y. Kim, Basic Clin. Pharmacol. Toxicol., 2014, 115, 282–290 CrossRef CAS PubMed.
- H. Schweikl, K. A. Hiller, A. Eckhardt, C. Bolay, G. Spagnuolo, T. Stempfl and G. Schmalz, Biomaterials, 2008, 29, 1377–1387 CrossRef CAS PubMed.
- O. Okamoto, S. Fujiwara, M. Abe and Y. Sato, Biochem. J., 1999, 337, 537 CrossRef CAS PubMed.
- U. Takeda, A. Utani, J. Wu, E. Adachi, H. Koseki, M. Taniguchi, T. Matsumoto, T. Ohashi, M. Sato and H. Shinkai, J. Invest. Dermatol., 2002, 119, 678–683 CrossRef CAS PubMed.
- S. Liu, J. Qiu, G. He, C. Geng, W. He, C. Liu, D. Cai, H. Pan and Q. Tian, J. Cancer, 2020, 11, 6288–6298 CrossRef CAS PubMed.
- D. Ye, Y. Wang, X. Deng, X. Zhou, D. Liu, B. Zhou, W. Zheng, X. Wang and L. Fang, Cell Death Dis., 2023, 14, 106 CrossRef CAS PubMed.
- J. Feng and L. Tang, Curr. Pharm. Des., 2014, 20, 6182–6190 CrossRef CAS PubMed.
- S. M. Ham, M. J. Song, H. S. Yoon, D. H. Lee, J. H. Chung and S. T. Lee, Int. J. Mol. Sci., 2023, 24, 12179 CrossRef CAS PubMed.
- C. Ucaryilmaz Metin and G. Ozcan, BMC Cancer, 2022, 22, 692 CrossRef CAS PubMed.
- S. Dorafshan, M. Razmi, S. Safaei, E. Gentilin, Z. Madjd and R. Ghods, Cancer Cell Int., 2022, 22, 315 CrossRef PubMed.
- Z. Zhao, C. C. Lee, S. Jiralerspong, R. C. Juyal, F. Lu, A. Baldini, F. Greenberg, C. T. Caskey and P. I. Patel, Hum. Mol. Genet., 1995, 4, 589–597 CrossRef CAS PubMed.
- J. Deng, J. Zhang, K. Gao, W. Yan, L. Zhou, Y. Jiang, J. Wang and Y. Wu, Neurochem. Res., 2022, 47, 3747–3760 CrossRef CAS PubMed.
- Y. Han, K. Xia and T. Su, Med. Sci. Monit., 2021, 27, e931238 CAS.
- E. M. Kim and O. Hwang, J. Neurochem., 2011, 116, 22–32 CrossRef CAS PubMed.
- P. Ershov, S. Poyarkov, Y. Konstantinova, E. Veselovsky and A. Makarova, Curr. Mol. Med., 2023, 23, 239–249 CrossRef CAS PubMed.
- T. Neill, L. Schaefer and R. V. Iozzo, Am. J. Pathol., 2012, 181, 380 CrossRef CAS PubMed.
- H. I. Lim and K. A. Hajjar, Int. J. Mol. Sci., 2021, 22, 6836 CrossRef CAS PubMed.
- M. V. Christensen, C. K. Høgdall, K. M. J. Umsen and E. V. S. Høgdall, Int. J. Oncol., 2018, 52, 5–18 CAS.
- E. M. Stone, T. A. Braun, S. R. Russell, M. H. Kuehn, A. J. Lotery, P. A. Moore, C. G. Eastman, T. L. Casavant and V. C. Sheffield, N. Engl. J. Med., 2004, 351, 346–353 CrossRef CAS PubMed.
- D. Markova, Y. Zou, F. Ringpfeil, T. Sasaki, G. Kostka, R. Timpl, J. Uitto and M. L. Chu, Am. J. Hum. Genet., 2003, 72, 998–1004 CrossRef CAS PubMed.
- R. Li, H. Wu, H. Jiang, Q. Wang, Z. Dou, H. Ma, S. Yan, C. Yuan, N. Yang and B. Kong, Oncol. Rep., 2020, 44, 2143–2151 CAS.
- S. Chakravarti, T. Magnuson, J. H. Lass, K. J. Jepsen, C. LaMantia and H. Carroll, J. Cell Biol., 1998, 141, 1277–1286 CrossRef CAS PubMed.
- O. Yamanaka, Y. Yuan, V. J. Coulson-Thomas, T. F. Gesteira, M. K. Call, Y. Zhang, J. Zhang, S. H. Chang, C. Xie, C. Y. Liu, S. Saika, J. V. Jester and W. W. Y. Kao, PLoS One, 2013, 8, e82730 CrossRef PubMed.
