Open Access Article
Open Access ArticleMicrofluidic mimicry of the Golgi-linked N-glycosylation machinery†
Florin N.
Isenrich
a,
Marie-Estelle
Losfeld
b,
Markus
Aebi
*b and
Andrew J.
deMello
 *a
*a
aInstitute for Chemical and Bioengineering, ETH Zurich, Zürich, 8093, Switzerland. E-mail: andrew.demello@chem.ethz.ch
bInstitute of Microbiology, ETH Zurich, Zürich, 8093, Switzerland. E-mail: markus.aebi@micro.biol.ethz.ch
First published on 20th March 2025
Abstract
The complexity of the eukaryotic glycosylation machinery hinders the development of cell-free protein glycosylation since in vitro methods struggle to simulate the natural environment of the glycosylation machinery. Microfluidic technologies have the potential to address this limitation due to their ability to control glycosylation parameters, such as enzyme/substrate concentrations and fluxes, in a rapid and precise manner. However, due to the complexity and sensitivity of the numerous components of the glycosylation machinery, very few “glycobiology-on-a-chip” systems have been proposed or reported in the literature. Herein, we describe the design, fabrication and proof-of-concept of a droplet-based microfluidic platform able to mimic N-linked glycan processing along the secretory pathway. Within a single microfluidic device, glycoproteins and glycosylation enzymes are encapsulated and incubated in water-in-oil droplets. Additional glycosylation enzymes are subsequently supplied to these droplets via picoinjection, allowing further glycoprotein processing in a user-defined manner. After system validation, the platform is used to perform two spatiotemporally separated consecutive enzymatic N-glycan modifications, mirroring the transition between the endoplasmic reticulum and early Golgi.
1. Introduction
Protein glycosylation occurs in all domains of life, with N-linked glycosylation being the most abundant type of glycosylation in eukaryotes.1–3 Such post-translational modifications on the surface of proteins are intimately involved in various processes, including protein folding and its quality control, cell–cell recognition or antibody binding/recognition.4–9 In eukaryotic cells, protein glycosylation is initiated in the endoplasmic reticulum (ER) by oligosaccharyltransferase (OST) transferring a pre-assembled oligosaccharide, GlcNAc2Man9Glc3, onto an asparagine residue of the Asn-X-Ser/Thr amino acid sequon of nascent polypeptides.10,11 This N-glycan chain is subsequently used and trimmed in the ER during protein quality control.5,6 In the Golgi, further saccharide modifications lead to a variety of different glycan structures covalently linked to proteins.8,12,13 These modifications are dependent on parameters such as substrate and enzyme concentrations, their fluxes through the glycosylation compartments as well as the protein's structure.12,14 The influence of protein structure leads to further complexity if multiple glycosylation sites exist. Here, the type and degree of the processing may vary between each site. The resulting site-specific heterogeneity requires the use of bespoke mass spectrometry methods for a detailed characterisation.12,14,15Since the glycosylation patterns of glycoproteins serve many functions in vivo, they are highly important in therapeutics. Specific forms of glycosylation have been shown to influence and control the effects of therapeutic proteins.16–19 Indeed, chemoenzymatic approaches have been developed to “glycoengineer” monoclonal antibodies towards more desirable glycan forms, with a focus on achieving homogeneous glycosylation patterns.20–23 For example, N-linked glycans have been enzymatically hydrolyzed and a drug-glycan conjugate subsequently attached.24 Nevertheless, substrate specificities of processing enzymes are not universal, and the aforementioned approaches primarily address homogeneous IgG glycosylation for antibody-drug conjugations. However, the presence of heterogeneous glycosylation patterns may be advantageous in certain applications, such as the development of glycoprotein or bioconjugate vaccines.16,25,26 Accordingly, and to better control glycosylation heterogeneity and thus leverage it in therapeutic applications a better understanding of the underlying mechanisms must be developed.
The traditional approach to controlling glycosylation heterogeneity involves genetic glycoengineering. However, this normally requires extensive cell-line engineering and is limited since such alterations may have unfavourable effects on the cell due to the importance of glycosylation in eukaryotic processes.16,25,26 In parallel, the complexity of the glycosylation machinery makes predictive network engineering demanding and the study of competing glycosylation reactions remains far from trivial. In both genetic and chemoenzymatic glycoengineering, an understanding of the glycosylation machinery and the enzyme kinetics involved is key to understanding and further developing approaches to yield glycoproteins with desired glycosylation patterns. To this end, alternative approaches and techniques able to investigate the processes and effects of glycosylation are required.16,26
In vivo glycosylation parameters, such as protein, saccharide and enzyme concentrations, their availabilities due to competing reactions, and the spatial and temporal separation of reactions between ER and different Golgi cisternae are difficult to control using conventional or “bulk” approaches. Microfluidic technologies, however, allow for the efficient control over reagent (protein and buffer) concentrations, facilitate reaction compartmentalization and enable control over fluxes between user-defined compartments.27 These features make microfluidic systems particularly interesting as tools to mimic the glycosylation machinery in vitro. Indeed, microfluidic technologies have previously been used to synthesize a variety of biomolecules including oligosaccharides and proteins.28–34
The earliest example of the use of microfluidics to mimic aspects of the Golgi apparatus was reported by Linhardt and co-workers in 2009.35 Using a digital microfluidic platform, heparin was enzymatically sulfonated by merging two reactant-containing source droplets. Unfortunately, the low throughput nature and structural complexity of digital microfluidic systems severely limited its application and adoption by others. Additionally, separation of the product from the reaction mixture was achieved by immobilizing the substrate (heparan sulfate) onto streptavidin functionalized magnetic nanoparticles. Although successful, immobilization of the reaction substrate significantly increased the complexity of the workflow. More recently, DeLisa and co-workers presented a continuous flow microfluidic system capable of performing cell-free protein synthesis of superfolder green fluorescent protein (sfGFP) and its subsequent glycosylation.36 In a first module, sfGFP was synthesized from plasmid DNA (encoding the acceptor protein) mixed with a crude yeast cell extract. The reaction product was then delivered to a second module, containing the bacterial OST enzyme, C. jejuni PglB, linked to the surface of the microfluidic channel. Here, a heptasaccharide is transferred from an undecaprenol-pyrophosphate-linked heptasaccharide glycan donor. A final module was then used to isolate the protein product via metal affinity capture. Whilst this study established the feasibility of glycosylation-on-a-chip (transferring an initial glycan to yield a glycoprotein) it should be noted that microfluidic systems have yet to be used to modify the glycosylation profile of glycoproteins. This is in large part due to a number of practical considerations. Of particular importance is the nature of the material used to form the microfluidic device itself. The most common material used to make microfluidic systems is polydimethylsiloxane (PDMS). Unfortunately, PDMS absorbs/adsorbs a wide range of chemical and biological molecules, including proteins.37–41 This is highly problematic when performing complex reactions or assays since macromolecular concentrations will vary in an uncontrollable manner in both space and time. The extent of biofouling within microfluidic systems is (macro)molecule-specific, posing a challenge when creating surface modification chemistries that can be “universally” employed to prevent biofouling.37–41 Indeed, since PDMS-based microfluidic systems are applied to a wide range of chemical/biological problems, involving a variety of small molecules and macromolecules, surface modification chemistries must be tailored on case by basis. To address this issue and create a robust and configurable platform for chip-based glycosylations, we describe the fabrication and testing of a PTFE-based microfluidic platform that employs ER and Golgi resident glycoenzymes to alter glycan structures of a model glycoprotein and generate distinct glycosylation patterns. Our microfluidic approach aims to control key parameters associated with the glycosylation machinery, including enzyme and substrate concentrations, temperature and retention times inside defined compartments. Specifically, enzymatic reactions are compartmentalized in water-in-oil droplets that can be incubated at elevated temperatures for defined periods of time. Passive mixing structures ensure proper mixing of the droplet contents, whilst the geometry of the incubation chambers used ensures a stable and uniform flow of droplets through the system. Additionally, by using picoinjectors, additional enzymes or reactants can be added to pre-formed droplets and subsequently incubated “on chip”. To demonstrate the efficacy of our platform for chip-based glycosylations, we enzymatically modify N-linked Man9GlcNac2 glycosylated yeast protein disulfide isomerase (PDI) by the glycosylation enzymes ER mannosidase I (ERMan I) and Golgi mannosidase I (GM I). Glycosylation patterns are then analyzed using tandem mass spectrometry (MS/MS), building on previous work by Hang et al.15 and Mathew et al.14 on the kinetics for these enzymes, we mimic the initial steps of mammalian glycosylation modifications between the ER and Golgi apparatus and use our microfluidic system to study the kinetics of the glycosylation machinery network.
