Open Access Article
Open Access ArticleExploring neuronal circuitry in neurodegenerative diseases: from traditional models to cutting-edge techniques
Chiara
Ausilio
abc,
Annachiara
Scalzone
a and
Paolo Antonio
Netti
 *abc
*abc
aCenter for Advanced Biomaterials for Healthcare@CRIB, Istituto Italiano di Tecnologia (IIT), 80125 Naples, Italy. E-mail: paolo.netti@iit.it
bDepartment of Chemical, Materials and Industrial Production Engineering (DICMAPI), University of Naples Federico II, P.le Tecchio 80, Naples 80125, Italy
cInterdisciplinary Research Centre on Biomaterials (CRIB), University of Naples Federico II, P.le Tecchio 80, Naples 80125, Italy
First published on 1st July 2025
Abstract
Current treatments of neurodegenerative diseases primarily address symptoms rather than halting pathology progression. This gap is due to the lack of effective methods for monitoring neural circuitry and dynamics over time. In this context, the development of in vitro models that more accurately replicate the human brain microenvironment has become essential. Traditional two-dimensional (2D) cell cultures, while providing valuable insights, fail to capture the intricate complexity of the human brain. Recent advancements in neuroscience spotlight the emergence of more sophisticated three-dimensional (3D) models, which can more faithfully recapitulate the intricacies of the brain. This review discusses the evolution of in vitro brain models, emphasizing the transition from traditional 2D cultures to sophisticated 3D systems, including neurospheroids, brain organoids, assembloids and micro-tissue engineered neuronal networks (micro-TENNs). We further highlight the emergence of brain-on-chip platforms, combining microfluidics with cell culture technologies to create precisely controlled environments mimicking the physiological conditions of the human brain. Furthermore, we discuss the application of 3D bioprinting technology enabling the generation of neural constructs with precise control over cell placement. Lastly, we delve into the potential of integrating brain organoids with 3D bioprinting technology, aiming to recapitulate the true three-dimensional complexity of the brain, thereby improving the physiological accuracy of brain models for advancing our understanding of neurodegenerative diseases.
Introduction
Neurodegenerative diseases significantly impact society by severely diminishing quality of life. Existing treatments mainly address symptoms rather than halting disease progression, largely due to our poor understanding of how brain diseases begin and develop. This gap in knowledge stems from the lack of methods to monitor neural circuitry dynamics over time.1 Consequently, the development of functional three-dimensional (3D) models in neuroscience is propelled by the imperative need to unravel the intricacies of the brain. Indeed, this sophisticated and fascinating organ embraces billions of electrically excitable neurons and supporting glial cells (astrocytes, microglia, and oligodendrocytes), working together to process and transport information and controlling several body functions.2 Moreover, the brain is able to dynamically respond, evolve and reshape its function in response to external and internal stimuli. This mechanism is mediated by synapses, connections between neurons, which change in strength and structure in response to experience or activity.3 This process, defined as synaptic plasticity, is critical for learning and memorizing and its dysregulation has been implicated in a range of neurological and psychiatric disorders. Driven by the complexity and the adaptive behaviour of the human brain, advances in cell culture techniques, tissue engineering and microfabrication have enabled the creation of increasingly sophisticated and biologically relevant in vitro systems. Several approaches have been exploited ranging from traditional two-dimensional (2D) cell cultures, which offer simplicity and ease of use, all the way up to more advanced 3D models that better emulate the brain cellular composition, architecture, and dynamics. Nevertheless, current methodologies fall short in facilitating a comprehensive correlation of in vitro and in vivo features and findings. Herein, we will briefly review the current state of art and limitations of 2D in vitro models for investigating neuronal behaviour. We will then report how 3D models, which more accurately replicate the architecture of the brain, provide enhanced opportunities to explore neuronal function and connectivity under both healthy and pathological conditions.4Finally, we will discuss the significant advancement in brain-on-chip (BoC) platforms, highlighting the potential of combining these models with cutting-edge manufacturing technology, such as 3D bioprinting. This integration aims to replicate the complex architecture of the brain in a highly controlled manner, thereby enhancing the biomimetic potential of neuro-hybrid systems.5
Cellular players in brain dynamics
The brain's complexity and functionality arise from the tuned interactions among its various cells and the surrounding extracellular components (Fig. 1B). Among these, the extracellular space (ECS), a narrow microenvironment enveloping all brain cells, plays a crucial role in modulating ionic diffusion and chemical communication, thereby enabling neuron excitability and the propagation of electrical signals. Neurons, as the primary signaling cells, communicate through a combination of electrical impulses and chemical synapses supporting rapid communication across neural networks.6 This signaling is further modulated by synaptic plasticity mechanisms, including long-term potentiation and depression, which are fundamental to learning and memory processes.7 Equally critical to brain function is the vast population of glial cells, making up approximately half of the cellular content in the mammalian central nervous system (CNS). Far from being passive supporters, glial cells actively interact with neurons through chemical signaling, mainly providing nutrition and tropic support.8,9 For instance, astrocytes play a crucial supporting role by maintaining the integrity of blood–brain barrier, regulating cerebral blood flow, and modulating synaptic transmission. They release gliotransmitters, influencing neuronal communication and contributing to synaptic plasticity and homeostatic balance.10 Oligodendrocytes, another major glial subtype, produce myelin sheaths that insulate axons, facilitating the rapid conduction of electrical impulses. Myelination is crucial not only for signal velocity but also for synchronizing neuronal firing across distant brain regions.11 Meanwhile, microglia act as brain's resident immune cells, constantly surveying the neuronal environment and responding to injury, infection, or pathological changes.12 The dynamic interactions between these cells within the brain are essential for maintaining homeostasis and adaptability, whether during development, learning, or injury.13–16Indeed, understanding how specific neural cell types contribute to brain homeostasis, provides critical insight into the pathogenesis of neurodegenerative diseases such as Alzheimer's disease (AD), Parkinson's disease (PD), and multiple sclerosis (MS), each of which arises from distinct cellular dysfunctions.
For instance, in AD, the pathological hallmarks include the extracellular deposition of β-amyloid (Aβ) plaques and the intracellular accumulation of hyperphosphorylated tau protein in neurofibrillary tangles (Fig. 1D). These aggregates disrupt synaptic communication and neuronal signaling, leading progressively to cell dysfunction and death, particularly in the hippocampus and cortical areas. Notably, astrocytes respond to Aβ deposition by becoming reactive, releasing pro-inflammatory cytokines (such as IL-1β and TNF-α) and reactive oxygen species that intensify synaptic dysfunction. Simultaneously, microglia initially exert a neuroprotective function by attempting to clear Aβ via receptors like TREM2 and CD36. However, chronic stimulation drives them toward a persistent pro-inflammatory state, marked by excessive cytokine secretion and oxidative stress, which exacerbates neuronal degeneration. Although oligodendrocytes are not primary targets in AD, their impaired function in response to inflammation may compromise axonal support and contribute indirectly to disease progression.
Similarly, in PD, the degeneration of dopaminergic neurons in the substantia nigra pars compacta is closely associated with the intracellular accumulation of misfolded α-synuclein in Lewy bodies (Fig. 1E). This disrupts intracellular trafficking, impairs mitochondrial function, and reduces dopamine release in basal ganglia circuits, ultimately producing characteristic motor symptoms. In this context, astrocytes display a reactive phenotype that limits their neurotrophic support and leads to impaired glutamate clearance, thereby increasing excitotoxic stress on neurons. At the same time, microglia become activated in response to α-synuclein and dying neurons, releasing inflammatory mediators that contribute to a neurotoxic environment and further dopaminergic cell loss. While the role of oligodendrocytes in PD is less well defined, evidence suggests that their dysfunction may be involved in white matter abnormalities and reduced metabolic support to neurons, potentially aggravating disease pathology.
In contrast, MS is primarily driven by chronic immune-mediated demyelination within the CNS. The autoimmune attack specifically targets oligodendrocytes, leading to the loss of myelin sheaths and resulting in impaired axonal conduction, neuronal damage, and functional deficits (Fig. 1F). As a consequence, microglia are rapidly recruited and participate in antigen presentation, phagocytosis of myelin debris, and amplification of inflammatory signals, contributing to lesion formation and chronic neuroinflammation. Simultaneously, astrocytes undergo hypertrophy and form glial scars that hinder axonal regeneration and remyelination. Their release of cytokines and extracellular matrix molecules further shapes the lesion microenvironment, often in ways that limit repair. Oligodendrocyte loss remains the central pathological event in MS, but their regenerative failure is compounded by the inflammatory environment and the inhibitory actions of reactive glial cells.
Taken together, these observations illustrate the disease-specific yet interconnected roles of neurons and glial cells across AD, PD, and MS. Understanding these complex interactions provides a robust foundation for investigating how cellular dysfunction contributes to neurological disorders. Recent advancements in in vitro platforms have significantly enhanced our ability to study the coordinated interplay between brain cells in neurological disorders. These advanced models replicate key aspects of brain architecture and function, facilitating detailed investigations of cellular interactions and underlined disease mechanisms.
