Open Access Article
Open Access ArticleVirus-based enzymatic nanoreactors with acid alpha-glucosidase for the potential treatment of Pompe disease†
Jordan
Molina-Solís
,
Pedro
Gama
,
Kanchan
Chauhan
 ,
Ana G.
Rodríguez-Hernández
and
Rafael
Vazquez-Duhalt
,
Ana G.
Rodríguez-Hernández
and
Rafael
Vazquez-Duhalt
 *
*
Centro de Nanociencias y Nanotecnología, Universidad Nacional Autónoma de México, Km 107 carretera Tijuana, Ensenada, Baja California 22860, Mexico. E-mail: rvd@ens.cnyn.unam.mx
First published on 24th January 2025
Abstract
Pompe disease is an inherited disorder originating from the enzyme acid alpha-glucosidase (GAA) deficiency that induces glycogen accumulation inside lysosomes. Enzyme replacement therapy (ERT) is the main treatment for Pompe disease. However, its effectivity is reduced by the immune response, enzyme proteolysis, and the enzyme difficulty in crossing the blood–brain barrier (BBB). This work aimed to design and synthesize enzymatic nanoreactors based on the enzyme confinement inside virus-like particles to improve the ERT for Pompe disease. The GAA confinement was performed by self-assembling the coat proteins of the brome mosaic virus in the presence of GAA. Then, nanoreactors were surface functionalized with a 6-phosphate mannose derivative using modified polyethylene glycol (PEG) to improve their capacity to cross the brain–blood barrier (BBB). The functionalized enzymatic nanoreactors showed a 1.6 times faster crossing rate than the unfunctionalized nanoreactors in an in vitro model of the BBB. In conclusion, enzymatic nanoreactors with GAA activity were successfully synthesized and functionalized with a 6-phosphate mannose derivative to improve the enzyme replacement therapy (ERT) and reduce the immunogenic response for Pompe disease treatment. The advantages of using virus-like particles as enzyme nanocarriers for treating Pompe disease are discussed.
1. Introduction
Pompe disease (PD) is an inherited neuromuscular disorder originating from a deficiency of the lysosomal enzyme acid-α-glucosidase (GAA), which hydrolyzes lysosomal glycogen to obtain free glucose-1-phosphate.1,2 Pompe disease is also known as glycogen storage disease type II (GSD II) or acid-maltase disease (AMD). A lack of GAA leads to glycogen accumulation in the lysosomes of cardiac, skeletal, and muscle cells and the central and peripheral nervous systems. Patients suffering from PD will present clinical manifestations such as muscular dystrophies, respiratory dysfunctions, cardiac dysfunctions, hepatomegaly, and cardiomegaly, among others.3,4 The most severe form is infantile-onset PD, presenting shortly after birth with symptoms of cardiomyopathy, respiratory failure, and skeletal muscle weakness.5 Pompe disease incidence is approximately 1 in 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 births.6
000 births.6
Enzyme replacement therapy (ERT) has been the standard of care for 15 years, slowing Pompe disease progression, particularly in cardiomyopathy, and improving survival rates. ERT involves the systemic delivery of exogenous therapeutic enzymes by the systemic route. In the case of Pompe disease, the treatment utilizes GAA, which was approved by the “Food and Drug Administration” (FDA) in 2008.7 Since then, it has been the primary treatment to combat PD, whose drawbacks are related to the activation of the immune system, the degradation of the enzyme by proteolysis, and the difficulties of the enzyme in crossing the brain blood–brain barrier.7–10 Thus, seeking and developing alternatives to overcome these obstacles is necessary.
Recently, the use of viral capsids or virus-like particles (VLPs) for ERT has been proposed.11 The encapsulation of the enzyme inside the VLPs shows several advantages when compared to the administration of exogenous free enzymes, such as protection against proteolysis, increasing the lifetime of the enzyme activity in the body, the possibility of ligand-functionalization to be recognized by specific cell receptors, and the reduction of immunogenic response. The encapsulation of enzymes into VLPs for different purposes has been recently reviewed.11–14 Specifically, for ERT, there are a few examples of using VLPs as enzyme nanocarriers in the literature. Glucocerebrosidase and α-galactosidase A, deficient enzymes in Gaucher15 and Fabry16 diseases, were encapsulated in VLPs from the cowpea chlorotic mottle virus (CCMV). The external nanoreactor surface was conjugated with PEG to improve the overall biodistribution and functionalized with a mannose group to enhance the targeting of diseased cells.17 On the other hand, virus-based enzymatic nanoreactors were proposed as an alternative for the ERT of classic galactosemia.18 Galactose-1-phosphate uridylyl-transferase (GALT) was encapsulated into plant virus capsids by a molecular self-assembly strategy. These virus-based GALT nanoreactors were effectively internalized into different human cell lines, such as hepatocytes, fibroblasts, and kidney cells, as demonstrated in vitro. Finally, the encapsulation of the enzyme β-glucocerebrosidase into VLPs from the brome mosaic virus (BMV) as a potential ERT for Gaucher disease was reported.15 The enzymatic nanoreactors were functionalized on their surface with PEG-mannose to be targeted to macrophages. The mannose-targeted enzymatic nanoreactors showed enhanced internalization into macrophage cells.15
In this work, the enzyme acid alpha-glucosidase (GAA) was encapsulated into VLP-based enzymatic nanoreactors for the ERT of Pompe disease. Nanoreactors with retained GAA enzyme activity were synthesized by optimizing the assembly of GAA with the BMV virus's coat proteins. Furthermore, nanoreactors were functionalized with a 6-phosphate mannose derivative to target lysosome delivery and improve their BBB-crossing capability (Fig. 1). The BBB delivery has been evaluated through an in vitro model demonstrating the potential of the developed nanoreactors in treating Pompe disease.