2. Materials & methods
Recombinant protein and enzyme production and purification
The recombinant glycoprotein protein disulfide isomerase (PDI) and glycoenzymes ERMan I and GM I were constructed, produced and purified as described in previously.14,15 Briefly, secreted PDI was transformed into DH10Bac E. coli cells (#10359016, Thermo Fisher Scientific, Switzerland) for bacmid expression. After isolation of bacmid DNA, sf21 cells (#11497013, Thermo Fisher Scientific, Switzerland) were transfected using Cellfectin™ II (Thermo Fisher Scientific, Switzerland) and the baculovirus stock harvested from the supernatant after 72 hours. Baculovirus stocks of N-terminally His8-tagged, secreted human glycoside hydrolases ERMan I (MAN1B1) and GM I (MAN1A2) were obtained from the glycoenzyme repository.14,42 Baculovirus stocks were amplified in sf21 cells to ensure high titers of infectious virus particles.Protein expression of PDI, ERMan I and GM I was achieved by infecting High-Five™ cells (#B855-02, Thermo Fisher Scientific, Switzerland) through the addition of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 v/v of the respective virus stock to the cell culture. 10 μM of the α-1,2-mannosidase inhibitor kifunensine (Sigma-Aldrich, Switzerland) was added during initial infection of High-Five™ cells with PDI baculovirus to yield a homogeneous PDI glycosylation pattern. Cells were pelleted after 48 hours and then flash-frozen using liquid nitrogen. After cell lysis, with 1% TritonX-100 (Carl Roth, Germany) in phosphate buffered saline solution (PBS, 135 mM NaCl, 2.5 mM KCl, 10 mM Na2HPO4, 1.75 mM KH2PO4, pH = 7.4), the His10-tagged PDI was purified using Protino® Ni-NTA agarose affinity chromatography (Machery-Nagel, Germany) (Fig. S1†).14 Following purification, PDI was immediately buffer exchanged to PBS (pH = 7.4). Protein concentration was determined using a Nano-Drop Spectrophotometer (Thermo Fisher Scientific, Switzerland). PDI was flash-frozen in liquid nitrogen and stored at −80 °C.
100 v/v of the respective virus stock to the cell culture. 10 μM of the α-1,2-mannosidase inhibitor kifunensine (Sigma-Aldrich, Switzerland) was added during initial infection of High-Five™ cells with PDI baculovirus to yield a homogeneous PDI glycosylation pattern. Cells were pelleted after 48 hours and then flash-frozen using liquid nitrogen. After cell lysis, with 1% TritonX-100 (Carl Roth, Germany) in phosphate buffered saline solution (PBS, 135 mM NaCl, 2.5 mM KCl, 10 mM Na2HPO4, 1.75 mM KH2PO4, pH = 7.4), the His10-tagged PDI was purified using Protino® Ni-NTA agarose affinity chromatography (Machery-Nagel, Germany) (Fig. S1†).14 Following purification, PDI was immediately buffer exchanged to PBS (pH = 7.4). Protein concentration was determined using a Nano-Drop Spectrophotometer (Thermo Fisher Scientific, Switzerland). PDI was flash-frozen in liquid nitrogen and stored at −80 °C.
The secreted glycosylation enzymes ERMan I and GM I were harvested from the infected High-Five™ supernatant after 72 hours and centrifuged for 10 minutes at 3500 rcf. The supernatant was then filtered using sterile 0.22 μm filters (TPP Techno Plastic Products, Switzerland) and incubated with 2% v/v Ni-NTA beads (Machery-Nagel, Germany) for 3 hours at 4 °C. As with PDI, glycosylation enzymes were purified over their His10-tag using identical Ni-NTA affinity chromatography protocols (Fig. S2†). After purification, ERMan I and GM I were immediately buffer exchanged to their respective activity buffers (Table S1†). Protein concentrations were determined using a Nano-Drop Spectrophotometer (Thermo Fisher Scientific, Switzerland). The purified glycosylation enzymes were diluted with glycerol (Sigma-Aldrich, Switzerland) (25% v/v final concentration), flash-frozen in liquid nitrogen and stored at −80 °C.