Exploring the dynamic role of brain extracellular space in health and disease
The formation of a functional nervous system requires neurons to respond to an array of biochemical, mechanical, and topographical signals within the brain's ECS. ECS is the narrow microenvironment that surrounds every brain cell and contains a solution that closely resembles cerebrospinal fluid with the addition of extracellular matrix (ECM) molecules. This latter is a dynamic intricate framework consisting of proteins and carbohydrates, mechanically and biochemically directing cell behavior.17 ECM can be broadly classified into two components: a structured phase and an amorphous one. The structured component consists of elastin, laminins, collagen which form an organized scaffold. In contrast, the less organized amorphous component is composed of hyaluronic acid (HA), proteoglycans, tenascins, link proteins, and glycoproteins, which fill the interstitial spaces.18 Among the proteins, fibronectin is directly involved in cell adhesion influencing cell migration, morphogenesis, and proliferation, while collagen type IV is the most abundant fibrous protein acting as a mechanical scaffold, providing an adhesive substrate for neurite outgrowth. Laminin facilitates neural migration and serves as a scaffold for axon guidance. Tenascins, occurring in ‘C’ and ‘R’ forms, can either promote or inhibit axon guidance. Additionally, HA, which is a simple glycosaminoglycan (GAG), interacts with cell surface receptors, impacting neural precursor cell migration and guiding axons. Finally, proteoglycans, core proteins often anchored to the apical membrane of endothelial cells, play a central role in inhibiting nerve growth, limiting plasticity, and fostering neural repair. Especially, chondroitin sulphate proteoglycan (CSPG) regulates neural stem cell proliferation and inhibits growth cone sprouting. Understanding the intricate composition of the ECM is not only essential for comprehending normal brain function, but also for unravelling the mechanisms underlying neurodegeneration.18–21Beyond its structural and biochemical roles, the ECS is a critical medium for ionic streaming, which underpins the electrical activity of neurons and forms an intercellular chemical communication channel. Ionic streaming involves the movement of key ions, including sodium (Na+), potassium (K+), calcium (Ca2+), and chloride (Cl−), through the ECS, creating ionic gradients essential for maintaining neuronal excitability and signal propagation. These ionic currents facilitate the generation and transmission of action potentials, allowing neurons to communicate rapidly and precisely. Additionally, the modulation of ion concentrations in the ECS is fundamental for synaptic plasticity, as changes in Ca2+ levels at synapses regulate neurotransmitter release and post-synaptic response. The interplay between the ECM and the ECS is particularly significant, as ECM molecules, such as CSPGs, can influence ion fluxes and synaptic function by modulating the availability of binding sites for ions and neurotransmitters. Emerging evidence highlights that the disruptions in the ionic balance of the ECS and the ECM's structural integrity are implicated in neurological disorders.19 For example, in AD, alterations in the ECM, particularly in the levels and sulfation patterns of proteoglycans and GAGs, such as chondroitin sulfate and heparan sulfate, can modulate Aβ aggregation kinetics and tau pathology.20 Here, the interactions between ECM and ECS create a dynamic microenvironment that modulates the diffusion and spread of Aβ and tau aggregates, thereby facilitating their accumulation and exacerbating neuroinflammation. This interplay not only promotes protein misfolding and aggregation but also contributes to the disruption of neuronal signaling and the inflammatory response, accelerating disease progression. Similarly, in MS, the ECM regulates the migration, proliferation, and differentiation of oligodendrocyte progenitor cells (OPCs), which are responsible for the myelination process.21 Alterations in the ECM components, such as the accumulation of HA, laminin, tenascins and CSPGs can create a non-permissive microenvironment for OPCs. This in turn, impedes their differentiation and leads to dysfunctional interactions between ECM and ECS, which hinders remyelination. These ECM–ECS interactions exacerbate the inflammatory response in MS by affecting oligodendrocyte migration and proliferation, which are crucial for repair after demyelination. On the other hand, in PD the alteration of CSPGs in the ECM has been shown to inhibit neurogenesis, reducing the formation of new neural connections and impeding repair mechanisms.22 These alterations in ECM influence the ECS environment, leading to impaired synaptic plasticity and neurogenesis, and creating barriers to the regeneration of dopaminergic neurons. The ECM–ECS interplay further exacerbates neuronal dysfunction, which contributes to the progressive neurodegeneration observed in PD.
Lastly, in complex and multifactorial psychiatric disorders like schizophrenia, changes in ECM such as abnormal levels of CSPGs can affect synaptic stabilization and plasticity.23 These changes affect neuronal connectivity by modifying the ECM–ECS interactions that regulate synaptic structure and function. Altered ECM components disrupt the balance of neuronal signaling, leading to impaired connectivity between brain regions, a hallmark feature of schizophrenia.
It is worth mentioning that during neurodegeneration, the complex interaction between neuronal dysfunction and the surrounding microenvironment is bidirectional. As healthy neurons are lost, they disrupt the supportive ECM structure, but recent research suggests that damaged neurons also actively remodel their microenvironment.24 This remodeling process often involves the dysregulation of growth factors and proteases, which lead to the degradation of ECM components. Understanding these mechanisms is crucial for elucidating how impaired neurons not only contribute to their own degeneration but also perpetuate the pathological changes in the ECM that further exacerbate disease progression.25 In AD, for example, altered expression of neuronal matrix metalloproteinases (MMPs) has been linked to the accumulation of Aβ plaques, which in turn contribute to a hostile environment that exacerbates neuronal damage.26 Similarly, in PD, the loss of dopaminergic neurons is associated with changes in the ECM, including reduced levels of laminin, which is crucial for neuronal survival and function.27 Finally, in amyotrophic lateral sclerosis (ALS), studies suggest that dysfunctional motor neurons can trigger the activation of astrocytes, glial cells that can remodel the ECM and contribute to neuroinflammation, further accelerating disease progression.28
Given the well-established role of the crosstalk between neuronal cells and their microenvironment in maintaining brain health and its impact on neurodegeneration, it is evident that targeting key ECM components presents promising opportunities for effective therapeutic intervention. Such strategies could indirectly modulate cellular responses and restore the microenvironment, potentially slowing disease progression. In this regard, the development of reliable in vitro brain models has been essential for elucidating the ECM role in disease progression, as well as for exploring the fundamental mechanisms of ionic streaming in neural signaling and identifying potential therapeutic targets.
Unveiling modern perspectives
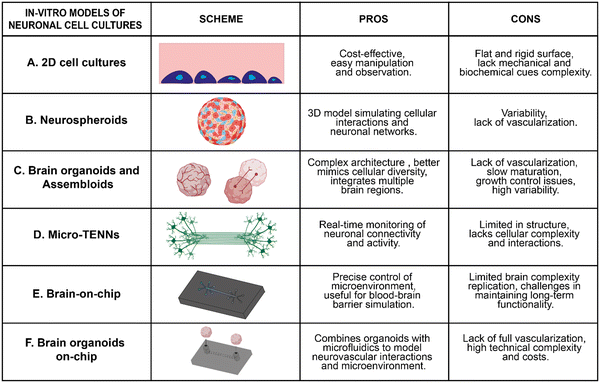
Driven by the intricacies of the human brain and the crucial role of its microenvironment, several strategies have been introduced to deeply investigate the architecture and connectivity of neuronal networks.29 Historically, the easy observation and manipulation of 2D cell cultures (Fig. 2A) has made them a widely used method to study cellular responses to physical, chemical and electrical signals, in a controlled environment.30 For instance, 2D cultures have been extensively used to explore the effects of substrate surface chemistry on neuronal behavior, revealing how positively and negatively charged surfaces, as well as protein-coated surfaces, impact neuronal viability, adhesion, and proliferation.31,32 Similarly, micro and nanostructures within these systems have shown how surface topography can dictate neuronal adhesion and modulate axon elongation and branching.33 Additionally, 2D platforms have demonstrated the potential of conductive materials to facilitate seamless integration between neurons and artificial devices by promoting cellular recognition of engineered interfaces as biologically compatible surfaces.34However, cellular behavior in 2D cultures often diverges significantly from that observed in the complex 3D microenvironment of the brain. In 2D systems, cells grow on flat, rigid substrates that fail to replicate the mechanical and biochemical cues present in native neural tissue. This artificial geometry alters cell polarity, reduces the formation of physiologically relevant cell–cell and cell–matrix interactions, and limits synaptic connectivity. To overcome these gaps, researchers have developed 3D models that better recapitulate brain architecture and function. These include self-organizing spheroids and brain organoids (BOs), microfluidic “brain-on-a-chip” devices, and other bioengineered neural tissues. Together, such models provide structural and functional advances for studying development, disease, and even cognition in controlled settings.35
For instance, neurospheroids (Fig. 2B) have emerged as robust platforms for high-throughput screening studies, particularly useful in disease modelling, drug testing as well as in the investigation of tumour invasion.36–39
Perhaps the most emblematic modern in vitro model is the human brain organoid (BO) (Fig. 2C). BO are 3D clusters of neural cells grown from stem cells that self-organize into brain-like structures.40–42 They can mimic aspects of early human brain development at cellular, structural, and functional levels. For instance, cerebral organoids have been shown to form discrete brain regions and even exhibit spontaneous neuronal electrical activity analogous to fetal brain patterns.43 These features make organoids powerful for disease modeling and personalized medicine. In this context, organoids derived from AD patients' iPSCs can develop hallmark Aβ plaques and tau tangles in vitro, recapitulating key disease pathology for drug testing.44–47 Apart from being reliable and valuable platforms for studying neurological disease, BOs could also serve as a scalable source for cell replacement therapy and facilitate the investigation of viral infection pathogenesis.48–50
In addition to BOs, researchers have developed brain assembloids, which are even more sophisticated models that combine multiple region-specific organoids to model inter-regional connectivity. For example, joining dorsal and ventral forebrain organoids allowed researchers to observe GABAergic interneurons migrating between regions, recreating developmental migration patterns.42,51 These platforms have also offered significant findings regarding tumour mechanisms, which could be further explored.52 Notably, brain glioblastoma cells by integrating tumour spheroids with cerebral organoids, enabling the observation of tumour infiltration in a human-like 3D context.53 This approach allows for the analysis of tumour–microenvironment interactions, response to therapies, and identification of molecular drivers of malignancy in a more physiologically relevant setting.
Such advances demonstrate how organoids provide more physiologically relevant neural models compared to conventional cell cultures.
