Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The effect of cyclic fluid perfusion on the proinflammatory tissue environment in osteoarthritis using equine joint-on-a-chip models†
Johannes Heidenberger‡
 a,
Eva I. Reihsab,
Jonathan Straussab,
Martin Frauenlobb,
Sinan Gültekinc,
Iris Gernerc,
Stefan Toegelad,
Peter Ertl
a,
Eva I. Reihsab,
Jonathan Straussab,
Martin Frauenlobb,
Sinan Gültekinc,
Iris Gernerc,
Stefan Toegelad,
Peter Ertl b,
Reinhard Windhagera,
Florien Jennerc and
Mario Rothbauer‡
b,
Reinhard Windhagera,
Florien Jennerc and
Mario Rothbauer‡ *ab
*ab
aDepartment of Orthopedics and Trauma Surgery, Karl Chiari Lab for Orthopaedic Biology, Medizinische Universität Wien, Währinger Gürtel 18-20, 1090 Vienna, Austria. E-mail: mario.rothbauer@muv.ac.at
bFaculty of Technical Chemistry, Technische Universität Wien, Getreidemarkt 9/163, 1060 Vienna, Austria. E-mail: mario.rothbauer@tuwien.ac.at
cDepartment for Small Animals and Horses, Centre for Equine Health and Research, Equine Surgery Unit, Veterinary Regenerative Medicine Laboratory, University of Veterinary Medicine Vienna, Veterinärplatz 1, 1210 Vienna, Austria
dLudwig Boltzmann Institute for Arthritis and Rehabilitation, Vienna, Austria
First published on 24th March 2025
Abstract
Osteoarthritis (OA) is a prevalent degenerative joint disorder characterized by cartilage degradation, chronic inflammation, and progressive joint dysfunction. Despite rising incidences driven by ageing and increasing obesity, potent treatments remain elusive, exacerbating the socioeconomic burden. OA pathogenesis involves an imbalance in extracellular matrix (ECM) turnover, mediated by inflammatory cytokines and matrix-degrading enzymes, leading to oxidative stress, chondrocyte apoptosis, and ECM degradation. Additionally, synovial inflammation (synovitis) plays a critical role in disease progression through molecular crosstalk with cartilage and other joint tissues. Existing in vitro and in vivo OA models face significant limitations in replicating human pathophysiology, particularly the complex interplay between joint tissues. Equine models, due to their anatomical and cellular similarities to humans, offer translational relevance but remain underutilized. This study aims to develop an advanced 3D coculture system using equine chondrocytes and synoviocytes to simulate tissue-level interactions and fluid mechanical forces involved in OA. By incorporating inflammatory stimuli and gravity-driven cyclic fluid actuation, this model enables the study of OA-related molecular interactions in both healthy and diseased conditions under dynamic fluid conditions. Findings from this research provide important insights into pathological tissue crosstalk. In turn, this can help to better understand underlying inflammatory pathways and the potential contribution of fluid flow as an influential factor on the tissue microenvironment.
Introduction
Osteoarthritis (OA), a leading musculoskeletal disorder affecting millions worldwide, is increasing in prevalence due to ageing populations, rising obesity rates, and changing activity patterns.1–3 This degenerative condition involves progressive cartilage breakdown and chronic inflammation, causing pain, stiffness, and swelling, with no curative treatments available.4 In healthy joints, chondrocytes balance ECM synthesis and degradation, but in OA, this balance is disrupted by inflammatory cytokines (e.g., IL-1β, IL-6, TNFα) and matrix-degrading enzymes (MMP1, MMP3, MMP13), leading to oxidative stress, a catabolic shift, cell apoptosis, and ECM loss.5,6OA affects not only cartilage but also other joint tissues, including the synovium. Synovitis, or inflammation of the synovium, is increasingly recognized as a key factor in OA pathogenesis. A recent transcriptomic study revealed significant molecular crosstalk between articular cartilage, meniscus, synovium, and subchondral bone, with the most frequent ligand–receptor pairings occurring between the synovium and cartilage.7 This highlights the synovium's active role in OA progression, as it secretes pro-inflammatory mediators that contribute to cartilage destruction and pain.8,9 During OA, the synovium becomes increasingly vascularized and inflamed, with cellular proliferation, hyperplasia, and fibrosis.10 The interplay between degraded cartilage products and synovial cells perpetuates inflammation, altering the joint environment and promoting disease progression.8 Interestingly, synoviocytes can also have protective effects on injured cartilage,11 underscoring the complexity of synovium–cartilage interactions in OA pathogenesis.12
Given the increasing number of OA patients, there is a critical need for more accurate models that replicate OA pathophysiology.13–17 Although in vivo models have significantly advanced our understanding of OA, the prevailing and widening translational gap highlights their limitations. The smaller joint size and differing biomechanical loads in small animals can produce distinct disease phenotypes that may not fully represent human OA.18 Moreover, small animal models, such as rodents, often exhibit exaggerated inflammatory responses and cytokine profiles compared to human OA. Genetic variability in these models also introduces challenges in translating findings to human populations. Equine models offer a promising alternative within the “One Health” framework, bridging human and veterinary medicine to address shared health challenges like osteoarthritis as a serious disease. Horses naturally develop OA and share key anatomical similarities with humans, including joint structure, cartilage thickness, and biomechanics, making them more clinically relevant than small animal models.18 Horses also allow for advanced diagnostic imaging, repeated synovial fluid sampling, arthroscopic procedures, and the collection of larger tissue samples from both healthy and OA-affected joints. Additionally, equine chondrocytes exhibit a response to IL-1β that is similar to human chondrocytes, further supporting their potential in translational OA research.19 By leveraging equine models, the One Health approach facilitates the study of OA in a species that bridges the gap between small animal models and humans, improving the translational potential of findings. This integration not only addresses the shared burden of OA across species but also fosters collaborative solutions for developing therapeutics and preventive strategies that benefit both human and veterinary medicine.
Current in vitro models include ex vivo approaches like explant cultures, which are prone to high inter-species, donor, tissue, and regional variability, and in vitro models such as 2D monolayers, micromass, organoid and 3D pellet cultures. While in vitro models are cost-effective and scalable, they face challenges in preserving relevant tissue phenotypes over extended experimental periods.20 Moreover, joint tissue models frequently focus on individual tissue types in isolated settings, failing to capture the complex tissue interactions that are crucial for joint tissue homeostasis. A variety of microfluidic approaches have been introduced for modelling different aspects of synovial joint (patho)biology, including inflammation and fibrotic remodelling.21–23 More recently, microphysiological systems that recapitulate tissue-level interactions of joints, as well as mechanical compressive loading, are emerging to better understand the role of cartilage mechanobiology in OA.24–27 Microfluidic systems that elaborate on the effects of surface shear stress on the synovial membrane mostly focus on the synovial lining.28 To address the limitations, we investigated the effects of fluid perfusion realized by gravity-driven fluid actuation by tilting in a synovial joint coculture system enabling the investigation of tissue-level crosstalk and concurrent fluid perfusion in potential OA in vitro models (see Fig. 1). The model integrates healthy chondrocytes and synoviocytes from equine origins, enabling the simulation of initially healthy environments that can be pathologically induced through the addition of IL-1β as pro-inflammatory mediator in OA. Our microfluidic coculture model applies gravity-driven fluid perfusion, providing controlled conditions to explore the potential effects of fluidic perfusion on cartilage and synovial tissue microenvironments. Although these mechanical forces are considerably lower than physiological joint pressures, the system enables an improved understanding of biochemical crosstalk, nutrient diffusion, and their combined roles in OA pathology. The first objective of this study is to compare conventional 2D cultures with advanced 3D coculture systems by analysing the morphology and gene expression of chondrocytes and synoviocytes in these settings. Subsequently, the indirect, soluble factor-mediated molecular interactions between synoviocytes and chondrocytes in coculture were analysed, to determine their impact on OA-related gene expression levels. As a final goal, the current study assessed the effects of gravity-driven fluid perfusion on both healthy and inflamed cocultures versus the respective monocultures to decipher the role of molecular tissue crosstalk in a fluid-actuated pro-inflammatory microenvironment.
