Open Access Article
Open Access ArticlePrecision single cell analysis to characterize host dependent antimicrobial response heterogeneity in physiological medium†
Ryuichiro
Abe‡
 a,
Jyong-Huei
Lee‡
a,
Jyong-Huei
Lee‡
 b,
Siew Mei
Chin‡
b,
Nikhil
Ram-Mohan
b,
Siew Mei
Chin‡
b,
Nikhil
Ram-Mohan
 a,
Kristel C.
Tjandra
a,
Kristel C.
Tjandra
 a,
April M.
Bobenchik
a,
April M.
Bobenchik
 c,
Kathleen E.
Mach
c,
Kathleen E.
Mach
 d,
Joseph C.
Liao
d,
Joseph C.
Liao
 de,
Pak Kin
Wong
*bf and
Samuel
Yang
*a
de,
Pak Kin
Wong
*bf and
Samuel
Yang
*a
aDepartment of Emergency Medicine, Stanford University School of Medicine, Stanford, CA, USA. E-mail: syang5@stanford.edu
bDepartment of Biomedical Engineering, The Pennsylvania State University, University Park, PA, USA. E-mail: pak@engr.psu.edu
cDepartment of Pathology and Laboratory Medicine, Penn State Health, Hershey, PA, USA
dDepartment of Urology, Stanford University School of Medicine, Palo Alto, CA, USA
eVeterans Affairs Palo Alto Health Care System, Palo Alto, CA, USA
fDepartment of Mechanical Engineering and Department of Surgery, The Pennsylvania State University, University Park, PA, USA
First published on 3rd February 2025
Abstract
Antimicrobial stewardship plays an essential role in combating the global health threat posed by multidrug-resistant pathogens. Phenotypic antimicrobial susceptibility testing (AST) is the gold standard for analyzing bacterial responses to antimicrobials. However, current AST techniques, which rely on end-point bulk measurements of bacterial growth under antimicrobial treatment in a broth solution, have limitations in resembling the physiological working environment and resolving heterogeneity in response kinetics within the population. In this study, we investigate the responses of uropathogenic bacteria under antimicrobial treatment in individual urine. Our results demonstrate substantial heterogeneity in time–kill kinetics in response to antimicrobials in a host-dependent manner. We also establish a microfluidic gel encapsulation platform for single cell imaging to rapidly resolve heterogeneous subpopulations in response to antimicrobials. The platform captures both bacterial growth and killing within the gel and enables medium exchange to assess the ability of surviving cells to resume growth after antimicrobial removal. Our study lays the foundation for a new generation of precision single cell analysis for personalizing antimicrobial treatment.
Introduction
Antimicrobial resistance represents an incessant global health threat that necessitates optimal approaches to therapy. Antimicrobial susceptibility testing (AST) is the current gold standard clinical microbiological test for predicting antimicrobial effect against the causative pathogen by determining the minimum inhibitory concentration (MIC) of antimicrobials in vitro. Traditional AST requires high inoculum of isolated colonies that is grown in culture with various concentrations of antimicrobials for 16–20 hours, followed by detection of cell proliferation. MIC results are then interpreted against valid clinical breakpoints to determine susceptibility and predict clinical therapeutic outcome. Clinical breakpoints used in AST interpretation is based on achievable unbound antimicrobial concentration in serum instead of the target site. However, many antimicrobials achieve much higher concentrations in other body fluids. For example, through glomerular filtration and tubular secretion, antimicrobial concentrations in urine have found to be 100–1000 times that achieved in serum.1 There is poor correlation between serum antimicrobial level and bacterial clearance from urine.2 In addition, AST is traditionally performed in standardized nutrient-rich media such as Mueller–Hinton II (MH2), where bacteria grow optimally, and antimicrobial activities are often highest. These conditions do not represent the working condition of antimicrobials in the urinary system. The physiology of the host infection environment