Furthermore, recent studies have successfully replicated the interconnectivity between different cortical areas by connecting neuronal ensembles with bundles of reciprocally extending axons, engineering the so-called micro-tissue engineered neural networks (micro-TENNs) (Fig. 2D).54–57 These systems typically feature a central core filled with ECM components (e.g. collagen, collagen–laminin, or fibrin), which provide essential support for axonal growth and function. An exterior agarose hydrogel shell encases the entire structure, providing a defined geometrical framework. Micro-TENNs have been largely used for studying growth processes and functional characteristics relevant to nervous system reconstruction, offering insights into neurological function and dysfunction, particularly in neurodegenerative diseases.58–60
Despite these achievements, model validation and limitations remain active areas of research. BOs, for example, still lack certain hallmarks of mature brains. They often do not develop the fully stratified cortical layers or gyrification seen in vivo, and their neuronal circuits are relatively rudimentary compared to an adult brain.43 Without blood vessels, organoids also face an oxygen/nutrient diffusion limit, causing cell death in their cores beyond ∼200–300 μm size.61,62 This inherent lack of vascularization not only limits organoid size but also makes it challenging to model late-onset neurodegenerative diseases that require long maturation.63 To address these issues, scientists are experimenting with vascularization strategies, such as embedding human endothelial cells into organoids to form capillary-like networks,64,65 or guided patterning approaches using bioengineering or genetic tools to induce layering and region-specific differentiation.43 Advances in genome engineering have also enabled more controllable organoids. For instance, introducing reporters or mutations to track and manipulate specific cell population. Looking ahead, researchers emphasize improving cross-talk between different tissues (for multi-organ interactions), applying spatiotemporal control over organoid development, and standardizing protocols to reduce variability.
Another modern paradigm is the integration of microfluidic technologies revolutionizing the development of BoC platforms (Fig. 2E).66–68 These devices use tiny engineered channels and chambers to provide a close-to-natural microenvironment with fluid perfusion, gradients, and real-time sensing. Moreover, BoC models enable fine-tuned control and observation of neuronal networks (e.g., applying shear flow or chemical gradients), bridging the gap between simplistic in vitro assays and the complex in vivo brain. BoC technology encompasses a variety of approaches, from brain-cells-on-chip (dissociated neurons in microfluidic networks) and brain-slices-on-chip (living brain tissue slices sustained on perfused chips) to brain organoids-on-chip (BOoC) (culturing mini-brains within microfluidic devices) (Fig. 2F).69–74 Despite different implementations, all such systems leverage microfluidics to maintain high cell viability over long periods and to incorporate sensors for continuous monitoring. For example, a BoC can integrate neuronal cells with engineered microenvironments and on-chip electrodes, yielding a structurally diverse, highly controlled model suitable for drug screening or toxicity tests. These chips also allow compartmentalization, such as modeling a blood–brain barrier by co-culturing brain endothelial cells with neurons and glia in adjacent channels.75 BoC technology has also advanced the accurate modeling of neurological diseases such as AD,PD and epilepsy, providing valuable insights into these conditions.76
Recent research has also highlighted the potential of interconnected BOs within these microfluidic platforms for investigating the roles of macroscopic circuits in the human brain. By interconnecting multiple organoids via engineered neural connections, it is possible to create complex and functional models for studying higher-order brain functions, such as memory, learning, and decision-making, potentially leading to a deeper understanding of neurological and psychiatric disorders.77 Indeed, the concept of “organoid intelligence” (OI) has been introduced, as BOs hold the potential to mimic key molecular and cellular aspects of learning, memory, and cognition.78 In this regard, Kagan et al. have embedded monolayers of cortical neurons in a real-time closed-loop environment using electrophysiological stimulation and recording.79 These cultures self-organized and rapidly adapted their activity, exhibiting goal-directed behavior within a simulated game environment. In this direction, ongoing challenges aim to develop biocomputing models through stimulus–response training and interfaces between BOs and computers.80 Future research will likely focus on assessing the learning capacity of BOs for computational purposes, utilizing biological learning patterns. The final goal of all these studies is to establish interfaces between organoids' models, including dissociated organoids, sliced organoids, and interconnected organoids, and computers, enabling supervised learning simulations through trained stimulus–response patterns.81,82 In this regard, high density-micro-electrode arrays (HD-MEAs), 3D bioelectronics, and flexible bioelectronics have been largely discussed for detailed exploration of BOs.83,84 Such findings underscore the remarkable progress in creating in vitro neuronal models that not only structurally resemble the brain but also perform complex, integrative functions. As these models continue to mature, they are poised to become indispensable for studying human neural development, neurodegenerative disease mechanisms, drug efficacy, and even the neural basis of behavior, all without relying on in vivo experiments.
Building brains: innovating the brain study with 3D bioprinting
While the abovementioned traditional 3D models have provided valuable insights into brain structure and function, they often lack the complexity and precision needed to fully replicate the intricate neural networks characteristic of the human brain. To address these limitations, recent studies emphasize the need for advanced and more controlled techniques to better mimic the complexity of the brain.85 In this scenario, 3D bioprinting offers a complementary strategy to engineer brain tissue by actively patterning cells and biomaterials into desired architectures.86 In 3D bioprinting, living cells (such as neurons, glia, or stem-cell-derived precursors) are mixed with a supportive hydrogel matrix to obtain a bioink, which can be printed to build a tissue construct. This technology grants extraordinary spatial control, by precisely placing multiple cell types in defined 3D arrangements, potentially recreating the layered structure of cortex or the organization of neural circuits.86 Unlike organoids, which rely on self-assembly and can be variable, bioprinting enables defined reproducible tissue geometry. For example, it is possible to print neuronal progenitors in a grid or a layered pattern and include supportive cells (astrocytes, oligodendrocytes) at strategic locations.87,88 Indeed, recent work has shown that bioprinting can produce complex neural tissue models that closely mimic the native brain's architecture and cell composition. These printed tissues are valuable for faithfully modeling brain function, screening drugs, and even exploring regenerative therapies, as they can be made patient-specific using iPSC-derived cells.More in details, several bioprinting strategies are being exploited for neural bioprinting. Extrusion-based printing uses a continuous flow of bioink extruded through a nozzle to directly write patterns on a substrate in a layer-by-layer fashion.89 While it allows for high cell densities and the obtainment of intricate structures, achieving uniform cell distribution within the bioprinted construct and maintaining its structural integrity remain ongoing challenges. Inkjet bioprinting deposits droplets of bioink containing cells with or without biomaterials onto a substrate in a precise manner, offering high printing speed and resolution at a lower cost. However, it has limitations in the viscosity range of bioinks.90 Lastly, laser-assisted bioprinting (LAB) uses laser pulses to propel cells and biomaterials from a donor slide to a receiving substrate, providing high precision and control over cell placement, though it involves a complex setup and potential thermal damage to cells.91 Alternative approach beyond these established methods have been developed and explored lately. Embedded bioprinting, for example, involves printing cells within a supportive hydrogel matrix (e.g. Pluronic F127, gelatin, Carbopol), which provides temporary structural support during the printing process and can be removed post-printing to yield delicate neural structures.92 This method offers high resolution and the ability to create complex structures geometries, but it requires careful optimization of the hydrogel properties to ensure optimal cell viability and functionality.
Apart from the selection of the bioprinting technique, also the bioink selection and optimization are crucial for the bioprinting process. In the context of in vitro brain models, bioinks for neural tissue engineering need to possess three main properties: (i) printability to ensure precise deposition of cells and biomaterials during the process of bioprinting; (ii) cytocompatibility, to ensure the bioink does not negatively impact neural cell viability, proliferation, and differentiation; and (iii) bio-instructiveness, which goes beyond simply mimicking the ECM, in fact it focuses on actively promoting desired cellular behaviors within the bioprinted construct.
Indeed, hydrogels must be carefully formulated: if the printed gel is too soft, the structure may collapse, but if it's too stiff, neurons cannot extend neurites or form synapses effectively.87,93 Early attempts often printed a supporting scaffold first (e.g. a 3D-printed polymer frame) and then seeded neurons onto it, but this indirect approach led to uneven cell distribution and physical barriers that impeded network formation between printed layers. However, new bioink strategies are overcoming these issues. For example, one recent study identified an optimal hydrogel formulation using fibrin as a base material, providing a biologically compatible, soft matrix for neurons, which was blended with hyaluronic acid to increase viscosity. This fibrin-based bioink has allowed printing human neural progenitor cells in a fibrin/gel construct differentiated into neurons that expressed mature markers (MAP2, NeuN), formed dense networks made of extensive connection characterized by synaptic puncta (vGlut1, Synapsin) observable within weeks.87
Advancements in bioinks specifically designed to mimic brain environment have further improved the structural integrity and signaling cues necessary for brain organoid constructs development. Hydrogels enriched with growth factors simulate the brain's ECM, promoting optimal cell viability and function. Recent research has focused on optimizing hydrogels like alginate, gelatine, collagen, fibrin, HA, and laminin to support 3D culture of neuronal cells and tissues.94 These hydrogels can be tailored to modulate their mechanical properties, promoting the differentiation of stem cells into specific neural lineages.95 Moreover, the use of multi-material bioprinting allows for the fabrication of tissues with distinct regions, closely mimicking the heterogeneity of brain. This enables the study of inter-regional communication and complex neurological processes with a new level of detail.
