Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Parallel dilution microfluidic device for enabling logarithmic concentration generation in molecular diagnostics†
Akira
Miyajima
*a,
Fumiya
Nishimura
a,
Daigo
Natsuhara
bc,
Yuka
Kiba
d,
Shunya
Okamoto
 a,
Moeto
Nagai
a,
Moeto
Nagai
 ae,
Tadashi
Yamamuro
f,
Masashi
Kitamura
d and
Takayuki
Shibata
ae,
Tadashi
Yamamuro
f,
Masashi
Kitamura
d and
Takayuki
Shibata
 *ae
*ae
aDepartment of Mechanical Engineering, Toyohashi University of Technology, 1-1 Hibarigaoka, Tempaku-cho, Toyohashi, Aichi 441-8580, Japan. E-mail: miyajima.akira.ci@tut.jp; shibata@me.tut.ac.jp
bInstitute for Advanced Research (IAR), Nagoya University, Furo-cho, Chikusa-ku, Nagoya, Aichi 464-8601, Japan
cDepartment of Materials Process Engineering, Nagoya University, Furo-cho, Chikusa-ku, Nagoya, Aichi 464-8603, Japan
dFaculty of Pharmacy and Pharmaceutical Sciences, Josai University, 1-1 Keyakidai, Sakado, Saitama 350-0295, Japan
eInstitute for Research on Next-generation Semiconductor and Sensing Science (IRES2), Toyohashi University of Technology, 1-1 Hibarigaoka, Tempaku-cho, Toyohashi, Aichi 441-8580, Japan
fNational Research Institute of Police Science, 6-3-1 Kashiwanoha, Kashiwa, Chiba 277-0882, Japan
First published on 4th June 2025
Abstract
In this study, we present a genetic diagnostic device with a four-stepwise logarithmic dilution capability for rapid and reliable detection of target nucleic acids in a single operation using the colorimetric loop-mediated isothermal amplification (LAMP) method. An innovative feature is the confluent point with differing microchannel heights ensuring the synchronized inflow of liquids while preventing backflow, even under large volumetric flow rate variations (10–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold). This enabled the independent generation of each dilution factor under constant pressure. Furthermore, an integrated asymmetric micromixer effectively mixed two liquids under laminar flow conditions, enabling simultaneous dispensing of the mixed solution at uniform concentrations into five microchambers for each dilution factor. Additionally, a permanent stop valve in the outlet of each microchamber prevented leakages, minimizing the waste of valuable samples and reagents. We demonstrate that diluted samples were accurately prepared at the intended logarithmic dilution factors in a single operation using purified cannabis seed DNA, achieving detection sensitivity similar to that of conventional turbidity-based LAMP assays. Moreover, we used crudely extracted cannabis resin DNA, which contains several gene amplification inhibitors, successfully detecting the target nucleic acids in a single test. Overall, this versatile device eliminates extensive manual sample preparation and has potential for on-site genetic testing in applications such as infectious disease detection, food safety, and illegal drug testing.
000-fold). This enabled the independent generation of each dilution factor under constant pressure. Furthermore, an integrated asymmetric micromixer effectively mixed two liquids under laminar flow conditions, enabling simultaneous dispensing of the mixed solution at uniform concentrations into five microchambers for each dilution factor. Additionally, a permanent stop valve in the outlet of each microchamber prevented leakages, minimizing the waste of valuable samples and reagents. We demonstrate that diluted samples were accurately prepared at the intended logarithmic dilution factors in a single operation using purified cannabis seed DNA, achieving detection sensitivity similar to that of conventional turbidity-based LAMP assays. Moreover, we used crudely extracted cannabis resin DNA, which contains several gene amplification inhibitors, successfully detecting the target nucleic acids in a single test. Overall, this versatile device eliminates extensive manual sample preparation and has potential for on-site genetic testing in applications such as infectious disease detection, food safety, and illegal drug testing.
Introduction
Generating dilutions of samples is essential in laboratory settings, including those for chemistry, biology, and clinical research.1,2 The primary purpose of serial dilutions is creating a series of solutions with decreasing concentrations. This is crucial for tasks such as determining the concentration of a substance, preparing standards for calibration, or facilitating experiments that require a wide range of concentrations. However, this process often involves human intervention that requires cumbersome manual pipetting. Precise concentration adjustment is critical in various fields such as medical diagnostics, drug development, and environmental monitoring. Nevertheless, the need for high-level expertise and technical skills introduces a risk of human error, compromising the reliability and reproducibility of experimental data. This underscores the need for innovative solutions to streamline and automate the concentration control, particularly when precise concentrations are essential for accurate and consistent results. Currently, robotic technology offers crucial advantages in precision, throughput, and safety for generating various dilution factors; however, challenges such as large sample volume requirements, high costs, complexity, and limited flexibility must be considered when opting for automated liquid handling.3,4To address these issues, lab-on-a-chip technology enables automated sample dilutions across several orders of magnitude, supporting applications such as drug screening, chemical toxicity analysis, chemotactic studies, and material synthesis.5–10 Microfluidic dilution generators manipulate small volumes of reagents, providing a platform that excels in precision, quantitation, and spatiotemporal gradient control. Table 1 summarizes the performance of previously developed dilution microfluidic devices, with the device proposed in this study included for reference. Whitesides group first used a tree-shaped microfluidic network to generate concentration gradients.11 Gradient generation is achieved through complete diffusive mixing of two liquids under laminar flow, using repeated splitting, mixing, and recombination of fluid streams. More complex gradient profiles can be generated as the number of inlets increases.12 Campbell et al.13 proposed a different tree-shaped microfluidic network that generates concentration gradients with the shape of any given monotonic function, using an alternative architecture comprising vertical serpentine channels of different lengths at the same stage. Additionally, there is a wide horizontal channel where an individual serpentine channel stream splits between the next-stage three serpentine channels, unlike the two channels in the previous design. Yusuf et al.14 simplified generating concentration gradients using significantly reduced microfluidic networks. The lengths of the serpentine channels in the network, equivalent to hydrodynamic resistance, were determined through computer optimization to achieve well-controlled volumetric mixing ratios at each branch. This provides highly precise linear concentration gradients at the outlet while simplifying the network design. Currently, tree-shaped microfluidic dilution devices are widely employed in fundamental studies related to chemotaxis,15 chemotherapy,16,17 and drug screening.18–21 Lee et al.22,23 proposed a combinatorial dilution device using a three-layer microfluidic network that generates seven combinations of three additive samples into a buffer solution. As a proof-of-concept, a combinatorial cytotoxicity test was performed using mitomycin C, doxorubicin, and 5-FU on MCF-7 human breast cancer cells. However, conventional concentration gradients are limited to a single order of magnitude and require approximately 20–30 min to reach steady-state conditions.14,24,25
| Design configuration | Flow control | Mixing | Liquid inlet/outlet | Concentration profile | Dilution range | Ref. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a SHM: Staggered-herringbone mixer. b P-ACE: Planar asymmetric contraction-and-expansion mixer. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tree-shape | Flow rate | Serpentine channel | 3-Input/9-output | Linear | One order | 11 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Flow rate | Serpentine channel | 3-Input/9-output or 24-output | Polynomial | One order | 12 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Periodic | One order | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Applied pressure | Serpentine channel | 2-Input/16-output | Linear | One order | 13 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parabolic | One order | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Logarithmic | Two orders | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Flow rate | Serpentine channel | 2-Input/6-output | Linear | One order | 14 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Ladder-shape | Flow rate | SHMa | 2-Input/9-output | Logarithmic | 2–103 | 26 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Flow rate | Tesla mixer | Linear | One order | 27 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Logarithmic | 101–104 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Flow rate | Serpentine channel | 2-Input/6-output | Linear | One order | 28 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Logarithmic | 2–102 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gaussian | One order | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Applied pressure | Diffusion-mixing | 2-Input/12-output | Logarithmic | 100–105 | 30 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Hybrid two-layer | Flow rate | Serpentine channel | 2-Input/18-output | Linear & Logarithmic | 101–103 | 29 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Serial format | Self-powered pumping | Expansion chambers | 4-Input/1-output | Logarithmic | 101–103 | 39 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parallel format | Applied pressure | P-ACEb | 2-Input/4-output | Logarithmic | 101–104 | Present work | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