has been shown to alter growth and expression of essential genes in various pathogens and consequently influence antimicrobial susceptibility.3–5 Moreover, AST, a growth based measurement, does not resolve antimicrobial tolerance. Tolerance is the ability of a bacterial population to survive a transient exposure to antimicrobials, even at concentrations far exceeding its MIC, and has been associated with treatment failure and relapse of many bacterial infections.6–8 A tolerant bacterial strain can have the same MIC as a susceptible strain but requires longer exposure to an antimicrobial, rather than higher concentration, to produce the same level of killing as a susceptible strain. Thus, traditional AST based on a static in vitro MIC value cannot provide insight into the kinetics of antimicrobial activity and does not mimic in vivo exposure dynamics at the effect site. Rapid accurate prediction of the pathogen's response to antimicrobial therapy to tailor treatment regimens that can provide optimal bacterial killing and prevent the amplification of resistance remains a critical gap in clinical care.Time–kill curves measure the bacterial response to antimicrobials over time, providing detailed information on both growth and killing kinetics, and offering more meaningful pharmacodynamic insights compared to static MIC values.9 Importantly, intercellular heterogeneity in antimicrobial response phenotypes can exist within a bacterial population. Despite a lack of consensus definitions, tolerance refers to a phenotypically, but non-genetically heritable, subpopulation of surviving cells, typically comprising less than 1% of the population. This subpopulation, which includes persister cells and viable but non-culturable (VBNC) cells, has been suggested to serve as evolutionary steppingstones towards resistance due to their increased mutation rate.10–14 Persister cells are phenotypic variants that survive antimicrobial exposure without replicating but resume growth after the antimicrobial is removed.15 In contrast, VBNC cells do not replicate post-antibiotic removal.16 The presence of these tolerant variants, implicated to cause treatment failure from relapse and emergence of antimicrobial resistance,17–20 can be characterized by a bimodal (or multimodal) time–kill curve that deviates from the simple decay expected from a uniform population.
In this study, we explored precision single cell analysis to capture both growth and killing kinetics in physiological medium, aiming to optimize antimicrobial selection and treatment duration. Time–kill curves offer a valuable tool for characterizing antimicrobial kinetics, distinguishing tolerant or variant subpopulations from susceptible phenotypes. We assessed their utility in evaluating the kinetics of common pathogens causing complicated urinary tract infections (UTIs) in response to frontline intravenous (IV) antimicrobials. Given the substantial differences in physiological conditions between urine and the standard MH2 broth typically used, we conducted our analyses in individual urine samples at physiologically relevant antimicrobial concentrations. Our aim was to examine inter-individual heterogeneity, dependent on the drug–pathogen–host combination, in both susceptible and resistant strain responses. Furthermore, to streamline the labor-intensive plating process involved in bulk time–kill analysis, we developed a microfluidic gel encapsulation platform for single cell microscopy, enabling the rapid resolution of intercellular response heterogeneity within a microbial population. Leveraging recent advancements in single cell imaging and microfluidic techniques for bacterial analysis,21–29 this platform offers a cost-effective solution for analyzing growth and killing kinetics at the single cell level. Additionally, the micrometer-thick gel layer is designed to facilitate rapid medium exchange, enabling the assessment of regrowth following antimicrobial removal to identify persistent subpopulations.
