DOI:
10.1039/D4MA01047G
(Paper)
Mater. Adv., 2025,
6, 2338-2355
Exploration of the antibacterial and photocatalytic properties of copper oxide (CuO) nanoparticles synthesized from Azadirachta indica leaf extract and electronic waste cable
Received
17th October 2024
, Accepted 2nd March 2025
First published on 3rd March 2025
Abstract
This study examined the antibacterial properties of CuO nanoparticles derived from Azadirachta indica leaf extract and waste Cu wire as a source of CuNO3. This environmentally friendly material efficiently facilitated the breakdown of industrial pollutants (dye and ciprofloxacin). The X-ray diffraction (XRD) analysis confirmed that the produced sample was CuO, with a crystallite size ranging from 5 to 92 nm, determined using the Sahadat–Scherrer, Monshi–Scherrer, Halder–Wagner, size-strain plot, and Williamson–Hall methods and the linear straight-line method of Scherrer's equation. A thermogravimetric analyzer confirmed three stages of weight loss (totaling 30.22%), while scanning electron microscopy revealed spherical-shaped NPs. The elemental composition (76.55% Cu and 23.45% O) was confirmed through energy-dispersive X-ray spectroscopy. Fourier transform infrared spectroscopic analysis, along with XRD, confirmed the presence of Cu–O bonding, indicating the generation of CuO NPs. The degradation percentages of CR dye (95%) and ciprofloxacin (80%) were estimated at a 20 ppm concentration (pH 7) using 0.1 g of catalyst, with degradation occurring over 120 min and 210 min, respectively. The computed rate constants, based on the Langmuir–Hinshelwood model equation, were 0.0198 min−1 for CR dye and 0.0047 min−1 for ciprofloxacin. The zones of inhibition for two Gram-positive bacteria were 27 mm (Staphylococcus aureus) and 14 mm (Bacillus megaterium), while for Gram-negative bacteria, the zones were 20 mm (Escherichia coli) and 13 mm (Salmonella typhi).
1. Introduction
Metal oxide nanoparticles have garnered significant interest due to their extensive applications in environmental, dental, food, agricultural, energy, pharmaceutical, electronic, catalytic, and biosensing fields.1 Copper oxide nanoparticles (CuO NPs) are potent anti-carcinogenic, antibacterial, catalytic, and coating agents that demonstrate intriguing activity across several biomedical sectors.2 Numerous synthesis techniques, including sol–gel, co-precipitation, hydrothermal,3 microwave irradiation,4 and microemulsion5 methods, facilitate the generation of these NPs. Chemical techniques often use surfactants or capping agents to regulate the growth and stability of CuO NPs.6 Moreover, CuO NPs can be produced by extracting plant components, such as flowers, fruits, roots, stems, leaves, seeds, and microbes (bacteria and fungi).7 CuO NPs exhibit effective antimicrobial properties and are used as disinfectants against healthcare-related infections in medical nanotechnology applications.2
The stability of nanoparticles depends on the synthesis process; as a result, nanoparticles generated from plant extracts tend to be more stable than those produced through other approaches (physical or chemical). Plant extracts contain biomolecules, such as aromatic compounds, proteins, vitamins, amino acids, saponins, polysaccharides, and enzymes, which assist in the environmentally friendly synthesis of nanoparticles.8 The secondary metabolites in plant extracts can function as reducing and stabilizing agents for the synthesized nanoparticles.9 Plants serve as reducing agents during the production of NPs, helping to eliminate harmful superfluous compounds.10 Therefore, the synthesis of CuO NPs utilizing neem (Azadirachta indica) leaves is promising. The neem tree (Azadirachta indica), which belongs to the Meliaceae family, is native to the Indian subcontinent. India is the world's largest producer of neem, and it is also native to Bangladesh, Pakistan, China, Myanmar, and Sri Lanka.11 Neem is also called a “wonder tree” due to its economic, environmental, medicinal, and ornamental value. It is found along roadsides and in the backyards of homes and offices. Moreover, several phytochemicals such as vitamins, terpenoids, alkaloids, minerals, amino acids, flavonoids, glycosides, and sterols are present in it.12,13 Due to secondary metabolites, when a reaction occurs between neem leaf extract and a metal salt, it reduces the salt and produces nanoparticles.14 A CuNO3 solution was used to carry out this research, which was synthesized using waste Cu wires as a source of Cu (58.3%).15 Due to cutting-edge technology, people use different types of electronic equipment to make their lives comfortable and fast. Therefore, repurposing electronic waste after its useful life can be an effective way to help protect the environment.
The aquatic environment has been negatively impacted by water pollution, and the amount of water for human consumption is so less (0.6%).16 The scarcity of water worldwide is exacerbated by industrial chemicals,17 the tanning industry, the textile industry,18 metal smelting, active pharmaceutical ingredients,19 the paper industry, electroplating, surfactants,20 and other factors. Polluted water from textile effluents (dyes) and antibiotics (ciprofloxacin) has adverse effects on the environment and living organisms. About 10%–25% of the dye is released from several industries and the varieties of commercial dyes are around 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 on the global market.21 In addition, antibiotic consumption has increased dramatically (200%) compared to the years 2000–2015, and during this period, the consumption was about 65%.22 This has happened due to the increasing drug consumption rate and the growing pharmaceutical industry, especially in Asian countries (China, Bangladesh, and India) that discharge pharmaceutical effluents in large amounts.23 In that regard, the green-synthesized nanoparticles were applied as efficient catalysts to treat wastewater (dye and antibiotics) effectively through the photocatalytic method for saving the environment and human beings from organic pollutants, antibiotics, and dyes.
000 on the global market.21 In addition, antibiotic consumption has increased dramatically (200%) compared to the years 2000–2015, and during this period, the consumption was about 65%.22 This has happened due to the increasing drug consumption rate and the growing pharmaceutical industry, especially in Asian countries (China, Bangladesh, and India) that discharge pharmaceutical effluents in large amounts.23 In that regard, the green-synthesized nanoparticles were applied as efficient catalysts to treat wastewater (dye and antibiotics) effectively through the photocatalytic method for saving the environment and human beings from organic pollutants, antibiotics, and dyes.
Based on the previous literature, no research studies have discussed the in-depth crystallographic information for plant-mediated and Cu wire-utilized CuO NPs and their effectiveness for all the microbes. Moreover, the green-synthesized CuO NPs showed efficient photocatalytic activity for the degradation of Congo red dye (CR dye) and ciprofloxacin, which has not been found in the earlier research work.
This study focuses on the green synthesis of CuO NPs using Azadirachta indica leaves and the emergence of CuO NP's antimicrobial activity against both Gram-positive (Staphylococcus aureus and Bacillus megaterium) and Gram-negative (Salmonella typhi and Escherichia coli) bacteria. Moreover, in this work, the CuO nanoparticles acted as effective catalysts to treat wastewater that contained CR dye and ciprofloxacin separately. Plant-mediated CuO NPs have been previously reported in the literature,24–26 but this research synthesized CuO NPs by combining leaf extract with waste Cu wires. Therefore, the main focus of this research is to offer detailed crystallographic insights into plant-mediated and waste Cu wire-synthesized CuO NPs, highlighting their effectiveness against microbes and their efficiency in degrading CR dye and ciprofloxacin.
2. Materials and method
2.1. Materials
The waste cable incapable of passing electricity was collected from the nearest cable shop in Dhaka, the capital of Bangladesh. Sodium hydroxide (NaOH; purity more than 99%), was used to maintain the pH, and nitric acid (purity 65%) was scored from E-Merck, Germany. Moreover, deionized water (DI) and ethanol were used to carry out the reaction. Further, Congo red dye (CR dye) and ciprofloxacin obtained from Shanghai Chemical Reagent, China, were used to examine the application of CuO NPs.