 | ||
| Fig. 1 Schematic diagram for functionalization of VLPs with the mannose-6-phosphate derivative and cyanine5 dye. | ||
2. Materials and methods
2.1. Reagents
Recombinant human lysosomal acid alpha-glucosidase (GAA) was purchased from R&D Systems (Minneapolis, MN). The human blood–brain barrier modeling kit (Cat. 8738) was obtained from ScienCell Research Laboratories (San Diego, CA). The kit includes human umbilical vein endothelial cells (HUVEC), human brain vascular pericytes (HBVP), and human astrocytes (HA). α-Man-6-phosphate-PEG3-azide (M6P-PEG3-N3) was obtained from Sussex Research Laboratories Inc. (Canada). (1R,8S,9s)-Bicyclo[6.1.0]non-4-yn-9-ylmethyl N-succinimidyl carbonate (BCN-NHS) was obtained from Sigma Aldrich (St. Louis, MO). The fluorescent dye, sulfo-cyanine5-NHS ester, was obtained from Lumiprobe Life Science Solutions (USA).2.2. BMV production and coat protein purification
The VLPs were produced and purified from the native virus, as previously reported.19 The native BMV virus was amplified through the infection of barley plants (Hordeum vulgare). Briefly, young barley leaves were inoculated with the BMV. Once the leaves showed the infection pattern, they were harvested and liquefied in a blender in a buffer containing 0.5 M sodium acetate and 80 mM magnesium acetate at pH 4.5. The resulting macerated material was filtered through cheesecloth, and then chloroform (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v) was added to the filtrate and mixed. The mixture was centrifuged at 10
1 v/v) was added to the filtrate and mixed. The mixture was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 4 °C, for 20 min. The upper aqueous phase containing the BMV virions was collected and stirred at 4 °C overnight. The aqueous mixture was then ultracentrifuged on a 5 mL sucrose cushion at 32
000 rpm, 4 °C, for 20 min. The upper aqueous phase containing the BMV virions was collected and stirred at 4 °C overnight. The aqueous mixture was then ultracentrifuged on a 5 mL sucrose cushion at 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 4 °C, for 2 h. After centrifugation, the supernatant was discarded, and a pellet containing the BMV virions was resuspended in a buffer containing 50 mM sodium acetate and 8 mM magnesium acetate at pH 4.5. Then, the BMV virion suspension was poured into a previously prepared centrifuge tube with a sucrose density gradient (10–40%) and centrifuged at 30
000 rpm, 4 °C, for 2 h. After centrifugation, the supernatant was discarded, and a pellet containing the BMV virions was resuspended in a buffer containing 50 mM sodium acetate and 8 mM magnesium acetate at pH 4.5. Then, the BMV virion suspension was poured into a previously prepared centrifuge tube with a sucrose density gradient (10–40%) and centrifuged at 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 4 °C, for 2 h. After centrifugation, the tube was examined under white light to confirm the presence of the blue phase containing the viruses. The blue phase was centrifuged at 32
000 rpm, 4 °C, for 2 h. After centrifugation, the tube was examined under white light to confirm the presence of the blue phase containing the viruses. The blue phase was centrifuged at 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 4 °C, for 3 h to concentrate the virions in the pellet.
000 rpm, 4 °C, for 3 h to concentrate the virions in the pellet.
Then, the BMV virions were disassembled by dialysis (12 kDa cutoff membrane) against the disassembly buffer (0.5 M CaCl2, 1 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, and 50 mM Tris–HCl, pH 7.4) for 24 h at 4 °C. Then, the resulting disassembled coat protein (CP) was centrifuged at 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 4 °C, for 8.5 h. The supernatant containing the CP was collected in 0.5 mL fractions, and the virus genetic material pellet was discarded. The CP purity of the fractions was estimated using the A280/A260 nm quotient. The best fractions (A280/A260 ≥ 1.5) were combined and stored.
000 rpm, 4 °C, for 8.5 h. The supernatant containing the CP was collected in 0.5 mL fractions, and the virus genetic material pellet was discarded. The CP purity of the fractions was estimated using the A280/A260 nm quotient. The best fractions (A280/A260 ≥ 1.5) were combined and stored.
2.3. Synthesis of enzymatic nanoreactors containing acid alpha-glucosidase (GAA)
The self-assembly process, based on the complementarity of electrostatic charges between the internal surface of the BMV capsid protein and the surface of the GAA molecule, is the mechanism for encapsulating enzyme molecules.15 Therefore, the electrostatic properties of the GAA enzyme and the BMV capsid protein were studied using Shrödinger® Maestro software (version 2019-1). The coordinates of the crystallographic structure of human GAA (PDB 5KZW) and the BMV capsid protein (PDB 1JS9) were obtained from the central repository of protein data (Protein Data Bank). The Poisson–Boltzmann equation was used to model the surface electrical charge distribution of the GAA enzyme and the BMV capsid protein to determine the GAA–CP electrostatic interactions for VLP formation.Enzymatic nanoreactors were formed by self-assembly procedure as follows: enzyme (GAA) and coat protein (CP) solutions at different molar proportions were dialyzed in assembly buffer solution (0.05 M NaCl; 0.05 M Tris; 0.01 M KCl, and 0.005 M MgCl2, at pH 7.2 adjusted with hydrochloric acid) for 12 h at −4 °C. Then, the mixture was dialyzed against an acidification buffer (0.05 M sodium acetate, 0.008 M magnesium acetate at pH 4.7, adjusted with acetic acid). The formation of nanoreactors (VLP–GAA) was monitored by DLS, identifying the appearance of hydrodynamic diameters similar to that of the native BMV. Once the formation of nanoreactors was confirmed, the samples were purified by centrifugation at 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 2 hours at 4 °C, and washed twice. After centrifugation, the supernatant was discarded, and the precipitate was resuspended in an acidification buffer.
000 rpm for 2 hours at 4 °C, and washed twice. After centrifugation, the supernatant was discarded, and the precipitate was resuspended in an acidification buffer.