Microfluidic platform design considerations
As described previously, the microfluidic platform comprises four functional modules that allow the encapsulation, mixing, reaction and incubation of substrate glycoprotein and glycosylation enzymes, and the controlled addition of secondary enzymes/reagents after user-defined time periods. Droplets are formed at a flow focusing geometry having a nozzle width of 30 μm and a height of 40 μm. Droplet reaction/incubation times are then defined using a series of constrictions and chambers that redistribute droplets repeatedly as they move along the flow path. This process results in droplet shuffling and provides for control of droplet incubation times without significant incubation time distributions.43 The developed platform contains two droplet incubation modules containing either 117 or 286 170 μm-high chambers in series. The first incubation module has a total volume of 23 μl, whereas the second incubation module has a volume of 60 μl. Introduction of additional enzymes or reagents into preformed droplets is achieved using a picoinjector that incorporates 1 M saltwater electrodes. The height of the picoinjection channel was set equal to the height of the droplet generation module (40 μm) to prevent backflow into the injection channel. Features in the rest of the picoinjection module were made to be 80 μm high, so as to limit any backpressure generated in the module.Microfluidic platform fabrication
Channel patterns were designed using AutoCAD® 2018 software (Autodesk, USA). Master molds were fabricated using a previously described protocol.44 Briefly, SU-8 photoresist layers (GM1070, Gersteltec, Switzerland) of variable thickness were spin coated on a single silicon wafer (Siegert Wafer, Germany). The layer for inlet channels was 40 μm thick, picoinjection layer 80 μm and the droplet incubation and inlet holes were 170 μm thick. Alignment of different layers was performed using a UV-KUB3 mask aligner (Kloe, France). After master mold fabrication, the entire wafer was exposed to chlorotrimethylsilane (Sigma-Aldrich, Switzerland) vapour for at least 1 hour to aid the removal of PDMS later in the fabrication process. PDMS microfluidic devices were fabricated using standard soft-lithographic techniques.44,45 Briefly, this involved casting a PDMS mixture made using a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 w/w ratio of base to curing agent (Elastosil RT 601 A/B, Ameba, Switzerland) onto the patterned silicon wafer and curing for at least 1 hour at 70 °C. The cured PDMS was then peeled off the mold, and individual devices were formed by dicing. 0.76 mm diameter inlet and outlet ports were created using a Shaft 20 catheter punch (Syneo, USA). The PDMS replicas were then plasma bonded onto 76 × 26 mm glass slides (Menzel-Glaser, Germany) using a Zepto air plasma (Diener electronic, Germany) and a 120 °C post-bake for 4 hours. Immediately after bonding, microfluidic channels were filled and incubated with tridecafluoro-1,1,2,2-tetrahydrooctyl-1-trichlorosilane (abcr, Germany) in HFE 7500 Novec oil (Interelec, Switzerland) for 5 minutes and then post-baked at 120 °C for at least 5 hours.
1 w/w ratio of base to curing agent (Elastosil RT 601 A/B, Ameba, Switzerland) onto the patterned silicon wafer and curing for at least 1 hour at 70 °C. The cured PDMS was then peeled off the mold, and individual devices were formed by dicing. 0.76 mm diameter inlet and outlet ports were created using a Shaft 20 catheter punch (Syneo, USA). The PDMS replicas were then plasma bonded onto 76 × 26 mm glass slides (Menzel-Glaser, Germany) using a Zepto air plasma (Diener electronic, Germany) and a 120 °C post-bake for 4 hours. Immediately after bonding, microfluidic channels were filled and incubated with tridecafluoro-1,1,2,2-tetrahydrooctyl-1-trichlorosilane (abcr, Germany) in HFE 7500 Novec oil (Interelec, Switzerland) for 5 minutes and then post-baked at 120 °C for at least 5 hours.
PTFE microfluidic devices were fabricated by adapting a previously described protocols.46,47 First, an uncured PDMS mixture (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 w/w base to curing agent) containing 40 μl saturated (200 mg ml−1 in EtOH) Pluronic F127 (Sigma-Aldrich, Switzerland) per 10 grams PDMS mixture was cast onto the SU-8 master mold. The PDMS-Pluronic mixture was then cured for at least 1 hour at 70 °C and then removed from the master mold. The cured PDMS replica was then heated for 2 minutes in a microwave oven at 700 W. Subsequently, a double-negative PDMS mold was fabricated by casting uncured PDMS (13
1 w/w base to curing agent) containing 40 μl saturated (200 mg ml−1 in EtOH) Pluronic F127 (Sigma-Aldrich, Switzerland) per 10 grams PDMS mixture was cast onto the SU-8 master mold. The PDMS-Pluronic mixture was then cured for at least 1 hour at 70 °C and then removed from the master mold. The cured PDMS replica was then heated for 2 minutes in a microwave oven at 700 W. Subsequently, a double-negative PDMS mold was fabricated by casting uncured PDMS (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 w/w base to curing agent) containing 40 μl saturated Pluronic™ F127 per 10 grams PDMS onto the initial Pluronic-containing PDMS block. This double-negative PDMS mold was cured for 90 minutes, cooled to room temperature over 30 minutes and subsequently removed from the initial PDMS negative mold. Embedding Pluronic™ F127 within the PDMS passivates the PDMS surfaces and reduces polymer movement from one PDMS mold into the double-negative mold during curing. This improves the subsequent separation of the two PDMS blocks. Such a strategy allowed the fabrication of microfluidic channel structures with low width to height ratios. Inlet and outlet ports were created in the double-negative PDMS mold using a Shaft 20 catheter punch (Syneo, USA). Metal pins were inserted into these holes and THV 500GZ PTFE pellets (3 M, Germany) melted onto the double-negative PDMS mold overnight in a Vacucenter VC50 vacuum oven (SalvisLab, Switzerland) at 200 °C. Next, the molten PTFE block was cooled to room temperature and the metal pins removed using household pliers. The PTFE block was then placed face down onto a flat THV 500GZ sheet that was spin coated from a 5% THV 221GZ PTFE solution in acetone (3 M, Germany). The two parts were pressed together with minimal pressure using a custom-made bonding device based on Ren et al.47 but with metal instead of glass plates. The complete PTFE device was thermally bonded at 115 °C for at least 2 hours. After bonding, hollow metal connections (Chuang Mei Wei Technology, China) having an outer diameter of 0.76 mm were connected to 0.56 mm ID PTFE tubing (Rotima, Switzerland) and inserted into the fully bonded PTFE device.
1 w/w base to curing agent) containing 40 μl saturated Pluronic™ F127 per 10 grams PDMS onto the initial Pluronic-containing PDMS block. This double-negative PDMS mold was cured for 90 minutes, cooled to room temperature over 30 minutes and subsequently removed from the initial PDMS negative mold. Embedding Pluronic™ F127 within the PDMS passivates the PDMS surfaces and reduces polymer movement from one PDMS mold into the double-negative mold during curing. This improves the subsequent separation of the two PDMS blocks. Such a strategy allowed the fabrication of microfluidic channel structures with low width to height ratios. Inlet and outlet ports were created in the double-negative PDMS mold using a Shaft 20 catheter punch (Syneo, USA). Metal pins were inserted into these holes and THV 500GZ PTFE pellets (3 M, Germany) melted onto the double-negative PDMS mold overnight in a Vacucenter VC50 vacuum oven (SalvisLab, Switzerland) at 200 °C. Next, the molten PTFE block was cooled to room temperature and the metal pins removed using household pliers. The PTFE block was then placed face down onto a flat THV 500GZ sheet that was spin coated from a 5% THV 221GZ PTFE solution in acetone (3 M, Germany). The two parts were pressed together with minimal pressure using a custom-made bonding device based on Ren et al.47 but with metal instead of glass plates. The complete PTFE device was thermally bonded at 115 °C for at least 2 hours. After bonding, hollow metal connections (Chuang Mei Wei Technology, China) having an outer diameter of 0.76 mm were connected to 0.56 mm ID PTFE tubing (Rotima, Switzerland) and inserted into the fully bonded PTFE device.