In disease modeling, bioprinted constructs offer a powerful tool for recreating complex neural environments that are essential for understanding the mechanisms underlying neurological disorders such as AD, PD, and MS.88 For example, in AD models, bioprinting allows spatially controlled deposition of neurons and glial cells within amyloid-rich matrices to mimic plaque formation and neuroinflammatory cascades. In PD, constructs enriched with dopaminergic neurons derived from iPSCs can reproduce key features of substantia nigra degeneration and enable the study of synaptic loss and α-synuclein aggregation. For MS, 3D printed constructs incorporating oligodendrocytes and myelinated axons provide an in vitro platform to investigate demyelination and remyelination processes under autoimmune-like conditions.96 These models enable more predictive screening of therapeutic compounds in physiologically relevant environments compared to abovementioned 3D model.97 In the table below (Table 1) are summarized recent articles reporting the use of bioprinting techniques for generating neural models; articles are categorized according to the exploited bioprinting technique and for each study the bioink formulation (cells and biomaterials), the application (healthy or disease) and any limitations encountered are reported. The fidelity of bioprinted models is continuously improving and bioprinting holds great promise for generating high-throughput, standardized process for producing physiologically accurate brain constructs, necessary for advancing our understanding of the nervous system and developing innovative treatments for neurological conditions. However, several key challenges remain in overcoming the limitations of each bioprinting technique, optimizing bioprinting parameters and optimizing bioinks to support high cell viability, differentiation, and functional integration, ensuring nutrient and oxygen supply through vascularization for the long-term viability of printed neural constructs. Equally important to note are the current limitations of 3D bioprinting in neuroscience. Maintaining viability and guiding maturation in thick, complex printed tissues remains challenging. Printed constructs with the lack of vasculature face the same diffusion limits as organoids, so researchers either print built-in microchannels or keep designs thin for now.86 Achieving the full cellular diversity of brain tissue including microglia, vasculature, and other components, in a single print is a complex challenge. Bioprinted models focus on specific subsets of cells such as neurons and astrocytes. Additionally, resolution limitations make it difficult to print extremely fine neural microarchitectures, like minuscule cortical columns, with current technology. Nevertheless, rapid progress is being made on all these points. A 2024 comprehensive review highlights that researchers are now systematically addressing such challenges by analyzing which printing methods work best for neural cells, formulating new bioinks tailored for neural stem cells, and even experimenting with stimuli-responsive materials to encourage proper organization over time.93 The field is moving towards creating brain-like constructs that incorporate multiple cell types and functionalities, possibly even printing supporting vasculature alongside neurons (using coaxial printing of endothelial cells, for example). However, challenges remain, such as limited printing resolution, the need for further optimization of bioinks for delicate stem and progenitor cells, and maintaining high cell viability during and after printing. Additionally, the lack of standardized post-printing maturation protocols and reproducibility across different platforms limits broader application.
| Bioprinting technique | Cells | Biomaterials | Application | Limitation | Ref. |
|---|---|---|---|---|---|
| Extrusion bioprinting | hiPSC-derived spinal neuronal progenitor cells (NPCs) and oligodendrocyte progenitor cells | Bioinks: Matrigel; gelatin, fibrin; gelatin methacrylate (GelMa), PI | Bioengineered spinal cord | Poor structural integrity and shape fidelity in low-viscosity inks, and potential cell damage from shear forces while extruding | 98 |
| Extrusion bioprinting | hiPSCs and endothelial cells | Bioink: HA, fibrinogen | Modelling neural network impairment | Limited vertical layering due to the softness of the gel, constrained tissue thickness for optimal neural network formation and lack of neuron orientation | 87 |
| Crosslinking buffer: CaCl2, transglutaminase, thrombin | |||||
| Extrusion embedded bioprinting | Mouse brain microvascular endothelial cells | Bioink: sodium alginate, collagen I | In vitro neurovascular unit models | Limited ability to recapitulate native architectural features and structural organisation | 99 |
| Mouse-glioma cells (GL261); undifferentiated rat pheochromocytoma cells (PC12); astrocyte | Support bath: gelatin, CaCl2 | ||||
| Extrusion-embedded bioprinting | Astrocytes and Neuroepithelial cells (NE-4C) | Bioink: GelMA, gelatin, alginate, PI | Resembling neural stem cell niche | Lack of vascularizion and perfusion needed for longer-term cultures | 100 |
| Support bath: GelMA, CaCl2, PI | |||||
| Microfluidic bioprinting | Cortical neurons and glial precursors derived from hiPSCs | Bioink: Matrigel and alginate | 3D models of the human nervous systems | Immature neuronal network; limited size, dimension and weight inhibit the easy handling of the samples | 101 |
| Microfluidic bioprinting | hiPSC-derived NPCs | Core Channel: fibrinogen, alginate, genipin, guggulsterone microspheres | 3D bioprinting of neural tissues to study hiPSC-derived NPC into dopaminergic neurons | Challenge of ensuring the differentiation of hiPSCs into the desired mature phenotypes | 102 |
| Coaxial channel: chitosan, CaCl2, thrombin | |||||
| Microfluidic bioprinting | NPCs-derived from both healthy and AD patient-derived hiPSCs | Bioink: fibrinogen, sodium alginate, genipin, PCL microspheres | 3D bioprinted models of AD | Further optimization of the bioink composition and printing parameters is needed to enhance the physiological relevance. The consistent and uniform distribution of microspheres within the bioink can be challengings | 103 |
| Inkjet bioprinting | Rat glial and retinal cells | Bioink: cell culture media | CNS grafts | Limited structural integrity and shape fidelity | 104 |
| Laser assisted bioprinting | Dorsal root ganglion (DRG) neuronal cells | Substrate: gelatin-coated quartz print ribbon | Understanding of neurophysiology and advance studies in neurodegenerative diseases and therapeutic interventions | Lower survival rate of primary DRG neurons due to the printing conditions (absence of cell culture medium and laser pulse) | 105 |
Synergistic advances: bioprinting and brain organoids
An exciting frontier in neuroscience involves integrating 3D bioprinting with BOs, offering unprecedented opportunities to better mimic the intricate architecture and cellular interactions of the human brain (Fig. 3).106Bioprinting and BOs offer complementary advantages for the fabrication of physiologically relevant neural models. While brain organoids recapitulate key aspects of early brain development, their intrinsic variability and limited spatial organization can hinder reproducibility. Bioprinting addresses these challenges by enabling controlled, reproducible tissue fabrication with precise spatial arrangement of different cell types and matrix components. This synergy facilitates the generation of complex, multi-regional brain models with enhanced structural fidelity and reproducibility, allowing for more accurate studies of neuronal connectivity, disease progression, and therapeutic screening in neurodegenerative conditions. Moreover, the combination of bioprinting and BOs supports the recreation of key features of brain architecture, including regional compartmentalization and cellular heterogeneity, through both spatial control and self-organization.107 Although the ability to fully replicate the brain's intricate cytoarchitecture remains under development, the integration of these technologies represents a significant advancement over conventional 3D in vitro models, offering unprecedented opportunities to model human brain function and pathology with higher physiological relevance.
Importantly, this synergy opens new possibilities for personalized medicine. Using patient-derived induced pluripotent stem cells (iPSCs), researchers can create individualized brain models to study patient-specific disease mechanisms, identifying biomarkers, and testing potential therapeutic interventions in an environments that closely resemble the patient's brain.108
To fully realize the potential of the coupling between bioprinting and BOs, the challenges and limitations of both techniques need to be overcome. For instance, keeping organoids evenly suspended in the printer's syringe is notoriously difficult. Without constant agitation, BOs quickly settle in low-viscosity bioinks and can clog the nozzle mid-print. Even the nozzle itself imposes a constraint: its diameter must be large enough to accommodate the organoids, which unavoidably limits printing resolution. To address these challenges, researchers are exploring solutions such as dynamic agitation systems and optimized bioink formulations that help maintain organoid suspension and prevent clogging.109,110 Incorporating neurovascular units to improve nutrient diffusion within bioprinted BOs will enable more realistic studies of neurovascular interaction, which are critical in conditions like stroke and MS. These approaches can help sustain the long-term viability and functionality of the constructs, making them more suitable for studying complex brain functions and diseases. Furthermore, combining bioprinting with advanced imaging techniques, such as two-photon microscopy and optogenetics, can provide real-time monitoring of neuronal activity and network formation. For instance, two-photon microscopy enables deep tissue imaging with high resolution, allowing researchers to visualize cellular processes within the BOs over time. Optogenetics, on the other hand, provides a method to manipulate neuronal activity with light, facilitating the study of functional neural circuits and their roles in different neurological conditions. Similarly, integration with real-time feedback systems and machine learning algorithms can optimize bioprinting protocols, enhancing the precision of cell placement and improving the outcomes of organoid maturation. Remarkably, these integrated systems have the potential to unlock the full capability of organoid intelligence. This breakthrough could lead to the creation of constructs with capabilities for memory and cognition, fundamentally redefining our understanding of the brain and opening unprecedented possibilities for innovation in medicine, artificial intelligence, and beyond.
Moreover, innovations such as multi-material printing for replicating brain heterogeneity, AI-guided print parameter optimization, and dynamic control systems for real-time cell monitoring are emerging as essential tools to bridge these gaps and enhance the synergy between organoids and bioprinting.111,112
In summary, the integration of 3D bioprinting and BOs offers transformative potential for neuroscience. Overcoming challenges such as vascularization and full regional specialization will not only enhance our understanding of brain development, function, and disease but also pave the way for groundbreaking therapies and regenerative strategies.
Conclusion and future perspectives
The rising prevalence of neurodegenerative diseases and the severe decline in quality of life they cause have a profound impact on society. This highlights an urgent need to develop in vitro models that more accurately mimic the human brain's microenvironment and functionality, offering a more effective approach than traditional assays for monitoring neural circuitry dynamics over time. Recent advancements in neuroscience emphasize the transition from simplified 2D cultures to more complex 3D models, which better replicate the complex architecture and dynamic interactions of the brain. In the last decades, the integration of microfluidic technologies has further enhanced these models, resulting in the development of BoC platforms. These platforms offer unparalleled precision in controlling the microenvironment, allowing for the manipulation of factors such as fluid flow, nutrient gradients, and cellular interactions. Despite these innovations, BoC models still fall short of fully recapitulating the complexity of in vivo brain architecture and function, underscoring the necessity for more sophisticated and controlled in vitro systems that can bridge the gap between current models and in vivo complexity. In this context, 3D models such as neurospheroids, BOs and assembloids, have offered a more realistic representation for studying neural development and connectivity. Similarly, microTENNs have been developed to replicate the interconnectivity between different cortical areas, enabling the study of growth processes and functional features relevant to nervous system reconstruction. However, while these systems represent major steps forward, they often lack essential features such as vascularization, full neuronal maturation, and scalable reproducibility, factors that are critical for accurately modeling chronic and late-onset neurodegenerative conditions. To overcome these limitations, recent research highlights the transformative potential of 3D bioprinting, which offers precise control over the spatial placement of neural cells and enables the creation of high-resolution functional neuronal constructs. Moreover, this technology provides the ability to integrate key supporting elements such as glial populations and even vascular precursors, enhancing both the structural fidelity and functional viability of engineered tissues. In doing so, their physiological relevance would be significantly increased.Importantly, the combination of 3D bioprinting with BOs stands out as a revolutionary leap forward, offering transformative potential for neuroscience. This synergy allows for unprecedented control over the spatial arrangement of cells, facilitating the creation of high-resolution, functional neuronal networks. Such innovation not only enhances the physiological relevance of in vitro brain models, but it is a paradigm shift that could fundamentally transform our understanding of the brain.