2.4. Transmission electron microscopy analysis of GAA nanoreactors
The characterization of the structure of the VLP–GAA was performed by transmission electron microscopy (TEM) (JEOL JEM-2010) operated at 200 kV, and the images were analyzed using Digital Micrograph 3 (Gatan, Inc. Pleasanton, CA). Ten μL of the nanoreactor suspension were placed on a copper grid for TEM, where it was allowed to incubate for one minute at room temperature, and then the drop was removed with blotting paper. Next, one more drop of 4 μL of 1% uranyl acetate was deposited on the grid and incubated under the same conditions, removing the uranyl acetate with blotting paper. Finally, the obtained micrographs were analyzed using ImageJ software (version 1.8.0) to determine the size and homogeneity of the produced nanoparticles.2.5. Catalytic characterization of GAA nanoreactors
Enzymatic activity assays of the synthesized VLP–GAA and the free GAA enzyme were performed following the protocol described by Siro and Lovgren,20 which uses the synthetic substrate p-nitrophenol α-D-glucoside (pNP-Glc) to estimate the catalytic activity. The data fitted the Michaelis–Menten model. The enzymatic reaction was monitored at 400 nm using a Nanodrop 2000c UV-vis spectrophotometer (Thermo Fisher Scientific). The rate constant (kcat) and the affinity constant (KM) were obtained from a non-linear Michaelis–Menten regression using CurveExpert Professional software (version 2.7.3.).The total protein was determined with the BioRad protein reagent and using a standard curve with bovine serum albumin (BSA). The GAA content in the nanoreactors was estimated utilizing a denaturing polyacrylamide gel electrophoresis assay (SDS-PAGE) and a densitometric analysis using ImageJ software.
2.6. Functionalization of VLP-GAA nanoreactors with PEG-6-phosphate-mannose
To prepare the targeted virus-like particles (VLP-M6P-Cy5), BCN-NHS and M6P-PEG3-N3 were mixed in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio and a 250-fold molar excess of VLPs for 60 min. The suspension of VLPs (1 mg mL−1, 0.5 mL) was mixed with the reaction mixture and stirred overnight at 4 °C. Excess reagents were removed the following day using Amicon filtration with 30 kDa cut-off filters and centrifuged at 4000 rpm for 15 minutes. The concentrated VLPs were then diluted in PBS (pH 7), and a 100-fold molar excess of the fluorescent dye CY5-NHS was added. This suspension was gently stirred for 2 hours. The VLPs were purified by three rounds of Amicon filtration and washing with PBS. The purified, targeted VLPs (VLP-M6P-Cy5) were stored at 4 °C for further analysis. For the non-targeted VLPs (VLP-Cy5), the VLPs were treated similarly, but only with a 100-fold molar excess of Cy5-NHS ester without the targeting ligand.
1 ratio and a 250-fold molar excess of VLPs for 60 min. The suspension of VLPs (1 mg mL−1, 0.5 mL) was mixed with the reaction mixture and stirred overnight at 4 °C. Excess reagents were removed the following day using Amicon filtration with 30 kDa cut-off filters and centrifuged at 4000 rpm for 15 minutes. The concentrated VLPs were then diluted in PBS (pH 7), and a 100-fold molar excess of the fluorescent dye CY5-NHS was added. This suspension was gently stirred for 2 hours. The VLPs were purified by three rounds of Amicon filtration and washing with PBS. The purified, targeted VLPs (VLP-M6P-Cy5) were stored at 4 °C for further analysis. For the non-targeted VLPs (VLP-Cy5), the VLPs were treated similarly, but only with a 100-fold molar excess of Cy5-NHS ester without the targeting ligand.
Zeta-potential and hydrodynamic diameter by dynamic light scattering (DLS) were measured with a Zetasizer Nanoseries (Nano-ZS, Malvern Instruments). The fluorescence was analyzed at room temperature using a Hitachi F-4700 spectrofluorometer with a 200 W Xe-lamp as an excitation source. The suspension of VLPs (200 μL) in PBS buffer (pH 7) at a concentration of 0.5 mg mL−1 was added to a fluorescence 96-well plate, and readings were obtained at λex = 600 nm and λem = 620–750 nm at a slit size of 10 nm for both excitation and emission.
2.7. Blood–brain barrier crossing in an in vitro model
The transfer of fluorescence-labeled nanoreactors through the blood–brain barrier (BBB) was estimated using the human blood–brain barrier modeling kit from ScienCell Research Laboratories and following the manufacturer's instructions. Cell lines were grown and maintained separately under 2D culture conditions until they reached 80% confluence (37 °C and 5% CO2 atmosphere), using the 2D-BBB co-culture medium basal supplemented with 5% fetal bovine serum, 1% epidermal growth factor, and 1% penicillin/streptomycin (v/v), all from ScienceCell™. HUVECs were cultured in 10-cm Petri dishes treated for cell culture and previously functionalized with fibronectin. The HBVP and HA cells were cultured in Petri dishes pre-functionalized with poly-L-lysine.Once the 3 cell lines reached 80% of confluence, the cells were washed with 1× PBS, lifted from the plate using 0.05% trypsin/EDTA (5 min at 37 °C and 5% CO2 atmosphere), and then neutralized with a supplemented basal medium.
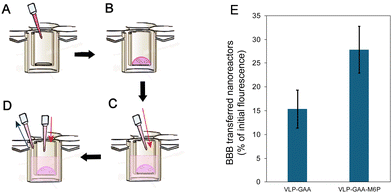
3D cultures with the three types of cell lines were performed in a 3D collagen matrix onto a transwell. The 3D scaffold was prepared by adding 75 μL of type I collagen solution (4 mg mL−1) to each well and mixed with a solution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v of 100 mM HEPES buffer and 2× PBS. Then, the indicated number of cells per well was added, gently mixed with a micropipette, and placed into the transwell insert (Fig. 5). The initial cell proportion was 7.5 × 105 HUVEC, 1.5 × 104 HBVP, and 3.6 × 104 HA per well. The cells/collagen mixture was then allowed to settle for 5 minutes and incubated at 37 °C and 5% CO2 for one hour. Finally, 750 μL of the 3D basal serum-free medium supplemented with a 1% 3D growth factor and 1% penicillin/streptomycin was added to each well. The medium was changed every 24 hours, and colonization of the entire scaffold took approximately 5 to 6 days to conduct the assay with labeled nanoreactors.