To generate concentration gradients of several orders of magnitude, serial dilution devices using straightforward ladder microfluidic network have been developed. Whitesides group achieved a dynamic range of 2–103 in ladder-shaped microfluidic dilution devices using 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dilution and 10 sequential stages of mixing and dilution.26 Staggered-herringbone mixers were embedded in the mixing microchannels to enhance the mixing efficiency. Kim et al.27 proposed a serial dilution device comprising a simple ladder microfluidic network and serpentine Tesla mixers capable of generating four-stepwise logarithmic concentrations (101–104) or three-stepwise linear concentrations (0.5, 0.2, and 0.1). However, computational fluid dynamics (CFD) simulations are necessary for the optimal design of the fluidic network in the two aforementioned devices, as they are essential for precisely estimating the hydrodynamic resistance of the micromixers. Lee et al.28 generalized a simple equivalent electrical circuit model to develop universal microfluidic concentration gradient devices without CFD simulations. The desired concentrations are achieved by controlling volumetric mixing ratios of two merging solutions in each stage, resulting in the generation of linear, logarithmic, and Gaussian gradients. For example, a 10-fold logarithmic gradient is obtained by the 9
1 dilution and 10 sequential stages of mixing and dilution.26 Staggered-herringbone mixers were embedded in the mixing microchannels to enhance the mixing efficiency. Kim et al.27 proposed a serial dilution device comprising a simple ladder microfluidic network and serpentine Tesla mixers capable of generating four-stepwise logarithmic concentrations (101–104) or three-stepwise linear concentrations (0.5, 0.2, and 0.1). However, computational fluid dynamics (CFD) simulations are necessary for the optimal design of the fluidic network in the two aforementioned devices, as they are essential for precisely estimating the hydrodynamic resistance of the micromixers. Lee et al.28 generalized a simple equivalent electrical circuit model to develop universal microfluidic concentration gradient devices without CFD simulations. The desired concentrations are achieved by controlling volumetric mixing ratios of two merging solutions in each stage, resulting in the generation of linear, logarithmic, and Gaussian gradients. For example, a 10-fold logarithmic gradient is obtained by the 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 volumetric mixing ratio of buffer and sample diluted from the previous step in each stage. Lee et al.29 also introduced a hybrid two-layer configuration combining serial and proportional microfluidic networks. This approach has the potential to significantly reduce the number of cascaded serial dilution stages required for 14 doses, by combining 4-order logarithmic and 4-point linear concentrations. As an alternative to passive micromixers embedded in mixing regions, Sugiura et al.30,31 proposed microfluidic networks consisting of thin fluidic-resistance and thick diffusion-mixing microchannels. This configuration enables short microchannel lengths and sufficient mixing of two liquids, generating precise concentration profiles over a wide range of flow rates. The serial dilution microfluidic device can generate logarithmic concentration profiles across six orders of magnitude with a relatively compact design. Currently, ladder-shaped microfluidic dilution devices capable of generating several orders of logarithmic concentrations are employed in fundamental studies related to cytotoxicity,27,32,33 dose–response,31,34 and cell proliferation assays,35 as well as chemotaxis,36 antibiotic susceptibility testing,37 and liposome nanoparticle synthesis.38 However, in serial dilutions, concentration errors in a stage propagate to the next, highlighting the importance of achieving complete mixing in each stage. Furthermore, many serial dilution steps can lead to a significant occupancy area, and generating a concentration gradient through continuous flow needs a relatively large amount of sample.
1 volumetric mixing ratio of buffer and sample diluted from the previous step in each stage. Lee et al.29 also introduced a hybrid two-layer configuration combining serial and proportional microfluidic networks. This approach has the potential to significantly reduce the number of cascaded serial dilution stages required for 14 doses, by combining 4-order logarithmic and 4-point linear concentrations. As an alternative to passive micromixers embedded in mixing regions, Sugiura et al.30,31 proposed microfluidic networks consisting of thin fluidic-resistance and thick diffusion-mixing microchannels. This configuration enables short microchannel lengths and sufficient mixing of two liquids, generating precise concentration profiles over a wide range of flow rates. The serial dilution microfluidic device can generate logarithmic concentration profiles across six orders of magnitude with a relatively compact design. Currently, ladder-shaped microfluidic dilution devices capable of generating several orders of logarithmic concentrations are employed in fundamental studies related to cytotoxicity,27,32,33 dose–response,31,34 and cell proliferation assays,35 as well as chemotaxis,36 antibiotic susceptibility testing,37 and liposome nanoparticle synthesis.38 However, in serial dilutions, concentration errors in a stage propagate to the next, highlighting the importance of achieving complete mixing in each stage. Furthermore, many serial dilution steps can lead to a significant occupancy area, and generating a concentration gradient through continuous flow needs a relatively large amount of sample.
Vloemans et al.39 proposed a dilution device incorporating hydrophobic valves and a self-powered pump, which acted as a passive driving source for liquid manipulation. This eliminated the need for external pumps, making the microfluidic platform suitable for point-of-care testing applications. However, the hydrophobic valves—composed of hydrophobically treated filter paper—must be manually positioned within the microfluidic channels, and both the top and bottom laminate films must be locally treated with hydrophobic and superhydrophobic solutions. These steps increase both labor requirements and device cost. Moreover, the dilution factor is limited to 1000-fold, which may be insufficient for applications requiring a wider dynamic range.
Previously, we developed a versatile microfluidic device for the multiplex detection of target nucleic acids based on the loop-mediated isothermal amplification (LAMP) method, which has been applied to crop disease detection,40 rapid identification of toxic plants,41 diagnosis of infectious diseases,42,43 food allergen detection,44 and foodborne pathogen detection.45 However, the task of adjusting the sample concentration to an appropriate level for genetic testing still relied on manual pipetting. Therefore, in this study, we present a microfluidic device designed for generating logarithmic concentrations in a parallel microchannel network for each dilution factor to reliably detect the target nucleic acids in a single operation. The DNA target was derived from cannabis (Cannabis sativa L.) because it is the most abused drug worldwide and is strictly regulated in many countries. Hence, developing simple and rapid testing methods for cannabis detection is crucial to addressing this global issue.46 While fundamental studies aiming for on-site detection of cannabis using LAMP have been reported,47,48 we aimed in this study to improve its practical applicability by integrating LAMP with the dilution microfluidic device.
Experimental
Design and fabrication of a two-heighted Y-junction for merging two liquids
In conventional T- or Y-junction for merging two liquids, if one reaches the confluent point first and flows into the downstream microchannel, air can become trapped between both liquids, disrupting the flow and hindering the subsequent entry of the other liquid. To address this issue, we designed a Y-junction wherein the microchannel width is narrowed just before the confluent point. This modification temporarily halts the liquid reaching first the confluent point through surface tension, allowing the other liquid to arrive at the confluent point. Once the interfaces of both liquids make contact, they simultaneously flow into the downstream microchannel. Consequently, we successfully controlled the timing of liquid merging. However, when the volumetric flow rate ratio of both liquids exceeded 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, backflow of the higher-flow rate liquid into the microchannel of the lower-flow rate liquid was observed (Fig. S1†).
1, backflow of the higher-flow rate liquid into the microchannel of the lower-flow rate liquid was observed (Fig. S1†).
Therefore, we propose a newly designed Y-junction with differing microchannel heights (hereinafter referred to as a “two-heighted Y-junction”) for the merging of two liquids with a significant difference in volume flow rates (Fig. 1a). By setting the microchannel height to 10 μm on the sample side (lower volumetric flow rate) and 90 μm on the dilution buffer side (higher volumetric flow rate), backflow was prevented by maintaining a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 flow velocity ratio during 10-fold dilution (with a 9
1 flow velocity ratio during 10-fold dilution (with a 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 volumetric flow rate ratio for buffer and sample, respectively). Furthermore, as previously mentioned, the inclusion of a constricted region (microchannel width: 32 μm) at the confluent point temporarily halts the liquid due to surface tension. The theoretical burst pressure was 12.7 kPa for water (ESI† S1 and Fig. S2); this design enables the exhaust of air from the other microchannel before the liquids come into contact, preventing air from getting trapped in the microchannel and allowing precise control of the timing of the merging process. Fig. 1b shows images of a polydimethylsiloxane (PDMS)-based two-heighted Y-junction acquired with a scanning electron microscope (GeminiSEM 560; ZEISS, Jena, Germany). The confluent point of the Y-junction with different microchannel heights (10 and 90 μm on the sample and dilution buffer sides, respectively) were successfully fabricated. Additionally, the width of the rectangular microchannel gradually narrowed from 200 μm as it approaches the confluent point to 32 μm at the constricted region.