Experimental
Tissue isolations & cell culture
The horses used in this study were mixed robust, warmblood and draft breeds (mixed sex incl. three mares, four geldings, and one stallion; age range: 5–24 years). They were euthanised for reasons unrelated to the study, including fractures (at sites distant from the articulatio genu), colic, pneumonia, and oral squamous cell carcinoma. Cartilage was harvested from the articulatio genu (knee joint). Joints were macroscopically assessed using the Osteoarthritis Research Society International (OARSI) scoring system and only joints with no macroscopically evident osteoarthritic changes (score 0) were included in this study.29 Equine chondrocytes and synoviocytes were isolated from the harvested cartilage and synovia through sequential digestion. For chondrocytes, a 0.1 mg mL−1 collagenase II solution was prepared by dissolving 5 mg of collagenase II powder (C2674, Sigma/Merck) in 5 mL DMEM (31885049, Gibco) with 1% Pen/Strep (P4333, Sigma) as 10× stock. Cartilage pieces were harvested sterilely and kept in sterile PBS with Ca and Mg (11580456, Gibco), cut into 1 mm3 pieces, and digested in a sterile beaker containing 0.1 mg mL−1 collagenase II solution. Digestion is performed on a magnetic stirrer in the incubator at 37 °C with slow stirring for 5–6 hours for adult cartilage. Synovial membrane tissue was carefully dissected to separate it from surrounding tissue and washed with PBS with Ca and Mg (Cat no.: 11580456, Gibco) containing 1% Pen/Strep (Cat no.: P4333, Sigma) to maintain sterility. Tissue pieces were enzymatically dissociated using Trypsin (10608924, Fisher Scientific) and transferred to a sterile Petri dish (83.3902, Sarstedt). A cell scraper (353085, Corning) was used to further process and dislodge cells if necessary.Harvested cells were suspended in complete DMEM and then seeded into sterile T-75 (90076, TPP®) culture flasks with 10 mL of prepared culture medium. After reaching 80% confluency, primary cells were cryostored in fetal bovine serum (FBS; Capricorn Scientific GmbH, #FBS-12A) containing 10% dimethylsulfoxide (DMSO) until cultivation. Equine chondrocytes and (fibroblast-like) synoviocytes were thawed by the addition of 1 mL of 2D culture medium (low glucose medium) (DMEM 1 g L−1D-glucose, Gibco™, #31885-023) with 10% FBS 1% anti–anti (Gibco™, #15240-062). Cultures were distributed into T-75 flasks (TPP®, #90076) with 10 mL of complete culture medium. The cells were then incubated at 37 °C and 5% CO2. The medium was changed every three days.
Microfabrication
The detailed chip designs of the mono- and coculture devices used in the current study as well as the detailed manufacturing protocols are described elsewhere (for detailed biochip dimensions see ESI† Fig. S1A).21 In brief, cleaned glass object slides (Paul Marienfeld, Germany) were used as the bottom layer for the microfluidic chips. These were first treated for 5 minutes in a 2% Hellmanex III (Hellma Analytics, Germany) solution in an ultrasonic bath to remove the coating. The slides were then treated in aqua dest. for 5 min to remove residual Hellmanex solution and finally with isopropanol for 5 min to assist drying. Both procedures were also performed in an ultrasonic bath. For the fabrication of the individual microfluidic chip layers a xurography method was used. First, 0.5 mm thick silicone sheets (MVQ Silicone) were cut into the different two-dimensional layers with a CAMM-1 GS-24 vinyl cutter (Roland, Germany) which was controlled by the Roland Studio software. For bonding the silicone layers to each other and to the glass slides, a plasma cleaner (Harrick Plasma, Plasma Cleaner PDC-002-CE) was used. The different layers and glass slides were treated for 2 minutes in the plasma cleaner. Next, the layers were positioned on top of each other and subsequently incubated at 80 °C for 10 minutes. After combining all 7 layers the final chip was again incubated at 80 °C overnight. For storage, individual devices were vacuum packaged in polymer pouches (Amazon Basics) and UV-treated for 60 min prior to use.Inflammatory stimulation
Dose titration of cytokine IL-1β was carried out to ensure consistent induction of an inflammatory response without detrimental effects on cell viability. Synoviocytes and chondrocytes cultured in 2D were exposed to increasing concentrations (0.5, 1.0, 10, 50 ng mL−1) of human recombinant IL-1β (BioLegend, #5794029) for 7 d. Cell viability was assessed using a calcein AM/ethidium bromide dye exclusion assay on single-cell resolution.On-chip cell culture
The mono- and coculture chips were sterilized in the laminar flow hood under UV light overnight directly before using them. The chondrocytes or synoviocytes were dissociated using TrypLE™ at 37 °C for 2 min. The cells were afterwards centrifuged for 8 minutes at 229 rcf and then resuspended in 2 ml starvation medium. The cells were then counted using a counting chamber and again centrifuged at 229 rcf for 8 minutes. The cells were then resuspended to reach a concentration of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per μL. To create the fibrin clots which form the organoids, 15 μL of cell suspension (12
000 cells per μL. To create the fibrin clots which form the organoids, 15 μL of cell suspension (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per μL) were pipetted to 30 μL of 100 mg mL−1 fibrinogen (TISSEEL, Baxter International) and mixed by pipetting up and down. Afterwards 15 μL of a 4 U mL−1 thrombin solution (Baxter International) were transferred into the lid of the Eppendorf tube (Safe-Lock Tubes 1.5 mL, Eppendorf AG, #0030120086) which held the cells and fibrinogen. The 15 μL of thrombin were then taken up, mixed five times and 45 μL were then transferred into the biochip through the hydrogel inlet port. Three chips were placed into a cassette (Quadriperm™, Sarstedt) holding four slots, with one slot left empty as a water reservoir to prevent evaporation. Polymerization occurred at 37 °C, 5% CO2 for 45 min.
000 cells per μL) were pipetted to 30 μL of 100 mg mL−1 fibrinogen (TISSEEL, Baxter International) and mixed by pipetting up and down. Afterwards 15 μL of a 4 U mL−1 thrombin solution (Baxter International) were transferred into the lid of the Eppendorf tube (Safe-Lock Tubes 1.5 mL, Eppendorf AG, #0030120086) which held the cells and fibrinogen. The 15 μL of thrombin were then taken up, mixed five times and 45 μL were then transferred into the biochip through the hydrogel inlet port. Three chips were placed into a cassette (Quadriperm™, Sarstedt) holding four slots, with one slot left empty as a water reservoir to prevent evaporation. Polymerization occurred at 37 °C, 5% CO2 for 45 min.