Despite these advances, significant gaps remain and filling them requires targeted engineering efforts. Based on the recent literature, several practical strategies can be proposed to advance current models: (i) functional vascularization through co-culture with endothelial cells and perfusion via microfluidic chips, vascular-like networks can be introduced into 3D constructs to enhance viability and long-term maturation; (ii) maturation and integration controlling delivery of morphogens and neurotrophic factors, use of matrix compositions mimicking developmental gradients, and patterned electrical stimulation can promote synaptogenesis and circuit maturation; (iii) sensor integration embedding flexible bioelectronics and high-density microelectrode arrays into 3D platforms will enable real-time, non-invasive electrophysiological readouts, essential for tracking network functionality and disease progression; (iv) protocol standardization harmonizing fabrication and characterization protocols, especially for bioink formulation, stem cell differentiation timelines, and quality control, will be essential to ensure reproducibility and facilitate regulatory adoption.
Looking forward, the continued convergence of BOs, BoC technologies and 3D bioprinting promises to deliver next-generation platforms that are predictive, personalized, and translationally relevant. The precision and control offered by bioprinting, coupled with the biological relevance of BOs, are likely to pave the way towards unraveling the complexities of the brain. As these technologies advance, we can anticipate significant breakthroughs in understanding brain function and dysfunction, leading to the development of more effective novel therapeutic strategies.
Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Author contributions
Chiara Ausilio: conceptualization, investigation, visualization, methodology, writing – original draft. Annachiara Scalzone: conceptualization, supervision, investigation, visualization, writing – original draft, writing – review & editing; Paolo A. Netti: conceptualization, funding acquisition, supervision, visualization, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge the support of the COMBO “A multiscale integrated approach to the study of the nervous system in health and disease” project (Project identification code PE00000006) funded under the National Recovery and Resilience Plan (NRRP), Mission 4 Component 2 Investment 1.3 of Italian Ministry of University and Research (MUR) funded by the European Union – NextGenerationEU. We also acknowledge the support of FIT4MEDROB: Fit for Medical Robotics – PNRR MUR (PNC0000007).References
- Ageing and health, https://www.who.int/news-room/fact-sheets/detail/ageing-and-health (accessed 2024-08-18).
- P. R. Laming, H. Kimelberg, S. Robinson, A. Salm, N. Hawrylak, C. Müller, B. Roots and K. Ng, Neuronal–Glial Interactions and Behaviour, Neurosci. Biobehav. Rev., 2000, 24(3), 295–340, DOI:10.1016/S0149-7634(99)00080-9.
- A. Citri and R. C. Malenka, Synaptic Plasticity: Multiple Forms, Functions, and Mechanisms, Neuropsychopharmacology, 2008, 33(1), 18–41, DOI:10.1038/sj.npp.1301559.
- S. Bang, S. Lee, N. Choi and H. N. Kim, Emerging Brain-Pathophysiology-Mimetic Platforms for Studying Neurodegenerative Diseases: Brain Organoids and Brains-on-a-Chip, Adv. Healthcare Mater., 2021, 10(12), 2002119, DOI:10.1002/adhm.202002119.
- P. Rawal, D. M. Tripathi, S. Ramakrishna and S. Kaur, Prospects for 3D Bioprinting of Organoids, Bio-Des. Manuf., 2021, 4(3), 627–640, DOI:10.1007/s42242-020-00124-1.
- I. B. Levitan and L. K. Kaczmarek, The Neuron: Cell and Molecular Biology, Oxford University Press, 4th edn, 2015, DOI:10.1093/med/9780199773893.001.0001.
- T. V. P. Bliss and G. L. Collingridge, A Synaptic Model of Memory: Long-Term Potentiation in the Hippocampus, Nature, 1993, 361(6407), 31–39, DOI:10.1038/361031a0.
- J. B. Zuchero and B. A. Barres, Glia in Mammalian Development and Disease, Development, 2015, 142(22), 3805–3809, DOI:10.1242/dev.129304.
- Y. Liu, X. Shen, Y. Zhang, X. Zheng, C. Cepeda, Y. Wang, S. Duan and X. Tong, Interactions of Glial Cells with Neuronal Synapses, from Astrocytes to Microglia and Oligodendrocyte Lineage Cells, Glia, 2023, 71(6), 1383–1401, DOI:10.1002/glia.24343.
- M. V. Sofroniew and H. V. Vinters, Astrocytes: Biology and Pathology, Acta Neuropathol., 2010, 119(1), 7–35, DOI:10.1007/s00401-009-0619-8.
- M. Bradl and H. Lassmann, Oligodendrocytes: Biology and Pathology, Acta Neuropathol., 2010, 119(1), 37–53, DOI:10.1007/s00401-009-0601-5.
- M. B. Graeber and W. J. Streit, Microglia: Biology and Pathology, Acta Neuropathol., 2010, 119(1), 89–105, DOI:10.1007/s00401-009-0622-0.
- N. Baumann, A. B.-V. Evercooren, C. Jacque and B. Zalc, Glial Biology and Disorders, Curr. Opin. Neurol., 1993, 6(1), 27 CAS.
- B. J. Andreone, M. Larhammar and J. W. Lewcock, Cell Death and Neurodegeneration, Cold Spring Harbor Perspect. Biol., 2020, 12(2), a036434, DOI:10.1101/cshperspect.a036434.
- S. Zhang, R. Meng, M. Jiang, H. Qing and J. Ni, Emerging Roles of Microglia in Blood-Brain Barrier Integrity in Agingand Neurodegeneration, CN, 2024, 22(7), 1189–1204, DOI:10.2174/1570159X21666230203103910.
- S.-A. Ahadiat and Z. Hosseinian, Astrocytes' Innate Role in Neurodegenerative Disorders, Bull. Natl. Res. Cent., 2023, 47(1), 105, DOI:10.1186/s42269-023-01083-0.
- E. R. Burnside and E. J. Bradbury, Review: Manipulating the Extracellular Matrix and Its Role in Brain and Spinal Cord Plasticity and Repair, Neuropathol. Appl. Neurobiol., 2014, 40(1), 26–59, DOI:10.1111/nan.12114.
- A. Soles, A. Selimovic, K. Sbrocco, F. Ghannoum, K. Hamel, E. L. Moncada, S. Gilliat and M. Cvetanovic, Extracellular Matrix Regulation in Physiology and in Brain Disease, Int. J. Mol. Sci., 2023, 24(8), 7049, DOI:10.3390/ijms24087049.
- T. Chen, Y. Dai, C. Hu, Z. Lin, S. Wang, J. Yang, L. Zeng, S. Li and W. Li, Cellular and Molecular Mechanisms of the Blood–Brain Barrier Dysfunction in Neurodegenerative Diseases, Fluids Barriers CNS, 2024, 21(1), 60, DOI:10.1186/s12987-024-00557-1.
- S. M. A. Naini and N. Soussi-Yanicostas, Heparan Sulfate as a Therapeutic Target in Tauopathies: Insights From Zebrafish, Front. Cell Dev. Biol., 2018, 6, 163, DOI:10.3389/fcell.2018.00163.
- S. Ghorbani and V. W. Yong, The Extracellular Matrix as Modifier of Neuroinflammation and Remyelination in Multiple Sclerosis, Brain, 2021, 144(7), 1958–1973, DOI:10.1093/brain/awab059.
- I. Jahan, M. Harun-Ur-Rashid, M. A. Islam, F. Sharmin, S. K. Al Jaouni, A. M. Kaki and S. Selim, Neuronal Plasticity and Its Role in Alzheimer's Disease and Parkinson's Disease, Neural Regener. Res., 2026, 21(1), 107–125, DOI:10.4103/NRR.NRR-D-24-01019.
- X.-L. Wu, Q.-J. Yan and F. Zhu, Abnormal Synaptic Plasticity and Impaired Cognition in Schizophrenia, World J. Psychiatry, 2022, 12(4), 541–557, DOI:10.5498/wjp.v12.i4.541.
- S. Samanta, L. Ylä-Outinen, V. K. Rangasami, S. Narkilahti and O. P. Oommen, Bidirectional Cell-Matrix Interaction Dictates Neuronal Network Formation in a Brain-Mimetic 3D Scaffold, Acta Biomater., 2022, 140, 314–323, DOI:10.1016/j.actbio.2021.12.010.
- T. Behl, G. Kaur, A. Sehgal, S. Bhardwaj, S. Singh, C. Buhas, C. Judea-Pusta, D. Uivarosan, M. A. Munteanu and S. Bungau, Multifaceted Role of Matrix Metalloproteinases in Neurodegenerative Diseases: Pathophysiological and Therapeutic Perspectives, Int. J. Mol. Sci., 2021, 22(3), 1413, DOI:10.3390/ijms22031413.
- N. D'Ambrosi, M. Cozzolino and S. Apolloni, The Contribution of Non-Neuronal Cells in Neurodegeneration: From Molecular Pathogenesis to Therapeutic Challenges, Cells, 2022, 11(2), 193, DOI:10.3390/cells11020193.
- W. A. Rike and S. Stern, Proteins and Transcriptional Dysregulation of the Brain Extracellular Matrix in Parkinson's Disease: A Systematic Review, Int. J. Mol. Sci., 2023, 24(8), 7435, DOI:10.3390/ijms24087435.
- M. Basso, S. Pozzi, M. Tortarolo, F. Fiordaliso, C. Bisighini, L. Pasetto, G. Spaltro, D. Lidonnici, F. Gensano, E. Battaglia, C. Bendotti and V. Bonetto, Mutant Copper-Zinc Superoxide Dismutase (SOD1) Induces Protein Secretion Pathway Alterations and Exosome Release in Astrocytes, J. Biol. Chem., 2013, 288(22), 15699–15711, DOI:10.1074/jbc.M112.425066.
- R. Rauti, N. Renous and B. M. Maoz, Mimicking the Brain Extracellular Matrix in Vitro : A Review of Current Methodologies and Challenges, Isr. J. Chem., 2020, 60(12), 1141–1151, DOI:10.1002/ijch.201900052.