1 volumetric flow rate ratio for buffer and sample, respectively). Furthermore, as previously mentioned, the inclusion of a constricted region (microchannel width: 32 μm) at the confluent point temporarily halts the liquid due to surface tension. The theoretical burst pressure was 12.7 kPa for water (ESI† S1 and Fig. S2); this design enables the exhaust of air from the other microchannel before the liquids come into contact, preventing air from getting trapped in the microchannel and allowing precise control of the timing of the merging process. Fig. 1b shows images of a polydimethylsiloxane (PDMS)-based two-heighted Y-junction acquired with a scanning electron microscope (GeminiSEM 560; ZEISS, Jena, Germany). The confluent point of the Y-junction with different microchannel heights (10 and 90 μm on the sample and dilution buffer sides, respectively) were successfully fabricated. Additionally, the width of the rectangular microchannel gradually narrowed from 200 μm as it approaches the confluent point to 32 μm at the constricted region.
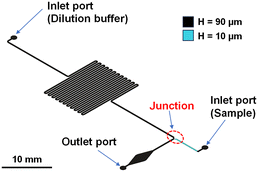
Fig. 2 shows a schematic diagram of the microfluidic device (microchannel width: 200 μm) designed to evaluate a 10-fold dilution factor of two liquids passing through the two-heighted Y-junction. The microchannel design was optimized so that under the same pressure for dilution buffer and sample, the flow resistance ratios from the inlet ports to the confluent point on both sides would achieve the volumetric flow rate ratio required for generating a 10-fold dilution factor. Similarly, the devices were designed for generating dilution factors from 100- to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold (Fig. S3†). Based on the theory describing flow resistance in a rectangular microchannel (ESI† S2), the theoretical volumetric flow rates of the dilution buffer and sample solutions for each dilution factor were calculated and are listed in Table S1.†
000-fold (Fig. S3†). Based on the theory describing flow resistance in a rectangular microchannel (ESI† S2), the theoretical volumetric flow rates of the dilution buffer and sample solutions for each dilution factor were calculated and are listed in Table S1.†
For the two-liquid merging experiments, pure and blue-colored (5.0 w/v%) waters were introduced as the dilution buffer through the left inlet and sample through the right inlet, respectively, using two pressure-controlled micropumps (Flow EZ™ 345 mbar; Fluigent SA, Le Kremlin-Bicêtre, France) at the same applied pressure of 3.5 kPa. The experimental procedure was as follows: first, pure water (dilution buffer) or colored water (sample) was aspirated into a 200 μL pipette tip using a pipette. After detaching the pipette tip from the pipette and connecting it to the pump, it was inserted into the inlet port of the device. The sample was then introduced first and temporarily halted at the confluent point, followed by the introduction of the dilution buffer. This sequence was necessary because the buffer, flowing through a lower-resistance microchannel, would otherwise reach the junction first and could not be stopped at the confluent point. To evaluate the dilution factor of the solution (diluted blue-colored water) across four orders of magnitude (10- to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold) in the device, 60 μL of each diluted solution was collected from the outlet port, and absorbance was measured 629 nm using a spectrophotometer (DS-11; DeNovix, Wilmington, DE, USA). For each dilution factor, absorbance measurements were performed thrice using 2 μL of solution per measurement, and the average value was calculated.
000-fold) in the device, 60 μL of each diluted solution was collected from the outlet port, and absorbance was measured 629 nm using a spectrophotometer (DS-11; DeNovix, Wilmington, DE, USA). For each dilution factor, absorbance measurements were performed thrice using 2 μL of solution per measurement, and the average value was calculated.
The PDMS-based microfluidic devices were fabricated through a soft lithography process briefly described as follows: first, a negative thick photoresist (SU-8 3010; MicroChem, Newton, MA, USA) was patterned on a 4 inch single-crystal silicon (Si) wafer (e-Prize, Yokohama, Japan) for the first layer with a thickness of 10 μm through a photolithography process. Next, the second layer with a thickness of 80 μm was patterned on the same Si wafer using a higher-viscosity negative thick photoresist (SU-8 3050) to form a mold with the microchannel heights of 10 and 90 μm on the sample and dilution buffer sides, respectively. Subsequently, the SU-8 master mold was replicated in PDMS (Silpot 184; Dow Corning Toray, Tokyo, Japan) after curing on a hot plate at 80 °C for 40 min. After peeling off the PDMS from the SU-8 master mold, circular holes for two inlet (with a diameter of 1.5 mm) and one outlet (with a diameter of 1.0 mm) ports were punched into the PDMS microfluidic device using a biopsy punch piercing tool (Kai Industries, Gifu, Japan). Finally, the microchannels were sealed with a white polyvinyl chloride (PVC) sheet (EB-235; Hikari, Osaka, Japan) using a silicone-based adhesive double-sided tape (no. 5303W; Nitto Denko, Osaka, Japan).
Design and fabrication of a microfluidic device for logarithmic concentration generation
Fig. 3 shows a schematic diagram of the proposed microfluidic device that generates logarithmic dilutions across four orders of magnitude (10- to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold) in a single operation. Notably, each dilution factor can be generated independently in a parallel microchannel network under constant pressure by employing the two-heighted Y-junction at the confluent points. Consequently, the flow resistance ratios of buffer and sample can be independently optimized for each dilution factor. Compared with conventional ladder-type logarithmic dilution devices,28 this approach significantly simplifies the microchannel design process. The detailed design of the four-stepwise logarithmic parallel dilution microfluidic device is described in Fig. S4,† and the designed flow resistance between the buffer and sample sides for each dilution factor is listed in Table S2.† Furthermore, downstream of the confluent point, both liquids in a laminar flow state are thoroughly mixed using a planar asymmetric contraction-and-expansion (P-ACE) micromixer49 embedded in a 15 mm long microchannel. The fully mixed solution then is evenly dispensed into five 3-μL microchambers. To ensure simultaneous filling, flow resistance is balanced by adjusting the length of the serpentine microchannels leading to each microchamber. Additionally, each microchamber outlet has a structure with a reduced cross-sectional area (width: 20 μm, height: 10 μm), hereinafter referred to as a permanent stop valve.42,44 This design facilitates the exhaust of air from the microchambers during filling, after which the liquid is halted at the permanent stop valve with a theoretical burst pressure of 9.5 kPa for water due to surface tension (ESI† S1). This prevents unnecessary waste of valuable samples and reagents.
000-fold) in a single operation. Notably, each dilution factor can be generated independently in a parallel microchannel network under constant pressure by employing the two-heighted Y-junction at the confluent points. Consequently, the flow resistance ratios of buffer and sample can be independently optimized for each dilution factor. Compared with conventional ladder-type logarithmic dilution devices,28 this approach significantly simplifies the microchannel design process. The detailed design of the four-stepwise logarithmic parallel dilution microfluidic device is described in Fig. S4,† and the designed flow resistance between the buffer and sample sides for each dilution factor is listed in Table S2.† Furthermore, downstream of the confluent point, both liquids in a laminar flow state are thoroughly mixed using a planar asymmetric contraction-and-expansion (P-ACE) micromixer49 embedded in a 15 mm long microchannel. The fully mixed solution then is evenly dispensed into five 3-μL microchambers. To ensure simultaneous filling, flow resistance is balanced by adjusting the length of the serpentine microchannels leading to each microchamber. Additionally, each microchamber outlet has a structure with a reduced cross-sectional area (width: 20 μm, height: 10 μm), hereinafter referred to as a permanent stop valve.42,44 This design facilitates the exhaust of air from the microchambers during filling, after which the liquid is halted at the permanent stop valve with a theoretical burst pressure of 9.5 kPa for water due to surface tension (ESI† S1). This prevents unnecessary waste of valuable samples and reagents.