Microfluidic platform characterization
Operational testing of the microfluidic system was performed using an Eclipse Ti-E inverted microscope (Nikon, Switzerland) equipped with a Plan Fluor 4×/0.13 objective (Nikon, Switzerland) or a Plan Fluor 10×/0.3 objective (Nikon, Switzerland) and a MotionPro Y5 high-speed camera (IDT Vision, USA). A Dino-Lite digital microscope (AnMo Electronics, Taiwan) was used to monitor droplets during enzymatic assays.Enzymatic reactions
All enzymatic assays were performed at 42 °C by placing the entire microfluidic device on a hot plate. Prior to use, the respective glycosylation enzymes ERMan I and/or GM I were diluted to 22.5 μg ml−1 in the respective enzyme activity buffer (Table S1†) and 2.25 % v/v glycerol. PDI (1 mg ml−1) and ERMan I (22.5 μg ml−1) and/or GM I (22.5 μg ml−1) were taken up in 0.56 mm ID PTFE tubing (Rotima AG, Switzerland) connected to 1 mL Hamilton glass syringes (Sigma-Aldrich, Switzerland) containing water (MicroPure™ UV/UF 0.2 μm, Thermo Scientific, Germany) as a supporting fluid. An air bubble between the sample and the supporting fluid in the PTFE tubing prevented contamination and dilution of the sample by the supporting fluid whilst allowing for complete sample consumption. For experiments involving only one glycosylation enzyme, an equivalent blank buffer solution was prepared accordingly.The oil phase, consisting of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of droplet generation oil (Bio-Rad Laboratories, Switzerland) and HFE 7500 Novec oil, was taken up in a 1 mL Hamilton glass syringe. Precision neMESYS syringe pumps (CETONI, Germany) were used to move all fluids and provide for a stable flow rate of 0.8 μl min−1 for each fluid. This resulted in an overall flowrate through the incubation chambers of 2.4 μl min−1 prior picoinjection and 3.2 μl min−1 post picoinjection. For picoinjection, a voltage of 60 V at 1 kHz was applied to the saltwater electrodes. Droplets were collected in 40 μl 100% trichloroacetic acid (500 μg in 227 μl H2O, Sigma-Aldrich, Switzerland) and 60 μl PBS. The aqueous phase was isolated from the oil phase using 1H,1H,2H,2H-perfluorooctanol (Apollo Scientific Ltd, UK). The recovered protein solution was precipitated in 15% v/v trichloroacetic acid for 10 minutes and pelleted for 5 minutes at 20′000 rcf and 4 °C. The resulting protein pellet was washed 3 times with acetone, air dried and stored at 20 °C as previously described.14
1 mixture of droplet generation oil (Bio-Rad Laboratories, Switzerland) and HFE 7500 Novec oil, was taken up in a 1 mL Hamilton glass syringe. Precision neMESYS syringe pumps (CETONI, Germany) were used to move all fluids and provide for a stable flow rate of 0.8 μl min−1 for each fluid. This resulted in an overall flowrate through the incubation chambers of 2.4 μl min−1 prior picoinjection and 3.2 μl min−1 post picoinjection. For picoinjection, a voltage of 60 V at 1 kHz was applied to the saltwater electrodes. Droplets were collected in 40 μl 100% trichloroacetic acid (500 μg in 227 μl H2O, Sigma-Aldrich, Switzerland) and 60 μl PBS. The aqueous phase was isolated from the oil phase using 1H,1H,2H,2H-perfluorooctanol (Apollo Scientific Ltd, UK). The recovered protein solution was precipitated in 15% v/v trichloroacetic acid for 10 minutes and pelleted for 5 minutes at 20′000 rcf and 4 °C. The resulting protein pellet was washed 3 times with acetone, air dried and stored at 20 °C as previously described.14
Mass spectrometry measurements and glycoform quantification
For MS analysis, precipitated protein pellets were resuspended in 400 μl of 8 M urea (Sigma-Aldrich, Switzerland). Sample was then processed according to a previously described protocol.14 Briefly, proteins were first reduced in 50 mM dithiothreitol (Sigma-Aldrich, Switzerland) followed by alkylation in 130 mM iodoacetic acid (Sigma-Aldrich, Switzerland) and 50 mM ammonium bicarbonate (Sigma-Aldrich, Switzerland) for 30 minutes at 37 °C to facilitate trypsin digestion using a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80 trypsin (Promega, Switzerland) to PDI weight ratio (overnight at 37 °C). Following tryptic digestion, the resulting peptides were desalted with 0.6 μL of Zip-Tip C18 resin (Milipore, Ireland) and dried until use. MS/MS measurements were performed using a calibrated Q Exactive mass spectrometer (Thermo Fisher Scientific, Switzerland) coupled to a Acquity UPLC M-Class system (Waters AG, Switzerland) with a PV-500 Picoview nanospray source (Sciex, USA).
80 trypsin (Promega, Switzerland) to PDI weight ratio (overnight at 37 °C). Following tryptic digestion, the resulting peptides were desalted with 0.6 μL of Zip-Tip C18 resin (Milipore, Ireland) and dried until use. MS/MS measurements were performed using a calibrated Q Exactive mass spectrometer (Thermo Fisher Scientific, Switzerland) coupled to a Acquity UPLC M-Class system (Waters AG, Switzerland) with a PV-500 Picoview nanospray source (Sciex, USA).
MS/MS data were analyzed using Xcalibur 4.0 software (Thermo Fisher Sceintific, Switzerland) as described previously.14,15 Spectral peak areas were defined manually and integrated. For simplicity, only the relative amounts of glycoforms on the glycosylation site 2 peptide are investigated in the current study. Additional information regarding the investigated glycosylation site 2 peptide is provided in Table S2† and in Mathew et al.14
Assessment of protein adsorption “on-chip”
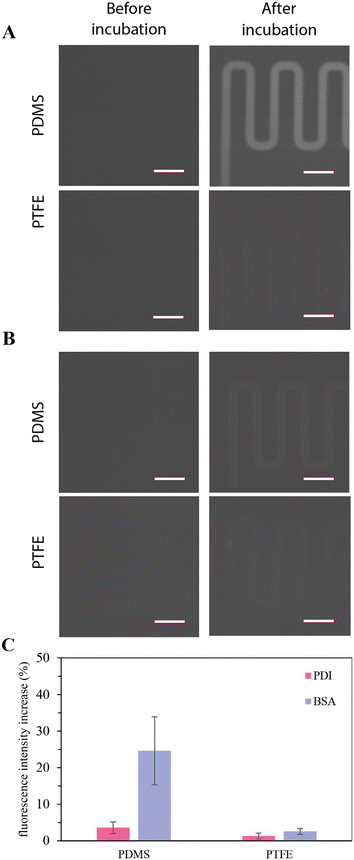
For experiments in PDMS and Teflon-based microfluidic devices PDI was expressed in High-Five™ cells using a baculovirus expression system described above. For the adsorption experiments PDI was not co-expressed with the α-1,2-mannosidase inhibitor kifunensine (Sigma-Aldrich, Switzerland).Recombinant PDI and commercially available bovine serum albumin (Sigma-Aldrich, Switzerland) were fluorescently labelled by incubating 15.9 μM PDI or BSA in PBS (pH = 7.4) with 100 molar equivalents of NHS modified Atto 488 in DMSO (ATTO-TEC, Germany) at 37 °C for 4 hours followed by four buffer exchanges to PBS (pH = 7.4).