2.2. Synthesis method
At first, the newly cut leaves (Azadirachta indica) were collected from the residential area of the Bangladesh Council of Scientific and Industrial Research (Dhaka, Bangladesh). The neem plant (Azadirachta indica) was identified based on its distinct characteristics such as pinnate leaves, serrated leaflets, and bitter taste, following standard botanical keys and literature.27,28 Moreover, previous work reported in the literature also used the neem plant (Azadirachta indica) to conduct the experiment.29,30 Then the collected leaves (Fig. 1) were dried at 60 °C after sun-drying to obtain their powder. An electric blender was used to powder the dried leaves. A required amount of dry powdered leaves was taken in a beaker with deionized water that was heated at 80 °C for 30 min. The mixture was filtered after it was cooled to room temperature. After cooling, the extract was segregated using Whatman filter paper and then the filtered extract was stored frozen for subsequent processes. CuNO3 was prepared using a waste electric wire from which the Cu-part was segregated and washed with water. The cleaned Cu part was fragmented and pure nitric acid was added drop by drop. Consequently, a reaction occurred, which is presented as eqn (1). Afterward, the CuNO3 solution was placed in a 250 mL beaker and a burette was used to add the leaf extract gradually. At that moment, the light blue color of the solution became dark green. Again, using a burette, a NaOH solution (2 M) was added dropwise into the mixture with continuous stirring to attain a pH of 12. During this time, the reaction temperature was 80 °C.31 The reaction was continued until the dark brown precipitate was formed. Afterward, the filtrate was cleaned with deionized water and ethanol three times and once, respectively. In the last step, the washed precipitate was dried at 80 °C and then ground using a mortar and pestle for characterization and application. The overall synthesis process of copper oxide nanoparticles (CuO NPs) is depicted in Fig. 2.| | | soild Cu wire + conc. HNO3 → CuNO3(solution) | (1) |
 |
| | Fig. 1 Fresh leaves of neem (Azadirachta indica). | |
 |
| | Fig. 2 Graphical representation of green-synthesized CuO NPs. | |
2.3. Characterization
The crystallographic phase was explored using a Rigaku SE model X-ray diffractometer with a CuKα radiation source at (1.5406 nm). During scanning, the temperature was 20 °C, and the range was 20 to 70° with steps of 0.01. During this period, the voltage and current were 50 kV and 40 mA, respectively. A standard reference (silicon) was deployed for calibration before analysis. To identify the functional groups of the green-synthesized CuO NPs, an IR-prestige 21 Fourier Transform Infrared Spectrometer (Shimadzu, Japan) was employed. The attenuated total reflection (ATR) approach assisted in assembling the FTIR data (wavenumber range: 400–4000 cm−1). During the generation of the FTIR spectrum, average scanning and spectral resolution were 30 scans and 4 cm−1, respectively. The thermal stability of green-synthesized CuO NPs was inspected using a thermogravimetric analyzer (PerkinElmer; TGA 8000) in the scanning range of 50–800 °C. Nitrogen gas was applied to maintain a neutral atmosphere inside the apparatus. The generated CuO NPs’ surface characteristics and chemical composition were analyzed using an SEM (scanning electron microscopy) and an EDX (energy-dispersive X-ray spectroscopy) device. During the analysis, the Zeiss machine (SEM) operated at 25 kV and the AMETEK US EDX at 15 kV. The synthesized nanoparticle was deployed as a catalyst and a U-2910 UV-vis spectrophotometer was used to determine the efficacy and degradation percentage of ciprofloxacin and CR dye. In this case, the scanning range was 450–550 nm (CR dye) and 200–400 nm (ciprofloxacin). Similar instruments were also used in the previous work reported in the literature to characterize the sample.32
2.4. Point of zero charge
A material has zero surface density when it is at the point of zero charge. First, 0.1 M of NaCl (30 mL) and a fixed amount of catalyst (0.02 g) were taken in a conical flask to explore the catalyst's point of zero charge and its initial pHi was kept under three conditions such as acidic (3, 5), neutral (7), and basic (9, 11) using HNO3 and NaOH. Then to agitate the solution and catalyst, the speed and time of the orbital shaker were 200 rpm and 30 min, respectively, and after this step, the pHf of the sample was noted to compute the pH value employing eqn (2):33
2.5. Photocatalytic functioning
An SEN TAI JM-500 halogen lamp (500 W) was installed in a box made of wooden material in which a beaker containing the synthesized sample with the solution of pollutants (dye/antibiotic) was placed to investigate the degradation percentage and capacity. A relative humidity of 60% and a temperature of 25 °C were maintained during the experiment. In addition, the halogen lamp was positioned at 0.14 meters from the solution. Eqn (3) and (4) are the mathematical formulas for computing the degradation percentage and capacity.34| |  | (3) |
| | 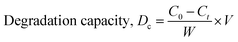 | (4) |
where C0 is the concentration of the sample at the initial stage, Ct is the concentration of the sample after time t, V is the solution's volume, and W is the catalyst's weight.
2.6. Test for antimicrobial efficacy
The tested bacterial strains were purchased from Thermo–Fisher Scientific Co. Ltd in lyophilized conditions and then sub-cultured in our laboratory for further applications. All the strains were reference strains containing a specific ATCC number (Escherichia coli – 11303, Salmonella typhi – 13311, Staphylococcus aureus – 9144, and Bacillus megaterium – 9885). The antimicrobial activity was explored by applying the typical agar well diffusion approach. Gram-positive and Gram-negative strains of bacteria were used in this research. The turbidity of the McFarland standard was 0.5 and fresh cultures (5 μL) were equally dispersed on the agar medium. The plates were set up to generate an agar well (6 mm) and the cork borer used for this investigation was sterile. Each well was filled with CuO NPs (50 μL of 200 μg mL−1). Kanamycin was employed as a reference standard, whereas DMSO was used as a solvent controller. To ensure homogeneous dispersion, 4 °C was upheld for the specimen at a fixed time (3 h). The zone of inhabitation was determined after completing incubation at 37 °C aerobically and 24 h of incubation time.
2.7. Statistical analysis
To compare the data, a one-way Analysis of Variance (ANOVA) test was executed using Microsoft Excel. A value of p < 0.05 was considered statistically significant, whereas a value of p > 0.05 was considered statistically non-significant.
3. Results and discussion
3.1. Crystallographic analysis of CuO NPs
Fig. 3 shows the X-ray diffractogram of CuO NPs generated via the green approach. To ensure that the synthesized product was CuO NPs, the obtained peaks in the X-ray diffractogram were correlated with the standard database (ICDD: #00-005-0661). The produced specimen was proven to be CuO NPs by the International Centre for Diffraction Data (ICDD). The outcome also aligned with the results previously reported in the literature. Table 1 exposes the peaks and planes of the generated CuO NPs with standard data.
 |
| | Fig. 3 Graphical representation of the XRD pattern of CuO NPs. | |
Table 1 Diffraction angles and planes of the generated CuO NPs with their references
| Diffraction angle (2θ) |
| CuO NPs |
Standard value card no: #00-005-0661 |
Planes (h k l) |
| 35.480 |
35.553 |
−1 1 1 |
| 38.669 |
38.730 |
1 1 1 |
| 48.446 |
48.761 |
−2 0 2 |
| 53.350 |
53.411 |
0 2 0 |
| 57.970 |
58.315 |
2 0 2 |
| 61.670 |
61.569 |
− 1 1 3 |
| 65.840 |
65.806 |
0 2 2 |
| 67.840 |
68.140 |
2 2 0 |
To compute the green-synthesized CuO NPs’ crystallographic parameters, eqn (5)–(11) were applied:35,36
| |  | (5) |
| |  | (6) |
| |  | (7) |
| |  | (8) |
| |  | (9) |
In the above-mentioned mathematical equations, θ is the angle of diffraction, K is Scherrer's constant and its value is 0.90, H1, H2, and H3 are the heights of the three strongest peaks, β is the Full width at Half maxima in radian, and h, k, l and a, b, c are the lattice parameters of crystals. The estimated values in the case of crystallographic parameters are enlisted in Table 2.
Table 2 Estimation of the crystallographic parameters
| Crystallographic parameters |
Deduced value |
| Crystallite size, Dc (nm) |
11.24 |
| Dislocation density (1015 lines per m2) |
0.0079 |
| Lattice parameters (Å) |
a = 4.71, b = 3.38, c = 5.05 |
| Microstrain |
0.53 |
| Crystallinity index |
1.84 |
To compute the preference growth, a specific plane (1 1 1) was compared to three additional planes (−1 1 1), (−2 0 2), and (0 0 2) and the relative intensity was considered (eqn (10)). Eqn (11) was employed to assess the preference growth of the (1 1 1) plane based on the relative intensities of the standard (RIst) and synthesized (RIs) samples. Similarly, the plane's (−1 1 1) preference growth was calculated against the (0 0 2), (1 1 1), and (−2 0 2) planes. In the same way, the preferential growth of the (0 2 0), and (2 2 0) planes was calculated, and all values are enlisted in Table 3. The estimated positive value of preference growth implied the thermodynamically favorable plane, whereas the negative value mentioned the thermodynamically unfavorable plane under the reaction condition.
| |  | (10) |
| |  | (11) |
Table 3 Calculation of the relative intensity and preferential growth
| Plane |
Other three planes |
Relative intensity |
Preferential growth |
| Standard, RIst |
Sample, RIs |
| 1 1 1 |
−1 1 1, 0 0 2, −2 0 2 |
0.55 |
1.50 |
1.73 |
| −1 1 1 |
0 0 2, 1 1 1, −2 0 2 |
0.58 |
0.45 |
−0.22 |
| 0 2 0 |
2 0 2, 0 2 2, 2 2 0 |
0.17 |
0.37 |
1.11 |
| 2 2 0 |
2 0 2, 0 2 2, 0 2 0 |
0.58 |
0.34 |
−0.38 |
3.2. Calculation of the crystallite size
The following model equations were applied from which the crystallite size, energy density, and strain were estimated.
3.2.4. Halder–Wagner method.
According to the Halder–Wagner method (HWM), the XRD reflection cannot be fully explained by either a Gaussian or Lorentzian function or a combination of both. This method introduced a new idea to focus on the cell and reciprocal lattice point.41 In this model, the reflection's tail follows the Lorentzian function. In contrast, the Gaussian function follows the peak. By using this combination, referred to as the Vigot function, eqn (16) precisely represents the broadening of the XRD peak.41
The integral breadth can be expressed as  , while a lattice plane spacing can be written as
, while a lattice plane spacing can be written as  . Eqn (17) represents the Halder–Wagnar approach in a mathematical context. Eqn (18) was created by swapping out the lattice plane and integral breadth to compute the strain and size of crystallites.42
. Eqn (17) represents the Halder–Wagnar approach in a mathematical context. Eqn (18) was created by swapping out the lattice plane and integral breadth to compute the strain and size of crystallites.42
| | 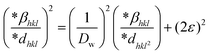 | (17) |
| |  | (18) |
By placing all values of  on the Y axis, and
on the Y axis, and  on the X-axis, an equation was formulated, which can be observed in Fig. 4(D). The size of the crystallite was determined from the generated slope, and the strain from the evolved intercept. In the case of the generated CuO, the crystallite size measured 5.05 nm and the strain was (1.5 × 10−5).
on the X-axis, an equation was formulated, which can be observed in Fig. 4(D). The size of the crystallite was determined from the generated slope, and the strain from the evolved intercept. In the case of the generated CuO, the crystallite size measured 5.05 nm and the strain was (1.5 × 10−5).