1 v/v of 100 mM HEPES buffer and 2× PBS. Then, the indicated number of cells per well was added, gently mixed with a micropipette, and placed into the transwell insert (Fig. 5). The initial cell proportion was 7.5 × 105 HUVEC, 1.5 × 104 HBVP, and 3.6 × 104 HA per well. The cells/collagen mixture was then allowed to settle for 5 minutes and incubated at 37 °C and 5% CO2 for one hour. Finally, 750 μL of the 3D basal serum-free medium supplemented with a 1% 3D growth factor and 1% penicillin/streptomycin was added to each well. The medium was changed every 24 hours, and colonization of the entire scaffold took approximately 5 to 6 days to conduct the assay with labeled nanoreactors.
The experiments to evaluate the nanoreactor transfer through the blood–brain barrier were carried out by adding 100 μL of the nanoreactor suspension (75 μg mL−1) into the transwell and incubating at 37 °C (in a 5% CO2 atmosphere). After 3 and 6 hours of incubation, 100 μL were removed from the transwell's outer well, and the fluorescence was measured using a spectrofluorometer. It is essential to add the same volume of the supplemented 3D basal medium into the transwell immediately after extraction.
2.8. Potential immune response by RawBlue cell activation
The immunogenic potential of nanoreactors was estimated using RAW-Blue cells (Invivogen, San Diego, CA). RAW-Blue cells were derived from the murine macrophage cell line RAW 264.7 and expressed a secreted embryonic alkaline phosphatase (SEAP) reporter gene under the control of two transcription factors, which play a central role in inflammation and immunity, NF-κB and AP-1 promoters. RAW-Blue cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) and 100 μg mL−1 Normocin. The cells (1 × 105) were incubated with 25 μg mL−1 of unmodified and M6P-modified nanoreactors for 12 h at 37 °C and 5% CO2. Supernatants were then collected, and SEAP production was evaluated based on the activity of alkaline phosphatase (AP) after adding 150 μL of quantity blue, and the absorbance was determined at 655 nm. As a positive control, 75 ng of purified lipopolysaccharides from Escherichia coli O111:B4 (Sigma-Aldrich) were used. The experiments were carried out in triplicate.3. Results and discussion
The lack or reduced acid alpha-glucosidase (GAA) activity is the origin of the Pompe disease. Virus-like nanoparticles have been explored as nanocarriers of enzymatic activity for biomedical purposes21–26 and specifically for ERT.11,15,18 To encapsulate the GAA inside BMV capsids by charge complementarity, first, an in silico analysis must be performed to determine the surface charge of the GAA enzyme. The surface charge analysis of the GAA enzyme showed a surface negative charge distribution (Fig. 2). On the opposite, the BMV capsid shows an internal side with a highly positive distribution of electrostatic charges.27 The DLS measurement of zeta potential of GAA at pH 7 showed a value of −8.1 ± 4.7 mV. Thus, enzyme encapsulation into BMV capsids by charge complementarity should be possible.The hydrodynamic diameter of the GAA molecule, determined by dynamic light scattering (DLS), is shown to be 6.238 nm (Fig. S1, ESI†). The BMV capsid comprises 60 asymmetric units made of 3 proteins (T = 3), for a total of 180 capsid proteins. An internal diameter of the BMV capsid of 22.6 nm is considered.27 Thus, the theoretical maximum capacity of GAA molecules inside the capsid is 24 GAA enzyme molecules per BMV capsid. Considering this, three different molar ratios (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 1
2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, and 1
10, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 VLP
20 VLP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GAA) were tested to synthesize enzymatic nanoreactors by self-assembly. The successful formation of enzymatic nanoreactors by the self-assembly method in all three molar ratios is shown in transmission electron microscopy (TEM) (Fig. 3). The nanoreactor preparations showed an icosahedral, quasi-spherical shape resembling the native BMV. The purified nanoreactors (VLP–GAA) showed an average hydrodynamic diameter of around 26 nm (Table 1), similar to the native virion.27,28 All nanoreactors are icosahedral particles of ∼28 nm in diameter. The DLS analysis detected no free GAA nor capsid protein, suggesting a complete encapsulation.
GAA) were tested to synthesize enzymatic nanoreactors by self-assembly. The successful formation of enzymatic nanoreactors by the self-assembly method in all three molar ratios is shown in transmission electron microscopy (TEM) (Fig. 3). The nanoreactor preparations showed an icosahedral, quasi-spherical shape resembling the native BMV. The purified nanoreactors (VLP–GAA) showed an average hydrodynamic diameter of around 26 nm (Table 1), similar to the native virion.27,28 All nanoreactors are icosahedral particles of ∼28 nm in diameter. The DLS analysis detected no free GAA nor capsid protein, suggesting a complete encapsulation.
 | ||
Fig. 3 Transmission electron microscopy (TEM) images of enzymatic nanoreactors (VLP–GAA). The different molar ratios of VLP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GAA are shown in the upper left corner of each image. GAA are shown in the upper left corner of each image. | ||
Nanoreactors VLP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GAA ratio GAA ratio |
Hydrodynamic diametera (nm) | GAA content in protein basisb (%) | CP/GAA molar ratio | GAA molecules per VLP capsid |
|---|---|---|---|---|
| a Determined by dynamic light scattering (DLS). b Determined by SDS-PAGE and image densitometry. | ||||
| BMV virion | 26 ± 7 | — | — | — |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
24 ± 8 | 8 | 62.4 | 2.9 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 10 |
26 ± 11 | 11 | 43.9 | 4.1 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 20 |
28 ± 10 | 21 | 20.4 | 8.8 |
The GAA content in the nanoreactors was estimated by SDS-PAGE and gel densitometry (Fig. S2, ESI†). The quantitative results are shown in Table 1. As expected, the greater the proportion of GAA, the greater the encapsulation.