For protein adsorption experiments, 50 μm wide and 40 μm high microfluidic channels were initially flushed with PBS (pH 7.4). This was then replaced by a previously prepared fluorescent protein solutions and incubated for five minutes. Subsequently, channels were flushed with 10 μl PBS (pH 7.4). Residual fluorescence originating from microchannels after flushing was quantified and compared to the pre-incubation background measurement. Fluorescence detection was performed using an Eclipse Ti-E inverted microscope (Nikon, Switzerland). Fluorescence emission was collected using a Plan Fluor 10×/0.3 objective (Nikon, Switzerland), filtered through 469/35 excitation and 525/39 emission filters (IDEX Health & Science, USA). Fluorescence emission was detected using an ORCA-flash 4.0 CMOS camera (Hamamatsu, Solothurn, Switzerland). μManager 1.4 software was used to control and automate fluorescence collection, while ImageJ software (National Institutes of Health, USA) was used for image processing and analysis.
3. Results
Microfluidic substrate
As discussed previously, the microfluidic platform aims to enable spatiotemporal control over protein concentrations by compartmentalizing enzymatic reactions in droplets and employing picoinjection to add additional enzymes, reactants, substrates, or buffers. A basic consideration in this regard is the choice of substrate material. Despite the widespread adoption of PDMS by the microfluidics community, biofouling of microfluidic channel surfaces will severely compromise the study of biochemical reaction networks. As expected, the use of PDMS-based microfluidic devices in preliminary experiments was characterized by the depletion (adsorption/absorption) of enzyme prior to droplet formation at the flow focusing geometry. Such biofouling impaired downstream enzymatic reactions in droplets (Fig. S3B–D†). To assess the likelihood and/or magnitude of biofouling, 15.9 μM fluorescently labelled protein disulfide isomerase (PDI) and bovine serum albumin (BSA) solutions were separately incubated in 50 μm × 40 μm cross-section microfluidic channels within PDMS and PTFE substrates for 5 minutes. Subsequently, channels were flushed with 10 μl PBS and residual fluorescence measured. As shown in Fig. 1A and C, Atto-488-labelled BSA remains on or in the microfluidic substrate after washing for PDMS (top) but not for PTFE (bottom). In contrast, only a slight increase in fluorescence after incubation with Atto-488-labelled PDI (and washing) was seen for the PDMS substrate (top), and even less for the PTFE substrate, indicating reduced biofouling (Fig. 1B and C).Microfluidic platform
Inside a microfluidic chip, substrate glycoprotein and glycosylation enzymes were co-encapsulated and subsequently incubated “on-chip” for an extended period of time of up to 30 minutes. As glycan hydrolysis is rarely a terminal reaction and further glycan processing by the same enzyme may occur over time, “off-chip” storage of reaction intermediates was limited. The addition of subsequent reaction mixtures was therefore implemented within the same microfluidic chip. A schematic of the entire microfluidic platform is presented in Fig. 2. It comprises four adaptable modules. In the first, droplets are generated at a flow focusing geometry with substrate and enzyme being co-encapsulated and passively mixed by chaotic advection within a winding channel section.46,48 Each reagent flux can be independently controlled by the user to regulate droplet payloads. The second module integrates a series of incubation chambers that ensures a stable flow of droplets over an extended incubation period. The average residence time of droplets in this incubation module is controlled by the inlet flow rates and the number of incubation chambers.43 In the current study, the total residence time of droplets in the incubation module was approximately 10 minutes. The third module contains a picoinjector and allows the controlled addition of small volumes of additional enzyme, substrate, or buffer to the droplets emerging from the first incubation module.49,50 Picoinjection works by flowing droplets past a channel containing a pressurized reagent stream. If a droplet is enveloped by a surfactant layer, this fluid stream will normally not enter the droplet. However, application of an electric field can be used to destabilize and breach the surfactant layer, allowing the reagent stream to enter the droplet over a short period of time. The process is highly robust and allows controlled addition of femtolitre–picolitre volumes at kilohertz rates. To reduce the probability of droplet fusion during picoinjection, a grounding electrode that acts as a shielding electrode is employed.50,51 The final module has a similar structure to the second module and provides for the controllable incubation of droplets for periods up to 20 minutes after picoinjection.Biosynthetic system
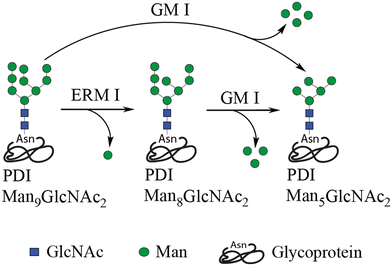
To prove principle, we focused our attention on glycan processing enzymes of the ER and early Golgi glycosylation machinery. While ERMan I hydrolyses one terminal Mannose on the N-linked glycan Man9GlcNAc2 down to Man8GlcNac2, GM I can hydrolyse the glycan further to yield Man5GlcNAc2,14,52 as shown in Fig. 3. Due to the fact that the ER and Golgi are separated in living systems, a combined mode of action is possible where ERMan I cleaves off a first mannose in the ER, with GM I subsequently trimming the glycan further within the Golgi apparatus. Additionally, it should be noted that prolonged incubation of Man8GlcNAc2 with ERMan I can lead to further mannose trimming in vitro.14First, the glycoprotein substrate (PDI) was expressed in presence of the α-1,2-mannosidase inhibitor (kifunensine) to yield homogeneously glycosylated PDI bearing the N-linked Man9GlcNAc2 structure. For simplicity, we focused our analysis on glycosylation site 2 of PDI. Enzymatic assays were performed entirely within the microfluidic device and followed by droplet collection and protein precipitation off-chip. Glycosylation patterns were then analysed by tandem mass spectrometry (MS/MS).