Materials and methods
Bacterial strains and healthy volunteer human urines for bulk time–kill curves
To investigate the bacterial responses to antimicrobials in human urines, Escherichia coli, Klebsiella pneumoniae, and Enterococcus faecalis were used as representative UTI causing species.30K. pneumoniae strains KP10 (AR_0010), KP12 (AR_0012), KP16 (AR_0016), KP120 (AR_0120), and KP153 (AR_0153) were obtained from The CDC & FDA Antimicrobial Resistance Isolate Bank. E. coli strains EC10789 (ATCC10789) and EC1427 (ATCC BAA-1427) and E. faecalis strains EF19432 (ATCC19432) and EF47077 (ATCC47077) were purchased from American Type Culture Collective (ATCC). MICs of antimicrobials to each strain and antimicrobial combination were measured by the broth microdilution method based on the CLSI guideline.31 Midstream urine was collected from healthy volunteers, who had not taken any antibiotics in the prior two weeks, after informed consent as per the IRB approved protocol (70759). Collected urine was then aliquoted and stored at −80 °C until further testing. Growth in MH2 broth was used as a control. Susceptible strains were tested in 2 healthy volunteers (urine 1 and urine 2) and resistant strains were tested in 3 healthy volunteers. Urine from individuals 1 and 2 were collected at two independent dates (urines 1 and 4, and 2 and 5). All healthy volunteer urines were confirmed to be sterile by culturing 50 μL of each urine on MH2 agar plate overnight at 37 °C.Bulk time–kill curves to assess bacterial response kinetics to antimicrobials in urine
Overnight cultures of K. pneumoniae and E. coli strains on MH2 agar, and E. faecalis strains on brain heart infusion (BHI) agar were used for estimating the time–kill kinetics. Colonies were suspended in phosphate-buffered saline (PBS) to achieve a turbidity of 0.5 McFarland standard and further diluted 10-fold in PBS to result in 1.5 × 107 CFU mL−1. Meropenem, ciprofloxacin, gentamicin, and ampicillin were each dissolved in PBS to concentrations of 320, 120, 220, and 5120 μg mL−1, respectively. 10 μL of bacterial suspension was inoculated in 80 μL of urine or MH2 broth and 10 μL of antimicrobial solution to reach the physiologically minimum concentrations of the antimicrobials in human urine under treatment. After 2, 4, 6, 8, and 24 hours of culturing at 37 °C, cells were taken, diluted in PBS buffer, and spotted on MH2 agar (K. pneumoniae and E. coli) or BHI agar (E. faecalis), and incubated at 37 °C overnight to determine the number of viable cells. The minimum urinary concentrations of meropenem,32 ciprofloxacin,33 and gentamicin34 in patients under antibiotic treatment for UTI were estimated based on previous studies. The concentration of ampicillin was between the thresholds of urinary concentration 3 to 4 h after 500 mg intramuscular injection, which is much higher than the lowest urinary concentration during treatment.35Microfluidic gel encapsulation
A microfluidic gel encapsulation technique was developed to embed bacteria in thin layers of micropatterned gel pads for evaluating individual bacterial responses to antimicrobials. We used a VLS 3.5 Desktop Series laser machining system to make an array of small wells with a diameter of 0.5 mm on a 5 μm ultra-thin double-sided tape (Dwell GM4095T, PET-base). We then attached the micropatterned tape to glass coverslips as a substrate for single cell microscopy. To implement gel encapsulation, 3% low-melting-point agarose (Fisher BioReagents, BP165-25) was dissolved in 1× PBS and heated to 65 °C to ensure complete dissolution. The gel was then carefully cooled to 37 °C or below before mixing with bacteria to prevent thermal damage to the cells. This mixture was then rapidly applied to the micropatterns. A microscope glass slide was gently used to scrape across the surface of the template to create thin micropatterned gel pads. Following this, we attached a laser-machined acrylic plate with a diameter of 1 cm to the coverslip to create a multiwell platform that is compatible with standard imaging setups and liquid handling equipment. Alternatively, thin layers of gel with embedded bacteria can be produced by centrifugation using a PDMS mold on a multiwell array (Fig. S1†). This method is particularly suitable for experiments requiring a large number of samples and diverse experimental conditions.Clinical samples
The microfluidic gel encapsulation platform was tested using clinical isolates from Penn State Milton S. Hershey Medical Center. These isolates, which included E. coli, Klebsiella oxytoca, K. pneumoniae, and Pseudomonas aeruginosa, were extracted from urine samples from patients with UTI or positive blood tube samples. All procedures were approved by the Institutional Review Boards of Penn State (STUDY00003415). The clinical isolates were subjected to varying concentrations of ciprofloxacin.We examined a total of 16 clinical isolates. Initially, we used a centrifuge operating at 5000 rpm for five minutes to effectively isolate the bacterial cultures from the samples. After isolation, the bacteria were pre-cultured in MH2 broth for one hour. Bacterial samples were then prepared to a concentration of ∼1.5 × 108 CFU ml−1 and quantified using a NanoDrop spectrophotometer. Antibiotics were diluted to the desired concentrations using MH2 broth. For the 96-well plate experiments, 100 μl of the appropriate growth medium was added to each well. The antibiotic solutions were dispensed into the corresponding wells, typically using a two-fold serial dilution to achieve a gradient of antibiotic concentrations. We inoculated each well with 10 μl of the standardized microbial suspension, ensuring that the final volume in each well was 200 μl. To prevent evaporation and contamination, we sealed the plates with gas permeable parafilm. The plates were incubated at 37 °C for 16–24 hours to allow for bacterial growth. After incubation, we examined the wells for microbial growth; the presence of a visible pellet or turbidity indicated growth. The MIC was determined as the lowest concentration of antibiotics that completely inhibited the growth of the organism. When using the NanoDrop spectrophotometer for quantification, we followed the same protocol as that used for the 96-well experiments. To establish a baseline, we recorded the initial measurements at 0 hours and compared them with those at 2 hours to determine the fold change. Significant changes in optical density were indicative of the response to each antibiotic concentration.