VLPs from the brome mosaic virus (BMV) were chosen because they show no cytotoxicity on different cell lines, including 293T human embryonic kidney cells,29 MOLT-4 lymphoblastic human leukemia cells, Rhe human B-cell lines,30 MDA-MB-231 breast tumoral cells,31 and HBE4 human bronchial epithelial cells.32
The enzymatic activity of the free enzyme and the different VLP–GAA nanoreactors was characterized (Table 2). The enzymatic assays were performed with different molar concentrations of the substrate p-nitrophenol α-D-glucoside (pNP-Glc) and monitored at 400 nm. Data were fitted with the Michaelis–Menten model, and the rate constant (kcat) was determined considering the GAA content in each nanoreactor (Table 1). As was expected, the catalytic activity constant (kcat) was lower for the nanoreactors when compared with the free enzyme. However, the enzyme encapsulation did not significantly affect the affinity constant (KM). On the other hand, among the nanoreactors, the preparation from the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 VLP/GAA ratio showed a higher activity rate constant (kcat). Thus, this preparation was used for further experiments. The nanoreactors showed one-third of the specific activity than the free enzyme. This activity is higher when compared with other enzymatic nanoreactors, where the enzymatic activity could be orders of magnitude lower than their respective free enzymes.33 Thus, the VLP–GAA nanoreactors seem to have low mass transfer limitations, as supported by the non-significative change of the KM (Table 2).
10 VLP/GAA ratio showed a higher activity rate constant (kcat). Thus, this preparation was used for further experiments. The nanoreactors showed one-third of the specific activity than the free enzyme. This activity is higher when compared with other enzymatic nanoreactors, where the enzymatic activity could be orders of magnitude lower than their respective free enzymes.33 Thus, the VLP–GAA nanoreactors seem to have low mass transfer limitations, as supported by the non-significative change of the KM (Table 2).
Preparation VLP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GAA ratio GAA ratio |
k cat (s−1) | K M (mM) | k cat/KM (s−1 mM−1) |
|---|---|---|---|
| a The rate constant kcat was determined considering the GAA content in the nanoreactors. | |||
| Free GAA | 356 | 0.16 | 2223 |
Nanoreactors 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
113 | 0.11 | 1032 |
Nanoreactors 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 10 |
116 | 0.12 | 964 |
Nanoreactors 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 20 |
25 | 0.10 | 247 |
The virus-based enzymatic nanoreactors have demonstrated better stability against proteolytic degradation, increasing their lifetime inside the body and reducing the needed doses.34,35 On the other hand, the presence of reactive groups on the nanoreactor surface facilitates conjugation with specific ligands targeted to specific cells or tissues.15,22,23
Enzymatic replacement therapy (ERT) for Pompe disease has two main challenges: a risk of immunological responses and a poor uptake in target tissues, and the inability to reach the central nervous system because of the blood–brain barrier (BBB).3 To contend with both challenges, VLP–GAA nanoreactors were functionalized with polyethylene glycol (PEG) containing mannose-6-phosphate (M6P) at the molecule end to reduce the immunological response and to facilitate the cell uptake and BBB crossing. Mannose-6-phosphate glycan targets the M6P receptors on early endosomes, a strategy often used for the targeted delivery of therapeutic enzymes for lysosomal diseases. The GAA targeting the cation-independent mannose 6-phosphate receptor (CI-M6PR) expressed on the cell surface enhances the efficiency of ERT. CI-M6PR, also known as insulin-like growth factor receptor (IGF2R), is a polyfunctional transmembrane protein with a high affinity for M6P.36,37 A modified glycoengineered enzyme with a synthetic oligosaccharide harboring mannose 6-phosphate (M6P) residues with a much-improved affinity for the CI-MPR and uptake by muscle cells showed more efficient glycogen clearance in vivo compared to the unmodified enzyme.38
On the other side, as with many drugs, GAA does not efficiently cross the BBB. Some receptor-mediated endocytosis and transcytosis strategies have been explored.39 Tran GAA fused with an anti-human transferrin receptor antibody enables transcytosis across the BBB and delivery into the central nervous system.40
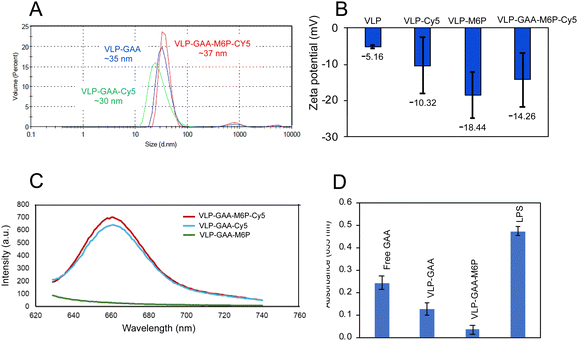
A schematic representation of the nanoreactor functionalization procedure is summarized in Fig. 1. Firstly, the azide-modified mannose-6-phosphate derivative was conjugated with bicyclo[6.1.0]non-4-yn-9-ylmethyl N-succinimidyl carbonate (BCN-NHS) ester via a copper-free click reaction at room temperature and then functionalized on the VLP surface by simple carbodiimide chemistry. Later, the targeted and non-targeted VLPs were labeled with the red-emitting dye Cyanine5 (Cy5) to track the BBB penetration by fluorescence analysis. The modified VLPs were analyzed through DLS and showed similar hydrodynamic diameters in all stages of modification. However, the surface charge varied after each modification compared to the unmodified VLPs (Fig. 4A and B). The fluorescence analysis showed the presence of Cy5 in the labeled VLPs, where an emission at 660 nm was observed after excitation at 600 nm, while VLP–M6P did not show any signal (Fig. 4C).