3.2.5. Williamson–Hall plot.
The equation of Scherrer does not consider the crystals’ lattice condition originating from the staking faults, line defect, point defect, lattice strain, area defect, grain boundary, etc., whereas real crystals invariably contain defects that induce strain.41,43Eqn (21) provides a comprehensive description of the overall broadening from the Williamson–Hall perspective considering the impact of strain and size. Stokes and Wilson formulated the following mathematical expression eqn (19) to estimate strain using peak broadening, with the value being influenced by the tan(θ) term.44
The following eqn (20) was derived by rearranging eqn (18) to calculate the strain:
| |  | (20) |
The crystal size can be calculated by the following eqn (21) based on the Scherrer eqn (12):
| |  | (21) |
The peak broadening of the sample was influenced by the size and strain, and the full width at half maxima (FWHM) can be expressed as follows:
| | | βtotal = βsize + βstrain | (22) |
Eqn (23) and (24) were obtained by substituting βsize and βstrain with new variables in eqn (22), resulting in the well-known Williamson–Hall equation:
| |  | (23) |
| |  | (24) |
The crystals are present in various environments, resulting in the segmentation of the Williamson–Hall equation into three distinct modified forms: the uniform deformation model (UDM), the uniform deformation energy density model (UDEDM), and the uniform stress deformation model (USDM).
3.2.5.1. Uniform deformation model (UDM).
Following the uniform deformation model (UDM) of the W–H model, the crystals within the powder sample are predicted to uphold identical strain, referred to as isotropic deformation of crystals. The conversion of eqn (24) gives rise to the following renowned eqn (25) by multiplying both sizes with cos(θ) within the uniform deformation model context to compute the crystallite size and strain in the crystals where ε is the strain, λ is the wavelength of the X-ray source, K is the Scherrer constant, D is the crystallite size, and βtotal is the full width at half maxima:45| |  | (25) |
The crystallite size and strain were computed considering the intercept and the slope of the equation by taking cos(θ)βtotal on the Y-axis and 4. sin(θ) on the X-axis in Fig. 5(A). After that, the generated equation and the standard linear equation (y = mx + c) were compared, from which the crystallite size was estimated. The crystallite size was determined to be 8.45 nm. The intrinsic strain was computed using the slope and a value was obtained, which was found to be 0.0004. Due to the negative value of intrinsic strain, it indicated the generation of compressive strain.46
 |
| | Fig. 5 Determination of the crystallite size using the uniform deformation model (A), uniform stress deformation model (B), uniform deformation energy density model (C), and size-strain plot method (D). | |
| |  | (27) |
Eqn (28) is a well-known representation of uniform stress deformation, established by placing eqn (27) in eqn (25):48
| |  | (28) |
The plot in Fig. 5(B) shows the relationship between  on the X-axis and cos(θ)βtotal on the Y-axis. Eqn (28) was compared with the straight-line equation to compute the stress value. Young's modulus value was 676.76 GPa.15 Additionally, the crystallite size became 8.45 nm taking into account the intercept of the equation and the estimated stress was 303 × 106 N m−2. The computed value of strain utilizing eqn (27) was found to be 4.47 × 10−4.
on the X-axis and cos(θ)βtotal on the Y-axis. Eqn (28) was compared with the straight-line equation to compute the stress value. Young's modulus value was 676.76 GPa.15 Additionally, the crystallite size became 8.45 nm taking into account the intercept of the equation and the estimated stress was 303 × 106 N m−2. The computed value of strain utilizing eqn (27) was found to be 4.47 × 10−4.
3.2.5.3. Uniform deformation energy density model (UDEDM).
The presence of defects in real crystals poses a challenge in achieving consistent stress and strain. To overcome this, the Williamson–Hall model was used. The uniform deformation energy density model (UDEDM) was established considering the novel idea of energy density along with the size and stress of crystallites. To evaluate the size and stress of crystallites as the anisotropic strain is uniform, a new concept known as energy density was included, leading to the development of the widely recognized uniform deformation energy density model (UDEDM).45eqn (29) and (30) illustrate the formulation of Hook's law concerning energy density:49| |  | (29) |
Or| | 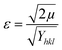 | (30) |
Eqn (30) was subjected to a transformation where the strain was substituted with the energy density, leading to the formulation of a distinct eqn (31) known as UDEDM:
| |  | (31) |
In Fig. 5(C), an equation was formed by plotting  on the X-axis and cos(θ)βtotal on the Y-axis, which was used to estimate the crystallite size and energy density. The resulting intercept of this calculation yielded a crystallite size of 8.45 nm. Moreover, the intercept also provided an energy density of 67.28 kJ m−3 from which the strain was estimated to be 0.00044.
on the X-axis and cos(θ)βtotal on the Y-axis, which was used to estimate the crystallite size and energy density. The resulting intercept of this calculation yielded a crystallite size of 8.45 nm. Moreover, the intercept also provided an energy density of 67.28 kJ m−3 from which the strain was estimated to be 0.00044.
3.3. FTIR analysis
Fig. 6(A) depicts the FTIR spectrum of Azadirachta indica leaf extract in which the wave number 3347 cm−1 was for the strong stretching band that related to the overlapping of the bending vibration of N–H of the amine group and OH stretching vibration of the phenolic group in the neem leaf extract. The peak at 1634 cm−1 was for C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching of the –COOH group in the extract. A similar result was observed in the earlier investigation.53 The bands at 661 and 551 cm−1 were for alkyl halide, whereas 599 cm−1 was assigned to the –OH stretching vibration. The previous exploration yielded similar results.54
O stretching of the –COOH group in the extract. A similar result was observed in the earlier investigation.53 The bands at 661 and 551 cm−1 were for alkyl halide, whereas 599 cm−1 was assigned to the –OH stretching vibration. The previous exploration yielded similar results.54
 |
| | Fig. 6 FTIR spectra of leaf extract (A) and CuO NPs (B). | |
The functional groups of the green-synthesized CuO NPs were obtained by analyzing the FTIR spectra visualized in Fig. 6(B). The absorption band at a low range of 489 to 695 cm−1 confirmed the vibration mode of Cu–O. Comparable forms of vibration mode were documented in the previous literature and the range of wavenumber was within 400–700 cm−1.55 The wavenumber of 927 cm−1 was the indicator of alkene (C–H and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond); related peaks were identified in the literature and the range was 841–937 cm−1.56 The wavenumber of 1061 cm−1 indicated the C–O stretching of phenols that was assured by the previous exploration, where the range was 1042–1076 cm−1.57 The absorption band at 1374 cm−1 (in the earlier exploration was 1381 cm−1) could be because of the alkene group's C–H stretching vibration or the esters’ O–C–O stretching.57 The peaks at 1488 cm−1 and 1587 cm−1 indicated the N–H groups of amines due to the biomolecules from leaf extract; this one was very close to previous outcomes (1518–1580 cm−1).58 The absorption band at 2358 cm−1 was assigned to the presence of environmental carbon dioxide which matched the earlier reports (2300–2385 cm−1) as well.59 The peak found at 3309 cm−1 was indicative of the stretching vibration of O–H that was found in the previous investigation (3264–3356 cm−1).60 The peak of 3577 cm−1 was very similar to the peak (3570 cm−1) of the previous study that was ascribed to the N–H stretching of amines involved in the stabilization of CuO NPs.61
C bond); related peaks were identified in the literature and the range was 841–937 cm−1.56 The wavenumber of 1061 cm−1 indicated the C–O stretching of phenols that was assured by the previous exploration, where the range was 1042–1076 cm−1.57 The absorption band at 1374 cm−1 (in the earlier exploration was 1381 cm−1) could be because of the alkene group's C–H stretching vibration or the esters’ O–C–O stretching.57 The peaks at 1488 cm−1 and 1587 cm−1 indicated the N–H groups of amines due to the biomolecules from leaf extract; this one was very close to previous outcomes (1518–1580 cm−1).58 The absorption band at 2358 cm−1 was assigned to the presence of environmental carbon dioxide which matched the earlier reports (2300–2385 cm−1) as well.59 The peak found at 3309 cm−1 was indicative of the stretching vibration of O–H that was found in the previous investigation (3264–3356 cm−1).60 The peak of 3577 cm−1 was very similar to the peak (3570 cm−1) of the previous study that was ascribed to the N–H stretching of amines involved in the stabilization of CuO NPs.61
3.4. Thermogravimetric analysis
The weight loss of green-synthesized CuO NPs during scanning through a thermogravimetric analyzer occurred in three stages, as graphically represented in Fig. 7. In the first stage (50–140 °C), 3.42% weight loss occurred on account of the evaporation of the molecules of water from the surface of the produced nanoparticles. Then, 16% weight loss (50–140 °C) was previously reported in the literature due to the same reason.62,63 The next two stages of weight loss occurred in the temperature range of 130–200 °C and 201–650 °C, respectively. The obtained weight loss was 4.08% and 6.72% as well. These occurred because of the combustion or decomposition of biomolecules on the surface of CuO NPs synthesized using the Azadirachta indica leaf extract. The same reason was also mentioned in the prior research investigation in which the weight loss became 17% (160–270 °C) and 48% (270–480 °C).62
 |
| | Fig. 7 Thermogravimetric analysis of CuO NPs. | |
3.5. Scanning electron microscopy
The morphological features of the green-synthesized CuO NPs were explored using an SEM, and the obtained image is represented in Fig. 8(A). The micrograph depicted the nanoparticles as spherical with an agglomeration state previously reported in the literature with a particle size of 50–100 nm.64,65 From the particle size distribution histogram visualized in Fig. 8(C), the computed size of the particle was 68.73 nm, which was within the range of earlier results. In addition, an energy-dispersive X-ray spectra (EDX) analyzer assured the chemical structure of the obtained product. Fig. 8(B) portrays the EDX graph of the CuO NPs, which revealed the weight percentages of 76.55% (copper) and 23.45% (oxygen) in the prepared nanoparticles that also supported the early exploration, where the weight percentages were 73.15% (copper) and 22.17% (oxygen).66 In the EDX spectrum, the peak for copper was so intense that confirmed the incredible crystalline particles of CuO NPs.