Single cell imaging and data analysis
The bacteria were pre-cultured in MHB broth for one hour prior to the experiment to ensure they were harvested during the exponential (log) growth phase, providing actively dividing cells for subsequent microfluidic experimentation. The microfluidic gel encapsulation platform was mounted on a Leica DMi8 epifluorescence microscope, equipped with an Okolab UNO-T-H temperature chamber to maintain a constant temperature at 37 °C for optimal bacterial growth. Both bright-field and fluorescence microscopy were performed using a Leica HCX PLAN APO 40×/0.85 objective lens. Bright-field images were captured with an exposure time of 15 ms. Fluorescence images for PI staining were captured using a Leica Y3 filter set, with an exposure time of 0.5 s.Automated image analysis was implemented using an ImageJ Macro to analyze the images, which identified the position of bacteria and captured morphological features, including area and perimeter. For precise calculations, images were converted to a 32-bit format, and background noise was subtracted using a rolling ball algorithm with a 10-pixel radius, assuming a lighter background. Image segmentation was performed using the particle analysis function in ImageJ by setting the threshold using the default method, and particles were analyzed by setting a minimum particle size of 0.02 pixels to exclude background noise. From this analysis, we extracted features for each bacterium, including area, mean intensity, XY coordinates, perimeter, aspect ratio, and solidity. We also analyzed the ratio of bacteria whose perimeter exceeds specific thresholds to compare the fold change of features relative to the initial time point. Additionally, individual cells were tracked using their XY coordinates to analyze single-cell growth and killing kinetics. The position of a cell between two time points was calculated using the Euclidean distance formula:
- d is the distance between the two points,
- (x1, y1) are the coordinates of the cell at the initial time point,
- (x2, y2) are the coordinates of the cell at the subsequent time point.
This method enabled accurate tracking of individual cells across different time points. Regions of interest (ROIs) were generated from bright-field images to enhance the analysis of fluorescence images and extract critical viability information indicated by the dye.
Statistical analysis
All assays were performed in triplicate unless specified otherwise. Data are presented as mean ± standard deviation. For our single cell growth analysis, we acquired three images per measurement, each capturing approximately 3000 bacteria.Results
Killing kinetics of susceptible strains depends on growth media
We first performed bulk time–kill analyses of bacteria. We compared the time–kill curves for carbapenem and/or ciprofloxacin susceptible K. pneumoniae and E. coli strains grown in MH2 broth and healthy volunteers' urines with physiologically minimum concentrations of meropenem (32 μg mL−1) and ciprofloxacin (12 μg mL−1) (Fig. 1), a concentration that is more than 64× the MIC for the strains tested. For meropenem, most bacteria were completely killed within 2–4 hours of treatment, except for KP10. This strain exhibited a tolerant subpopulation (<1%) that could survive even after 8 hours of treatment, while the majority of the population was killed within 2 hours of treatment. In urine 2, the surviving population after 8 hours of treatment was larger than in MH2 or urine 1. When prolonging the meropenem treatment of KP10 to 20 hours, similar to the standard AST, no survivors were observed in replicates with MH2 (0/14) (Table S1†). In contrast, 3 out of 28 (10.7%) replicates in urine samples treated with meropenem and 2 out of 24 (8.3%) replicates in urine samples treated with ciprofloxacin showed colony formation after treatment. These results suggest that the working media, particularly urine in a host-dependent manner, can contribute to heterogeneity in bacterial susceptibility to antimicrobials.Heterogeneities in susceptibility were also observed in other antimicrobials in these strains. When treated with ciprofloxacin, the time–kill curves revealed bacterial responses distinct from those observed with meropenem. KP10, which exhibited a tolerant subpopulation to meropenem, was killed sharply by ciprofloxacin under all tested conditions. In contrast, KP16 and EC10789, which were killed sharply by meropenem, exhibited surviving subpopulations against ciprofloxacin after 6–8 hours treatment. Notably, urine 2 exhibited a substantially larger surviving subpopulation compared to urine 1 and MH2. In contrast, ECBAA1427 was completely killed by both agents within 4 hours.