The potential immune response was evaluated on RAW Blue cells. This cell line is a commonly used model of mouse macrophages for the study of potential immunogenic response. RAW-Blue cells express a secreted inducible alkaline phosphatase (SEAP) reporter gene to monitor the activation of the NF-κB transcription factors. The immunological response to RawBlue macrophage cells with 100 μg mL−1 of free enzymes or nanoreactors compared with those with 75 ng mL−1 of the bacterial lipopolysaccharide (LPS). The GAA encapsulation into a BMV capsid reduces the immunological response when compared with the free enzyme (Fig. 4D). It is known that the BMV capsids show a low immunological response.31 Furthermore, the nanoreactor functionalization with polyethylene glycol conjugated with mannose-6-phosphate (M6P–PEG) showed even lower immunological response (Fig. 4D).
Drug delivery crossing the blood–brain barrier (BBB) is still a big challenge. Nanoparticle functionalization with insulin,41,42 dopamine,43 and especially with transferrin has demonstrated an enhanced BBB crossing. Transferrin functionalization proceeds by transcytosis using the transferrin receptors (D and E).44,45 The mannose-6-phosphate glycan's crucial role in lysosomal targeting has been elucidated in studies on lysosome dysfunction, such as Pompe disease.46 Mannose-6-phosphate47,48 has also enhanced the BBB crossing. VLPs, in addition to protecting the cargo, can be functionalized with any of these ligands. Functionalized VLP-GAA with PEG molecules conjugated with mannose-6-phosphate was assayed for BBB crossing in an in vitro model (Fig. 1).
The labeled nanoreactors were assayed in the 3D cellular model that consists of a three-layer cell 3D culture, including co-cultures of endothelial cells, brain vascular pericytes, and human astrocytes in a collagen I matrix. The cultures are performed in transwell plates, as shown in Fig. 5. Transwell plates consist of two chambers separated by a semipermeable membrane, where the BBB cell culture model is cultured. This in vitro system allows researchers to evaluate the BBB crossing of different metabolites in an organized manner.49–51 The three cell lines are cultured in an ordered manner inside the internal well (Fig. 5B). Then, the nanoreactor suspension is placed over the cell layer. After incubation, the medium from the outside well is taken for fluorescence analysis (Fig. 5D).
The proportion of nanoreactors that crossed the in vitro model of the BBB was higher when the nanoreactors were functionalized with the mannose-6-phosphate moiety (Fig. 5E).
Among the diseases caused by the deficiency or absence of enzymatic activity, Pompe disease, although considered rare,52 is still significant and severe. The “classic infantile” form of the disease is more frequently described, inducing severe cardiomegaly, hepatomegaly, hypotonia from birth, and early death. On the other hand, the “non-classic infantile” form manifests from the first year but with slower progression and less severe cardiomyopathy. The “late” form manifests in childhood, youth, or adulthood, between the second and sixth decades of life, with slowly progressive myopathy, generally without cardiomyopathy.53
The ERT for Pompe disease requires lifelong administration of recombinant human GAA (rhGAA). The doses needed have been recently revised,53 and recommended 40 mg kg−1 every two weeks, instead of the previously recommended 20 mg kg−1, which is relatively high compared to other lysosomal storage diseases. These doses achieve an effective glycogen clearance in the heart and skeletal muscles, thus reducing cardiomyopathy.54 However, it is less effective for respiratory or neurological affections.55
4. Conclusions
Virus-based enzymatic nanoreactors are an exciting alternative for enzyme replacement therapy (ERT), as in the case of Pompe disease. Acid alpha-glucosidase (GAA) was efficiently encapsulated inside BMV capsids. The enzymatic nanoreactors showed high catalytic activity and reduced immunogenicity. The GAA nanoreactors were functionalized with polyethylene glycol carrying a mannose-6-phosphate moiety at the molecule end to target specific cell receptors. The functionalization improved the BBB crossing in an in vitro 3D cell culture. Thus, lower doses and better treatment efficiency are expected because of the enzyme protection that increases its lifetime and the specific targeting. Developing clinically relevant animal models for Pompe disease will allow the evaluation of VLP–GAA–M6P enzymatic nanoreactors in ERT.Author contributions
J. M. S., P. G., and A. G. R. H.: investigation and data curation. K. C.: investigation, data curation, and funding acquisition. R. V. D.: conceptualization, methodology, data curation, writing – original draft preparation, supervision, and funding acquisition.Data availability
The data that support the findings are also available upon request from the corresponding author.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the National Council of Humanities, Sciences, and Technologies (grant CBF 2023-2024-253).References
- K. Kok, C. Kuo, R. E. Katzy, L. T. Lelieveld, L. Wu, V. Roig-Zamboni, G. A. van der Marel, J. D. C. Codée, G. Sulzenbacher, G. J. Davies, H. S. Overkleeft, J. M. F. G. Aerts and M. Artola, J. Am. Chem. Soc., 2022, 144, 14819–14827 CrossRef CAS PubMed.
- D. Stevens, S. Milani-Nejad and T. Mozaffar, Curr. Treat. Options Neurol., 2022, 24, 573–588 CrossRef PubMed.
- Z. Unnisa, J. K. Yoon, J. W. Schindler, C. Mason and N. P. van Til, Biomedicines, 2022, 10, 302 CrossRef CAS PubMed.
- C. I. van Capelle, L. P. F. Winkel, M. L. C. Hagemans, S. K. Shapira, W. F. M. Arts, P. A. van Doorn, W. C. J. Hop, A. J. J. Reuser and A. T. van der Ploeg, Neuromuscular Disord., 2008, 18, 447–452 CrossRef CAS PubMed.