 |
| | Fig. 8 SEM micrographs (A), EDX spectra (B), and histogram analysis (C) of CuO NPs. | |
3.6. Point of zero charge
A pH higher than the pHZPC value indicates a negatively charged catalyst surface, while a pH lower than the pHZPC value suggests a positively charged surface. The point of intersection was obtained after plotting the values of ΔpH and pHi on the Y-axis, as well as the X-axis, respectively (visualized in Fig. 9). The pHZPC value generated was 8, higher than the pH 7 of the collective trial with the maximum degradation percentage. Congo red is an azo dye that carries a negative charge. Therefore, the negatively charged CR dye and the positively charged catalyst experienced strong electrostatic forces leading to a maximum degradation percentage.67
 |
| | Fig. 9 Estimation of the point of zero charge of CuO NPs. | |
3.7. Photocatalytic activity of CuO
3.7.1. Impact of time variation.
With the time variation, the degradation percentage and capacity were changed significantly. The rate of degradation increased progressively with the increasing time, but at a certain time, the percentage became eternal. This happened due to the saturation of the catalyst's active site.68 The maximum degradation was 95% at 120 min and the minimum was 74% at 30 min. The same incident happened for the degradation capacity. The maximum degradation capacity value was 7.6 mg g−1 at 120 min. The degradation percentage and capacity with time intervals are graphically represented in Fig. 10(A).
 |
| | Fig. 10 Assessment of the degradation capacity and degradation percentage of the CR dye based on time variation (A), catalyst dosage variation (B), distinct dye concentrations (C), and various pH limits (D). | |
3.7.2. Implications of various catalyst dosages.
The photocatalytic activity relies on the catalyst amount depicted in Fig. 10(B). To see the impact of the catalyst's dosages on the degradation capacity and percentage, 0.025–0.3 g catalyst was taken. Nearly, 95% was the highest degradation percentage due to 0.1 g catalyst. Furthermore, the degradation capacity came down substantially. The observed situation in photodegradation that followed can be explained by the photocatalyst particles clustering together with the increasing amount of the catalyst. As a result, it lessens the light that reaches the catalyst's active sites.69 The maximum degradation capacity was 26 mg g−1 because of the 0.025 g catalyst. To carry out the next two parameters, 0.1 g catalyst and 120 min were taken as fixed.
3.7.3. Influence of distinct concentrations of the dye.
Fig. 10(C) represents the impact of several dye concentrations (5–40 ppm) on the degradation capacity and percentage. The degradation percentage escalated minimally due to the variation in dye concentration, but at 40 ppm, the percentage dropped modestly and became 89%. As the dye concentration increased, so did the amount of dye that remained unabsorbed by the solution. Consequently, reduced light penetration through the solution may inhibit the light from reaching the photocatalyst surface. Thereby, the degradation percentage falls due to the diminishing of the concentration of radicals (˙OH) on the surface.70 The highest value was approximately 95% when the dye concentration was 20 ppm. On the contrary, the degradation capacity climbed sharply and the maximum capacity was 14 mg g−1 at 40 ppm concentration.
3.7.4. Consequences of various pH limits.
Beneath the light source, the solution's pH impacts the photodegradation percentage and capacity, as illustrated in Fig. 10(D). The maximum degradation was 95% at a medium pH level (pH 7). Both the degradation capacity and percentage were topmost at the intermediate level (pH 7) rather than higher (pH 9) and lower (pH 5) levels. The maximum degradation capacity was 7.6 mg g−1 at pH 7. Consequently, the photochemical reaction that was initiated by the synthesized CuO NPs was profoundly influenced by surplus hydrogen and hydroxyl ions.71 Moreover, the immediate interaction between the CuO catalyst and the CR dye molecules is significantly slower in acidic environments.72 So, the ideal pH was determined to be 7 because it did not require extra acid or base like pH 5 and pH 9.
3.8. Ciprofloxacin degradation
The degradation of ciprofloxacin across several time intervals was noted (visualized in Fig. 11). In this case, 0.1 g of CuO catalyst and 20 ppm of 40 mL ciprofloxacin solution were taken to evaluate the percentage and capacity of degradation. The maximum and minimum degradation were 80%, and 60% at 210 min and 60 min, respectively. At the same time, the degradation capacity was maximum at 210 min and the value was 6.46 mg g−1.
 |
| | Fig. 11 Assessment of the degradation percentage and capacity of ciprofloxacin based on several time intervals. | |
3.9. Kinetic studies
A well-known model equation (Langmuir–Hinshelwood) was used to compute the dye degradation rate. According to the Langmuir–Hinshelwood model equation, the mathematical correlation between the concentration considering time t (min) and the degradation rate can be revealed using eqn (34):73| |  | (34) |
In eqn (33), Kr is the rate of reaction and Kae is the adsorption equilibrium constant. The reaction is the expression of a pseudo-first-order reaction. After abandoning the denominator and considering C0 (t = 0) negligible because of its small value, the newly formed eqn (35) is given as follows:74
| |  | (35) |
After plotting  on the Y-axis and time (t, min) on the X-axis, the generated slope is the indicator of the apparent first-order rate constant (Kapp). The obtained rate constant from the graph (visualized in Fig. 12) for the green-synthesized CuO was 0.0198 min−1 (CR dye) and 0.0047 min−1 (ciprofloxacin).
on the Y-axis and time (t, min) on the X-axis, the generated slope is the indicator of the apparent first-order rate constant (Kapp). The obtained rate constant from the graph (visualized in Fig. 12) for the green-synthesized CuO was 0.0198 min−1 (CR dye) and 0.0047 min−1 (ciprofloxacin).
 |
| | Fig. 12 Kinetics of the photocatalytic degradation of CR dye (A) and ciprofloxacin (B) using CuO NPs as photocatalysts. | |
3.10. Recyclability and stability of photocatalysts
To examine the longevity of the generated catalyst, A recycling test for the degradation of CR dye and ciprofloxacin was carried out. To evaluate the reusability in the case of the degradation of CR dye, 0.1 g catalyst and 20 ppm of 40 mL dye solution were taken in a beaker and placed under a halogen lamp for 120 min. In the case of ciprofloxacin, the time was 210 min and other parameters were similar. Three cycles were executed and after completing every cycle, the catalyst was decanted and washed three times with water. After oven drying at 80 °C, the catalyst was used for the subsequent cycles. Gradually, the degradation percentage and capacity dropped because of the reduction of the catalyst's active site after every cycle. Fig. 13 depicts the recyclability and stability of green-synthesized Ag2O NPs. ANOVA tests revealed that the p-values due to degradation percentage were 0.015% (CR dye) and 0.007% (Ciprofloxacin) during the reusability test of the catalyst. Therefore, in this case, the degradation percentage was statistically significant because the p-value was smaller than 0.05. Furthermore, the p-values for degradation capacity were 0.20% (CR dye) and 0.32% (Ciprofloxacin); both were greater than 0.05, and hence, the degradation capacity was statistically insignificant.
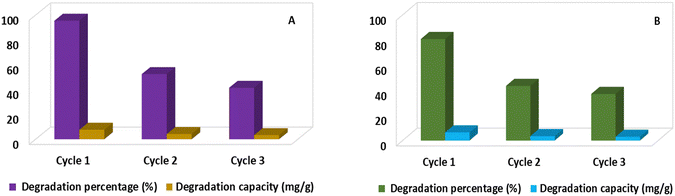 |
| | Fig. 13 Recyclability test of Ag2O NPs for the degradation of CR dye (A) and ciprofloxacin (B). | |
3.11. Scavenging experiment
To observe the active site of the catalyst during the breakdown of CR dye and ciprofloxacin, radical scavenging tests were performed using EDTA (ethylene diamine tetraacetic acid), DPPH (2,2-diphenyl-1-1-picrylhydrazyl) and IPA (isopropyl alcohol). DPPH and IPA were for free and hydroxyl radicals, respectively, while EDTA was for scavenging holes.75 Due to the addition of the scavenger, the degradation percentage of CR dye (95%) and ciprofloxacin (80%) dropped, which is portrayed in Fig. 14. The degradation of ciprofloxacin was 31% (DPPH), 44% (IPA), and 35% (EDTA), while that of CR dye was 36% (DPPH), 50% (IPA), and 11% (EDTA). The scavenging experiments gave the same outcome as the prior exploration.76,77
 |
| | Fig. 14 Scavenger test for CR dye (A) and ciprofloxacin (B). | |
3.12. Mechanism of the photocatalytic decomposition of pollutants
Numerous earlier studies have demonstrated the utilization of CuO nanoparticles as photocatalysts to disintegrate both industrial and pharmaceutical pollutants. As CuO nanoparticles become exposed to light, they generate hole (h+)–electron (e−) pairs. This occurs when electrons move from the valence band to the conduction band.78 When electrons (e−) and holes (h+) are created, the electrons and holes act as reducing agents and oxidizing agents, respectively. The photo-generated electrons and holes are confined to the CuO surface during photocatalysis. The electron interacts with the adsorbed oxygen and it generates superoxide radicals. The generated superoxide radical can act as a reductant or an oxidant.79 The potent oxidant (hydroxyl radical) is yielded due to the interaction of adsorbed water and holes. Several contaminants may be degraded during the photocatalysis of CuO because, at the time of photocatalysis, it generates numerous reactive molecules (eCB−, h+VB, OH˙, O2˙−, H2O2, O2, etc.) with byproducts (CO2 and H2O).80 The dye degradation step during photocatalysis is depicted in Fig. 16.