The next step was to add 400 μL of 3D culture medium (low glucose medium with 1% FBS, 1% anti–anti antibiotic/-mycotic mix, 1% ITS (insulin–transferrin–selenium, Gibco™, #41400-045), 1% NEAA (MEM NEAA, Gibco™, #11140-035) and 0.1 mM ascorbic acid) with (inflamed group) or without (control group) addition of 10 ng mL−1 interleukin-1β (IL-1β) to the biochips. To complete the preparation of the cassette for incubation, the fourth slot, previously left empty, was filled with 1× PBS until it was half full. Furthermore, a nonwoven wound dressing was inserted into the PBS to absorb the liquid and function as a sponge. Finally, the ports of the chips were sealed with small strips of PCR foil (MicroAmp™ Optical Adhesive Film, Applied Biosystems™, #4311971). The chips were incubated at 37 °C and 5% CO2 for 10 days. All cassettes containing microfluidic chips were closed with parafilm on three sides from day 7 on to minimize evaporation and create a comparable setting between static and dynamic chips. The medium was changed on days three and seven for mono- and coculture chips with 350 μL of 3D culture medium. Samples were harvested on day three and seven for monocultures and on day three, seven, eight, nine and ten for co-cultures for qPCR.
Gravity-driven fluid perfusion
To examine the influence of cyclic perfusion on the gene expression of matrix-related genes and inflammation markers, chondrocytes and synoviocytes-containing biochip organoids were exposed to cyclic bi-directional fluid flow. Starting on day 7 post-seeding, fluid flow was initiated by placing healthy and inflamed cartilage and synovial biochips on a tilting plate (PMR-30 Mini Rocker-Shaker, Grant-bio) operating at tilting speed of 1 Hz to create fluid actuation inside the biochips's microchannel next to the hydrogel compartment and provide the 3D organoids with a more dynamic fluidic microenvironment. Samples were harvested, analogous to static cultures, after 10 days of culture (including 3 days of actuation from day 7 on).RNA isolation, cDNA synthesis and RT-qPCR analysis
RNA was isolated using the innuPREP RNA Mini kit 2.0 (IST Innuscreen GmbH, #845-KS-2040250). For 2D cultures, cells were lysed in 350 μL lysis buffer, while 3D cultures were excised, washed in PBS with Ca and Mg (Gibco, #11580456), and lysed in lysis buffer with 1% β-mercaptoethanol (Sigma, #M6250). cDNA synthesis used the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, #4368814) with 10× RT buffer, dNTP mix, random primers, and MultiScribe™ Reverse Transcriptase, run on a QuantStudio 3 thermocycler (Applied Biosystems). qPCR analysis used the manufacturer's protocol using PowerTrack SYBR Green Master Mix (Applied Biosystems, #A46109; see Tables 1 and 2 for details on master mix compositions and primer sequences) with SDHA as a housekeeping gene which showed a coefficient of variation (Cv) of 3.27% in chondrocytes and 3.92% in synoviocytes, ensuring reliable normalization in all conditions used in this study. Each well contained 19 μL master mix and 1 μL cDNA, loaded into MicroAmp® Fast 96-Well Reaction Plates (Applied Biosystems, #4346907), sealed with optical adhesive film, and spun down before PCR cycling (95 °C denaturation, 60 °C annealing, and elongation).| Component | Volume per reaction in μL per well |
|---|---|
| Molecular grade H2O | 5.8 |
| SYBR Green MM | 10 |
| Forward primer | 1.6 |
| Reverse primer | 1.6 |
| Total | 19 |
| cDNA sample | 1 |
| Final volume | 20 |
| Gene | Protein | Primer orientation | Sequence |
|---|---|---|---|
| ACAN | Aggrecan | Forward | TGGGAGAGCAGATGTCAGTG |
| Reverse | GTTCTGGAGGCTGGGATGTA | ||
| CDH11 | Cadherin-11 | Forward | TCAGAACAGCCCTTCCCAAC |
| Reverse | ATCTGGTATACGCTCTGCGG | ||
| COL1A1 | Collagen type I, alpha 1 | Forward | TGGACGCCATCAAGGTCTTC |
| Reverse | GGCCACCATACTCGAACTGG | ||
| COL2A1 | Collagen type II, alpha 1 | Forward | ACGAGCACCTCTTTTGCCTT |
| Reverse | TTCGTGTCTGTTCTCAGGGC | ||
| COL3A1 | Collagen type III, alpha 1 | Forward | TGCTCCCATCTTGGTCAGTC |
| Reverse | GGAATCTCTGGGTTGGGACA | ||
| COMP | Cartilage oligomeric matrix protein | Forward | GGTGCGGCTGCTATGGAA |
| Reverse | CCAGCTCAGGGCCCTCAT | ||
| IL6 | Interleukin-6 | Forward | ATGGCAGAAAAAGACGGATG |
| Reverse | GGGTCAGGGGTGGTTACTTC | ||
| CXCL8 | Interleukin-8 | Forward | GCCACACTGCGAAAACTCAG |
| Reverse | CCCACTTTGTATGGGGGTTC | ||
| MMP1 | Matrix metalloproteinase-1 | Forward | CAGTGCCTTCAGAAACACGA |
| Reverse | GCTTCCCAGTCACTTTCAGC | ||
| MMP3 | Matrix metalloproteinase-3 | Forward | TGTGGAGGTGATGCACAAATC |
| Reverse | GCATGCCAGGAAATGTAGTGAA | ||
| SDHA | Succinate dehydrogenase complex flavoprotein subunit A | Forward | CAGTTCCACCCTACAGGCAT |
| Reverse | CTCCATGAACCTTTCGCCCT | ||
| SOX9 | Transcription factor SOX-9 | Forward | GAGGAAGTCGGTGAAGAACG |
| Reverse | GTTGGGGGAGATGTGTGTCT | ||
| SPARC | Secreted protein acidic and rich in cysteine | Forward | AGAATGAGAAGCGCCTGGAG |
| Reverse | TGCACGGGGAAGATGTACAT | ||
| COL10A1 | Collagen type X alpha 1 chain | Forward | AACGGCACCCCAGTAATGTA |
| Reverse | GAGGAGTACAGGCCATTCGA | ||
| SSP1 | Osteopontin | Forward | GCCGAGGTGACAGTTTGAGT |
| Reverse | ATGTCCTTGCTTTCCACAGG | ||
| COX2 | Cyclooxygenase-2 | Forward | CGAGTGGTTCTCCCCATAGA |
| Reverse | GGCCACGAGAGTTGTCTGAT | ||
| RUNX2 | Runt-related transcription factor 2 | Forward | GACGAGGCAAGAGTTTCACC |
| Reverse | GGGGTCCATCCACTGTAACT | ||
| TNFA | Tumor necrosis factor | Forward | ATGTGTGACCTGGACAACGG |
| Reverse | CCAGTGAGTTCTGGAAGCCC |
CFD simulations
Computational fluid dynamics (CFD) simulations were performed using COMSOL Multiphysics 6.1 to evaluate the theoretical wall shear stress, relative gravity-driven pressure difference, culture media supply, and molecular compound release in a monoculture chip design involving chondrocytes and synoviocytes based on the tilting motion, thus gravity-driven flow. For wall shear stress analysis, a 3D model employing the Brinkman equation was used to simulate time-dependent Darcy fluid flow in porous media. The fluid was assumed to have the properties of water at 25 °C and 1 atm, with gel porosity (ε) set at 0.98 and permeability (κ) at 10−11 m2, based on literature values.30 To study culture media supply and molecular compound release, a mass transfer, with the fluid's properties matching those of water at 25 °C and 1 atm, a gel porosity (ε) of 0.98, and a diffusion coefficient (D) of 10−9 m2 s−1 for both fluid and gel, as cited in literature.1 For the media supply simulation, inflow and reservoir geometries were assigned a concentration of 1 mol m−3, while the channel and gel were initialized at 0 mol m−3. For molecular compound release, the concentration settings were inverted. Boundary conditions included no-slip walls. Mesh refinement varied based on the simulation duration: extra-fine for short-term studies (1 s), fine for medium-term studies (up to 6 s), and coarse for long-term studies (3600 s) to optimize computational efficiency. Convergence plots and mesh geometries can be seen in ESI† Fig. S1B–D. This model assumed ideal convection and time-dependent transport of diluted species model in porous media was applied.Data visualization and statistical analysis