The PDMS-based parallel dilution microfluidic devices were fabricated through a soft lithography process, as described previously. However, there is a difference: initially, the SU-8 master mold with microchannel heights of 10 and 90 μm on the sample and dilution buffer sides, respectively, was patterned on a Si wafer using a two-step photolithography process. Next, to create deep localized microchamber structures of up to 1 mm in depth suitable for colorimetric LAMP assays, the SU-8 mold and pieces of wax of 2.7 mg (Ferris File-A-Wax; Freeman Manufacturing & Supply, Avon, OH, USA) were simultaneously subjected to a low-pressure air plasma surface treatment in a plasma asher (JPA300; J-Science Lab, Kyoto, Japan) at 150 W for 3 min. Then, the pieces of wax were positioned at the center of each SU-8 microchamber pattern. Subsequently, a reflow process was conducted by heating the mold on a hotplate at 115 °C for 3 min (EC1200-N; AS ONE, Osaka, Japan) (Fig. S5†). The SU-8 master mold, featuring semi-elliptical wax structures, was replicated using PDMS and cured on a hot plate at 80 °C for 40 min. After detaching the PDMS from the SU-8 master mold, circular holes with 1.5 and 1.0 mm in diameter for the inlet and outlet ports, respectively, were created in the PDMS device using a biopsy punch tool. The microchambers and microchannels on the PDMS surface were finally sealed with a white PVC sheet utilizing adhesive double-sided tape. The detailed fabrication process, including the wax reflow process, is further described in our previous study.44
Fig. 4a shows an example of the fabricated parallel dilution microfluidic device with a size of approximately 40 × 70 mm. In the experiment, blue-colored water (10.0 v/v%) was first introduced as the sample through the right inlet port at 7.0 kPa using a pressure-controlled micropump. After the blue-colored water was temporarily pinned at all the two-heighted Y-junctions for each dilution factor, yellow-colored water (1.0 v/v%) was introduced as the dilution buffer through the left inlet port at the same pressure. The image was automatically stitched using high-magnification images acquired with a digital microscope (BZ-X810; Keyence, Osaka, Japan). To quantitatively evaluate the dilution factors, RGB values were analyzed using the ImageJ software package (version 1.54d; National Institutes of Health, Bethesda, MD, USA). A circular measurement area of 1 mm2 was defined at the center of the microchamber. The dilution performance was evaluated using the R/G ratio, which provided the most quantitative representation of dilution levels. A higher R/G value indicates a greater concentration of the yellow-colored dilution buffer. Fig. 4b shows the mixing behavior of two liquids in the asymmetric micromixer, observed using an inverted microscope (ECLIPSE Ti2; Nikon, Tokyo, Japan) equipped with a complementary metal-oxide semiconductor (CMOS) camera (DS-Ri2; Nikon, Tokyo, Japan). Before reaching the entrance of the micromixer, both liquids exhibited a steady laminar flow profile. However, they were effectively mixed after passing through the 15 mm long micromixer.
Operating procedure for the colorimetric LAMP assay in the dilution microfluidic device
The method for conducting a colorimetric LAMP assay within a parallel dilution microfluidic device for the simultaneous detection of DNA samples generated across four orders of magnitude (10–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold) in a single operation is described in this section. LAMP primer sets for detecting cannabis were designed for the fiber-type tetrahydrocannabinolic acid (THCA) synthase gene (DDBJ/EMBL/GenBank database accession no. AB212830).47 Tetrahydrocannabinol (THC), the decarboxylated form of THCA, is the primary cannabinoid responsible for the strong psychoactive effects. In this study, two types of DNA samples—purified and crude—were extracted from cannabis seeds and resin to serve two distinct experimental purposes: (1) purified DNA was used to generate a standard curve for quantitative analysis, and (2) crude DNA was used to evaluate the effect of LAMP inhibitors in dilution devices. Cannabis seeds were purchase from Uchida Wakanyaku Co., Ltd. (Tokyo, Japan) and cannabis resin was issued by the Narcotics Control Department (Kanto-Shin'etsu Regional Bureau of Health and Welfare, Ministry of Health, Labor and Welfare, Tokyo, Japan) in accordance with official procedures. To obtain the standard curve for a LAMP assay using DNA samples diluted 10–10
000-fold) in a single operation is described in this section. LAMP primer sets for detecting cannabis were designed for the fiber-type tetrahydrocannabinolic acid (THCA) synthase gene (DDBJ/EMBL/GenBank database accession no. AB212830).47 Tetrahydrocannabinol (THC), the decarboxylated form of THCA, is the primary cannabinoid responsible for the strong psychoactive effects. In this study, two types of DNA samples—purified and crude—were extracted from cannabis seeds and resin to serve two distinct experimental purposes: (1) purified DNA was used to generate a standard curve for quantitative analysis, and (2) crude DNA was used to evaluate the effect of LAMP inhibitors in dilution devices. Cannabis seeds were purchase from Uchida Wakanyaku Co., Ltd. (Tokyo, Japan) and cannabis resin was issued by the Narcotics Control Department (Kanto-Shin'etsu Regional Bureau of Health and Welfare, Ministry of Health, Labor and Welfare, Tokyo, Japan) in accordance with official procedures. To obtain the standard curve for a LAMP assay using DNA samples diluted 10–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold in a single operation, high-quality genomic DNA was extracted from cannabis seed (10 mg) using a FavorPrep Plant Genomic DNA Extraction Mini Kit (Chiyoda Science, Tokyo, Japan). To investigate the effect of LAMP inhibitors in DNA samples, total genomic DNA was extracted from cannabis resin (10 mg) using a Kaneka Easy DNA Extraction Kit version 2 (Kaneka, Tokyo, Japan) according to the manufacturer's manual. DNA concentration was quantified using Quantus Fluorometer (Promega, Madison, WI, USA) and QuantiFluor ONE dsDNA System (Promega), and adjusted to a final concentration of 10 ng μL−1 (1.04 × 104 copies per μL) for purified cannabis seed and 8 ng μL−1 (8.33 × 103 copies per μL) for crudely extracted cannabis resin.
000-fold in a single operation, high-quality genomic DNA was extracted from cannabis seed (10 mg) using a FavorPrep Plant Genomic DNA Extraction Mini Kit (Chiyoda Science, Tokyo, Japan). To investigate the effect of LAMP inhibitors in DNA samples, total genomic DNA was extracted from cannabis resin (10 mg) using a Kaneka Easy DNA Extraction Kit version 2 (Kaneka, Tokyo, Japan) according to the manufacturer's manual. DNA concentration was quantified using Quantus Fluorometer (Promega, Madison, WI, USA) and QuantiFluor ONE dsDNA System (Promega), and adjusted to a final concentration of 10 ng μL−1 (1.04 × 104 copies per μL) for purified cannabis seed and 8 ng μL−1 (8.33 × 103 copies per μL) for crudely extracted cannabis resin.
Hydroxynaphthol blue (HNB; FUJIFILM Wako Pure Chemical, Osaka, Japan) was used as a colorimetric indicator to signal LAMP reactions, with a positive reaction indicated by a color shift from violet to sky blue.43 Each microchamber's final HNB concentration was adjusted to 150 μM within the blend of sample and LAMP reagents. The LAMP reactions employed the Loopamp® DNA amplification kit (Eiken Chemical, Tokyo, Japan), which includes a 2× reaction mix and thermostable Bst polymerase. The final mixture volume was 112.5 μL, comprising the LAMP reagents and primer sets (1.6 μM of inner primers (FIP and BIP), 0.2 μM of outer primers (F3 and B3), and 0.8 μM of loop primers (LF and LB)) except for the DNA sample, which was prepared for colorimetric LAMP assays in the dilution device (Table S3†).
For comparative purposes in the cannabis seed LAMP assays, conventional off-chip LAMP assays were conducted using a 25 μL reaction mixture in 0.2 mL PCR tubes with a real-time turbidimeter (LoopampEXIA; Eiken Chemical, Tokyo, Japan). The Loopamp® DNA amplification kit was employed to perform the LAMP reactions (Table S4†). Additionally, for comparative purposes in the cannabis resin LAMP assays, conventional real-time fluorescent LAMP assays were conducted using a 25 μL reaction mixture with a Thermal Cycler Dice Real Time System Lite (Takara Bio, Shiga, Japan). The LAMP reactions employed the Loopamp® DNA amplification kit, which includes a 2× reaction mix and Bst polymerase. A 25-μM EvaGreen® dye (20× in water; Biotium, Fremont, CA, USA) was used as a fluorescent indicator (Table S5†). Furthermore, real-time PCR was also conducted in a final volume of 25 μL containing Taq 2× Master Mix (New England Biolabs, Ipswich, MA, USA), primers, EvaGreen® dye, and genomic DNA as template with the Thermal Cycler Dice Real Time System Lite (Table S6†). The PCR primers targeting the chloroplast TrnH-TrnL intragenic spacer regions were designed according to a previous study.50 Real-time PCR conditions were as follows: initial denaturation at 95 °C for 10 min, followed by 45 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s, and extension at 70 °C for 30 s.
To obtain DNA amplification curves from time-dependent color changes—shifting from violet (negative) to sky blue (positive)—in each reaction microchamber during the colorimetric LAMP assays, time-lapse images of the dilution device were captured and analyzed using a homebuilt imaging apparatus (Fig. S6†). This setup included a CMOS camera (STC-MCS163U3V; Omron Sentech, Kanagawa, Japan) with a fixed focal lens (M117FM35; Tamron, Saitama, Japan) and an LED ring light (LED-R48; Arms System, Tokyo, Japan) to ensure even lighting. Images of the device were taken every 30 s using a Python macro program, and the colorimetric LAMP assay proceeded at 63 °C for 60 min for cannabis seed and resin in a hot-water bath (TB-1NC; AS ONE, Osaka, Japan). After the LAMP assay, the time-dependent color change between positive (sky blue) and negative (violet) reactions in each microchamber was automatically analyzed using a homebuilt hue-based analysis software.43 The hue angle change (Δh) in the CIELAB color space was used as an indicator of gene amplification progression. For visualization purposes, the color saturation of the images presented later as experimental results was enhanced to improve the distinction between positive and negative microchambers. However, unprocessed raw images were used for the quantitative colorimetric analysis.