- N. McBrien, M. Cornell and A. Gentle, Invest. Ophthalmol. Visual Sci., 2001, 42, 2179–2187 CAS.
- X. Chen, X. Li, X. Hu, F. Jiang, Y. Shen, R. Xu, L. Wu, P. Wei and X. Shen, Front Oncol., 2020, 10, 533667 Search PubMed.
- Y. Zhao, F. Zhao, L. Zong, P. Zhang, L. Guan, J. Zhang, D. Wang, J. Wang, W. Chai, L. Lan, Q. Li, B. Han, L. Yang, X. Jin, W. Yang, X. Hu, X. Wang, N. Li, Y. Li, C. Petit, J. Wang, H. Y. J. Wang and Q. Wang, PLoS One, 2013, 8, e69549 CrossRef CAS PubMed.
- T. Yoshida, T. Akatsuka and K. Imanaka-Yoshida, Cell Adhes. Migr., 2015, 9, 96–104 CrossRef CAS PubMed.
- M. Shi, X. He, W. Wei, J. Wang, T. Zhang and X. Shen, Apoptosis, 2015, 20, 843–857 CrossRef CAS PubMed.
- H. Kuivaniemi and G. Tromp, Gene, 2019, 707, 151–171 CrossRef CAS PubMed.
- N. Dwivedi, N. Shukla, K. M. Prathima, M. Das and S. K. Dhar, Sci. Rep., 2023, 13, 13899 CrossRef CAS PubMed.
- S. R. Lamandé and J. F. Bateman, Matrix Biol., 2018, 71–72, 348–367 CrossRef PubMed.
- J. Qiao, C.-Y. Fang, S.-X. Chen, X.-Q. Wang, S.-J. Cui, X.-H. Liu, Y.-H. Jiang, J. Wang, Y. Zhang, P.-Y. Yang, F. Liu, J. Qiao, C.-Y. Fang, S.-X. Chen, X.-Q. Wang, S.-J. Cui, X.-H. Liu, Y.-H. Jiang, J. Wang, Y. Zhang, P.-Y. Yang and F. Liu, Oncotarget, 2015, 6, 29929–29946 CrossRef PubMed.
- H. Devos, J. Zoidakis, M. G. Roubelakis, A. Latosinska and A. Vlahou, Int. J. Mol. Sci., 2023, 24, 10004 CrossRef CAS PubMed.
- J. C. Marini, A. Forlino, H. P. Bächinger, N. J. Bishop, P. H. Byers, A. De Paepe, F. Fassier, N. Fratzl-Zelman, K. M. Kozloff, D. Krakow, K. Montpetit and O. Semler, Nat. Rev. Dis. Primers, 2017, 3, 17052 CrossRef PubMed.
- X. Li, X. Sun, C. Kan, B. Chen, N. Qu, N. Hou, Y. Liu and F. Han, Pathol., Res. Pract., 2022, 236, 154013 CrossRef CAS PubMed.
- J. A. N. Meester, G. Vandeweyer, I. Pintelon, M. Lammens, L. Van Hoorick, S. De Belder, K. Waitzman, L. Young, L. W. Markham, J. Vogt, J. Richer, L. M. Beauchesne, S. Unger, A. Superti-Furga, M. Prsa, R. Dhillon, E. Reyniers, H. C. DIetz, W. Wuyts, G. Mortier, A. Verstraeten, L. Van Laer and B. L. Loeys, Genet. Med., 2017, 19, 386–395 CrossRef CAS PubMed.
- H. Sun, X. Wang, Y. Zhang, X. Che, Z. Liu, L. Zhang, C. Qiu, Q. Lv and J. Jiang, Arch. Gynecol. Obstet., 2016, 293, 429–438 CrossRef CAS PubMed.
- V. Casagrande, M. Federici and R. Menghini, Acta Diabetol., 2021, 58, 1587–1594 CrossRef CAS PubMed.
- W. T. Lee, P. Y. Wu, Y. M. Cheng and Y. F. Huang, Int. J. Mol. Sci., 2024, 25, 3191 CrossRef CAS PubMed.
- C. Kannabiran and G. K. Klintworth, Hum. Mutat., 2006, 27, 615–625 CrossRef CAS PubMed.
- A. Corona and G. C. Blobe, Cell Signal, 2021, 84, 110028 CrossRef CAS PubMed.
- J. M. Łopacińska, J. Emnéus and M. Dufva, PLoS One, 2013, 8, e53107 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ma01191k |
| This journal is © The Royal Society of Chemistry 2025 |