Statistical analysis and visualisation were performed using GraphPad Prism 8.0.1. After the normality assessment, the nonparametric Mann–Whitney test was used to assess the statistical significance level (*p < 0.05 and **p < 0.01).Results and discussion
Primary equine chondrocytes and synoviocytes exhibit improved physiology in 3D on-chip culture and react to IL-1β-treatment with OA-like hallmarks
To characterize any alterations in morphology and gene expression profiles of the cartilage and synovial organoid models, equine chondrocytes and synoviocytes were cultured as 2D monolayer or 3D on-chip conditions for seven days. Culture in a 3D versus 2D environment, significantly improved the chondrogenic phenotype of primary chondrocytes by significantly (p = 0.0238) decreasing the expression of dedifferentiation marker COL1A1 by approx. 100-fold, (Fig. 2A and B). In addition, chondrocytes in 3D culture displayed a more physiological spherical morphology. Similarly, synoviocytes cultured in the 3D biochip showed significantly higher expression of COL3A1 compared to 2D culture. For COL1A1 no significant change was observed, but a trend toward downregulation in 3D culture was evident. Regarding their morphology, synoviocytes arranged themselves in a spindle form in 2D culture, while they became more elongated in 3D culture. Using immunofluorescence and immunohistochemical stainings, the pericellular localization of collagen type 2 in equine cartilage and 3D on-chip cartilage organoids was confirmed (ESI† Fig. S2). To initially rule out that the 10 ng mL−1 IL-1β exposure was negatively influencing organoid viability, we next screened conventional synovial and chondrocyte 2D cultures viability for extended exposure to increasing concentrations of cytokine up to 50 ng mL−1. ESI† Fig. S3 shows the data on the analysis of the calcein AM/ethidium bromide dye exclusion assay on single-cell resolution for 7 days of cultivation with no observable viability decline even up to 50 ng mL−1 (above 90% of calcein-AM positive primary chondrocytes and synoviocytes). The lack of cell necrosis for these elevated interleukin levels was in line with previous studies.31Stimulation of on-chip chondrocyte organoids with 10 ng mL−1 IL-1β over 10 days resulted in significant increase of inflammatory markers MMP1 (p = 0.0238), MMP3 (p = 0.0238), IL6 (p = 0.0238) and CXCL8 (p = 0.0357), and a non-significant (p = 0.5476) downregulation of COL2A1 while ACAN stayed on comparable levels (Fig. 3A and B). Similarly, analogous inflammatory stimulation of on-chip synoviocyte organoids resulted, in a significant upregulation of MMP1 (p = 0.0238), MMP3 (p = 0.0238), IL6 (p = 0.0238) and CXCL8 (p = 0.0238) and a significant downregulation of COL1A1 (p = 0.0238). The expression of CDH11, on the other hand, seemed to not be affected by the IL-1β treatment (Fig. 3C and D). Notably, the pro-inflammatory responsiveness of synovial organoids was overall higher than of the cartilage organoids.
Effect of cyclic fluid perfusion on healthy 3D monoculture gene expression
Next, 3D monocultures of synoviocytes and chondrocytes were exposed to cyclic fluid actuation. To validate the design's functionality, computational fluid dynamics (CFD) simulations were performed for up to 3600 seconds. The system utilized a maximum tilting angle of 4°, height differentials, and a reservoir centre-to-centre distance of 2 cm, resulting in inlet pressures oscillating between 12.7 Pa and −12.7 Pa (Fig. 4A). Driven by a rocking motion at 1 Hz, the sinusoidal inlet flow produced a maximum hydrostatic pressure of 6.35 Pa on the gel interface at 0.5-second intervals (see also Fig. S5A†). The hydrostatic pressure peaked at 0.25 seconds, governing both shear stress and fluid velocity within the media supply channel. This process generated a maximum wall shear stress of 2.2 dyn cm−2 at the 3D cell culture interface (Fig. 4B) and a maximum channel velocity of 7.2 cm s−1 (Fig. 4C), respectively. Further simulation analysis of media supply revealed the establishment of a concentration gradient across the gel perpendicular to the media channel (Fig. 4D). After one hour, the media successfully diffused across the entire gel width (0.9 cm), increasing concentration at the distal end to 0.02 M. Time-dependent animations illustrating changes in velocity (Video S1†), pressure (Video S2†), and concentration dynamics for media supply (Video S3†) and molecular compound release (Video S4†) further supported these findings. More detailed CFD simulations were conducted to get a better understanding of the impact of tilting angle on wall shear stress, hydrostatic pressure changes and molecule diffusion dynamics the on-chip organoids are being subjected to during gravity-driven perfusion.As shown in ESI† Fig. S4, variations of the tilting plate angle from 0 up to 4° (see ESI† Fig. S5A and B) resulted in a linear increase in both peak wall shear stress as well as hydrostatic pressure generation inside the hydrogel compartment of up to 2 dyn cm−2 and 6.5 Pa, respectively. Notably, the perfusion of nutrients from the microchannels into the organoids also increased gradually with the increase of the tilting angle (see ESI† Fig. S6C–F). CFD simulations demonstrated that tilting the system at 4° generates low-level hydrostatic pressures (∼6.35 Pa) and shear stresses (∼2.2 dyn cm−2). These pressure regimes are significantly below physiological joint loading conditions, suggesting that the beneficial cellular responses observed are primarily due to improved diffusion of nutrients and metabolites rather than meaningful mechanotransductive signalling. When organoid 3D monocultures were next subjected to cyclic fluid actuation at a maximum 4° tilting angle of the rocking system, chondrocyte and synoviocyte organoids exhibited distinct alterations in gene expression. Cyclic fluid flow application significantly reduced COL1A1 expression (ESI† Fig. S6; p = 0.0087 and p = 0.0022) for chondrocytes and synoviocytes. In chondrocytes, COL2A1 expression (p = 0.9372) showed a trend toward downregulation, while synoviocytes exhibited a similar trend in COL3A1 expression (p = 0.1797). Interestingly, the expression of ACAN (p = 0.5587) and CDH11 (p = 0.1320) remained largely unaffected by fluid flow. Notably, MMP1 and MMP3 were significantly upregulated (p = 0.0260 and p = 0.0022) in chondrocytes exposed to fluid flow, indicating heightened matrix degradation activity. Conversely, synoviocytes experienced a marked downregulation in MMP1 (p = 0.0087), MMP3 (p = 0.0152), and IL6 (p = 0.0087) expression under cyclic fluid stimulation, suggesting a dampening of pro-inflammatory responses.