We further performed time–kill analysis of ampicillin- and gentamicin-susceptible strains of E. faecalis, EF19432 and EF47077, against these antimicrobials in healthy urine. Interestingly, both strains with MICs of 1 μg mL−1 ampicillin showed incomplete killing after 24 hours of treatment with 512 μg mL−1 of ampicillin (Fig. 2). In contrast, both strains had MICs of <0.25 μg mL−1 for gentamicin and were killed when treated with 22 μg mL−1 of the antimicrobial. There was distinguishable heterogeneity in the killing kinetics of EF47077 in the two urines and MH2, with fastest killing, under 2 hours, in urine 2 and slowest in MH2. Subjecting the two strains to both ampicillin and gentamicin had no additive effect on EF19432 but hastened the killing of EF47077 to under 2 hours, highlighting the value of time–kill analysis in resolving the combinatorial effects of the antibiotics on the bacteria. These results further support the notion that the working media can influence the heterogeneity in bacterial susceptibility to antimicrobial.
Killing kinetics of resistance strains depends on growth media
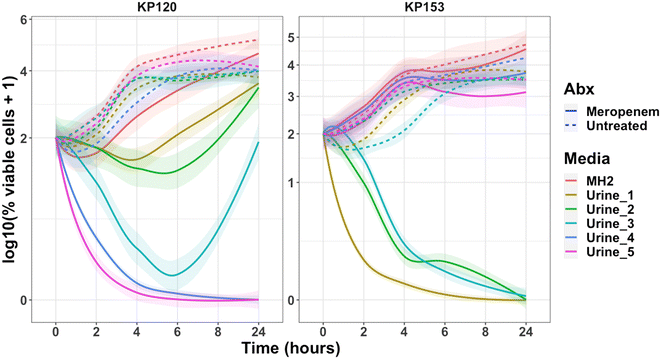
Heterogeneity was also evident in the resistant strains, KP120 and KP153 with known MICs of 32 μg mL−1 for meropenem (Fig. 3). As expected, both strains grew in all urines and MH2 without antimicrobial. However, resistance to meropenem was delayed in urines 1 and 2. The majority (99%) of KP120 was killed within 4 hours in urine 3, but growth resumed after 8 hours. Remarkably, at the strain's MIC of 32 μg mL−1, KP120 was completely cleared within 4 hours in urines 4 and 5, and KP153 was completely cleared in urines 1, 2, and 3, suggesting that the antimicrobial would be effective against this resistant strain in these individuals on those specific dates. Notably, the strain grew normally in urines 4 and 5 in the absence of the antimicrobial, suggesting the presence of unknown urine factor(s) that may synergize with the potency of meropenem. Such intraindividual and interindividual heterogeneity, and discrepancy in responses between MH2 and urine, underscore the complexity of host–pathogen–antimicrobial interactions and warrant precision measurement techniques for guiding antimicrobial treatment.Microfluidic gel encapsulation platform
Bulk time–kill analysis relies on plate counting to quantify colony-forming units, but the process is cumbersome and slow due to the colony-forming step. Moreover, bulk time–kill analysis is unable to detect VBNC subpopulations and distinguish between persisters and VBNC. To overcome these limitations, we devised a microfluidic gel encapsulation platform for single cell growth and killing analysis (Fig. 4A). The design concept focuses on trapping bacteria in a thin layer of low-temperature agarose gel, which enables single cell time-lapse microscopy. The thin gel structure is created by using a glass slide to spread and scrape the gel into laser-machined micropatterns, forming isolated thin gel pads that facilitate image analysis.The fabrication process begins with laser machining ultra-thin tape (5 μm) to create a micropatterned array consisting of circles with a diameter of 0.5 mm on a coverslip (Fig. 4B). The minimal thickness of the tape ensures that all bacteria are on the same focal plane for imaging. Additionally, the thin gel layer allows for easy reagent loading, washing, and medium exchange. Fig. 4C illustrates the process of spreading the gel with a glass slide. After gel loading, a laser-machined acrylic plate is attached to the coverslips to form a multiwell array for parallel processing. Each well has dimensions of 3 mm in height and 10 mm in diameter. The acrylic well can hold up to 235 μL of culture medium, while the volume of a single gel cylinder in the micropatterned well is 0.98 nL. Fig. 4D shows the assembled multiwell platform, which is designed for measuring single cell growth and killing kinetics. To accommodate a larger number of samples, we also developed an alternative method to create thin gel layers with embedded bacteria using centrifugation (Fig. S1†).
Microfluidic gel encapsulation platform for single cell AST