- C. Kuschel, H. Steuer, A. N. Maurer, B. Kanzok, R. Stoop and B. Angres, Cell Adhesion Profiling Using Extracellular Matrix Protein Microarrays, BioTechniques, 2006, 40(4), 523–531, DOI:10.2144/000112134.
- C. Ausilio, C. Lubrano, A. Mariano and F. Santoro, Negatively-Charged Supported Lipid Bilayers Regulate Neuronal Adhesion and Outgrowth, RSC Adv., 2022, 12(47), 30270–30277, 10.1039/D2RA05147H.
- T. Marques-Almeida, C. Ribeiro, I. Irastorza, P. Miranda-Azpiazu, I. Torres-Alemán, U. Silvan and S. Lanceros-Méndez, Electroactive Materials Surface Charge Impacts Neuron Viability and Maturation in 2D Cultures, ACS Appl. Mater. Interfaces, 2023, 15(26), 31206–31213, DOI:10.1021/acsami.3c04055.
- L. Matino, A. Mariano, C. Ausilio, R. Garg, T. Cohen-Karni and F. Santoro, Modulation of Early Stage Neuronal Outgrowth through Out-of-Plane Graphene, Nano Lett., 2022, 22(21), 8633–8640, DOI:10.1021/acs.nanolett.2c03171.
- A. Mariano, C. Lubrano, U. Bruno, C. Ausilio, N. B. Dinger and F. Santoro, Advances in Cell-Conductive Polymer Biointerfaces and Role of the Plasma Membrane, Chem. Rev., 2022, 122(4), 4552–4580, DOI:10.1021/acs.chemrev.1c00363.
- E. G. Z. Centeno, H. Cimarosti and A. Bithell, 2D versus 3D Human Induced Pluripotent Stem Cell-Derived Cultures for Neurodegenerative Disease Modelling, Mol. Neurodegener., 2018, 13(1), 27, DOI:10.1186/s13024-018-0258-4.
- G. Joseph, R. P. Orme, T. Kyriacou, R. A. Fricker and P. Roach, Effects of Surface Chemistry Interaction on Primary Neural Stem Cell Neurosphere Responses, ACS Omega, 2021, 6(30), 19901–19910, DOI:10.1021/acsomega.1c02796.
- C. E. Strong, J. Zhang, M. Carrasco, S. Kundu, M. Boutin, H. D. Vishwasrao, J. Liu, A. Medina, Y.-C. Chen, K. Wilson, E. M. Lee and M. Ferrer, Functional Brain Region-Specific Neural Spheroids for Modeling Neurological Diseases and Therapeutics Screening, Commun. Biol., 2023, 6(1), 1211, DOI:10.1038/s42003-023-05582-8.
- J. Guyon, L. Andrique, N. Pujol, G. V. Røsland, G. Recher, A. Bikfalvi and T. Daubon, A 3D Spheroid Model for Glioblastoma, J. Visualized Exp., 2020, 158, 60998, DOI:10.3791/60998.
- H. Y. Shin, K.-S. Han, H. W. Park, Y. H. Hong, Y. Kim, H. E. Moon, K. W. Park, H. R. Park, C. J. Lee, K. Lee, S. J. Kim, M. S. Heo, S.-H. Park, D. G. Kim and S. H. Paek, Tumor Spheroids of an Aggressive Form of Central Neurocytoma Have Transit-Amplifying Progenitor Characteristics with Enhanced EGFR and Tumor Stem Cell Signaling, Exp. Neurobiol., 2021, 30(2), 120–143, DOI:10.5607/en21004.
- L. Smirnova and T. Hartung, The Promise and Potential of Brain Organoids, Adv. Healthcare Mater., 2024, 2302745, DOI:10.1002/adhm.202302745.
- Y. Li, P.-M. Zeng, J. Wu and Z.-G. Luo, Advances and Applications of Brain Organoids, Neurosci. Bull., 2023, 39(11), 1703–1716, DOI:10.1007/s12264-023-01065-2.
- I. Pereira, M. J. Lopez-Martinez and J. Samitier, Advances in Current in Vitro Models on Neurodegenerative Diseases, Front. Bioeng. Biotechnol., 2023, 11, 1260397, DOI:10.3389/fbioe.2023.1260397.
- T. Lokai, B. Albin, K. Qubbaj, A. P. Tiwari, P. Adhikari and I. H. Yang, A Review on Current Brain Organoid Technologies from a Biomedical Engineering Perspective, Exp. Neurol., 2023, 367, 114461, DOI:10.1016/j.expneurol.2023.114461.
- H. Kim, S. Kang, B. Cho, S. An, Y. Kim and J. Kim, Parkinson's Disease Modeling Using Directly Converted 3D Induced Dopaminergic Neuron Organoids and Assembloids, Adv. Sci., 2025, 12(14), 2412548, DOI:10.1002/advs.202412548.
- E. Frattini, G. Faustini, G. Lopez, E. V. Carsana, M. Tosi, I. Trezzi, M. Magni, G. Soldà, L. Straniero, D. Facchi, M. Samarani, M. Martá-Ariza, C. M. G. De Luca, E. Vezzoli, A. Pittaro, A. Stepanyan, R. Silipigni, I. Rosety, J. C. Schwamborn, S. P. Sardi, F. Moda, S. Corti, G. P. Comi, F. Blandini, N. X. Tritsch, M. Bortolozzi, S. Ferrero, F. M. Cribiù, T. Wisniewski, R. Asselta, M. Aureli, A. Bellucci and A. Di Fonzo, Lewy Pathology Formation in Patient-Derived GBA1 Parkinson's Disease Midbrain Organoids, Brain, 2025, 148(4), 1242–1257, DOI:10.1093/brain/awae365.
- N. Daviaud, T. Mehta, W. Holzman, A. McDermott and S. A. Sadiq, Impaired Myelination in Multiple Sclerosis Organoids: P21 Links Oligodendrocyte Dysfunction to Disease Subtype, bioRxiv, 2025, preprint, DOI:10.1101/2025.01.08.631924.
- W. K. Raja, A. E. Mungenast, Y.-T. Lin, T. Ko, F. Abdurrob, J. Seo and L.-H. Tsai, Self-Organizing 3D Human Neural Tissue Derived from Induced Pluripotent Stem Cells Recapitulate Alzheimer's Disease Phenotypes, PLoS One, 2016, 11(9), e0161969, DOI:10.1371/journal.pone.0161969.
- S. Kim and M.-Y. Chang, Application of Human Brain Organoids—Opportunities and Challenges in Modeling Human Brain Development and Neurodevelopmental Diseases, Int. J. Mol. Sci., 2023, 24(15), 12528, DOI:10.3390/ijms241512528.
- W. Fan, K. M. Christian, H. Song and G. Ming, Applications of Brain Organoids for Infectious Diseases, J. Mol. Biol., 2022, 434(3), 167243, DOI:10.1016/j.jmb.2021.167243.
- J.-Q. Zhou, L.-H. Zeng, C.-T. Li, D.-H. He, H.-D. Zhao, Y.-N. Xu, Z.-T. Jin and C. Gao, Brain Organoids Are New Tool for Drug Screening of Neurological Diseases, Neural Regener. Res., 2023, 18(12), 2639–2645, DOI:10.4103/1673-5374.367983.
- Y. Miura, M.-Y. Li, O. Revah, S.-J. Yoon, G. Narazaki and S. P. Paşca, Engineering Brain Assembloids to Interrogate Human Neural Circuits, Nat. Protoc., 2022, 17(1), 15–35, DOI:10.1038/s41596-021-00632-z.
- J. Wen, F. Liu, Q. Cheng, N. Weygant, X. Liang, F. Fan, C. Li, L. Zhang and Z. Liu, Applications of Organoid Technology to Brain Tumors, CNS Neurosci. Ther., 2023, 29(10), 2725–2743, DOI:10.1111/cns.14272.
- T. G. Krieger, S. M. Tirier, J. Park, K. Jechow, T. Eisemann, H. Peterziel, P. Angel, R. Eils and C. Conrad, Modeling Glioblastoma Invasion Using Human Brain Organoids and Single-Cell Transcriptomics, Neurooncology, 2020, 22(8), 1138–1149, DOI:10.1093/neuonc/noaa091.
- W. A. Anderson, A. Bosak, H. T. Hogberg, T. Hartung and M. J. Moore, Advances in 3D Neuronal Microphysiological Systems: Towards a Functional Nervous System on a Chip, In Vitro Cell. Dev. Biol.: Anim., 2021, 57(2), 191–206, DOI:10.1007/s11626-020-00532-8.
- D. K. Cullen, W. J. Gordián-Vélez, L. A. Struzyna, D. Jgamadze, J. Lim, K. L. Wofford, K. D. Browne and H. I. Chen, Bundled Three-Dimensional Human Axon Tracts Derived from Brain Organoids, iScience, 2019, 21, 57–67, DOI:10.1016/j.isci.2019.10.004.
- L. A. Struzyna, J. A. Wolf, C. J. Mietus, D. O. Adewole, H. I. Chen, D. H. Smith and D. K. Cullen, Rebuilding Brain Circuitry with Living Micro-Tissue Engineered Neural Networks, Tissue Eng., Part A, 2015, 21(21–22), 2744–2756, DOI:10.1089/ten.tea.2014.0557.
- T. Kirihara, Z. Luo, S. Y. A. Chow, R. Misawa, J. Kawada, S. Shibata, F. Khoyratee, C. A. Vollette, V. Volz, T. Levi, T. Fujii and Y. Ikeuchi, A Human Induced Pluripotent Stem Cell-Derived Tissue Model of a Cerebral Tract Connecting Two Cortical Regions, iScience, 2019, 14, 301–311, DOI:10.1016/j.isci.2019.03.012.
- T. Marinov, H. A. L. Sánchez, L. Yuchi, D. O. Adewole, D. K. Cullen and R. H. Kraft, A Computational Model of Bidirectional Axonal Growth in Micro-Tissue Engineered Neuronal Networks (Micro-TENNs), In Silico Biol., 2020, 13(3–4), 85–99, DOI:10.3233/ISB-180172.