Results and discussion
Performance of a two-heighted Y-junction for the merging of two liquids
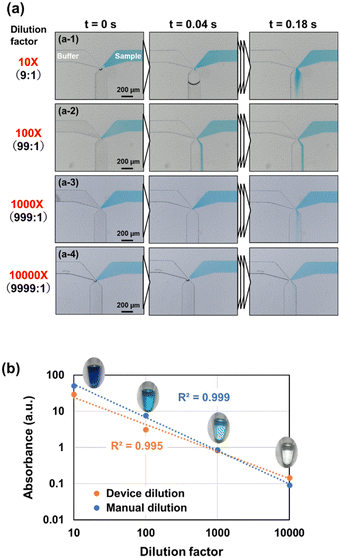
Fig. 5a shows the flow behavior of two liquids passing through the two-heighted Y-junction at dilutions from 10–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold within the dilution devices, as described in Fig. 2 and S3.† For generating a 10-fold dilution (a-1) with a volumetric flow rate ratio of 9
000-fold within the dilution devices, as described in Fig. 2 and S3.† For generating a 10-fold dilution (a-1) with a volumetric flow rate ratio of 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 between buffer and sample, pure (dilution buffer) and blue-colored (sample) waters were introduced into a microchannel, both at 3.5 kPa. The blue-colored water was temporarily pinned at the confluence point due to surface tension. When the pure water arrived, the menisci of both liquids came into contact, allowing them to flow simultaneously into the downstream microchannel under laminar flow conditions. At this point, no backflow of the higher-flow rate pure water into the microchannel of the blue-colored water—with the lower flow rate—was observed, likely due to the 1
1 between buffer and sample, pure (dilution buffer) and blue-colored (sample) waters were introduced into a microchannel, both at 3.5 kPa. The blue-colored water was temporarily pinned at the confluence point due to surface tension. When the pure water arrived, the menisci of both liquids came into contact, allowing them to flow simultaneously into the downstream microchannel under laminar flow conditions. At this point, no backflow of the higher-flow rate pure water into the microchannel of the blue-colored water—with the lower flow rate—was observed, likely due to the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 flow velocity ratio. Notably, the proposed two-heighted Y-junction performed effectively even under 100-fold (99
1 flow velocity ratio. Notably, the proposed two-heighted Y-junction performed effectively even under 100-fold (99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 flow rate ratio) (a-2), 1000-fold (999
1 flow rate ratio) (a-2), 1000-fold (999![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 flow rate ratio) (a-3), and 10
1 flow rate ratio) (a-3), and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold (9999
000-fold (9999![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 flow rate ratio) (a-4) dilutions. In all cases, it effectively controlled the timing of the inflow of both liquids while suppressing backflow. The flow behavior of two liquids at the two-heighted Y-junction at various dilution factors (10–10
1 flow rate ratio) (a-4) dilutions. In all cases, it effectively controlled the timing of the inflow of both liquids while suppressing backflow. The flow behavior of two liquids at the two-heighted Y-junction at various dilution factors (10–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold) is shown in Video S1.† This efficacity is attributed to the significantly higher flow resistance in the sample microchannel, which has a reduced height of 10 μm. Theoretically, flow resistance increases in proportion to the third power of the microchannel height.42 Thus, the flow resistance differs by approximately 1000-fold between the upstream and downstream regions of the confluence point. Consequently, both liquids preferentially inflow into the downstream microchannel below the confluence point.
000-fold) is shown in Video S1.† This efficacity is attributed to the significantly higher flow resistance in the sample microchannel, which has a reduced height of 10 μm. Theoretically, flow resistance increases in proportion to the third power of the microchannel height.42 Thus, the flow resistance differs by approximately 1000-fold between the upstream and downstream regions of the confluence point. Consequently, both liquids preferentially inflow into the downstream microchannel below the confluence point.
Fig. 5b shows the absorbance measurements for the diluted solutions generated at each dilution factor. The horizontal and vertical axes represent the dilution factor and absorbance on a log–log scale, respectively. Mean and standard deviation (SD) values calculated from five experimental replicates for the device-based dilution, and a single replicate for the manual dilution, are summarized in Table S7.† For comparison, the absorbance values of manually diluted solutions are also plotted. The diluted solutions generated by the device exhibited linearity with a high coefficient of determination (R2 > 0.99). Additionally, data agreed with those of the manually diluted solutions, demonstrating that the dilution device accurately generated four-stepwise logarithmic dilutions across four orders of magnitude (10–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold).
000-fold).
Performance of the microfluidic device for logarithmic concentration generation
Fig. 6a shows representative experimental results evaluating the dilution factors in the developed device. Yellow-colored (1 v/v%) and blue-colored (10 v/v%) waters were introduced into the 90- and 10 μm-high microchannel left and right inlet ports, respectively, at 7.0 kPa. For a 10-fold dilution, the yellow- and blue colored waters in the dilution buffer and sample sides, respectively, were mixed at a volumetric flow rate ratio of 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, turning the water color to green. As the dilution increased from 100- to 10
1, turning the water color to green. As the dilution increased from 100- to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold, the mixed solution progressively shifted to a more yellowish color, reflecting the increasing dominance of the yellow-colored water in the mixture. Furthermore, no visible differences in color were observed between the five microchambers at each dilution factor because the liquids were effectively mixed in the asymmetric micromixer before being dispensed into the microchambers (Fig. 4b). By comparing the experimental result with those obtained from introducing into the device manually mixed solutions (10–10
000-fold, the mixed solution progressively shifted to a more yellowish color, reflecting the increasing dominance of the yellow-colored water in the mixture. Furthermore, no visible differences in color were observed between the five microchambers at each dilution factor because the liquids were effectively mixed in the asymmetric micromixer before being dispensed into the microchambers (Fig. 4b). By comparing the experimental result with those obtained from introducing into the device manually mixed solutions (10–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold dilutions) (Fig. 6b), it seems that the color at each dilution factor were similar. Conversely, there were significant differences in color between the five microchambers at each dilution factor in the device without the micromixer (Fig. 6c). These results confirm that the micromixer is crucial for achieving uniform dispensing of the mixed solution into microchambers.
000-fold dilutions) (Fig. 6b), it seems that the color at each dilution factor were similar. Conversely, there were significant differences in color between the five microchambers at each dilution factor in the device without the micromixer (Fig. 6c). These results confirm that the micromixer is crucial for achieving uniform dispensing of the mixed solution into microchambers.
Fig. S7a† shows the results of the device-based dilutions with the micromixer, while Fig. S7b† shows the results of the manually prepared dilutions. It is shown that the RGB value ratios at each dilution factor are similar. Fig. 6d shows a comparison of the R/G values between the device-based and manually prepared dilutions. Mean and SD values calculated from five experimental replicates for both the device-based and manual dilutions are summarized in Table S8.† The graph indicates that the slope of the approximation line is approximately 0.9 with a high coefficient of determination (R2 > 0.99), suggesting that the dilution factors of both methods are well aligned. Notably, no significant difference in the R/G values was observed between the 1000 and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold device dilutions. This is likely due to the yellow-colored water becoming excessively dominant relative to the blue-colored water. However, a t-test revealed a statistically significant difference in the R/G values between the 1000 and 10
000-fold device dilutions. This is likely due to the yellow-colored water becoming excessively dominant relative to the blue-colored water. However, a t-test revealed a statistically significant difference in the R/G values between the 1000 and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold dilutions generated by the device (p < 0.01).
000-fold dilutions generated by the device (p < 0.01).