The following eqn (36)–(43) are the expressions of the reaction during photocatalysis, and Fig. 15 depicts the dye/antibiotic degradation mechanism.79 Furthermore, the outcome of this exploration was compared to the previously reported results in the literature (enlisted in Table 5).
| | | CuO NPs + hν → CuO NPs(eCB−) + h+VB | (36) |
| | | OH + reactant → Oxidised product | (38) |
| | | HO2˙ + HO2˙ → H2O2 + O2 | (41) |
| | | H2O2 + eCB− → OH˙ + OH− | (42) |
| | | Reactant + O2 → degradation products | (43) |
 |
| | Fig. 15 Schematic of the dye degradation mechanism by CuO NPs. | |
 |
| | Fig. 16 Dye degradation step during photocatalysis. | |
Table 5 Comparison chart revealing the degradation of pollutants
| Name of catalyst |
Synthesis approach |
Crystallite size (nm) |
Source of light |
Name of the pollutant |
Degradation percentage (%) |
Ref. |
| Abbreviations: GM – green method, HL – halogen lamp, SL – sunlight. |
| CuO |
GM |
12.24 |
HL |
Congo red dye |
95.4 |
This work |
| CuO |
GM |
— |
HL |
Ciprofloxacin |
80 |
This study |
| CuO |
Sol–gel method |
17 |
SL |
Methylene blue dye |
94.07 |
81
|
| CuO |
GM |
20 |
SL |
Methylene blue dye |
92 |
82
|
| CuO |
Biogenic method |
— |
UV-vis light |
Rhodamine B |
65 |
83
|
| CuO |
GM |
8.42–9.42 |
— |
Methyl orange |
96.4 |
84
|
| CuO/Fe2O3 |
GM |
— |
UV-vis light |
Tetracycline |
88 |
85
|
| CuO/ZnO |
Hydrothermal method |
— |
SL |
Ciprofloxacin |
93 |
86
|
3.13. Antimicrobial activities of green-synthesized CuO NPs
Nano-based remedies have been extensively researched to prepare new medications for detecting and treating illnesses. Similar to copper oxide nanoparticles (CuO NPs), metal oxide nanoparticles have been extensively explored for their antimicrobial features against several pathogenic bacteria. According to previous investigations, CuO NPs can inhibit several infections because of their identical antimicrobial activity. Fig. 17 portrays the antimicrobial activity of green-synthesized CuO NPs obtained from the leaves of the medicinal plant Azadirachta indica against Gram-negative and Gram-positive bacteria. In this case, kanamycin was used as the standard. Gram-positive (S. aureus and B. megaterium) and Gram-negative (E. coli and S. typhi) bacteria were manipulated to assess the antimicrobial activity of the generated product. The diameter of the zone of inhibition for the standard was 29 mm and 25 mm for S. aureus and E. coli, respectively. In contrast, for B. megaterium and S. typhi, the inhibition zone's diameter was similar (22 mm) to that of the standard. The maximum zone of inhibition was 27 mm for S. aureus, whereas the minimum was 13 mm in the case of S. typhi for green-synthesized CuO NPs. In the case of B. megaterium, and E. coli, the inhibitory active zone of CuO NPs was 14 mm, and 20 mm, respectively. Therefore, the highest active zone of CuO NPs was 27 mm for S. aureus (Gram-positive bacteria), which was nearly the standard. Several researchers also looked at CuO NPs’ antibacterial properties, which are included in Table 6. The ANOVA tests revealed that the p-value was 0.03 in the inhibition zone and the p-value was smaller than 0.05, and hence, it was statistically significant.
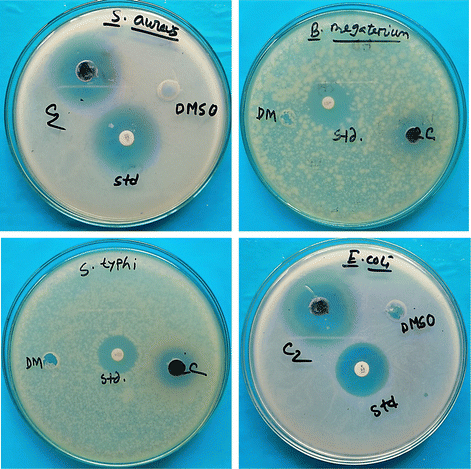 |
| | Fig. 17 Antimicrobial effectiveness of CuO NPs against Gram-negative and Gram-positive bacteria. | |
Table 6 Comparative visualization of the antimicrobial activity, showing the inhibition zone of the synthesized CuO NPs versus previous studies
| Nanoparticles |
Source of plant |
Zone of inhibition (mm) |
Ref. |
|
E. coli
|
S. aureus
|
S. typhi
|
B. megaterium
|
| CuO |
Azadirachta indica
|
20 |
27 |
13 |
14 |
This study |
| CuO |
Bifurcaria bifurcate
|
— |
16 |
— |
— |
87
|
| CuO |
Phyllanthus amarus
|
24 |
28 |
— |
— |
88
|
| CuO |
Aerva javanica
|
5 |
9 |
— |
— |
89
|
| CuO |
Nilgirianthus ciliatus
|
13 |
15 |
— |
— |
90
|
| CuO |
Ocimum tenuiflorum
|
14 |
12 |
— |
— |
91
|
| CuO |
Pterocarpus marsupium
|
10 |
11 |
— |
— |
92
|
3.14. Mechanism of the antimicrobial activity of CuO NPs
Reactive oxygen species are produced from green-synthesized CuO NPs, which traverse the cell membrane of the bacteria and then penetrate the membrane of the cell. Then, the penetrated CuO NPs inhibit the raising of the cells of bacteria; consequently, it causes cell death.93 Moreover, hydroxyl radicals and superoxide are released from CuO NPs, which also causes bacteria cell death.94 The generated reactive oxygen species (ROS) from CuO NPs can damage DNA, protein, mitochondria's functionality, and the stability of ribosomes.95 In this way, green-synthesized CuO NPs prevent the growth of both Gram-positive and Gram-negative bacteria. Fig. 18 depicts the mechanism of the antimicrobial capacity of green-synthesized CuO NPs.
 |
| | Fig. 18 Illustration of the function of CuO NPs in antimicrobial capacity. | |
3.15. FTIR analysis of the catalyst after photocatalytic activity
There were no additional peaks of CuO NPs after dipping in CR dye and ciprofloxacin. Fig. 19 reveals that even after the photocatalytic experiment was over (degrading the dye and ciprofloxacin), the functional groups of CuO NPs remained the same as those of the raw catalysts. Therefore, the photocatalytic activity had no impact on the CuO NPs.
 |
| | Fig. 19 FTIR spectra of CuO after the degradation of CR dye (A) and ciprofloxacin (B). | |
3.16. XRD analysis of the catalyst after photocatalysis
The X-ray diffractogram of CuO after photocatalysis in both ciprofloxacin and dye is displayed in Fig. 20. However, there was no change in the peak position compared to the X-ray diffractogram of CuO before photocatalysis (Fig. 2), which indicated that after the completion of the photocatalysis process, the catalyst's crystal structure remained stable.
 |
| | Fig. 20 X-ray diffractogram of CuO after the degradation of dye and ciprofloxacin. | |
3.17. Discussion
CuO NPs were synthesized using the leaf extract of Azadirachta indica, a common plant with excellent medicinal value. Besides, a waste electric Cu wire was used for the synthesis process. Therefore, the overall synthesis process was eco-friendly through the proper utilization of waste and leaf extract. The activity of this nano-sized (less than 100 nm) product was outstanding as a catalyst because it was able to degrade the CR dye (95.4%) and ciprofloxacin (80%). These two pollutants cause serious water pollution and create a scarcity of fresh water. Table 5 reveals the comparison of this product as a catalyst with previous explorations. Moreover, the green-synthesized CuO NP was able to inhibit the growth of Gram-positive (S. aureus and B. megaterium) and Gram-negative (E. coli and S. typhi) bacteria. Table 6 compares the inhibition zone of this product with that reported in previous studies.
3.18. Socio-economic importance
This research project focused on sustainable development goals (SDGs) towards 3Rs – reduce, reuse, and recycle for waste management to save the environment. Due to swift technological advancement and the usage of technological items in every sector, the amount of waste Cu-wires has increased with no bounds. Hence, the proper utilization of electronic waste can help to achieve the SDGs. Consequently, green-synthesized CuO NPs are beneficial to society by promoting eco-friendly production through the utilization of leaf extract and waste cable wires, combating antimicrobial resistance and enabling water pollution remediation, contributing to public health and environmental sustainability. Moreover, the cost of production for green-synthesized CuO NPs is relatively low due to the use of readily available Azadirachta indica leaf extract and the utilization of waste cable Cu wires. However, the scalability of the green synthesis process, the long-term stability of CuO NPs, and real-world applicability in complex wastewater remain unexplored due to the lack of large-scale production and application facilities.