Microfluidic mimicking of the glycosylation machinery
As described, we first validated our microfluidic platform by performing the enzymatic reactions shown in Fig. 3 within a first-generation microfluidic device shown in Fig. S4.† First, PDI was co-encapsulated with either ERMan I or GM I at the flow focusing geometry, while a blank buffer solution was used in the picoinjector leading to a total incubation time of 18 minutes. The resulting glycosylation patterns for glycosylation site 2 of PDI are shown in Fig. 4A. As expected, incubating PDI with ERMan I inside droplets yielded Man8GlcNAc2 as the dominant N-linked glycan. The incubation of PDI and GM I yielded a more heterogeneous glycosylation pattern, with a slow trimming of the first mannose, but accelerated trimming of further mannoses. As a control and to exclude any effects that picoinjection may have on enzyme reactivity, droplets containing the PDI substrate without glycosylation enzymes were picoinjected with ERMan I and GM I. The resulting glycosylation pattern shown in Fig. 4B(left) indicates that picoinjection has a negligible effect on enzyme activity. The slightly reduced processing of Man9GlcNAc2 by ERMan I can be explained by the shorter incubation time of 8 minutes due to the absence of enzyme in the droplets during the first incubation module. We subsequently used the final microfluidic device shown in Fig. 2 to mimic the early steps of N-linked glycan processing by the glycosylation machinery. In this proof-of-concept, we investigated the sequential actions of ERMan I and the Golgi resident GM I. Compartmentalization inside droplets ensured their spatial and temporal separation, with droplets containing the PDI substrate and ERMan I being initially incubated on-chip for 10 minutes. The subsequent addition of GM I being achieved by picoinjection was followed by the second incubation for 20 minutes. As shown in Fig. 4C, the combined and sequential action of the two glycosylation enzymes greatly enhances the hydrolysis of Man9GlcNAc2, with Man8GlcNAc2 and Man5GlcNAc2 being the dominant hydrolysis products. These findings suggest increased reaction kinetics of GM I for the conversion of Man7GlcNAc2 to Man6GlcNAc2 and Man5GlcNAc2, which is in good agreement with recent investigations into the reaction kinetics of the employed glycosylation enzymes.144. Discussion
In the current study, we have presented a droplet-based microfluidic platform able to mimic the transition between the ER and early Golgi by controlling the spatiotemporal separation of enzymatic reactions with ER and Golgi resident mannosidases. When mimicking mammalian glycosylation, it is important to consider that glycosylation enzymes are sensitive to environmental conditions and produced at relatively low concentrations in cell culture. This dictates that enzymes should only minimally interact with substrate material of the microfluidic device to avoid their depletion. Any biofouling alters reaction composition in the microfluidic system, which in turn alters or prevents enzymatic reactions. The use of droplet-based microfluidics addresses this issue in part by encapsulating enzymatic reactions and minimizing the interaction between enzyme and microfluidic channel surfaces. However, prior to droplet formation both enzyme and substrate contact the microfluidic channel walls. Such residual biofouling can have severe effects, especially or when processing sensitive enzymes or working with low enzyme concentrations. Accordingly, PDMS is poorly suited for use in such investigations due to its propensity to biofoul. Instead, we fabricated of PTFE-based microfluidic devices, provides access to a far wider range of glycosylation reactions. Although the developed microfluidic platform is able to mimic aspects of the early Golgi-linked glycosylation machinery, the capacity to control molecular fluxes spatially and temporally, means that it can be easily adapted to investigate other parts of the glycosylation machinery and more complex enzymatic networks. For example, the current platform could be extended to incorporate further enzymatic reactions such as those involving glycosyl transferases. Picoinjection could be used to supply reaction droplets with glycosyl transferase (GnT I) and its corresponding saccharide substrate (UDP-GlcNAc). Here, the attachment of a first GlcNAc to the N-linked glycan is crucial in forming hybrid and complex N-linked glycans.53–55 Picoinjection of kifunensine at varying concentrations could then be used to inhibit glycan trimming by preceding α-1,2-mannosidases. Additionally, enzymes could be removed from droplets using the enzyme's Strep II tag, available in the employed expression system of the glycoenzyme repository.42 Here, magnetic nanoparticles functionalized to bind the Strep II tag could be picoinjected into the droplets, with enzyme–nanoparticle complexes subsequently being removed through the use of magnetic fields and asymmetric droplet splitting as described by Choi and co-workers.56 Alternatively, enzyme nanoparticle conjugations could be used to perform glycosylation reactions. From an analytical standpoint, further insight would be gained through expanding the analyzed glycosylation patterns to other glycosylation sites. In the current study, we focused our analysis on glycosylation site 2 of PDI. Extension to the other four glycosylation sites of PDI would likely yield additional information about surface-specific enzyme–protein interactions.12,14 Finally, our work can also be seen in light of recent advances where microfluidic systems have been used to synthesize glycoproteins.36 For example, by combining our work with such advances, one could possibly synthesize glycoproteins in vitro and subsequently study their processing within an integrated microfluidic platform. Finally, enzyme-functionalized magnetic nanoparticles could be co-encapsulated with the reactants in our microfluidic platform. Using asymmetric droplet splitting, one could then also regenerate the precious enzymes for further experiments.Abbreviations
| Asn | Asparagine |
| BSA | Bovine serum albumin |
| ER | Endoplasmic reticulum |
| ERMan I | ER mannosidase I |
| Glc | Glucose |
| GlcNAc | N-Acetylglucosamine |
| GM I | Golgi mannosidase I |
| Man | Mannose |
| MS | Mass spectrometry |
| MS/MS | Tandem mass spectrometry |
| OST | Oligosaccharyltransferase |
| PBS | Phosphate buffered saline |
| PDI | Protein disulfide isomerase |
| PDMS | Polydimethylsiloxane |
| PTFE | Polytetrafluorethylene |
| Ser | Serine |
| sfGFP | Superfolder green fluorescent protein |
| Thr | Threonine |
Data availability
Data supporting this article have been included as part of the ESI.† Further data will be shared on reasonable request to the corresponding author.Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
This work was partially supported by ETH Zürich. The authors would like to thank the Functional Genomics Center Zürich and specifically Dr. Chia-wei Lin for assistance with mass spectrometry analysis. We would also like to acknowledge prior work performed by Dr. Corina Mathew on the biosynthetic system. We also acknowledge Professor Donald Jarvis and Professor Kelley Moremen who developed the “Glycoenzyme repository” from which the initial expression constructs for the enzymes ERMan I and GM I were obtained.References
- M. Abu-Qarn, J. Eichler and N. Sharon, Not just for Eukarya anymore: protein glycosylation in Bacteria and Archaea, Curr. Opin. Struct. Biol., 2008, 18(5), 544–550 CrossRef CAS PubMed.
- K. T. Schjoldager, Y. Narimatsu, H. J. Joshi and H. Clausen, Global view of human protein glycosylation pathways and functions, Nat. Rev. Mol. Cell Biol., 2020, 21(12), 729–749 CrossRef CAS PubMed.
- D. F. Zielinska, F. Gnad, K. Schropp, J. R. Wisniewski and M. Mann, Mapping N-Glycosylation Sites across Seven Evolutionarily Distant Species Reveals a Divergent Substrate Proteome Despite a Common Core Machinery, Mol. Cell, 2012, 46(4), 542–548 CrossRef CAS PubMed.