- M. Kamudzandu, M. Köse-Dunn, M. G. Evans, R. A. Fricker and P. Roach, A Micro-Fabricated in Vitro Complex Neuronal Circuit Platform, Biomed. Phys. Eng. Express, 2019, 5(4), 045016, DOI:10.1088/2057-1976/ab2307.
- H. R. Karimian, K. J. Pollard and M. J. Moore, Kordjamshidi, P. Semantic Segmentation of Microengineered Neural Tissues, in 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), IEEE, Berlin, Germany, 2019, pp. 955–960, DOI:10.1109/EMBC.2019.8856378.
- I. Kelava and M. A. Lancaster, Dishing out Mini-Brains: Current Progress and Future Prospects in Brain Organoid Research, Dev. Biol., 2016, 420(2), 199–209, DOI:10.1016/j.ydbio.2016.06.037.
- R. M. Eglen and T. Reisine, Human iPS Cell-Derived Patient Tissues and 3D Cell Culture Part 2: Spheroids, Organoids, and Disease Modeling, SLAS Technol., 2019, 24(1), 18–27, DOI:10.1177/2472630318803275.
- J. Gopalakrishnan, The Emergence of Stem Cell-Based Brain Organoids: Trends and Challenges, BioEssays, 2019, 41(8), 1900011, DOI:10.1002/bies.201900011.
- M. Rizzuti, V. Melzi, L. Brambilla, L. Quetti, L. Sali, L. Ottoboni, M. Meneri, A. Ratti, F. Verde, N. Ticozzi, G. P. Comi, S. Corti and E. Abati, Shaping the Neurovascular Unit Exploiting Human Brain Organoids, Mol. Neurobiol., 2024, 61(9), 6642–6657, DOI:10.1007/s12035-024-03998-9.
- M. T. Pham, K. M. Pollock, M. D. Rose, W. A. Cary, H. R. Stewart, P. Zhou, J. A. Nolta and B. Waldau, Generation of Human Vascularized Brain Organoids, NeuroReport, 2018, 29(7), 588–593, DOI:10.1097/WNR.0000000000001014.
- B. M. Maoz, Brain-on-a-Chip: Characterizing the next Generation of Advanced in Vitro Platforms for Modeling the Central Nervous System, APL Bioeng., 2021, 5(3), 030902, DOI:10.1063/5.0055812.
- Z. Wang, Y. Zhang, Z. Li, H. Wang, N. Li and Y. Deng, Microfluidic Brain-on-a-Chip: From Key Technology to System Integration and Application, Small, 2023, 19(52), 2304427, DOI:10.1002/smll.202304427.
- P. Herreros, L. M. Ballesteros-Esteban, M. F. Laguna, I. Leyva, I. Sendiña-Nadal and M. Holgado, Neuronal Circuits on a Chip for Biological Network Monitoring, Biotechnol. J., 2021, 16(7), 2000355, DOI:10.1002/biot.202000355.
- S. Zhang, Z. Wan and R. D. Kamm, Vascularized Organoids on a Chip: Strategies for Engineering Organoids with Functional Vasculature, Lab Chip, 2021, 21(3), 473–488, 10.1039/D0LC01186J.
- A.-N. Cho, Y. Jin, Y. An, J. Kim, Y. S. Choi, J. S. Lee, J. Kim, W.-Y. Choi, D.-J. Koo, W. Yu, G.-E. Chang, D.-Y. Kim, S.-H. Jo, J. Kim, S.-Y. Kim, Y.-G. Kim, J. Y. Kim, N. Choi, E. Cheong, Y.-J. Kim, H. S. Je, H.-C. Kang and S.-W. Cho, Microfluidic Device with Brain Extracellular Matrix Promotes Structural and Functional Maturation of Human Brain Organoids, Nat. Commun., 2021, 12(1), 4730, DOI:10.1038/s41467-021-24775-5.
- M. A. M. Jahromi, A. Abdoli, M. Rahmanian, H. Bardania, M. Bayandori, S. M. M. Basri, A. Kalbasi, A. R. Aref, M. Karimi and M. R. Hamblin, Microfluidic Brain-on-a-Chip: Perspectives for Mimicking Neural System Disorders, Mol. Neurobiol., 2019, 56(12), 8489–8512, DOI:10.1007/s12035-019-01653-2.
- N. Shettigar, C. S. Suh and D. Banerjee, On Developing a Novel Brain-On-Chip Platform for Enhanced Control and Design of 3D Neural Circuit Informational Dynamics, in Dynamics, Vibration, and Control, American Society of Mechanical Engineers, Portland, Oregon, USA, 2024, vol. 5, p. V005T07A037, DOI:10.1115/IMECE2024-147442.
- K. Gomez, V. R. Yarmey, H. Mane and A. San-Miguel, Microfluidic and Computational Tools for Neurodegeneration Studies, Annu. Rev. Chem. Biomol. Eng., 2025, 16(1), 195–216, DOI:10.1146/annurev-chembioeng-082223-054547.
- H. Castiglione, P.-A. Vigneron, C. Baquerre, F. Yates, J. Rontard and T. Honegger, Human Brain Organoids-on-Chip: Advances, Challenges, and Perspectives for Preclinical Applications, Pharmaceutics, 2022, 14(11), 2301, DOI:10.3390/pharmaceutics14112301.
- P. A. Libet, L. Y. Polynkin, M. R. Saridis, E. V. Yakovlev, S. A. Korsakova, A. B. Salmina, A. S. Averchuk, N. A. Rozanova and S. O. Yurchenko, A Four-Channel Microfluidic Model of the Blood–Brain and Blood–Cerebrospinal Fluid Barriers: Fluid Dynamics Analysis, Micro Nano Syst. Lett., 2024, 12(1), 28, DOI:10.1186/s40486-024-00219-9.
- L. Amirifar, A. Shamloo, R. Nasiri, N. R. De Barros, Z. Z. Wang, B. D. Unluturk, A. Libanori, O. Ievglevskyi, S. E. Diltemiz, S. Sances, I. Balasingham, S. K. Seidlits and N. Ashammakhi, Brain-on-a-Chip: Recent Advances in Design and Techniques for Microfluidic Models of the Brain in Health and Disease, Biomaterials, 2022, 285, 121531, DOI:10.1016/j.biomaterials.2022.121531.
- T. Osaki, T. Duenki, S. Y. A. Chow, Y. Ikegami, R. Beaubois, T. Levi, N. Nakagawa-Tamagawa, Y. Hirano and Y. Ikeuchi, Complex Activity and Short-Term Plasticity of Human Cerebral Organoids Reciprocally Connected with Axons, Nat. Commun., 2024, 15(1), 2945, DOI:10.1038/s41467-024-46787-7.
- L. Smirnova, B. S. Caffo, D. H. Gracias, Q. Huang, I. E. M. Pantoja, B. Tang, D. J. Zack, C. A. Berlinicke, J. L. Boyd, T. D. Harris, E. C. Johnson, B. J. Kagan, J. Kahn, A. R. Muotri, B. L. Paulhamus, J. C. Schwamborn, J. Plotkin, A. S. Szalay, J. T. Vogelstein, P. F. Worley and T. Hartung, Organoid Intelligence (OI): The New Frontier in Biocomputing and Intelligence-in-a-Dish, Front. Sci., 2023, 1, 1017235, DOI:10.3389/fsci.2023.1017235.
- B. J. Kagan, A. C. Kitchen, N. T. Tran, F. Habibollahi, M. Khajehnejad, B. J. Parker, A. Bhat, B. Rollo, A. Razi and K. J. Friston, In Vitro Neurons Learn and Exhibit Sentience When Embodied in a Simulated Game-World, Neuron, 2022, 110(23), 3952–3969.e8, DOI:10.1016/j.neuron.2022.09.001.
- I. E. M. Pantoja, L. Smirnova, A. R. Muotri, K. J. Wahlin, J. Kahn, J. L. Boyd, D. H. Gracias, T. D. Harris, T. Cohen-Karni, B. S. Caffo, A. S. Szalay, F. Han, D. J. Zack, R. Etienne-Cummings, A. Akwaboah, J. C. Romero, D.-M. Alam El Din, J. D. Plotkin, B. L. Paulhamus, E. C. Johnson, F. Gilbert, J. L. Curley, B. Cappiello, J. C. Schwamborn, E. J. Hill, P. Roach, D. Tornero, C. Krall, R. Parri, F. Sillé, A. Levchenko, R. E. Jabbour, B. J. Kagan, C. A. Berlinicke, Q. Huang, A. Maertens, K. Herrmann, K. Tsaioun, R. Dastgheyb, C. W. Habela, J. T. Vogelstein and T. Hartung, First Organoid Intelligence (OI) Workshop to Form an OI Community, Front. Artif. Intell., 2023, 6, 1116870, DOI:10.3389/frai.2023.1116870.
- C. A. Trujillo, R. Gao, P. D. Negraes, J. Gu, J. Buchanan, S. Preissl, A. Wang, W. Wu, G. G. Haddad, I. A. Chaim, A. Domissy, M. Vandenberghe, A. Devor, G. W. Yeo, B. Voytek and A. R. Muotri, Complex Oscillatory Waves Emerging from Cortical Organoids Model Early Human Brain Network Development, Cell Stem Cell, 2019, 25(4), 558–569.e7, DOI:10.1016/j.stem.2019.08.002.
- T. Sharf, T. Van Der Molen, S. M. K. Glasauer, E. Guzman, A. P. Buccino, G. Luna, Z. Cheng, M. Audouard, K. G. Ranasinghe, K. Kudo, S. S. Nagarajan, K. R. Tovar, L. R. Petzold, A. Hierlemann, P. K. Hansma and K. S. Kosik, Functional Neuronal Circuitry and Oscillatory Dynamics in Human Brain Organoids, Nat. Commun., 2022, 13(1), 4403, DOI:10.1038/s41467-022-32115-4.
- K. Tasnim and J. Liu, Emerging Bioelectronics for Brain Organoid Electrophysiology, J. Mol. Biol., 2022, 434(3), 167165, DOI:10.1016/j.jmb.2021.167165.