- A. L. Roger, R. Sethi, M. L. Huston, E. Scarrow, J. Bao-Dai, E. Lai, D. D. Biswas, L. E. Haddad, L. M. Strickland, P. S. Kishnani and M. K. ElMallah, Expert Opin. Biol. Ther., 2022, 22, 1117–1135 CrossRef PubMed.
- K. S. Park, Mol. Genet. Metab. Rep., 2021, 27, 100734 CAS.
- J.-A. Lim, H. Yi, F. Gao, N. Raben, P. S. Kishnani and B. Sun, Mol. Ther. – Methods Clin. Dev., 2019, 12, 233–245 CrossRef CAS PubMed.
- F. Puzzo, P. Colella, M. G. Biferi, D. Bali, K. Nicole, P. Vidal, F. Collaud, M. Simon-Sola, S. Charles, C. Leborgne, A. Meliani, M. Cohen-Tannoudji, B. Gjata, P. Sellier, P. Wittenberghe, L. van, A. Vignaud, F. Boisgerault, M. Barkats, P. Laforet and F. Mingozzi, Sci. Transl. Med., 2017, 9, 1–27 CAS.
- G. Ronzitti, F. Collaud, P. Laforet and F. Mingozzi, Ann. Transl. Med., 2019, 7, 287 CrossRef CAS PubMed.
- U. Cagin, F. Puzzo, M. J. Gómez, M. Moya-Nilges, P. Sellier, C. Abad, L. van Wittenberghe, N. Daniele, N. Guerchet, B. Gjata, F. Collaud, S. Charles, M. S. Sola, O. Boyer, J. Krijnse-Locker, G. Ronzitti, P. Colella and F. Mingozzi, Mol. Ther., 2020, 28, 17 CrossRef.
- O. González-Davis, M. V. Villagrana-Escareño, M. A. Trujillo, P. Gama, K. Chauhan and R. Vazquez-Duhalt, Virology, 2023, 580, 73–87 CrossRef PubMed.
- R. Koyani, J. Pérez-Robles, R. D. Cadena-Nava and R. Vazquez-Duhalt, Nanotechnol. Rev., 2017, 6, 405–419 CAS.
- J. W. Wilkerson, S.-O. Yang, P. J. Funk, S. K. Stanley and B. C. Bundy, New Biotechnol., 2018, 44, 59–63 CrossRef CAS.
- M. Uchida, E. Manzo, D. Echeveria, S. Jiménez and L. Lovell, Curr. Opin. Virol., 2022, 52, 250–257 CrossRef CAS PubMed.
- K. Chauhan, C. N. Olivares-Medina, M. V. Villagrana-Escareño, K. O. Juárez-Moreno, R. D. Cadena-Nava, A. G. Rodríguez-Hernández and R. Vazquez-Duhalt, ChemMedChem, 2022, 17, e202200384 CrossRef CAS PubMed.
- D. F. M. Vervoort, Protein-engineered Cowpea Chlorotic Mottle Virus-like particles as a drug delivery platform. Biomedical Engineering, PhD thesis, Eindhoven University of Technology, 2021 Search PubMed.
- E. J. M. Oudmaijer, Stabilized Cowpea chlorotic mottle virus as a platform for targeted enzyme replacement therapy, PhD thesis, Eindhoven University of Technology, 2021 Search PubMed.
- P. Gama, R. D. Cadena-Nava, K. Juarez-Moreno, J. Pérez-Robles and R. Vazquez-Duhalt, ChemMedChem, 2021, 16, 1438–1445 CrossRef CAS PubMed.
- R. D. Cadena-Nava, M. Comas-Garcia, R. F. Garmann, A. L. N. Rao, C. M. Knobler and W. M. Gelbart, J. Virol., 2012, 86, 3318–3326 CrossRef CAS PubMed.
- M. R. Siro and T. Lovgren, Acta Chem. Scand., 1978, 32, 447–451 CrossRef CAS PubMed.
- M. Cayetano-Cruz, C. F. Coffeen, J. Valadez-García, C. Montiel and I. Bustos-Jaimes, Virus Res., 2018, 255, 1–9 CrossRef CAS PubMed.
- K. Chauhan, J. M. Hernández-Meza, A. G. Rodriguez-Hernández, K. Juarez-Moreno, P. Sengar and R. Vazquez-Duhalt, J. Nanobiotechol., 2018, 16, 17 CrossRef PubMed.
- A. Tapia-Moreno, K. Juarez-Moreno, O. Gonzalez-Davis, R. D. Cadena-Nava and R. Vazquez-Duhalt, Biotechnol. J., 2017, 12, 1600706 CrossRef PubMed.
- D. Cardinale, N. Carette and T. Michon, Trends Biotechnol., 2012, 30, 369–376 CrossRef CAS PubMed.
- O. González-Davis, K. Chauhan and R. Vazquez-Duhalt, Biocatalytic nanoreactors for medical purposes, in Pharmaceutical Biocatalysis, ed. Grundwald, P., Pan Stanford Publishing Pte. Ltd., Singapore, 2019, ch. 18, pp. 637–671 Search PubMed.
- J. L. Mejía-Méndez, R. Vazquez-Duhalt, L. R. Hernández, E. Sánchez-Arreola and H. Bach, Int. J. Mol. Sci., 2022, 23, 8579 CrossRef PubMed.
- R. W. Lucas, S. B. Larson and A. McPherson, J. Mol. Biol., 2002, 317, 95–108 CrossRef CAS PubMed.
- A. Strugała, J. Jagielski, K. Kamel, G. Nowaczyk, M. Radom, M. Figlerowicz and A. Urbanowicz, Int. J. Mol. Sci., 2021, 22, 1–15 Search PubMed.
- P. Gama, P. Juarez, A. G. Rodríguez-Hernández and R. Vazquez-Duhalt, Biotechnol. J., 2023, 18, 2300199 CrossRef CAS PubMed.
- F. Villanueva-Flores, A. Pastor, L. A. Palomares and A. Huerta-Saquero, Pharmaceutics, 2023, 15, 2260 CrossRef CAS PubMed.