4. Conclusion
CuO NPs have been successfully synthesized using the leaf extract of Azadirachta indica (a ubiquitous medicinal plant), which was ensured by correlating the XRD data with the standard ICDD data. The crystallite size of CuO NPs was less than 100 nm (nano size), as validated by all of the model equations. The thermogravimetric analysis revealed a total of 30.22% weight loss of the synthesized CuO NPs. The particle size was 68.72 nm, which was computed through histogram analysis. This study showed that CuO NPs effectively degraded ciprofloxacin and CR dye and remained catalytically effective after three reuse cycles. The maximum degradation was 80% (ciprofloxacin) and 95% (CR dye). The Azadirachta indica leaf extract-mediated CuO NPs effectively inhibited the growth of Gram-negative and Gram-positive bacteria and the widest zone of inhibition was 27 mm (S. aureus). These research findings reveal that green-synthesized CuO NPs have the potential activity to inhibit bacterial growth and treat wastewater. Therefore, future research may investigate advanced characterization techniques, expand studies on the antibacterial spectrum, and analyze detailed degradation mechanisms. Optimizing synthesis parameters and biocompatibility, testing real wastewater samples, and scaling up eco-friendly synthesis for industrial use hold great potential.
Author contributions
Sumaya Tabassum synthesized the product, performed the experiment, and wrote the draft and original manuscript. Md. Sahadat Hossain conceived the experiment, assisted in writing the final manuscript, and analyzed the data. Md. Abu Saeed assisted in writing the draft of the manuscript. Samina Ahmed supervised the overall work and managed the required facilities.
Data availability
Data will be made available upon request.
Conflicts of interest
There are no conflicts to declare.
Acknowledgements
The authors are grateful to the Bangladesh Council of Scientific and Industrial Research (BCSIR) for their support through the R&D project (ref. no. 39.02.00000.011.14.134.2021/900; Date: 30.12.2021). The authors also appreciate the financial support from the Ministry of Science and Technology (MOST), Bangladesh, through the R&D project. Sumaya Tabassum is also grateful to the BCSIR for approving Professor Nurul Afsar Khan's post-graduate fellowship.
References
- R. Javed, N. ul Ain, A. Gul, M. Arslan Ahmad, W. Guo, Q. Ao and S. Tian, IET Nanobiotechnol., 2022, 16, 171–189 CrossRef PubMed.
- M. E. Grigore, E. R. Biscu, A. M. Holban, M. C. Gestal and A. M. Grumezescu, Pharmaceuticals, 2016, 9, 75 CrossRef PubMed.
- D. Bharti and A. Bharati, Luminescence, 2017, 32(3), 317–320 CrossRef CAS PubMed.
- K. N’Konou, M. Haris, Y. Lare, M. Baneto, K. Napo and P. Torchio, Phys. Status Solidi, 2016, 253(2), 260–266 CrossRef.
- N. A. Mirgane, V. S. Shivankar, S. B. Kotwal, G. C. Wadhawa and M. C. Sonawale, Mater. Today: Proc., 2021, 37(2), 886–889 CAS.
- T. H. Tran and V. T. Nguyen, Int. Scholarly Res. Not., 2014, 856592 Search PubMed.
- M. Shamsuddin and N. Nordin, Mal. J. Fund. Appl. Sci, 2019, 15, 218–224 CrossRef.
- M. Sepahvand, F. Buazar and M. Sayahi, Appl. Organomet. Chem., 2020, 34, 1–11 CrossRef.
- K. Ganesan, V. K. Jothi, R. Arulmozhi and A. Natarajan, Mater. Adv., 2021, 124–142 Search PubMed.
- B. Malaikozhundan, R. Krishnamoorthi, J. Vinodhini, K. S. N. Nambi and S. Palanisamy, Inorg. Chem. Commun., 2022, 144, 109843 CrossRef CAS.
- R. Manimaran, Ind. Crops Prod., 2023, 204, 117304 CrossRef CAS.
- S. Srivastava, B. Agrawal, A. Kumar and A. Pandey, J. Sci. Res., 2020, 64, 285–290 Search PubMed.
- I. Ujah, C. Nsude, O. Ani, U. Alozieuwa, A. Okwor and I. Okpako, GSC Biol. Pharm. Sci., 2021, 14(02), 165–171 CrossRef CAS.
- F. Moradnia, S. Taghavi Fardood, A. Ramazani, B. Min, S. W. Joo and R. S. Varma, J. Cleaner Prod., 2021, 288, 125632 CrossRef CAS.
- Md Sahadat Hossain and S. Ahmed, Results Eng., 2023, 20, 101630 CrossRef CAS.
- K. Vikrant and K.-H. Kim, Chem. Eng. J., 2019, 358, 264–282 CrossRef CAS.
- O. Golovko, S. Örn, M. Sörengård, K. Frieberg, W. Nassazzi, F. Y. Lai and L. Ahrens, Sci. Total Environ., 2021, 754, 142122 CrossRef CAS PubMed.
- S. Vigneshwaran, P. Sirajudheen, C. P. Nabeena and S. Meenakshi, Colloids Surf., A, 2021, 611, 125789 CrossRef CAS.
- R. Lindberg, S. Namazkar, S. Lage, M. Östman, Z. Gojkovic, C. Funk, F. Gentili and M. Tysklind, Chemosphere, 2021, 271, 129763 CrossRef CAS PubMed.
- P. Y. Nguyen, G. Carvalho, M. A. M. Reis and A. Oehmen, Water Res., 2021, 188, 116446 CrossRef CAS PubMed.
- S. Khan, S. Khan and A. M. Asiri, Sci. Rep., 2016, 6, 35107 CrossRef PubMed.
- E. Klein, T. Boeckel, E. Martinez, S. Pant, S. Gandra, S. Levin, H. Goossens and R. Laxminarayan, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 201717295 Search PubMed.
- M. S. U. Rehman, N. Rashid, M. Ashfaq, A. Saif, N. Ahmad and J.-I. Han, Chemosphere, 2015, 138, 1045–1055 CrossRef CAS PubMed.
- D. M. Nzilu, E. S. Madivoli, D. S. Makhanu, S. I. Wanakai, G. K. Kiprono and P. G. Kareru, Sci. Rep., 2023, 13, 14030 CrossRef CAS PubMed.
- B. Malaikozhundan, V. N. Lakshmi and R. Krishnamoorthi, Mater. Today Commun., 2022, 33, 104348 CrossRef CAS.
- S. Faisal, N. S. Al-Radadi, H. Jan, Abdullah, S. A. Shah, S. Shah, M. Rizwan, Z. Afsheen, Z. Hussain, M. N. Uddin, M. Idrees and N. Bibi, Coatings, 2021, 11, 849 CrossRef CAS.
- I. V. S. Reddy and N. Palagani, Int. J. Econ. Plant, 2021, 9(1), 0437d Search PubMed.
- A. Verma and M. S. Mehata, J. Radiat. Res. Appl. Sci., 2016, 9, 109–115 CAS.
- S. Kumari, A. Patlolla and P. Madhusudhanachary, Micromachines, 2022, 13, 1416 CrossRef PubMed.
- N. S. Alharbi and N. S. Alsubhi, J. Nanomater., 2023, 2023, 9916777 Search PubMed.
- W. Andualem, F. Sabir, E. Tsegaye, H. Belay and B. Gonfa, J. Nanotechnol., 2020, 2932434 CAS.
- S. Tabassum, M. Sahadat Hossain, Md. M. H. Sachchu, M. Uddin and S. Ahmed, New J. Chem., 2025, 49, 1301–1313 RSC.
- N. S. Pinky, M. Mobarak, S. Mustafi, M. Rahman, A. Nahar, T. Saha and N. Bahadur, Arabian J. Chem., 2023, 16, 105080 CrossRef CAS.
- M. Kawsar, M. S. Hossain, S. Tabassum, N. Mohammed Bahadur and S. Ahmed, RSC Adv., 2024, 14, 11570–11583 RSC.
- Md Sahadat Hossain, S. Sarkar, S. Tarannum, S. M. Tuntun, M. Mahmud, M. Bin Mobarak and S. Ahmed, J. Saudi Chem. Soc., 2023, 27, 101769 CrossRef CAS.
- Md Sahadat Hossain and S. Ahmed, J. Saudi Chem. Soc., 2023, 27, 101649 CrossRef CAS.
- A. Monshi, M. R. Foroughi and M. R. Monshi, World J. Nano Sci. Eng., 2012, 02, 154–160 CrossRef.
- M. Sahadat Hossain and S. Ahmed, RSC Adv., 2022, 12, 25096–25105 RSC.
- M. Mobarak, M. Sahadat Hossain, F. Chowdhury and S. Ahmed, Arabian J. Chem., 2022, 15, 104117 CrossRef.
- Md Sahadat Hossain, S. A. Jahan and S. Ahmed, Results Chem., 2023, 5, 100822 CrossRef CAS.
- D. Nath, F. Singh and R. Das, Mater. Chem. Phys., 2020, 239, 122021 CrossRef CAS.
- Md. S. Saadi, I. Ahmed, A. Khazaal, K. Jasim and K. H. Harbbi, NeuroQuantology, 2022, 20, 199–204 CrossRef.
- S. Hossain, M. Mahmud, M. B. Mobarak, S. Sultana, Md. A. A. Shaikh and S. Ahmed, Chem. Pap., 2022, 76, 7245–7251 CrossRef.
- N. Jahan Tamanna, Md. S. Hossain, N. Mohammed Bahadur and S. Ahmed, Results Chem., 2024, 7, 101313 CrossRef CAS.
- V. Mote, Y. Purushotham and B. Dole, J. Theor. Appl., 2012, 6, 6 CrossRef.
- S. Tabassum, Md Sahadat Hossain, M. Bin Mobarak, F. Nigar and S. Ahmed, Arabian J. Chem., 2024, 17, 105901 CrossRef CAS.
- Y. Canchanya Huaman, F. Mayta Armas, J. Pomalaya Velasco, Y. Bendezu, J. Guerra and J. Ramos Guivar, Nanomaterials, 2021, 11, 2311 CrossRef CAS PubMed.