- J. W. Dennis, K. S. Lau, M. Demetriou and I. R. Nabi, Adaptive Regulation at the Cell Surface by N-Glycosylation, Traffic, 2009, 10(11), 1569–1578 CrossRef CAS.
- A. Helenius and M. Aebi, Roles of N-linked glycans in the endoplasmic reticulum, Annu. Rev. Biochem., 2004, 73, 1019–1049 CrossRef CAS PubMed.
- C. A. Jakob, P. Burda, J. Roth and M. Aebi, Degradation of misfolded endoplasmic reticulum glycoproteins in Saccharomyces cerevisiae is determined by a specific oligosaccharide structure, J. Cell Biol., 1998, 142(5), 1223–1233 CrossRef CAS PubMed.
- A. Varki, Biological roles of oligosaccharides: all of the theories are correct, Glycobiology, 1993, 3(2), 97–130 CrossRef CAS PubMed.
- A. Varki, R. D. Cummings, J. D. Esko, P. Stanley, G. W. Hart and M. Aebi, et al., Essentials of Glycobiology, 3rd edn, 2017 Search PubMed.
- Y. Mimura, T. Katoh, R. Saldova, R. O'Flaherty, T. Izumi and Y. Mimura-Kimura, et al., Glycosylation engineering of therapeutic IgG antibodies: challenges for the safety, functionality and efficacy, Protein Cell, 2018, 9(1), 47–62 CrossRef CAS PubMed.
- D. J. Kelleher and R. Gilmore, An evolving view of the eukaryotic oligosaccharyltransferase, Glycobiology, 2006, 16(4), 47R–62R CrossRef CAS PubMed.
- R. Wild, J. Kowal, J. Eyring, E. M. Ngwa, M. Aebi and K. P. Locher, Structure of the yeast oligosaccharyltransferase complex gives insight into eukaryotic N-glycosylation, Science, 2018, 359(6375), 545–549 CrossRef CAS PubMed.
- M. E. Losfeld, E. Scibona, C. W. Lin, T. K. Villiger, R. Gauss and M. Morbidelli, et al., Influence of protein/glycan interaction on site-specific glycan heterogeneity, FASEB J., 2017, 31(10), 4623–4635 CrossRef CAS PubMed.
- K. W. Moremen, M. Tiemeyer and A. V. Nairn, Vertebrate protein glycosylation: diversity, synthesis and function, Nat. Rev. Mol. Cell Biol., 2012, 13(7), 448–462 CrossRef CAS PubMed.
- C. Mathew, R. G. Weiss, C. Giese, C. W. Lin, M. E. Losfeld and R. Glockshuber, et al., Glycan-protein interactions determine kinetics of N-glycan remodeling, RSC Chem. Biol., 2021, 2(3), 917–931 CAS.
- I. Hang, C. W. Lin, O. C. Grant, S. Fleurkens, T. K. Villiger and M. Soos, et al., Analysis of site-specific N-glycan remodeling in the endoplasmic reticulum and the Golgi, Glycobiology, 2015, 25(12), 1335–1349 CrossRef CAS PubMed.
- M. J. Buettner, S. R. Shah, C. T. Saeui, R. Ariss and K. J. Yarema, Improving Immunotherapy Through Glycodesign, Front. Immunol., 2018, 9, 2485 Search PubMed.
- S. Elliott, T. Lorenzini, S. Asher, K. Aoki, D. Brankow and L. Buck, et al., Enhancement of therapeutic protein in vivo activities through glycoengineering, Nat. Biotechnol., 2003, 21(4), 414–421 CAS.
- C. Ferrara, S. Grau, C. Jager, P. Sondermann, P. Brunker and I. Waldhauer, et al., Unique carbohydrate-carbohydrate interactions are required for high affinity binding between Fc gamma RIII and antibodies lacking core fucose, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(31), 12669–12674 CrossRef CAS PubMed.
- P. Sondermann, R. Huber, V. Oosthuizen and U. Jacob, The 3.2-angstrom crystal structure of the human IgG1 Fc fragment-Fc gamma RIII complex, Nature, 2000, 406(6793), 267–273 CrossRef CAS PubMed.
- R. M. Anthony, F. Nimmerjahn, D. J. Ashline, V. N. Reinhold, J. C. Paulson and J. V. Ravetch, Recapitulation of IVIG anti-inflammatory activity with a recombinant IgG fc, Science, 2008, 320(5874), 373–376 CAS.
- J. P. Giddens, J. V. Lomino, D. J. DiLillo, J. V. Ravetch and L. X. Wang, Site-selective chemoenzymatic glycoengineering of Fab and Fc glycans of a therapeutic antibody, Proc. Natl. Acad. Sci. U. S. A., 2018, 115(47), 12023–12027 CAS.
- F. Higel, A. Seidl, F. Sorgel and W. Friess, N-glycosylation heterogeneity and the influence on structure, function and pharmacokinetics of monoclonal antibodies and Fc fusion proteins, Eur. J. Pharm. Biopharm., 2016, 100, 94–100 CrossRef CAS PubMed.
- W. Huang, J. Giddens, S. Q. Fan, C. Toonstra and L. X. Wang, Chemoenzymatic Glycoengineering of Intact IgG Antibodies for Gain of Functions, J. Am. Chem. Soc., 2012, 134(29), 12308–12318 CrossRef CAS PubMed.
- F. Tang, L. X. Wang and W. Huang, Chemoenzymatic synthesis of glycoengineered IgG antibodies and glycosite-specific antibody-drug conjugates, Nat. Protoc., 2017, 12(8), 1702–1721 CrossRef CAS PubMed.
- R. Donini, S. M. Haslam and C. Kontoravdi, Glycoengineering Chinese hamster ovary cells: a short history, Biochem. Soc. Trans., 2021, 49(2), 915–931 CrossRef CAS PubMed.
- B. Ma, X. Y. Guan, Y. H. Li, S. Y. Shang, J. Li and Z. P. Tan, Protein Glycoengineering: An Approach for Improving Protein Properties, Front. Chem., 2020, 8, 622 CrossRef CAS PubMed.
- O. J. Dressler, I. S. X. Casadevall and A. J. deMello, Chemical and Biological Dynamics Using Droplet-Based Microfluidics, Annu. Rev. Anal. Chem., 2017, 10(1), 1–24 CrossRef PubMed.
- M. H. Ayoubi-Joshaghani, H. Dianat-Moghadam, K. Seidi, A. Jahanban-Esfahalan, P. Zare and R. Jahanban-Esfahlan, Cell-free protein synthesis: The transition from batch reactions to minimal cells and microfluidic devices, Biotechnol. Bioeng., 2020, 117(4), 1204–1229 CrossRef CAS PubMed.
- J. Y. Ma, Y. C. Wang and J. Liu, Biomaterials Meet Microfluidics: From Synthesis Technologies to Biological Applications, Micromachines, 2017, 8(8), 255 CrossRef PubMed.