Colorimetric LAMP assay in the parallel dilution microfluidic device
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold). The filling times for the microchambers differed depending on the flow resistance: 1 min 45 s for the lowest resistance (10
000-fold). The filling times for the microchambers differed depending on the flow resistance: 1 min 45 s for the lowest resistance (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold dilution) and 3 min 23 s for the highest resistance (10-fold dilution) (Video S2†). Here, the filling times of the microchambers at dilution factors of 10
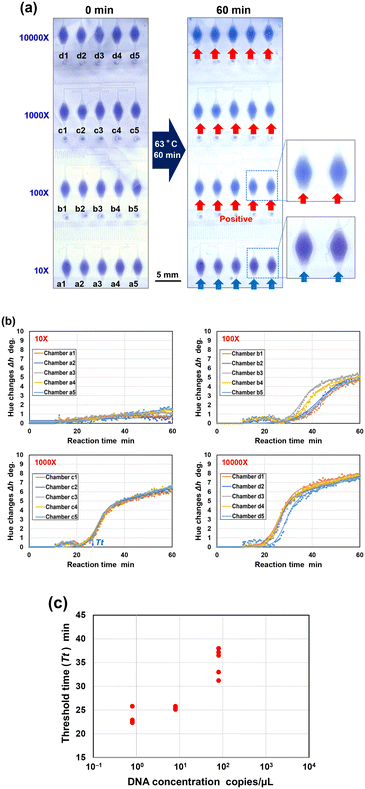
000-fold dilution) and 3 min 23 s for the highest resistance (10-fold dilution) (Video S2†). Here, the filling times of the microchambers at dilution factors of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold (1 min 45 s) and 1000-fold (1 min 42 s) were similar, as the flow resistance from the confluent point to the inlet of each microchamber was significantly greater than that from the inlet port to the confluent point. Consequently, the total flow resistance from each inlet port to the inlet of each microchamber was similar. As expected, the permanent stop valves at the outlet ports of the microchambers successfully prevented liquid outflow from previously filled microchambers. This design effectively reduced waste of samples and reagents. Once all microchambers were filled, all inlet ports and air exhaust outlet ports were sealed with adhesive double-sided tape. The device was then placed in a hot-water bath to facilitate the amplification of targeted nucleic acids via the LAMP reaction under stable isothermal conditions at 63 °C for 60 min. At dilution factors of 10–1000-fold, all reaction chambers showed a color change from violet to sky blue, indicating a positive reaction. In contrast, at a dilution factor of 10
000-fold (1 min 45 s) and 1000-fold (1 min 42 s) were similar, as the flow resistance from the confluent point to the inlet of each microchamber was significantly greater than that from the inlet port to the confluent point. Consequently, the total flow resistance from each inlet port to the inlet of each microchamber was similar. As expected, the permanent stop valves at the outlet ports of the microchambers successfully prevented liquid outflow from previously filled microchambers. This design effectively reduced waste of samples and reagents. Once all microchambers were filled, all inlet ports and air exhaust outlet ports were sealed with adhesive double-sided tape. The device was then placed in a hot-water bath to facilitate the amplification of targeted nucleic acids via the LAMP reaction under stable isothermal conditions at 63 °C for 60 min. At dilution factors of 10–1000-fold, all reaction chambers showed a color change from violet to sky blue, indicating a positive reaction. In contrast, at a dilution factor of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold, only the second and fourth reaction chambers showed a positive reaction.
000-fold, only the second and fourth reaction chambers showed a positive reaction.
Fig. 7b shows the DNA amplification curves obtained at each dilution factor using a home-built analysis software,43 where the hue angle changes (Δh) were plotted as a function of reaction time. As the dilution factor increased, the start of the gene amplification reaction was delayed and its variability between reaction chambers became more pronounced. The threshold time (Tt value) was determined from the DNA amplification curves of five reaction chambers at each dilution factor. Tt values were automatically calculated as the peak value of the second derivative of each DNA amplification curve to quantitatively estimate the reaction time required for DNA amplification.43
Fig. 7c shows a standard curve obtained by plotting the calculated Tt values (red solid circles) against DNA concentrations. Detailed data are listed in Tables S9 and S10.† A pronounced negative correlation (R2 = 0.952) was found for the detection of specific DNA within the dilution devices, except for data from the 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold dilution. These results indicate that samples were prepared at the correct dilution factors in the device in a single operation. The 10
000-fold dilution. These results indicate that samples were prepared at the correct dilution factors in the device in a single operation. The 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold dilution (DNA concentration: 1 pg μL−1; 1.04 copies per μL) likely reached the limit of detection (LOD) of the dilution device, resulting in a higher number of negative reaction chambers and Tt values, even in positive reaction chambers. Nevertheless, the experimental Tt values agreed with those from conventional turbidity-based LAMP assays (blue solid circles). Additionally, as the conventional LAMP method yielded a positive reaction only in one of three experiments, the 10
000-fold dilution (DNA concentration: 1 pg μL−1; 1.04 copies per μL) likely reached the limit of detection (LOD) of the dilution device, resulting in a higher number of negative reaction chambers and Tt values, even in positive reaction chambers. Nevertheless, the experimental Tt values agreed with those from conventional turbidity-based LAMP assays (blue solid circles). Additionally, as the conventional LAMP method yielded a positive reaction only in one of three experiments, the 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold dilution can be considered the LOD of cannabis seed.
000-fold dilution can be considered the LOD of cannabis seed.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold, indicating a positive reaction. In contrast, at a 10-fold dilution, no color change was observed in any of the five microchambers, resulting in false-negative results. This suggest that the crude DNA sample was not properly diluted, leading to the inhibition of the gene amplification reaction, likely due to the relatively high concentration of LAMP inhibitors in the sample. Fig. 8b presents the DNA amplification curves obtained at each dilution factor. No gene amplification was observed at 10-fold dilution, whereas gene amplification was detected at 100-fold dilution. However, there the DNA amplification curves varied across the five microchambers. When the dilution factor was further increased to 1000- and 10
000-fold, indicating a positive reaction. In contrast, at a 10-fold dilution, no color change was observed in any of the five microchambers, resulting in false-negative results. This suggest that the crude DNA sample was not properly diluted, leading to the inhibition of the gene amplification reaction, likely due to the relatively high concentration of LAMP inhibitors in the sample. Fig. 8b presents the DNA amplification curves obtained at each dilution factor. No gene amplification was observed at 10-fold dilution, whereas gene amplification was detected at 100-fold dilution. However, there the DNA amplification curves varied across the five microchambers. When the dilution factor was further increased to 1000- and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold, the DNA amplification curves became more distinctive, and the variability among the five microchambers decreased. Compared to the DNA amplification curve of cannabis seed DNA at 10
000-fold, the DNA amplification curves became more distinctive, and the variability among the five microchambers decreased. Compared to the DNA amplification curve of cannabis seed DNA at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold dilution (Fig. 7b), the detection sensitivity of cannabis resin DNA is higher. This is likely because the cannabis resin used in this study contained more target THCA sequences in its genome per unit weight than those in cannabis seed. Fig. 8c shows the calculated Tt values plotted against DNA concentrations. Detailed data are listed in Table S11.† The results are contrary to those obtained using purified cannabis seed DNA (Fig. 7c). When crudely extracted DNA was used as the sample, Tt values decreased as the dilution factor increased (i.e., as DNA concentration decreased). This suggests that the effect of LAMP inhibitors on the gene amplification reaction was more significant than the impact of reduced DNA concentration. Cannabis resin contains many impurities such as polyphenolic compounds that inhibit DNA amplification; thus, the amplification efficiency can be improved by diluting the crudely extracted DNA solution.51 The optimal dilution condition was found to be 1000-fold, where the variability in Tt values was the smallest. In contrast, at a 10
000-fold dilution (Fig. 7b), the detection sensitivity of cannabis resin DNA is higher. This is likely because the cannabis resin used in this study contained more target THCA sequences in its genome per unit weight than those in cannabis seed. Fig. 8c shows the calculated Tt values plotted against DNA concentrations. Detailed data are listed in Table S11.† The results are contrary to those obtained using purified cannabis seed DNA (Fig. 7c). When crudely extracted DNA was used as the sample, Tt values decreased as the dilution factor increased (i.e., as DNA concentration decreased). This suggests that the effect of LAMP inhibitors on the gene amplification reaction was more significant than the impact of reduced DNA concentration. Cannabis resin contains many impurities such as polyphenolic compounds that inhibit DNA amplification; thus, the amplification efficiency can be improved by diluting the crudely extracted DNA solution.51 The optimal dilution condition was found to be 1000-fold, where the variability in Tt values was the smallest. In contrast, at a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold dilution (0.8 pg μL−1; 0.83 copies per μL), the DNA concentration approached the LOD of cannabis resin DNA, leading to greater variability in Tt values.
000-fold dilution (0.8 pg μL−1; 0.83 copies per μL), the DNA concentration approached the LOD of cannabis resin DNA, leading to greater variability in Tt values.
The results of real-time PCR and real-time fluorescent LAMP are compared in Table 2, and the DNA amplification curves are presented in Fig. S8.† In real-time PCR, a positive reaction was observed only at the 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold dilution, indicating that PCR is highly sensible to inhibiting substances. In contrast, for the fluorescent LAMP, only the 10-fold dilution condition yielded a negative result, showing good agreement with the results from the dilution device. These findings demonstrate that even when crudely extracted DNA has a significant amount of amplification inhibitors, the proposed dilution device enables the reliable detection of the target nucleic acids by generating the appropriate dilution factor in a single operation. In the future studies, the effectiveness of the proposed dilution device will be further demonstrated using various samples that may contain inhibitors of gene amplification reactions, such as nucleic acid samples crudely extracted from blood, saliva, urine, environmental water, and food extracts.