Impact of cyclic flow on IL-1β-treated 3D monoculture organoids
In the subsequent experiments, the impact of cyclic gravity-driven fluid perfusion on inflamed 3D monocultures of chondrocytes and synoviocytes was evaluated to assess their potential modulatory effects in amplifying a pro-inflammatory microenvironment. To induce inflammation, both cell types were cultured from day 0 with the continuous addition of 10 ng mL−1 IL-1β cytokine solution. At day seven, one group of biochips was subjected to cyclic fluid flow via a tilting platform operating at 1 Hz up to three days, while a control group remained static to serve as a reference for another 3 days of culture. Application of cyclic fluid flow on IL-1β-treated 3D monoculture, resulted in a non-significant (p = 0.200) downregulation of COL1A1 in both cell types (Fig. 5). Chondrocytes exhibited a non-significant upregulation of COL2A1 (p = 0.100) and CXCL8 (p = 0.400) and downregulation of MMP1 (p = 0.400) and IL6 (p = 0.200) expression. Synoviocytes did not display any downregulation of COL3A1 (p = 0.400) and CDH11 (p = 0.700) as well as inflammation marker levels.Coculture of chondrocytes and synoviocytes leads to upregulation of pro-inflammatory gene level, which can be further exacerbated by cyclic flow
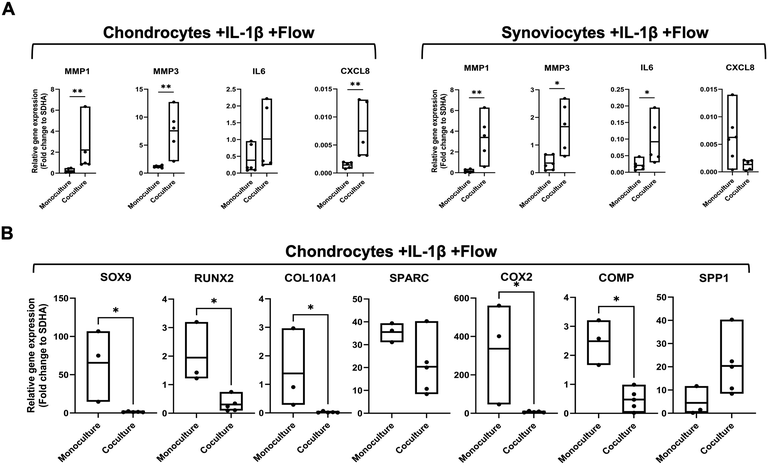
To examine the influence of cellular reciprocity between chondrocytes and synoviocytes on the pro-inflammatory gene expression profiles, a microfluidic biochip with two adjacent organoid compartments was used to analyze the effect of soluble molecular crosstalk compared to monoculture organoids. For this, chondrocyte and synoviocyte organoids were cultured for ten days in monoculture and coculture microfluidic chips (Fig. 6). The coculture chips maintained the proximity of the two cell-type organoids without direct physical contact, yet they shared the same culture medium, enabling the exchange of signalling molecules between them. Indirect 3D coculture of chondrocytes and synoviocytes in chips stimulated with IL-1β resulted in a non-significant but visible reduction of anabolic markers including COL2A1 (p = 0.662) and ACAN (p = 0.930) in chondrocytes but no further effects on matrix-related genes in either cell type compared to monoculture (see ESI† Fig. S7). As shown in Fig. 6A catabolic marker genes, including MMP1 and MMP3 expression, increased significantly in chondrocytes (MMP1 p = 0.0043 MMP3 p = 0.0043) and synoviocytes (MMP1 p = 0.0043 MMP3 p = 0.0173) in comparison to untreated controls. Additionally, chondrocytes significantly upregulated the expression of CXCL8 (Mann–Whitney t-test, p = 0.0079), while synoviocytes increased IL6 by 4.5-fold (p = 0.0173). Next, we investigated the reactiveness of such cocultures to further stimulation with 10 ng mL−1 IL-1β as a pro-inflammatory OA-like trigger, as shown in Fig. 6B. While all marker genes exhibited an elevated response in chondral coculture organoids, IL6 gene expression increased significantly by 10.2-fold (p = 0.0079). Similarly, synoviocytes showed a marked pro-inflammatory profile with a 203-fold increase in IL6 expression (p = 0.0079), along with elevated levels of MMP1 and CXCL8, which increased by 8.9-fold (p = 0.0079) and 218-fold (p = 0.0079), respectively.These results underline the role of intercellular crosstalk in triggering a pro-inflammatory cascade, significantly amplifying catabolic gene expression in both synovial and chondral organoids, especially when further stimulated by pro-inflammatory cytokines. Next, the effects of cyclic perfusion on inflamed chondrocytes and synoviocytes cocultured in a biochip 3D system over 10 days was investigated (Fig. 7). In chondrocyte biochip organoids, application of fluid flow over three days led to a significant reduction in MMP3 (p = 0.0317) and CXCL8 (p = 0.0159) expression, alongside a general downtrend in other pro-inflammatory markers (Fig. 7A). Similarly, synoviocyte biochip cultures showed significant decreases in MMP1 (p = 0.0159), MMP3 (p = 0.0164), and CXCL8 (p = 0.0068) expression, with a visible reduction trend for IL6 (p = 0.222) as illustrated in Fig. 7B. Moreover, a comparative analysis of matrix relating gene markers for chondrocyte cocultures subjected to cyclic fluid flow in the absence of IL-1β (ESI† Fig. S8) also showed an anabolic response with upregulation of COL2A1 (p < 0.05) and downregulation of COL1A1 (p < 0.05), similar to chondrocyte monoculture responses (see also Fig. 5A). In a final comparison, the effect of cyclic flow was investigated for synovial and chondral organoid cocultures in comparison to inflamed and perfused monoculture organoids.
As shown in Fig. 8, chondral biochip cultures in the dynamic coculture regimen retained significantly higher catabolic gene expression levels with 8.9-fold MMP1 (p = 0.0357) and 4.7-fold IL6 (p = 0.025) expression level increase from 1.8 ± 1.4 to 16.9 ± 12.2 and 5.7 ± 6.7 to 26.7 ± 10.9 fold, respectively. In contrast, synovial cocultures did not show any additional exacerbating effect of cyclic perfusion for MMP1 (p = 0.57) and IL6 (p = 0.25) expression levels. Further analysis of an extended panel of chondrogenic marker genes for chondral organoids, as depicted in Fig. 8B, confirmed significant downregulation of SOX9 (p = 0.0357), which is essential for maintaining the chondrogenic phenotype,32,33 COL10A1 (p = 0.0357), a hypertrophic chondrocyte marker,33 and COMP (p = 0.0357), a structural cartilage matrix component.34 Additionally, RUNX2 (p = 0.0357), an osteoblast differentiation marker,35 and COX2 (p = 0.0357), linked to inflammatory responses,36 showed reduced expression, which may reflect an altered cellular differentiation and inflammatory response in the coculture setting. Similar pro-inflammatory profiles typical to responses of OA-patient-derived synovial membranes were observable in cocultured synovial organoids subjected to the dynamic culture environment as shown in ESI† Fig. S9 with upregulation of TNFA (p = 0.0357) and COMP (p = 0.0357). Our overall findings on the tissue-specific response profiles of cocultured chondro–synovial organoids suggest that applying cyclic fluid flow can act as a modulating factor in synovial joint organoid coculture models. Notably, the cyclic fluid actuation method in the proposed joint-on-a-chip system can be sufficient to trigger a pro-chondrogenic response in healthy specimens while it can modulate and even exacerbate proinflammatory responses within an inflammatory cell microenvironment with a tissue-specific response profile for both chondrocyte and synovial organoids.