- Q. Huang, B. Tang, J. C. Romero, Y. Yang, S. K. Elsayed, G. Pahapale, T.-J. Lee, I. E. Morales Pantoja, F. Han, C. Berlinicke, T. Xiang, M. Solazzo, T. Hartung, Z. Qin, B. S. Caffo, L. Smirnova and D. H. Gracias, Shell Microelectrode Arrays (MEAs) for Brain Organoids, Sci. Adv., 2022, 8(33), eabq5031, DOI:10.1126/sciadv.abq5031.
- Y. Song, X. Su, K. F. Firouzian, Y. Fang, T. Zhang and W. Sun, Engineering of Brain-like Tissue Constructs via 3D Cell-Printing Technology, Biofabrication, 2020, 12(3), 035016, DOI:10.1088/1758-5090/ab7d76.
- X. Bocheng and R. França, Innovative 3D Bioprinting Approaches for Advancing Brain Science and Medicine: A Literature Review, Biomed. Phys. Eng. Express, 2024, 10(6), 062002, DOI:10.1088/2057-1976/ad795c.
- Y. Yan, X. Li, Y. Gao, S. Mathivanan, L. Kong, Y. Tao, Y. Dong, X. Li, A. Bhattacharyya, X. Zhao and S.-C. Zhang, 3D Bioprinting of Human Neural Tissues with Functional Connectivity, Cell Stem Cell, 2024, 31(2), 260–274.e7, DOI:10.1016/j.stem.2023.12.009.
- A. Orr, F. Kalantarnia, S. Nazir, B. Bolandi, D. Alderson, K. O'Grady, M. Hoorfar, L. M. Julian and S. M. Willerth, Recent Advances in 3D Bioprinted Neural Models: A Systematic Review on the Applications to Drug Discovery, Adv. Drug Delivery Rev., 2025, 218, 115524, DOI:10.1016/j.addr.2025.115524.
- S. Noh, K. Kim, J.-I. Kim, J. H. Shin and H.-W. Kang, Direct-Write Printing for Producing Biomimetic Patterns with Self-Aligned Neurites, Addit. Manuf., 2020, 32, 101072, DOI:10.1016/j.addma.2020.101072.
- S. Ilkhanizadeh, A. Teixeira and O. Hermanson, Inkjet Printing of Macromolecules on Hydrogels to Steer Neural Stem Cell Differentiation, Biomaterials, 2007, 28(27), 3936–3943, DOI:10.1016/j.biomaterials.2007.05.018.
- L. Koch, A. Deiwick, J. Soriano and B. Chichkov, Laser Bioprinting of Human iPSC-Derived Neural Stem Cells and Neurons: Effect on Cell Survival, Multipotency, Differentiation, and Neuronal Activity, Int. J. Bioprint., 2023, 9(2), 672, DOI:10.18063/ijb.v9i2.672.
- M. Ö. Öztürk-Öncel, B. H. Leal-Martínez, R. F. Monteiro, M. E. Gomes and R. M. A. Domingues, A Dive into the Bath: Embedded 3D Bioprinting of Freeform in Vitro Models, Biomater. Sci., 2023, 11(16), 5462–5473, 10.1039/D3BM00626C.
- C. K. Bektas, J. Luo, B. Conley, K.-P. N. Le and K.-B. Lee, 3D Bioprinting Approaches for Enhancing Stem Cell-Based Neural Tissue Regeneration, Acta Biomater., 2025, 193, 20–48, DOI:10.1016/j.actbio.2025.01.006.
- P. Layrolle, P. Payoux and S. Chavanas, Message in a Scaffold: Natural Biomaterials for Three-Dimensional (3D) Bioprinting of Human Brain Organoids, Biomolecules, 2022, 13(1), 25, DOI:10.3390/biom13010025.
- A. Patel and K. Mequanint, Hydrogel Biomaterials, in Biomedical Engineering – Frontiers and Challenges, ed. R. Fazel, InTech, 2011, DOI:10.5772/24856.
- D. Espinosa-Hoyos, A. Jagielska, K. A. Homan, H. Du, T. Busbee, D. G. Anderson, N. X. Fang, J. A. Lewis and K. J. Van Vliet, Engineered 3D-Printed Artificial Axons, Sci. Rep., 2018, 8(1), 478, DOI:10.1038/s41598-017-18744-6.
- C. Whitehouse, N. Corbett and J. Brownlees, 3D Models of Neurodegeneration: Implementation in Drug Discovery, Trends Pharmacol. Sci., 2023, 44(4), 208–221, DOI:10.1016/j.tips.2023.01.005.
- D. Joung, V. Truong, C. C. Neitzke, S. Guo, P. J. Walsh, J. R. Monat, F. Meng, S. H. Park, J. R. Dutton, A. M. Parr and M. C. McAlpine, 3D Printed Stem-Cell Derived Neural Progenitors Generate Spinal Cord Scaffolds, Adv. Funct. Mater., 2018, 28(39), 1801850, DOI:10.1002/adfm.201801850.
- S. Wang, L. Bai, X. Hu, S. Yao, Z. Hao, J. Zhou, X. Li, H. Lu, J. He, L. Wang and D. Li, 3D Bioprinting of Neurovascular Tissue Modeling with Collagen-Based Low-Viscosity Composites, Adv. Healthcare Mater., 2023, 12(25), 2300004, DOI:10.1002/adhm.202300004.
- Y.-C. E. Li, Y. A. Jodat, R. Samanipour, G. Zorzi, K. Zhu, M. Hirano, K. Chang, A. Arnaout, S. Hassan, N. Matharu, A. Khademhosseini, M. Hoorfar and S. R. Shin, Toward a Neurospheroid Niche Model: Optimizing Embedded 3D Bioprinting for Fabrication of Neurospheroid Brain-like Co-Culture Constructs, Biofabrication, 2020, 13(1), 015014, DOI:10.1088/1758-5090/abc1be.
- F. Salaris, C. Colosi, C. Brighi, A. Soloperto, V. De Turris, M. C. Benedetti, S. Ghirga, M. Rosito, S. Di Angelantonio and A. Rosa, 3D Bioprinted Human Cortical Neural Constructs Derived from Induced Pluripotent Stem Cells, J. Clin. Med., 2019, 8(10), 1595, DOI:10.3390/jcm8101595.
- R. Sharma, I. P. M. Smits, L. De La Vega, C. Lee and S. M. Willerth, 3D Bioprinting Pluripotent Stem Cell Derived Neural Tissues Using a Novel Fibrin Bioink Containing Drug Releasing Microspheres, Front. Bioeng. Biotechnol., 2020, 8, 57, DOI:10.3389/fbioe.2020.00057.
- C. Benwood, J. Walters-Shumka, K. Scheck and S. M. Willerth, 3D Bioprinting Patient-Derived Induced Pluripotent Stem Cell Models of Alzheimer's Disease Using a Smart Bioink, Bioelectron. Med., 2023, 9(1), 10, DOI:10.1186/s42234-023-00112-7.
- B. Lorber, W.-K. Hsiao, I. M. Hutchings and K. R. Martin, Adult Rat Retinal Ganglion Cells and Glia Can Be Printed by Piezoelectric Inkjet Printing, Biofabrication, 2013, 6(1), 015001, DOI:10.1088/1758-5082/6/1/015001.
- J. L. Curley, S. C. Sklare, D. A. Bowser, J. Saksena, M. J. Moore and D. B. Chrisey, Isolated Node Engineering of Neuronal Systems Using Laser Direct Write, Biofabrication, 2016, 8(1), 015013, DOI:10.1088/1758-5090/8/1/015013.
- H. Lee, Engineering In Vitro Models: Bioprinting of Organoids with Artificial Intelligence, Cyborg Bionic Syst., 2023, 4, 0018, DOI:10.34133/cbsystems.0018.
- D. Banerjee, Y. P. Singh, P. Datta, V. Ozbolat, A. O'Donnell, M. Yeo and I. T. Ozbolat, Strategies for 3D Bioprinting of Spheroids: A Comprehensive Review, Biomaterials, 2022, 291, 121881, DOI:10.1016/j.biomaterials.2022.121881.
- C. C. Clark, K. M. Yoo, H. Sivakumar, K. Strumpf, A. W. Laxton, S. B. Tatter, R. E. Strowd and A. Skardal, Immersion Bioprinting of Hyaluronan and Collagen Bioink-Supported 3D Patient-Derived Brain Tumor Organoids, Biomed. Mater., 2023, 18(1), 015014, DOI:10.1088/1748-605X/aca05d.
- D. Takagi, W. Lin, T. Matsumoto, H. Yaginuma, N. Hemmi, S. Hatada and M. Seo, High-Precision Three-Dimensional Inkjet Technology for Live Cell Bioprinting, Int. J. Bioprint., 2019, 5(2), 208, DOI:10.18063/ijb.v5i2.208.
- P. N. Bernal, M. Bouwmeester, J. Madrid-Wolff, M. Falandt, S. Florczak, N. G. Rodriguez, Y. Li, G. Größbacher, R. Samsom, M. Van Wolferen, L. J. W. Van Der Laan, P. Delrot, D. Loterie, J. Malda, C. Moser, B. Spee and R. Levato, Volumetric Bioprinting of Organoids and Optically Tuned Hydrogels to Build Liver-Like Metabolic Biofactories, Adv. Mater., 2022, 34(15), 2110054, DOI:10.1002/adma.202110054.
- Z. Zhang, X. Zhou, Y. Fang, Z. Xiong and T. Zhang, AI-Driven 3D Bioprinting for Regenerative Medicine: From Bench to Bedside, Bioact. Mater., 2025, 45, 201–230, DOI:10.1016/j.bioactmat.2024.11.021.
- G. Lee, S. J. Kim and J.-K. Park, Bioprinting of Heterogeneous and Multilayered Cell-Hydrogel Constructs Using Continuous Multi-Material Printing and Aerosol-Based Crosslinking, STAR Protoc., 2022, 3(2), 101303, DOI:10.1016/j.xpro.2022.101303.
| This journal is © The Royal Society of Chemistry 2025 |