000-fold dilution, indicating that PCR is highly sensible to inhibiting substances. In contrast, for the fluorescent LAMP, only the 10-fold dilution condition yielded a negative result, showing good agreement with the results from the dilution device. These findings demonstrate that even when crudely extracted DNA has a significant amount of amplification inhibitors, the proposed dilution device enables the reliable detection of the target nucleic acids by generating the appropriate dilution factor in a single operation. In the future studies, the effectiveness of the proposed dilution device will be further demonstrated using various samples that may contain inhibitors of gene amplification reactions, such as nucleic acid samples crudely extracted from blood, saliva, urine, environmental water, and food extracts.
| Dilution factor | Real-time PCR | Real-time fluorescent LAMP | ||
|---|---|---|---|---|
| C t value | T t value (min) | |||
| Mean value | SD | Mean value | SD | |
| 10× | n.d. | n.d. | n.d. | n.d. |
| 100× | n.d. | n.d. | 22.0 | 0.8 |
| 1000× | n.d. | n.d. | 24.4 | 0.1 |
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000× 000× |
24.7 | 0.7 | 27.2 | 1.2 |
Conclusions
In this study, we developed a genetic diagnostic device capable of performing four-stepwise logarithmic dilutions for rapid and reliable detection of target nucleic acids in a single operation using the colorimetric LAMP method. We proposed a confluent point with a two-layer structure and microchannel heights of 90- and 10 μm, which facilitated the synchronization of the inflow timing of two liquids and prevented backflow, even when the volumetric flow rate ratio differed significantly (10- to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold). By optimizing the microchannel design, we developed a device capable of generating logarithmic dilutions and dispensing the mixed solution in parallel into five microchambers at each dilution factor. Consequently, compared with conventional ladder-type logarithmic dilution devices that require complex microchannel networks, we developed a parallel flow path configuration that generates each dilution under constant pressure, significantly simplifying the overall device design. Additionally, an asymmetric micromixer installed downstream of the merging section efficiently mixed both liquids under laminar flow conditions, ensuring an uniform concentration before simultaneously dispensing them into five microchambers of 3 μL each for gene amplification. Furthermore, to minimize the waste of valuable samples and reagents, a narrowed microchannel structure implemented at the outlet of the microchambers effectively prevented liquid leakage due to surface tension. We performed LAMP assays using purified cannabis seed DNA as a test sample on the fabricated PDMS-based four-stepwise logarithmic parallel dilution microfluidic device. The standard curve obtained from the device indicated that its detection sensitivity was equivalent to that of conventional turbidity-based LAMP assays. Moreover, we used the device to detect crudely extracted cannabis resin DNA, demonstrating reliable detection of target nucleic acids, even in the presence of a significant amount of gene amplification inhibitors, by automatically generating the appropriate dilution factor in a single operation. The proposed dilution device eliminates extensive and multiple manual procedures in molecular diagnostics. However, the connection procedure between the device and the pump remains somewhat laborious. Therefore, to further improve usability, future work will explore an integrated design wherein reagent and sample reservoirs are built into the device inlets, allowing for direct pressure-driven liquid delivery without the need for manual connection to an external pump.
000-fold). By optimizing the microchannel design, we developed a device capable of generating logarithmic dilutions and dispensing the mixed solution in parallel into five microchambers at each dilution factor. Consequently, compared with conventional ladder-type logarithmic dilution devices that require complex microchannel networks, we developed a parallel flow path configuration that generates each dilution under constant pressure, significantly simplifying the overall device design. Additionally, an asymmetric micromixer installed downstream of the merging section efficiently mixed both liquids under laminar flow conditions, ensuring an uniform concentration before simultaneously dispensing them into five microchambers of 3 μL each for gene amplification. Furthermore, to minimize the waste of valuable samples and reagents, a narrowed microchannel structure implemented at the outlet of the microchambers effectively prevented liquid leakage due to surface tension. We performed LAMP assays using purified cannabis seed DNA as a test sample on the fabricated PDMS-based four-stepwise logarithmic parallel dilution microfluidic device. The standard curve obtained from the device indicated that its detection sensitivity was equivalent to that of conventional turbidity-based LAMP assays. Moreover, we used the device to detect crudely extracted cannabis resin DNA, demonstrating reliable detection of target nucleic acids, even in the presence of a significant amount of gene amplification inhibitors, by automatically generating the appropriate dilution factor in a single operation. The proposed dilution device eliminates extensive and multiple manual procedures in molecular diagnostics. However, the connection procedure between the device and the pump remains somewhat laborious. Therefore, to further improve usability, future work will explore an integrated design wherein reagent and sample reservoirs are built into the device inlets, allowing for direct pressure-driven liquid delivery without the need for manual connection to an external pump.
Furthermore, the device, with multiple reaction chambers at each dilution factor, has the potential for simultaneous detection of multiple nucleic acid targets (DNA/RNA) by varying the primers or combinations of pre-spotted primers in each reaction chamber. Therefore, this device can be applied to on-site multiplexed genetic detection of a wide range of infectious agents, including viruses, bacteria, fungi, and parasites in human, animal, and plant samples. Additionally, it has applications in detecting foodborne pathogens, allergens, and illegal substances. Future research will further validate the device's effectiveness using practical samples that commonly contain amplification inhibitors, including target nucleic acid samples (DNA/RNA) crudely extracted from clinical specimens (blood, saliva, and urine), as well as environmental and food-related samples. Furthermore, we will focus on optimizing the device design to reduce its size and shorten the dilution process time, thereby enhancing its practical utility. Additionally, we aim to develop a portable system integrating a heating element for LAMP reactions and a CMOS camera for colorimetric analysis.
Data availability
The authors confirm that the data supporting the findings of this study are available within the main article and ESI.†Author contributions
Akira Miyajima; methodology, investigation, writing – original draft preparation, Fumiya Nishimura; methodology, investigation, Daigo Natsuhara; methodology, investigation, writing – review and editing, Yuka Kiba; methodology, investigation, Shunya Okamoto; writing – review and editing, supervision, Moeto Nagai; writing – review and editing, supervision, Tadashi Yamamuro; methodology, writing – review and editing, supervision, Masashi Kitamura; methodology, writing – review and editing, supervision, Takayuki Shibata; conceptualization, methodology, writing – review and editing, supervision, project administration, funding acquisition. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare no conflicts of interests.Acknowledgements
This research was partially supported by JSPS KAKENHI Grant Number JP24K00776. We would like to thank Editage (http://www.editage.com) for English language editing.References
- D. C. Florian, M. Odziomek, C. L. Ock, H. Chen and S. A. Guelcher, Sci. Rep., 2020, 10, 13663, DOI:10.1038/s41598-020-70465-5.
- M. A. Torres-Acosta, G. J. Lye and D. Dikicioglu, Biochem. Eng. J., 2022, 188, 108713, DOI:10.1016/j.bej.2022.108713.
- P. Dettinger, T. Kull, G. Arekatla, N. Ahmed, Y. Zhang, F. Schneiter, A. Wehling, D. Schirmacher, S. Kawamura, D. Loeffler and T. Schroeder, Nat. Commun., 2022, 13, 2999, DOI:10.1038/s41467-022-30643-7.
- K. Thurow, Anal. Bioanal. Chem., 2023, 415, 5057–5066, DOI:10.1007/s00216-023-04727-2.
- T. M. Keenan and A. Folch, Lab Chip, 2008, 8, 34–57, 10.1039/B711887B.
- K. W. Oh, K. Lee, B. Ahn and E. P. Furlani, Lab Chip, 2012, 12, 515–545, 10.1039/C2LC20799K.
- N.-T. Nguyen, S. A. M. Shaegh, N. Kashaninejad and D.-T. Phan, Adv. Drug Delivery Rev., 2013, 65, 1403–1419, DOI:10.1016/j.addr.2013.05.008.
- A. G. G. Toh, Z. P. Wang, C. Yang and N.-T. Nguyen, Microfluid. Nanofluid., 2014, 16, 1–18, DOI:10.1007/s10404-013-1236-3.
- X. Wang, Z. Liu and Y. Pang, RSC Adv., 2017, 7, 29966–29984, 10.1039/C7RA04494A.
- C. Y. Lee and L. M. Fu, Sens. Actuators, B, 2018, 259, 677–702, DOI:10.1016/j.snb.2017.12.034.
- N. L. Jeon, S. K. W. Dertinger, D. T. Chiu, I. S. Choi, A. D. Stroock and G. M. Whitesides, Langmuir, 2000, 16, 8311–8316, DOI:10.1021/la000600b.