The equine primary organoids in this study demonstrated morphology and functionality comparable to previously reported human models, underscoring the potential of a ‘One Health’ approach. This approach leverages healthy, non-OA equine 3D cultures to model joint physiology, addressing the challenges of the limited availability of non-arthritic human tissue samples. Upon inflammatory stimulation with IL-1β, a key cytokine in OA progression, chondrocytes exhibited reduced ECM marker expression and significant upregulation of inflammatory markers and matrix metalloproteinases, reflecting a shift toward a catabolic and inflammatory state. Similarly, synoviocytes responded to IL-1β with a pronounced inflammatory profile, including significant increases in MMP1, MMP3, IL6, and CXCL8 expression, along with a marked downregulation of COL1A1. Synoviocytes exhibited a more robust pro-inflammatory response compared to chondrocytes, highlighting the critical role of the intimal synovium in amplifying inflammation during OA progression.
Synoviocytes and chondrocytes both play a role in joint inflammation, but synoviocytes are generally more responsive to inflammatory stimuli while also showing a stronger secretory activity. Chou et al., for example, found higher expression of key inflammatory cytokines in synoviocytes compared to chondrocytes.37 Their study detected IL1B, TNF, IL6, IL15, and IL18 in 5–36% of synoviocytes but only in less than 1% of chondrocytes, indicating a more inflammatory profile of the intima synovialis over cartilage tissue. Moreover, in damaged cartilage regions, synoviocytes exhibited expression levels of these cytokines that were up to 291 times higher than those observed in chondrocytes. Furthermore, they identified inflammatory macrophages and dendritic cells as the main producers of IL-1β, which is a classic pathogenic cytokine in OA.37 Additionally, in a study by Fu et al., synoviocytes exhibited a heightened pro-inflammatory response when exposed to IL-18, producing higher levels of inflammatory mediators compared to chondrocytes.38 Also, synovial cells originating from inflamed synovial membranes demonstrate greater NF-κB activity and cytokine release upon stimulation than chondrocytes, indicating a more robust and/or pronounced inflammatory response,39 which confirms the comparative data on our synovial and cartilage biochip organoids tissue-specific pro-inflammatory response patterns.
Similar tissue-specific responses were also confirmed in the current investigation into the effects of cyclic fluid-perfusion of the hydrogel compartment on healthy chondrocyte and synoviocyte organoids in both mono- and coculture biochip systems. 3D chondrocytes displayed an upregulation of MMP1 and MMP3 under the cyclic fluid perfusion regime, consistent with their susceptibility to fluid shear and overloading to potentially contribute to OA progression.40,41
Conversely, synoviocytes, key drivers of inflammation in osteoarthritic synovial fluid,42 responded by downregulating MMP1, MMP3, and IL6, suggesting a potentially protective role of fluid perfusion on healthy synovial tissues. These contrasting responses emphasize how dynamic culture conditions inside microfluidic joint microsystems can differentially affect the cartilage and synovial tissue organoids, influencing joint homeostasis and, consequently, also, the potential dynamics of OA progression. Our results also demonstrated that cyclic fluid flow of the equine OA model mitigated pro-inflammatory responses in both inflamed chondral and synovial monocultures, aligning with previous in vivo studies showing reduced inflammatory mediators after mechanical stimulation, such as treadmill exercise in OA models.43 Notably, both cell types exhibited a downregulation trend for COL1A1 under fluid flow, suggesting that dynamic culture conditions in the coculture biochips can modulate matrix-related responses and potentially influence fibrotic remodelling in joint tissues. The indirect proportionality between synovial and cartilage gene responses supports our earlier findings on fibrotic remodelling in rheumatic chondro–synovial models.21
Our coculture biochip platform has enabled the investigation of tissue reciprocity in pro-inflammatory OA progression using perfused in vitro organoids derived from healthy equine tissues within a single microfluidic system. This innovative approach reveals unprecedented insights into the molecular interplay between cartilage and synovial tissues under cyclic fluid actuation by tilting. We hypothesised that indirect tissue interactions in our dynamic OA model would intensify pro-inflammatory responses. Our results support the hypothesis that cyclic fluid perfusion indirectly influences gene expression changes, most likely due to the enchanced and dynamic cellular microenvironment. This corroborates the outcomes of previous studies on the effects of fluid perfusion on synovial and chondrocyte studies at various orders of magnitudes below an overloading regime of about 20 dyn cm−2.28,44,45 While inflamed monocultures in our current study exhibited pronounced inflammatory and catabolic responses, these cytokine-induced effects were partially mitigated over time. However, in perfused cocultures, the tissue crosstalk dramatically amplified inflammatory and catabolic pathways in the chondral but not the synovial organoid, potentially underscoring the synergistic impact of molecular signalling and crosstalk in OA pathogenesis. These findings suggest that the interaction between synovial and chondral tissues, driven by cyclic fluid flow, can play an important role in tissue homeostasis and, in turn, disease progression. The observed downregulation trends in pro-inflammatory markers over time also highlight that dynamic cultivation conditions can play an important role in joint homeostasis and pathological progression of arthritic phenotypes using in vitro 3D joint-on-a-chip technologies. Utilizing the concepts used in mechanical bioreactors and lab-on-a-chip systems for decades, the inclusion of on-chip dynamic culture conditions, including fluid perfusion, shear stress, hydrostatic loading, as well as mechanical loading and shearing, will serve as vital modulatory factors in advanced joint-on-a-chip systems for osteoarthritis.
Limitations of the current study using the coculture chip design as an equine OA model of synovial joints include i) indirect contact of synovial with cartilage organoids allowing only secretory exchange and ii) the uniformity of the applied fluid mechanical forces, which may not account for distinct differences in both the types of mechanical stresses applied as well as the orders of magnitude. The synovial membrane in situ is subjected to shear and tensile strains, whereas cartilage is predominantly exposed to compressive loading and mechanical surface shearing.46 To enhance the biomimetic nature of our biochip approach, the hydrogel chamber geometries should be adapted to account for different mechanical pressure regimes in addition to cyclic perfusion. As suggested by studies from Occhetta and Paggi on cartilage in microfluidic joint-on-a-chip models, this will allow for the inclusion of a broader set of mechanical stressors.24,26,47–49 In line with the design implementations demonstrated by Paggi and coworkers for chondrocyte 3D monocultures the combination of tissue-specific and variable biomechanical actuation regimes (i.e., 5–12% straining24 or 10–30% (ref. 25)) by microfluidic actuator design constraints in a single joint on a chip platform will enhance the physiological relevance of biochips regarding joint mechanobiology. In this study, we examined fluid perfusion conditions at approximately 2 dyn cm−2 and 12 Pa. Although these values are most probably at the lower range of the mechanical force ranges (i.e., wall shear stress) reported in previous studies for healthy (2–10 dyn cm−2) and overloaded joint conditions (10 dyn cm−2),50 the modulating effects of the cyclic fluid flow remain apparent. From a mechanical viewpoint, in vivo joint compression forces are higher and can reach approximately 1700 N for a typical hind limb51 (e.g., ∼129 kPa for a 500 kg horse with a hind hoof measuring 14 cm in length and 12 cm in width). Achieving such biomimetic forces will require more advanced biochip architectures and manufacturing processes to replace hydrogel approaches with decellularized tissue scaffolding to better approximate the biomechanical parameters of the extracellular matrix.