We first evaluated the microfluidic gel encapsulation platform for measuring bacterial growth. We tested 16 clinical isolates obtained from the Clinical Microbiology Laboratory (Fig. 5A). These clinical isolates were simultaneously tested against different concentrations of ciprofloxacin, taking advantage of the multiplexity of the microwell design. The cells were imaged using brightfield microscopy and analyzed with ImageJ to identify bacteria and automatically extract growth-dependent features, such as area or perimeter. In this experiment, the increase in perimeter after one hour was normalized to the initial value to quantify bacterial growth. For comparison, we also conducted the experiment using a 96-well plate format with both two-hour and overnight culture, resembling standard bulk AST. The growth of bacteria after two-hour culture was detected using a NanoDrop spectrophotometer (Fig. 5B and C). Among the clinical isolates tested, the observed trends showed consistency across the gel encapsulation platform, 96-well plate with NanoDrop, and overnight culture approaches. The MIC values were in general agreement within one to two-fold dilutions of each other, supporting the use of the microfluidic gel encapsulation platform for rapid single cell analysis.Microfluidic gel encapsulation platform for precision single cell analysis
The microfluidic gel encapsulation platform can resolve the growth kinetics of individual cells. We analyzed the response of a uropathogenic E. coli (EC137) exposed to ciprofloxacin. The strain is susceptible to ciprofloxacin with an MIC of 2 μg mL−1 (Fig. 6A). In this experiment, the clinical isolate was cultured in MHB and harvested at the logarithmic phase. A bacterial sample of ∼1 × 108 CFU ml−1 (measured using a NanoDrop) was prepared by centrifuging the culture and then resuspending it in media. As shown in Fig. 6B, notable bacterial growth was observed in the absence of ciprofloxacin. At 2 μg mL−1 (1× MIC), most bacteria exhibited filamentous morphology and ceased replication. Bacterial filamentation is a marker of cellular stress and is indicative of an induced SOS response to DNA damage or interrupted DNA replication.36,37 At concentrations of 4 and 8 mg L−1, the growth of bacteria were largely inhibited. Some cells were stained with propidium iodide (PI), despite ciprofloxacin not directly affecting the bacterial cell membrane integrity.38Fig. 6C depicts the fold increase in the perimeter and area of single cells across different concentrations of ciprofloxacin, underscoring the intercellular heterogeneity in response to the antimicrobial. Similar results were also obtained by K. pneumoniae (Fig. S2†). | ||
| Fig. 6 Single cell response of uropathogenic E. coli to different concentrations of ciprofloxacin. (A) Growth measurements obtained using the microfluidic gel encapsulation platform under the same cultivate condition as Fig. 5. The bacteria were incubated in varying concentrations of ciprofloxacin dissolved in MHB at 37 °C. (B) Brightfield and fluorescence images illustrating the response of E. coli (EC137) exposed to ciprofloxacin. (C) Changes in the perimeter and area of individual bacteria after 90 minutes. Red dots indicated dead cells, as determined by PI staining. Values were normalized to the initial measurement. | ||
In addition to analyzing a fixed time point, similar to traditional AST, the microfluidic gel encapsulation platform also allows time-lapse microscopy to study the growth and killing kinetics of single cells (Fig. 7A and S3–S5†). To demonstrate this capability, we expose bacteria to varying concentrations of ciprofloxacin while tracking them within the same imaging field at four different time points. In the absence of ciprofloxacin, substantial cell growth with large increases in both perimeter and area was observed (Fig. 7B and C). At 2 μg mL−1 (1× MIC), a small percentage of cells duplicated within 60 minutes; however, beyond this point, only slight elongation without replication was observed. At higher concentrations (20 μg mL−1 and 128 μg mL−1), bacterial growth was effectively inhibited, with minimal changes in morphology. These results demonstrate that the microfluidic gel encapsulation platform can reveal concentration-dependent kinetics of bacterial responses to antibiotics at the single cell level.