- T. W. Murphy, J. Y. Sheng, L. B. Naler, X. Y. Feng and C. Lu, On-chip manufacturing of synthetic proteins for point-of-care therapeutics, Microsyst. Nanoeng., 2019, 5, 13 Search PubMed.
- Y. Ono, M. Kitajima, S. Daikoku, T. Shiroya, S. Nishihara and Y. Kanie, et al., Sequential enzymatic glycosyltransfer reactions on a microfluidic device: Synthesis of a glycosaminoglycan linkage region tetrasaccharide, Lab Chip, 2008, 8(12), 2168–2173 CAS.
- F. Pinnock and S. Daniel, Small tools for sweet challenges: advances in microfluidic technologies for glycan synthesis, Anal. Bioanal. Chem., 2022, 414, 5139–5163 CAS.
- M. Weiss, J. P. Frohnmayer, L. T. Benk, B. Haller, J. W. Janiesch and T. Heitkamp, et al., Sequential bottom-up assembly of mechanically stabilized synthetic cells by microfluidics, Nat. Mater., 2018, 17(1), 89 CAS.
- A. T. Woolley, D. Hadley, P. Landre, A. J. deMello, R. A. Mathies and M. A. Northrup, Functional integration of PCR amplification and capillary electrophoresis in a microfabricated DNA analysis device, Anal. Chem., 1996, 68(23), 4081–4086 CAS.
- J. G. Martin, M. Gupta, Y. M. Xu, S. Akella, J. Liu and J. S. Dordick, et al., Toward an Artificial Golgi: Redesigning the Biological Activities of Heparan Sulfate on a Digital Microfluidic Chip, J. Am. Chem. Soc., 2009, 131(31), 11041–11048 CAS.
- A. K. Aquino, Z. A. Manzer, S. Daniel and M. P. DeLisa, Glycosylation-on-a-Chip: A Flow-Based Microfluidic System for Cell-Free Glycoprotein Biosynthesis, Front. Mol. Biosci., 2021, 8, 782905 CAS.
- D. Belder and M. Ludwig, Surface modification in microchip electrophoresis, Electrophoresis, 2003, 24(21), 3595–3606 CrossRef CAS.
- A. Gokaltun, Y. B. Kang, M. L. Yarmush, O. B. Usta and A. Asatekin, Simple Surface Modification of Poly(dimethylsiloxane) via Surface Segregating Smart Polymers for Biomicrofluidics, Sci. Rep., 2019, 9, 7377 CrossRef PubMed.
- A. Gokaltun, M. L. Yarmush, A. Asatekin and O. B. Usta, Recent advances in nonbiofouling PDMS surface modification strategies applicable to microfluidic technology, Technology., 2017, 5(1), 1–12 CrossRef PubMed.
- M. W. Toepke and D. J. Beebe, PDMS absorption of small molecules and consequences in microfluidic applications, Lab Chip, 2006, 6(12), 1484–1486 RSC.
- T. Robinson, Y. Schaerli, R. Wootton, F. Hollfelder, C. Dunsby and G. Baldwin, et al., Removal of background signals from fluorescence thermometry measurements in PDMS microchannels using fluorescence lifetime imaging, Lab Chip, 2009, 9(23), 3437–3441 RSC.
- K. W. Moremen, A. Ramiah, M. Stuart, J. Steel, L. Meng and F. Forouhar, et al., Expression system for structural and functional studies of human glycosylation enzymes, Nat. Chem. Biol., 2018, 14(2), 156 CrossRef CAS.
- L. Frenz, K. Blank, E. Brouzes and A. D. Griffiths, Reliable microfluidic on-chip incubation of droplets in delay-lines, Lab Chip, 2009, 9(10), 1344–1348 RSC.
- Y. N. Xia and G. M. Whitesides, Soft lithography, Annu. Rev. Mater. Sci., 1998, 28, 153–184 CrossRef CAS.
- T. Moragues, D. Arguijo, T. Beneyton, C. Modavi, K. Simutis and A. R. Abate, et al., Droplet-based microfluidics, Nat. Rev. Methods Primers, 2023, 3(1), 32 CrossRef CAS.
- D. Hess, V. Dockalova, P. Kokkonen, D. Bednar, J. Damborsky and A. DeMello, et al., Exploring mechanism of enzyme catalysis by on-chip transient kinetics coupled with global data analysis and molecular modeling, Chem, 2021, 7(4), 1066–1079 CAS.
- K. N. Ren, W. Dai, J. H. Zhou, J. Su and H. K. Wu, Whole-Teflon microfluidic chips, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(20), 8162–8166 CAS.
- H. Song, J. D. Tice and R. F. Ismagilov, A microfluidic system for controlling reaction networks in time, Angew. Chem., Int. Ed., 2003, 42(7), 768–772 CrossRef CAS PubMed.
- A. R. Abate, T. Hung, P. Mary, J. J. Agresti and D. A. Weitz, High-throughput injection with microfluidics using picoinjectors, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(45), 19163–19166 CAS.
- A. Sciambi and A. R. Abate, Generating electric fields in PDMS microfluidic devices with salt water electrodes, Lab Chip, 2014, 14(15), 2605–2609 RSC.
- B. O'Donovan, D. J. Eastburn and A. R. Abate, Electrode-free picoinjection of microfluidic drops, Lab Chip, 2012, 12(20), 4029–4032 RSC.
- A. Lal, P. Pang, S. Kalelkar, P. A. Romero, A. Herscovics and K. W. Moremen, Substrate specificities of recombinant murine Golgi alpha 1,2-mannosidases IA and IB and comparison with endoplasmic reticulum and Golgi processing alpha 1,2-mannosidases, Glycobiology, 1998, 8(10), 981–995 CrossRef CAS PubMed.
- E. Ioffe and P. Stanley, Mice lacking N-acetylglucosaminyltransferase I activity die at mid-gestation, revealing an essential role for complex or hybrid N-linked carbohydrates, Proc. Natl. Acad. Sci. U. S. A., 1994, 91(2), 728–732 CrossRef CAS PubMed.
- H. Schachter, biosynthetic controls that determine the branching and microheterogeneity of protein-bound oligosaccharides, Biochemistry and Cell Biology-Biochimie Et Biologie Cellulaire., 1986, 64(3), 163–181 CAS.
- H. Schachter, Complex N-glycans: the story of the “yellow brick road”, Glycoconjugate J., 2014, 31(1), 1–5 CrossRef CAS PubMed.
- N. Choi, J. Lee, J. Ko, J. H. Jeon, G. Rhie and A. J. Demello, et al., Integrated SERS-Based Microdroplet Platform for the Automated Immunoassay of F1 Antigens in Yersinia pestis, Anal. Chem., 2017, 89(16), 8413–8420 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5lc00005j |
| This journal is © The Royal Society of Chemistry 2025 |