Regarding inflammatory milieu, the investigation of lower levels of cytokine stimuli (i.e., using pg-instead of ng-levels) found in OA-patient synovial fluids and a broader variety of cytokine types (i.e., adipokines) will allow the current in vitro joint-on-a-chip models to be a better representation of the actual inflammatory environment found in vivo.52 Future bioanalytical investigation on the structure–function relationship of various tissue OA and their secretional changes of in vitro joint models will provide a more complex and detailed understanding of dynamic culture conditions at the protein level and, more importantly, the activation of signalling pathways connected to OA. Lastly, the use of equine cells as healthy organoid models is advantageous as human material is scarce. Notably, equine samples, nonetheless, may require further methodological optimizations and validations for protein techniques, including Western blot, Luminex, and ELISA assays. This may include the development of custom-made anti-horse antibodies in larger multi-national project frameworks since no commercial panels or anti-horse antibodies are currently available. This constitutes a disadvantage of equine cells over human and murine cells in advancing studies on the onset and progression modelling of different OA pathotypes of in vitro musculoskeletal research.
Conclusions
This study establishes equine joint-on-a-chip models as a powerful tool for OA research, demonstrating the potential of advanced 3D culture systems combined with biomechanical stimulation to replicate the intricate physiology and pathology of joint tissues. By preserving a native phenotype and incorporating tissue-level interactions, our microfluidic platform successfully models the inflammatory and catabolic mechanisms central to osteoarthritis. Dynamic culture conditions using cyclic fluid perfusion appear capable of modulating inflammatory profiles indirectly, predominantly through the potential improvement of nutrient diffusion and waste clearance. The low mechanical forces applied suggest limited physiological relevance to direct mechanobiological effects, underscoring the need for enhanced mechanical loading conditions (shear and loading) in future biochip studies.Our findings indicate that cyclic fluid perfusion is likely influencing cellular pathways predominantly through indirect biochemical mechanisms, notably enhanced nutrient diffusion, metabolite clearance, and soluble molecular crosstalk between tissues rather than through direct mechanical signalling. Understanding the nuanced and indirect effects of such dynamic culture conditions is important for accurately interpreting the biological responses observed in joint-on-a-chip systems. Future studies employing this platform will more clearly delineate between mechanical and biochemical contributions to better approximate physiological conditions relevant to OA.
Ethics statement
Primary cells were isolated from the waste material of non-arthritic horses euthanized for unrelated reasons. According to the University of Veterinary Medicine Vienna's “Good Scientific Practice. Ethics in Science and Research” regulations, in vitro cell culture studies do not require Institutional Ethics Committee approval. The tissue was obtained solely for diagnostic, therapeutic, or approved research purposes. Owner consent for sample collection, analysis, and data publication was secured per standard procedures.Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.Author contributions
JH, methodology, formal analysis, investigation, data curation, writing – original draft, writing – review and editing, visualization; EIR, methodology, formal analysis, investigation, data curation, writing – original draft, writing – review and editing; JS, formal analysis, investigation, data curation, writing – original draft, writing – review and editing, visualization; MF, methodology, formal analysis, investigation, data curation, writing – original draft, writing – review and editing, visualization; IG, investigation, data curation, writing – original draft, writing – review and editing, visualization; SG, methodology, formal analysis, investigation, data curation, writing – original draft, writing – review and editing; FJ, data curation, writing – original draft, writing – review and editing, visualization, supervision, project administration; PE, writing – original draft, writing – review and editing, RW, writing – original draft, writing – review and editing, visualization, supervision, project administration, funding acquisition; ST, writing – original draft, writing – review and editing, visualization, supervision; MR, methodology, data curation, writing – original draft, writing – review and editing, supervision, project administration.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge TU Wien Bibliothek for financial support through its Open Access Funding Programme. Fig. 1 was created in BioRender. Toegel, S. (2025) https://BioRender.com/y628302. ChatGPT v4 (OpenAI) and Grammarly Pro word-Plugin (Grammarly) were used for language editing and manuscript corrections.Notes and references
- R. Shumnalieva, G. Kotov and S. Monov, Life, 2023, 13, 1650 CrossRef CAS PubMed.
- G. Kuş, Z. Yasaci, C. Boz and E. Türkmen, Am. J. Phys. Med. Rehabil., 2023, 102, 901–906 Search PubMed.
- S. Safiri, A. A. Kolahi, E. Smith, C. Hill, D. Bettampadi, A. Mohammad, D. Mansournia, A. Hoy, M. Sepidarkish, A. Almasi-Hashiani, G. Collins, J. Kaufman, M. Qorbani, M. Moradi-Lakeh, A. D. Woolf, F. Guillemin, L. March and M. Cross, Ann. Rheum. Dis., 2020, 79, 819–828 Search PubMed.
- A. Eitner, G. O. Hofmann and H. G. Schaible, Front. Mol. Neurosci., 2017, 10, 349 Search PubMed.
- M. Di Francesco, A. Fragassi, M. Pannuzzo, M. Ferreira, S. Brahmachari and P. Decuzzi, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2022, 14, e1780 CAS.
- C. Jia, X. Li, J. Pan, H. Ma, D. Wu, H. Lu, W. Wang, X. Zhang and X. Yi, Oxid. Med. Cell. Longevity, 2022, 2022, 1135827 Search PubMed.
- M. Wang, J. Shen, H. Jin, H.-J. Im, J. Sandy and D. Chen, Ann. N. Y. Acad. Sci., 2011, 1240, 61–69 CrossRef CAS PubMed.
- B. Chen, Y. Sun, G. Xu, J. Jiang, W. Zhang, C. Wu, P. Xue and Z. Cui, Exp. Ther. Med., 2024, 27(5) DOI:10.3892/ETM.2024.12490.
- N. Fahy, E. Farrell, T. Ritter, A. E. Ryan and J. M. Murphy, Tissue Eng., Part B, 2015, 21, 55–66 Search PubMed.
- K. Thavaratnam, E. Gracey, A. Ratneswaran, S. Vohra, S. Lively, S. Dupont, J. Rockel, R. Gandhi, D. Elewaut and M. Kapoor, Osteoarthr. Cartil., 2022, 30, S118 Search PubMed.
- C. M. Lee, J. D. Kisiday, C. W. McIlwraith, A. J. Grodzinsky and D. D. Frisbie, BMC Musculoskeletal Disord., 2013, 14 DOI:10.1186/1471-2474-14-54.
- Z. Li, Z. Huang and L. Bai, Front. Cell Dev. Biol., 2021, 9, 720477 Search PubMed.
- Z. Lin, Z. Li, E. N. Li, X. Li, C. J. Del Duke, H. Shen, T. Hao, B. O'Donnell, B. A. Bunnell, S. B. Goodman, P. G. Alexander, R. S. Tuan and H. Lin, Front. Bioeng. Biotechnol., 2019, 7 DOI:10.3389/FBIOE.2019.00411.
- C. Mondadori, R. Visone, M. Rasponi, A. Redaelli, M. Moretti and S. Lopa, Osteoarthr. Cartil., 2018, 26, S122 CrossRef.
- M. Rothbauer, G. Höll, C. Eilenberger, S. R. A. Kratz, B. Farooq, P. Schuller, I. Olmos Calvo, R. A. Byrne, B. Meyer, B. Niederreiter, S. Küpcü, F. Sevelda, J. Holinka, O. Hayden, S. F. Tedde, H. P. Kiener and P. Ertl, Lab Chip, 2020, 20, 1461–1471 RSC.
- I. M. Oliveira, M. R. Carvalho, D. C. Fernandes, C. M. Abreu, F. R. Maia, H. Pereira, D. Caballero, S. C. Kundu, R. L. Reis and J. M. Oliveira, J. Mater. Chem. B, 2021, 9, 4211–4218 RSC.
- J. Rosser, B. Bachmann, C. Jordan, I. Ribitsch, E. Haltmayer, S. Gueltekin, S. Junttila, B. Galik, A. Gyenesei, B. Haddadi, M. Harasek, M. Egerbacher, P. Ertl and F. Jenner, Mater. Today Bio, 2019, 4 DOI:10.1016/J.MTBIO.2019.100023.