- P. Dey and R. Das, J. Mater. Eng. Perform., 2021, 30, 652–660 CrossRef CAS.
- N. Tayade and M. Tirpude, Bull. Mater. Sci., 2023, 46, 20 CrossRef CAS.
- Y. Prabhu, K. Rao, S. S. Kumar and B. Kumari, World J. Nano Sci. Eng., 2014, 04, 21–28 CrossRef CAS.
- H. Heryanto, D. Tahir and M. Mahmud, J. Phys.: Conf. Ser., 2019, 1317, 012052 CrossRef.
- H. Cheema, V. Yadav, R. S. Maurya, V. Yadav, A. Kumar, N. Sharma, P. A. Alvi and U. Kumar, J. Mater. Sci.: Mater. Electron., 2021, 32, 23578–23600 CrossRef CAS.
- H. A. Widatalla, L. F. Yassin, A. A. Alrasheid, S. A. R. Ahmed, M. O. Widdatallah, S. H. Eltilib and A. A. Mohamed, Nanoscale Adv., 2022, 4, 911–915 RSC.
- M. J. Islam, N. Khatun, R. H. Bhuiyan, S. Sultana, M. A. A. Shaikh, M. N. A. Bitu, F. Chowdhury and S. Islam, RSC Adv., 2023, 13, 19164–19172 RSC.
- A. Fouda, S. Salah Salem, A. Wassel, M. Hamza and T. Shaheen, Heliyon, 2020, 6, e04896 CrossRef CAS PubMed.
-
H. Selim, A. A. Nada, M. El-Sayed, H. Rm, E. R. Souaya and K. Mf.
- S. Alamdari, M. Sasani Ghamsari, C. Lee, W. Han, H.-H. Park, M. Jafar Tafreshi, H. Afarideh and M. Ara, Appl. Sci., 2020, 10, 3620 CrossRef CAS.
- M. Din, F. Arshad, A. Rani, A. Aihetasham, M. Mukhtar and H. Mehmood, Optoelectron. Adv. Mater., 2017, 9, 41–48 Search PubMed.
- J. Schott, C.-L. Do-Thanh, W. Shan, N. Puskar, S. Dai and S. Mahurin, Green Chem. Eng., 2021, 2(4), 392–401 CrossRef.
- D. S. Kumar, S. Sivaranjani, A. Mariam, B. Ravikumar, S. Pandiarajan, C. Veeralakshmi, J. Muthurulandi and M. Maaza, J. Semicond., 2018, 39(3) DOI:10.1088/1674-4926/39/3/032001.
- N. A. Thamer, N. Q. Muftin and S. N. Al-Rubae’i, Asian J. Chem., 2018, 30, 1559–1563 CrossRef CAS.
- H. C. A. Murthy, T. Desalegn, K. B. Tan, S. Ghotekar, M. W. Alam, B. Ruthramurthy, K. Chan, S. Fakrudeen, A. Kumar M R and C. R. Ravikumar, Results Chem., 2021, 3, 100141 CrossRef.
- M. Nasrollahzadeh, S. M. Sajadi, A. Rostami-Vartooni and S. M. Hussin, J. Colloid Interface Sci., 2016, 466, 113–119 CrossRef CAS PubMed.
- Z. Alhalili, Arabian J. Chem., 2022, 15, 103739 CrossRef CAS.
- I. Khaldari, M. R. Naghavi and E. Motamedi, RSC Adv., 2021, 11, 3346–3353 RSC.
- S. Sukumar, A. Rudrasenan and D. Nambiar, ACS Omega, 2020, 5(2), 1040–1051 CrossRef CAS PubMed.
- E. Abdelkader, N. Laouedj and A. Bekka, J. Chem. Eng. Process Technol., 2011, 2, 1–9 Search PubMed.
- E. Al-Farraj and E. Abdelrahman, ACS Omega, 2024, 9(4), 4870–4880 CrossRef CAS PubMed.
- F. Aisien, A. Amenaghawon and E. Ekpenisi, J. Eng. Appl. Sci., 2014, 9, 11–16 Search PubMed.
- M. Siddique, R. Khan, A. Khan and R. Farooq, J. Chem. Soc. Pak., 2014, 38, 37–43 Search PubMed.
- M. Mallikarjunaswamy, L. Ranganatha, R. Ramu, U. Gowda and N. Ganganagappa, J. Mater. Sci.: Mater. Electron., 2020, 31, 1004–1021 CrossRef.
- J. Shang, Y. Jiang, X. Qin, B. Zhao and X. Li, Front. Environ. Sci., 2021, 9, 783313 CrossRef.
- A. V. Petukhov, Chem. Phys. Lett., 1997, 277, 539–544 CrossRef CAS.
- E. Kusvuran, A. Samil, O. Atanur and O. Erbatur, Appl. Catal., B, 2005, 58, 211–216 CrossRef CAS.
- H. N. Jayasimha, K. G. Chandrappa, S. P. Fakrudeen and D. Kumar V G, Sens. Int., 2023, 5, 100254 CrossRef.
- A. Muthuvel, M. Jothibas and C. Manoharan, Nanotechnol. Environ. Eng., 2020, 5, 14 CrossRef CAS.
- Md. K. Alam, Md Sahadat Hossain, S. Tabassum, N. M. Bahadur and S. Ahmed, Open Ceram., 2024, 19, 100625 CrossRef CAS.
- J. Tang, J. Durrant and D. Klug, J. Am. Chem. Soc., 2008, 130, 13885–13891 CrossRef CAS PubMed.
- U. Gaya and A. Abdullah, J. Photochem. Photobiol., C, 2008, 9, 1–12 CrossRef CAS.
- L. Gemachu and A. Berhanu, Green Chem. Lett. Rev., 2024, 17(1), 2293841 CrossRef.
- B. Arunkumar, s Jeyakumar and M. Jothibas, Optik, 2019, 168, 414–425 Search PubMed.
- K. Dulta, G. K. Ağçeli, P. Chauhan, R. Jasrotia, G. Chandan, P. K. Chauhan and J. O. Ighalo, Sustainable Environ. Res., 2022, 32, 2 CrossRef CAS.
- M. B. Zaman, R. Poolla, P. Singh and T. Gudipati, Environ. Nanotechnol., Monit. Manage., 2020, 14, 100346 Search PubMed.
- S. Sharma, K. Kumar, N. Thakur, S. Chauhan and M. Chauhan, J. Environ. Chem. Eng., 2021, 9, 105395 CrossRef CAS.
- S. Kaushal, A. Kumar, H. Bains and P. P. Singh, Environ. Sci. Pollut. Res., 2023, 30, 37092–37104 CrossRef CAS PubMed.
- K. Bano, S. Kaushal, B. Lal, S. Joshi, R. Kumar and P. Singh, Environ. Nanotechnol., Monit. Manage., 2023, 20, 100863 CAS.
- Y. Abboud, T. Saffaj, A. Chagraoui, E. B. Abdeslam, K. Brouzi, O. Tanane and B. Ihssane, Appl. Nanosci., 2014, 4, 571–576 CrossRef CAS.
- N. Acharyulu, R. S. D. O. R. S. Dubey, P. Kollu, V. Swaminadham, R. Kalyani and S. V. N. Pammi, Int. J. Eng. Res. Sci. Technol., 2014, 3, 639 Search PubMed.
- G. Afzal, A. Jamal, S. Kiran, G. Mustafa, T. Mehmood, F. Ahmad, S. Saeed, A. Ali, N. Naz, S. Zehra, S. Khalil and S. Dawood, J. Anim. Plant Sci., 2022, 32(2), 547–553 CAS.
- R. Rajamma, S. Nair, F. Khadar and B. Baskaran, IET Nanobiotechnol., 2020, 14(9), 749–907 CrossRef PubMed.
- N. C. Joshi and Y. Prakash, Asian J. Pharm. Clin. Res., 2019, 12(8), 288–291 CAS.
- G. Sharma, J. Microb. Biochem. Technol., 2015, 7(3), 140–144 Search PubMed.
- D. Das, B. C. Nath, P. Phukon and S. K. Dolui, Colloids Surf., B, 2013, 101, 430–433 CrossRef CAS PubMed.
- P. Sutradhar, M. Saha and D. Maiti, J. Nanostruct. Chem., 2014, 4, 86 CrossRef.
- S. A. Akintelu, A. S. Folorunso, F. A. Folorunso and A. K. Oyebamiji, Heliyon, 2020, 6, e04508 CrossRef PubMed.
|
| This journal is © The Royal Society of Chemistry 2025 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article a,
Md.
Sahadat Hossain
a,
Md.
Sahadat Hossain
 *a,
Md.
Abu Saeed
c,
Md. Najem
Uddin
*a,
Md.
Abu Saeed
c,
Md. Najem
Uddin
 b and
Samina
Ahmed
b and
Samina
Ahmed
 *a
*a
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 on the global market.21 In addition, antibiotic consumption has increased dramatically (200%) compared to the years 2000–2015, and during this period, the consumption was about 65%.22 This has happened due to the increasing drug consumption rate and the growing pharmaceutical industry, especially in Asian countries (China, Bangladesh, and India) that discharge pharmaceutical effluents in large amounts.23 In that regard, the green-synthesized nanoparticles were applied as efficient catalysts to treat wastewater (dye and antibiotics) effectively through the photocatalytic method for saving the environment and human beings from organic pollutants, antibiotics, and dyes.
000 on the global market.21 In addition, antibiotic consumption has increased dramatically (200%) compared to the years 2000–2015, and during this period, the consumption was about 65%.22 This has happened due to the increasing drug consumption rate and the growing pharmaceutical industry, especially in Asian countries (China, Bangladesh, and India) that discharge pharmaceutical effluents in large amounts.23 In that regard, the green-synthesized nanoparticles were applied as efficient catalysts to treat wastewater (dye and antibiotics) effectively through the photocatalytic method for saving the environment and human beings from organic pollutants, antibiotics, and dyes.