- A. Nuñez-Rivera, P. G. J. Fournier, D. L. Arellano, A. G. Rodriguez-Hernandez, R. Vazquez-Duhalt and R. D. Cadena-Nava, Beilstein J. Nanotechnol., 2020, 11, 372–382 CrossRef PubMed.
- B. Jung, A. L. N. Rao and B. Anvari, ACS Nano, 2011, 5, 1243–1252 CrossRef CAS PubMed.
- K. Chauhan, A. Zárate-Romero, P. Sengar, C. Medrano and R. Vazquez-Duhalt, ChemCatChem, 2021, 13, 3732–3748 CrossRef CAS.
- J. D. Fiedler, S. D. Brown, J. L. Lau and M. G. Finn, Angew. Chem., Int. Ed., 2010, 49, 9648–9651 CrossRef CAS.
- L. Sánchez-Sánchez, A. Tapia-Moreno, K. Juarez-Moreno, D. P. Patterson, R. D. Cadena-Nava, T. Douglas and R. Vazquez-Duhalt, J. Nanobiotechnol., 2015, 13, 66 CrossRef PubMed.
- S. Kornfeld, Annu. Rev. Biochem., 1992, 61, 307–330 CrossRef CAS PubMed.
- R. J. Desnick and E. H. Schuchman, Nat. Rev. Genet., 2002, 3, 954–966 CrossRef CAS PubMed.
- Y. Zhu, J.-L. Jiang, N. K. Gumlaw, J. Zhang, S. D. Bercury, R. J. Ziegler, K. Lee, M. Kudo, W. M. Canfield, T. Edmunds, C. Jiang, R. J. Mattaliano and S. H. Cheng, Mol. Ther., 2009, 17, 954–963 CrossRef CAS PubMed.
- Z. Zhou, G. L. Austin, R. Shaffer, D. D. Armstrong and M. S. Gentry, Trends Mol. Med., 2019, 25, 1094–1109 CrossRef CAS PubMed.
- S. Tanaka, A. Yoshioka, M. Kinoshita, S. Kida, A. Imakiire, S. Takenaka, H. Morimoto, R. Yamamoto, K. Minami, H. Sonoda, K. Takahashi and T. Hirato, Mol. Genet. Metab., 2020, 129, S150–S151 CrossRef.
- O. Betzer, M. Shilo, R. Opochinsky, E. Barnoy, M. Motiei, E. Okun, G. Yadid and R. Popovtzer, Nanomedicine, 2017, 12, 1533–1546 CrossRef CAS.
- M. Shilo, M. Motiei, P. Hana and R. Popovtzer, Nanoscale, 2014, 6, 2146–2152 RSC.
- D. A. Gonzalez-Carter, Z. Y. Ong, C. M. McGilvery, I. E. Dunlop, D. T. Dexter and A. E. Porter, Nanomedicine, 2019, 15, 1–11 CrossRef CAS PubMed.
- M. S. Thomsen, K. B. Johnsen, K. Kucharz, M. Lauritzen and T. Moos, Pharmaceutics, 2022, 14, 2237 CrossRef CAS PubMed.
- K. B. Johnsen, A. Burkhart, L. B. Thomsen, T. L. Andresen and T. Moos, Prog. Neurobiol., 2019, 181, 101665 CrossRef CAS PubMed.
- J. Seo and D. B. Oh, Anim. Cells Syst., 2022, 26, 84–91 CrossRef CAS.
- A. Urayama, J. H. Grubb, W. S. Sly and W. A. Banks, Mol. Ther., 2008, 16, 1261–1266 CrossRef CAS PubMed.
- H. J. Byeon, L. Q. Thao, S. Lee, S. Y. Min, E. S. Lee, B. S. Shin, H.-G. Choi and Y. S. Youn, J. Controlled Release, 2016, 225, 301–313 CrossRef CAS PubMed.
- K. Hatherell, P.-O. Couraud, I. A. Romero, B. Weksler and G. J. Pilkington, J. Neurosci. Methods, 2011, 199, 223–229 CrossRef PubMed.
- E. Urich, C. Patsch, S. Aigner, M. Graf, R. Iacone and P.-O. Freskgård, Sci. Rep., 2013, 3, 1500 CrossRef.
- M. Pervin, K. Unno, A. Nakagawa, Y. Takahashi, K. Iguchi, H. Yamamoto, M. Hoshino, A. Hara, A. Takagaki, F. Nanjo, A. Minami, S. Imai and Y. Nakamura, Biochem. Biophys. Rep., 2017, 9, 180–186 Search PubMed.
- L. Kohler, R. Puertollano and N. Raben, Neurotherapeutics, 2018, 15, 928–942 CrossRef CAS.
- D. D. Koeberl, L. E. Case, A. Desai, E. C. Smith, C. Walters, S. Han, B. L. Thurberg, S. P. Young, D. Bali and P. S. Kishnani, Mol. Gen. Metab., 2020, 129, 67–72 CrossRef CAS PubMed.
- I. A. M. Ditters, H. H. Huidekoper, M. E. Kruijshaar, D. Rizopoulos, A. Hahn, T. E. Mongini, F. Labarthe, M. Tardieu, B. Chabrol, A. Brassier, R. Parini, G. Parenti, N. A. M. E. van der Beek, A. T. van der Ploeg and J. M. P. van den Hout, Lancet Child Adolesc. Health, 2022, 6, 28–37 CrossRef CAS PubMed.
- B. J. Ebbink, E. Poelman, F. K. Aarsen, I. Plug, L. Régal, C. Muentjes, N. A. M. E. V. D. Beek, M. H. Lequin, A. T. van Der Ploeg and J. M. P. V. D. Hout, Dev. Med. Child Neurol., 2018, 60, 579–586 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ma00943f |
| This journal is © The Royal Society of Chemistry 2025 |