- T. J. O'Brien, F. Hollinshead and L. R. Goodrich, Extracell. Vesicles Circ. Nucleic Acids, 2023, 4, 151–169 CrossRef PubMed.
- C. Little and M. Smith, Curr. Rheumatol. Rev., 2008, 4, 175–182 CrossRef CAS.
- P. J. Cope, K. Ourradi, Y. Li and M. Sharif, Osteoarthr. Cartil., 2019, 27, 230 CrossRef CAS PubMed.
- M. Rothbauer, R. A. Byrne, S. Schobesberger, I. Olmos Calvo, A. Fischer, E. I. Reihs, S. Spitz, B. Bachmann, F. Sevelda, J. Holinka, W. Holnthoner, H. Redl, S. Toegel, R. Windhager, H. P. Kiener and P. Ertl, Lab Chip, 2021, 21, 4128–4143 RSC.
- M. J. Makarczyk, S. Hines, H. Yagi, Z. A. Li, A. M. Aguglia, J. Zbikowski, A. M. Padget, Q. Gao, B. A. Bunnell, S. B. Goodman and H. Lin, Biomolecules, 2023, 13(2) DOI:10.3390/BIOM13020384.
- Z. Li, Z. Lin, S. Liu, H. Yagi, X. Zhang, L. Yocum, M. Romero-Lopez, C. Rhee, M. J. Makarcyzk, I. Yu, E. N. Li, M. R. Fritch, Q. Gao, K. B. Goh, B. O'Donnell, T. Hao, P. G. Alexander, B. Mahadik, J. P. Fisher, S. B. Goodman, B. A. Bunnell, R. S. Tuan and H. Lin, Adv. Sci., 2022, 9(21) DOI:10.1002/ADVS.202105909.
- C. A. Paggi, B. Venzac, M. Karperien, J. C. H. Leijten and S. Le Gac, Sens. Actuators, B, 2020, 315, 127917 CrossRef CAS.
- P. Occhetta, A. Mainardi, E. Votta, Q. Vallmajo-Martin, M. Ehrbar, I. Martin, A. Barbero and M. Rasponi, Nat. Biomed. Eng., 2019, 3, 545–557 CAS.
- C. A. Paggi, J. Hendriks, M. Karperien and S. Le Gac, Lab Chip, 2022, 22, 1815–1828 RSC.
- C. L. Thompson, T. Hopkins, C. Bevan, H. R. C. Screen, K. T. Wright and M. M. Knight, Biomed. Mater., 2023, 18(6) DOI:10.1088/1748-605X/ACF976.
- S. Piluso, Y. Li, L. M. Texeira, P. Padmanaban, J. Rouwkema, J. Leijten, R. van Weeren, M. Karperien and J. Malda, J. Cartil. Jt. Preserv., 2025, 100233 Search PubMed.
- C. W. McIlwraith, D. D. Frisbie, C. E. Kawcak, C. J. Fuller, M. Hurtig and A. Cruz, Osteoarthr. Cartil., 2010, 18(Suppl 3), S93–S105 CrossRef PubMed.
- C. Olaizola-Rodrigo, S. Palma-Florez, T. Ranđelović, C. Bayona, M. Ashrafi, J. Samitier, A. Lagunas, M. Mir, M. Doblaré, I. Ochoa, R. Monge and S. Oliván, Lab Chip, 2024, 24, 2094–2106 CAS.
- J. R. Anderson, M. M. Phelan, L. Foddy, P. D. Clegg and M. J. Peffers, J. Proteome Res., 2020, 19, 3652–3667 CAS.
- H. Song and K. H. Park, Semin. Cancer Biol., 2020, 67, 12–23 CAS.
- A. Haseeb, R. Kc, M. Angelozzi, C. de Charleroy, D. Rux, R. J. Tower, L. Yao, R. P. da Silva, M. Pacifici, L. Qin and V. Lefebvre, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2019152118 CAS.
- J. Cui and J. Zhang, Int. J. Mol. Sci., 2022, 23, 9253 CAS.
- Y. Mori-Akiyama, H. Akiyama, D. H. Rowitch and B. De Crombrugghe, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 9360–9365 CAS.
- Y. Y. Chow and K. Y. Chin, Mediators Inflammation, 2020, 2020, 8293921 Search PubMed.
- C. H. Chou, V. Jain, J. Gibson, D. E. Attarian, C. A. Haraden, C. B. Yohn, R. M. Laberge, S. Gregory and V. B. Kraus, Sci. Rep., 2020, 10(1) DOI:10.1038/s41598-020-67730-y.
- Z. Fu, P. Liu, D. Yang, F. Wang, L. Yuan, Z. Lin and J. Jiang, Int. J. Mol. Med., 2012, 30, 805–810 CAS.
- M. L. Libke, D. J. Cunningham, B. D. Furman, J. S. Yi, J. M. Brunger, V. B. Kraus, F. Guilak, A. L. McNulty and S. A. Olson, Sci. Rep., 2024, 14 DOI:10.1038/s41598-024-71964-5.
- P. Wang, P. P. Guan, C. Guo, F. Zhu, K. Konstantopoulos and Z. Y. Wang, FASEB J., 2013, 27, 4664 CAS.
- F. Zhu, P. Wang, N. H. Lee, M. B. Goldring and K. Konstantopoulos, PLoS One, 2010, 5, e15174 CAS.
- C. R. Scanzello and S. R. Goldring, Bone, 2012, 51, 249–257 CAS.
- L. Chen, Y. Lou, Z. Pan, X. Cao, L. Zhang, C. Zhu and J. Liang, Biochem. Biophys. Res. Commun., 2020, 523, 117–122 CAS.
- J. Zhao, Y. Xia and J. He, Sci. Rep., 2024, 14(1) DOI:10.1038/s41598-024-78676-w.
- E. G. Estell, L. A. Murphy, A. M. Silverstein, A. R. Tan, R. P. Shah, G. A. Ateshian and C. T. Hung, J. Biomech., 2017, 60, 91 Search PubMed.
- Q. Liu, X. Hu, X. Zhang, X. Duan, P. Yang, F. Zhao and Y. Ao, Sci. Rep., 2016, 6 DOI:10.1038/srep37268.
- C. Palma, S. Piazza, R. Visone, R. Ringom, U. Björklund, A. Bermejo Gómez, M. Rasponi and P. Occhetta, Adv. Healthcare Mater., 2024, 13, 2401187 CrossRef CAS PubMed.
- P. Occhetta, A. Mainardi, E. Votta, Q. Vallmajo-Martin, M. Ehrbar, I. Martin, A. Barbero and M. Rasponi, Nat. Biomed. Eng., 2019, 3(7), 545–557 CrossRef CAS PubMed.
- V. Peitso, Z. Sarmadian, J. Henriques, E. Lauwers, C. A. Paggi and A. Mobasheri, Curr. Protoc., 2024, 4, e70079 CrossRef CAS PubMed.
- T. Hodgkinson, I. N. Amado, F. J. O'Brien and O. D. Kennedy, APL Bioeng., 2022, 6, 11501 CrossRef CAS.
- S. O. Schuurman, W. Kersten and W. A. Weijs, J. Anat., 2003, 202, 355 CrossRef PubMed.
- A. L. McNulty, N. E. Rothfusz, H. A. Leddy and F. Guilak, J. Orthop. Res., 2013, 31, 1039 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4lc01078g |
| ‡ Authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2025 |