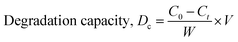








 and cos
and cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ were placed on the X and Y axes, respectively, from which a straight line was evolved. The originated straight line visualized in Fig. 4(A) was correlated with the original straight-line equation (y = mx + c). The computed crystallite size of the synthesized CuO was 92.43 nm.
θ were placed on the X and Y axes, respectively, from which a straight line was evolved. The originated straight line visualized in Fig. 4(A) was correlated with the original straight-line equation (y = mx + c). The computed crystallite size of the synthesized CuO was 92.43 nm.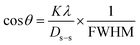
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ on the Y-axis and
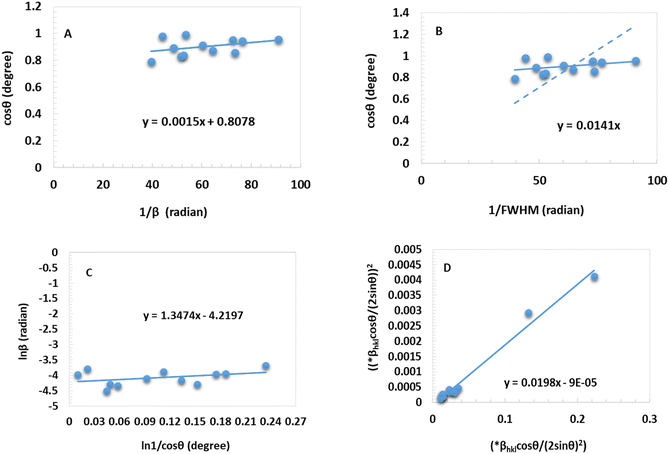
θ on the Y-axis and  on the X-axis that was straight. Another straight line was formed with a zero intercept to ensure it goes through the origin. The Sahadat–Scherrer model, which was utilized to estimate the crystallite size of synthesized CuO, is depicted in Fig. 4(B). The slope of the line was used to calculate the crystallite size of the generated CuO, which came out to be 9.83 nm.
on the X-axis that was straight. Another straight line was formed with a zero intercept to ensure it goes through the origin. The Sahadat–Scherrer model, which was utilized to estimate the crystallite size of synthesized CuO, is depicted in Fig. 4(B). The slope of the line was used to calculate the crystallite size of the generated CuO, which came out to be 9.83 nm.

 on the X-axis and ln
on the X-axis and ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β on the Y-axis, resulting in the formation of a linear equation, and the slope of the constructed equation was used to estimate the average size of the crystal. The resulting equation was then compared to the standard linear equation and the intercept was equal to
β on the Y-axis, resulting in the formation of a linear equation, and the slope of the constructed equation was used to estimate the average size of the crystal. The resulting equation was then compared to the standard linear equation and the intercept was equal to  . The calculated crystallite size of CuO was 9.43 nm by employing the Monshi–Scherrer method (MSM).
. The calculated crystallite size of CuO was 9.43 nm by employing the Monshi–Scherrer method (MSM). , while a lattice plane spacing can be written as
, while a lattice plane spacing can be written as  . Eqn (17) represents the Halder–Wagnar approach in a mathematical context. Eqn (18) was created by swapping out the lattice plane and integral breadth to compute the strain and size of crystallites.42
. Eqn (17) represents the Halder–Wagnar approach in a mathematical context. Eqn (18) was created by swapping out the lattice plane and integral breadth to compute the strain and size of crystallites.42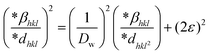

 on the Y axis, and
on the Y axis, and  on the X-axis, an equation was formulated, which can be observed in Fig. 4(D). The size of the crystallite was determined from the generated slope, and the strain from the evolved intercept. In the case of the generated CuO, the crystallite size measured 5.05 nm and the strain was (1.5 × 10−5).
on the X-axis, an equation was formulated, which can be observed in Fig. 4(D). The size of the crystallite was determined from the generated slope, and the strain from the evolved intercept. In the case of the generated CuO, the crystallite size measured 5.05 nm and the strain was (1.5 × 10−5).






 on the X-axis and cos(θ)βtotal on the Y-axis. Eqn (28) was compared with the straight-line equation to compute the stress value. Young's modulus value was 676.76 GPa.15 Additionally, the crystallite size became 8.45 nm taking into account the intercept of the equation and the estimated stress was 303 × 106 N m−2. The computed value of strain utilizing eqn (27) was found to be 4.47 × 10−4.
on the X-axis and cos(θ)βtotal on the Y-axis. Eqn (28) was compared with the straight-line equation to compute the stress value. Young's modulus value was 676.76 GPa.15 Additionally, the crystallite size became 8.45 nm taking into account the intercept of the equation and the estimated stress was 303 × 106 N m−2. The computed value of strain utilizing eqn (27) was found to be 4.47 × 10−4.
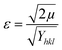

 on the X-axis and cos(θ)βtotal on the Y-axis, which was used to estimate the crystallite size and energy density. The resulting intercept of this calculation yielded a crystallite size of 8.45 nm. Moreover, the intercept also provided an energy density of 67.28 kJ m−3 from which the strain was estimated to be 0.00044.
on the X-axis and cos(θ)βtotal on the Y-axis, which was used to estimate the crystallite size and energy density. The resulting intercept of this calculation yielded a crystallite size of 8.45 nm. Moreover, the intercept also provided an energy density of 67.28 kJ m−3 from which the strain was estimated to be 0.00044.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos
cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ)2 on the Y-axis and (dhklβhkl
θ)2 on the Y-axis and (dhklβhkl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos
cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ) on the X-axis, as depicted in Fig. 5(D). The resulting equation was correlated with a straight-line equation. The slope of the line represents the crystallite size measurement, which was calculated to be 7.00 nm. Furthermore, the line's intercept allowed for the strain's estimation, resulting in a value of 0.02. The computed crystallite sizes using several model equations are enlisted in Table 4.
θ) on the X-axis, as depicted in Fig. 5(D). The resulting equation was correlated with a straight-line equation. The slope of the line represents the crystallite size measurement, which was calculated to be 7.00 nm. Furthermore, the line's intercept allowed for the strain's estimation, resulting in a value of 0.02. The computed crystallite sizes using several model equations are enlisted in Table 4.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching of the –COOH group in the extract. A similar result was observed in the earlier investigation.53 The bands at 661 and 551 cm−1 were for alkyl halide, whereas 599 cm−1 was assigned to the –OH stretching vibration. The previous exploration yielded similar results.54
O stretching of the –COOH group in the extract. A similar result was observed in the earlier investigation.53 The bands at 661 and 551 cm−1 were for alkyl halide, whereas 599 cm−1 was assigned to the –OH stretching vibration. The previous exploration yielded similar results.54
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond); related peaks were identified in the literature and the range was 841–937 cm−1.56 The wavenumber of 1061 cm−1 indicated the C–O stretching of phenols that was assured by the previous exploration, where the range was 1042–1076 cm−1.57 The absorption band at 1374 cm−1 (in the earlier exploration was 1381 cm−1) could be because of the alkene group's C–H stretching vibration or the esters’ O–C–O stretching.57 The peaks at 1488 cm−1 and 1587 cm−1 indicated the N–H groups of amines due to the biomolecules from leaf extract; this one was very close to previous outcomes (1518–1580 cm−1).58 The absorption band at 2358 cm−1 was assigned to the presence of environmental carbon dioxide which matched the earlier reports (2300–2385 cm−1) as well.59 The peak found at 3309 cm−1 was indicative of the stretching vibration of O–H that was found in the previous investigation (3264–3356 cm−1).60 The peak of 3577 cm−1 was very similar to the peak (3570 cm−1) of the previous study that was ascribed to the N–H stretching of amines involved in the stabilization of CuO NPs.61
C bond); related peaks were identified in the literature and the range was 841–937 cm−1.56 The wavenumber of 1061 cm−1 indicated the C–O stretching of phenols that was assured by the previous exploration, where the range was 1042–1076 cm−1.57 The absorption band at 1374 cm−1 (in the earlier exploration was 1381 cm−1) could be because of the alkene group's C–H stretching vibration or the esters’ O–C–O stretching.57 The peaks at 1488 cm−1 and 1587 cm−1 indicated the N–H groups of amines due to the biomolecules from leaf extract; this one was very close to previous outcomes (1518–1580 cm−1).58 The absorption band at 2358 cm−1 was assigned to the presence of environmental carbon dioxide which matched the earlier reports (2300–2385 cm−1) as well.59 The peak found at 3309 cm−1 was indicative of the stretching vibration of O–H that was found in the previous investigation (3264–3356 cm−1).60 The peak of 3577 cm−1 was very similar to the peak (3570 cm−1) of the previous study that was ascribed to the N–H stretching of amines involved in the stabilization of CuO NPs.61


 on the Y-axis and time (t, min) on the X-axis, the generated slope is the indicator of the apparent first-order rate constant (Kapp). The obtained rate constant from the graph (visualized in Fig. 12) for the green-synthesized CuO was 0.0198 min−1 (CR dye) and 0.0047 min−1 (ciprofloxacin).
on the Y-axis and time (t, min) on the X-axis, the generated slope is the indicator of the apparent first-order rate constant (Kapp). The obtained rate constant from the graph (visualized in Fig. 12) for the green-synthesized CuO was 0.0198 min−1 (CR dye) and 0.0047 min−1 (ciprofloxacin).