- S. K. W. Dertinger, D. T. Chiu, N. L. Jeon and G. M. Whitesides, Anal. Chem., 2001, 73, 1240–1246, DOI:10.1021/ac001132d.
- K. Campbell and A. Groisman, Lab Chip, 2007, 7, 264–272, 10.1039/b610011b.
- H. A. Yusuf, S. J. Baldock, R. W. Barber, P. R. Fielden, N. J. Goddard, S. Mohr and B. J. T. Brown, Lab Chip, 2009, 9, 1882–1889, 10.1039/B823101J.
- N. L. Jeon, H. Baskaran, S. K. W. Dertinger, G. M. Whitesides, L. Van De Water and M. Toner, Nat. Biotechnol., 2002, 20, 826–830, DOI:10.1038/nbt712.
- D. Liu, L. Wang, R. Zhong, B. Li, N. Ye, X. Liu and B. Lin, J. Biotechnol., 2007, 131, 286–292, DOI:10.1016/j.jbiotec.2007.06.014.
- W. Siyan, Y. Feng, Z. Lichuan, W. Jiarui, W. Yingyan, J. Li, L. Bingcheng and W. Qi, J. Pharm. Biomed. Anal., 2009, 49, 806–810, DOI:10.1016/j.jpba.2008.12.021.
- N. Ye, J. Qin, W. Shi, X. Liu and B. Lin, Lab Chip, 2007, 7, 1696–1704, 10.1039/B711513J.
- J. Ruan, L. Wang, M. Xu, D. Cui, X. Zhou and D. Liu, Mater. Sci. Eng., C, 2009, 29, 674–679, DOI:10.1016/j.jpba.2008.12.021.
- J. Kim, D. Taylor, N. Agrawal, H. Wang, H. Kim, A. Han, K. Rege and A. Jayaraman, Lab Chip, 2012, 12, 1813–1822, 10.1039/C2LC21202A.
- W. Lim and S. Park, Molecules, 2018, 23, 3355, DOI:10.3390/molecules23123355.
- K. Lee, C. Kim, G. Jung, T. S. Kim, J. Y. Kang and K. W. Oh, Microfluid. Nanofluid., 2010, 8, 677–685, DOI:10.1007/s10404-009-0500-z.
- K. Lee, C. Kim, Y. Kim, B. Ahn, J. Bang, J. Kim, R. Panchapakesan, Y. K. Yoon, J. Y. Kang and K. W. Oh, Microfluid. Nanofluid., 2011, 11, 75–86, DOI:10.1007/s10404-011-0775-8.
- Y. Tang, M. Gan, Y. Xie, X. Li and L. Chen, Lab Chip, 2014, 14, 1162–1167, 10.1039/C3LC51332G.
- J. T. S. Fernandes, S. Tenreiro, A. Gameiro, V. Chu, T. F. Outeiro and J. P. Conde, Lab Chip, 2014, 14, 3949–3957, 10.1039/C4LC00756E.
- X. Jiang, J. M. K. Ng, A. D. Stroock, S. K. W. Dertinger and G. M. Whitesides, J. Am. Chem. Soc., 2003, 125, 5294–5295, DOI:10.1021/ja034566+.
- C. Kim, K. Lee, J. H. Kim, K. S. Shin, K.-J. Lee, T. S. Kim and J. Y. Kang, Lab Chip, 2008, 8, 473–479, 10.1039/B714536E.
- K. Lee, C. Kim, B. Ahn, R. Panchapakesan, A. R. Full, L. Nordee, J. Y. Kang and K. W. Oh, Lab Chip, 2009, 9, 709–717, 10.1039/B813582G.
- K. Lee, C. Kim, Y. Kim, K. Jung, B. Ahn, J. Y. Kang and K. W. Oh, Biomed. Microdevices, 2010, 12, 297–309, DOI:10.1007/s10544-009-9385-6.
- K. Hattori, S. Sugiura and T. Kanamori, Lab Chip, 2009, 9, 1763–1772, 10.1039/B816995K.
- S. Sugiura, K. Hattori and T. Kanamori, Anal. Chem., 2010, 82, 8278–8282, DOI:10.1021/ac1017666.
- M. Hosokawa, T. Hayashi, T. Mori, T. Yoshino, S. Nakasono and T. Matsunaga, Anal. Chem., 2011, 83, 3648–3654, DOI:10.1021/ac2000225.
- G. Zheng, L. Lu, Y. Yang, J. Wei, B. Han, Q. Zhang and Y. Wang, Anal. Chem., 2018, 90, 13280–13289, DOI:10.1021/acs.analchem.8b02597.
- C.-Y. Chen, A. M. Wo and D.-S. Jong, Lab Chip, 2012, 12, 794–801, 10.1039/C1LC20548J.
- P. Occhetta, M. Centola, B. Tonnarelli, A. Redaelli, I. Martin and M. Rasponi, Sci. Rep., 2015, 5, 10288, DOI:10.1038/srep10288.
- M. R. Stehnach, R. J. Henshaw, S. A. Floge and J. S. Guasto, eLife, 2023, 12, e85348, DOI:10.7554/eLife.85348.
- A. V. Nguyen, M. Yaghoobi, M. Azizi, M. Davaritouchaee, K. W. Simpson and A. Abbaspourrad, Commun. Eng., 2023, 2, 15, DOI:10.1038/s44172-023-00064-5.
- H. Shi, Z. Xie, Y. Cao, Y. Zhao, C. Zhang, Z. Chen, N. M. Reis and Z. Liu, Chem. Eng. Sci., 2022, 263, 118080, DOI:10.1016/j.ces.2022.118080.
- D. Vloemans, A. Pieters, F. D. Dosso and J. Lammertyn, Lab Chip, 2024, 24, 2791–2801, 10.1039/D4LC00195H.
- D. Natsuhara, K. Takishita, K. Tanaka, A. Kage, R. Suzuki, Y. Mizukami, N. Saka, M. Nagai and T. Shibata, Micromachines, 2020, 11, 540, DOI:10.3390/mi11060540.
- S. Misawa, D. Natsuhara, Y. Kiba, T. Yamamuro, R. Suzuki, T. Shibata and M. Kitamura, Forensic Toxicol., 2021, 39, 259–265, DOI:10.1007/s11419-020-00557-4.
- D. Natsuhara, R. Saito, H. Aonuma, T. Sakurai, S. Okamoto, M. Nagai, H. Kanuka and T. Shibata, Lab Chip, 2021, 21, 4779–4790, 10.1039/D1LC00829C.
- D. Natsuhara, A. Miyajima, T. Bussho, S. Okamoto, M. Nagai, M. Ihira and T. Shibata, Analyst, 2024, 149, 3335–3345, 10.1039/D4AN00215F.
- D. Natsuhara, S. Misawa, R. Saito, K. Shirai, S. Okamoto, M. Nagai, M. Kitamura and T. Shibata, Sci. Rep., 2022, 12, 12852, DOI:10.1038/s41598-022-16945-2.
- D. Natsuhara, Y. Kiba, R. Saito, S. Okamoto, M. Nagai, Y. Yamauchi, M. Kitamura and T. Shibata, RSC Adv., 2024, 14, 22606–22617, 10.1039/D4RA04055D.
- United Nations Office on Drug and Crime, World Drug Report 2024, Contemporary Issues on Drugs, https://www.unodc.org/documents/data-and-analysis/WDR_2024/WDR24_Contemporary_issues.pdf, (accessed April 2025).
- M. Kitamura, M. Aragane, K. Nakamura, K. Watanabe and Y. Sasaki, Biol. Pharm. Bull., 2016, 39, 1144–1149, DOI:10.1248/bpb.b16-00090.
- M. Kitamura, M. Aragane, K. Nakamura, T. Adachi, K. Watanabe and Y. Sasaki, Biol. Pharm. Bull., 2018, 41, 1303–1306, DOI:10.1248/bpb.b18-00272.
- D. Natsuhara, R. Saito, S. Okamoto, M. Nagai and T. Shibata, Micromachines, 2022, 13, 1386, DOI:10.3390/mi13091386.
- C. E. Johnson, A. Premasuthan, J. S. Satkoski Trask and S. Kanthaswamy, J. Forensic Sci., 2013, 58, 486–490, DOI:10.1111/1556-4029.12055.
- T. Yamamuro, Y. T. Iwata, H. Segawa, K. Kuwayama, K. Tsujikawa, T. Kanamori and H. Inoue, Forensic Sci. Int., 2018, 287, 176–182, DOI:10.1016/j.forsciint.2018.03.044.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5lc00356c |
| This journal is © The Royal Society of Chemistry 2025 |