Under optimal growth conditions (i.e., rich media at 37 °C), the bacteria exhibited a maximum doubling time of ∼30 minutes during the exponential phase. In the microfluidic gel encapsulation platform, we observed a slight delay in the growth rate at the beginning of the experiment. This initial lag phase lasted approximately one duplication cycle (∼30 minutes) and may have resulted from the transition to the new solid-phase environment and potential thermal or physical stress introduced during the gel loading procedure. However, following the lag phase, the bacterial growth rate returned to the normal rate of approximately 30 minutes per doubling. This observation supports that the thin micropatterned gel pads in the gel encapsulation platform do not impose inherent limitations (e.g., nutrient diffusion or oxygen availability) on bacterial growth and are well-suited for analyzing bacterial responses to antimicrobial treatment.
Microfluidic gel encapsulation platform for fluid exchange
To assess the ability of surviving bacteria to resume growth after the removal of antibiotics, a fluid exchange step is necessary. Given that the gel encapsulating the bacteria is only 5 μm thick, fluid exchange can be easily performed on the microfluidic gel encapsulation platform. To demonstrate this capability, we conducted a single cell analysis experiment using K. pneumoniae (ATCC 11296) exposed to meropenem at the MIC of the strain (0.03 μg mL−1) for one hour. After antibiotic treatment, the broth containing meropenem was replaced with fresh medium, and viable cells were monitored during an additional hour of recovery incubation period (Fig. 8A). At the beginning of the experiment, PI was introduced to detect dead cells. As expected, the majority of bacteria were killed by meropenem at the MIC and stained by PI. In the analyzed field of view, we observed 95 bacteria, out of which 82 were stained by PI, indicating cell death. Conversely, 13 bacteria were not stained by PI and showed no signs of growth or elongation. These cells appeared to tolerate meropenem during the duration of antimicrobial treatment (dotted circles in Fig. 8B).To investigate if these viable bacteria could resume growth after the removal of the antimicrobial, a fluid exchange step was performed. Utilizing time-lapse microscopy and automated image analysis, we monitored the growth curve representing changes in the area of individual bacteria (Fig. 8C). Among the cells that were not stained by PI, three bacteria resumed growth within one hour of antimicrobial removal, consistent with the behavior of persister cells, while the remaining 10 cells could represent either VBNC cells or persister cells requiring longer recovery times. These results highlight the capability of the microfluidic gel encapsulation platform for single cell time–kill curve analysis and medium exchange, enabling the distinction of different cell fates. Future studies analyzing a larger number of bacteria and a broader range of conditions will be valuable for deciphering the differences between VBNC cells, persisters, and other dormant cells.
Microfluidic gel encapsulation platform for resolving bacterial response to antimicrobial in urine samples
We performed single cell analyses of uropathogenic E. coli (EC 137), a ciprofloxacin-susceptible strain, at 37 °C in MHB and in urine samples from three healthy volunteers (Fig. 9 and S7†). Using the MIC of ciprofloxacin (2 μg mL−1) as a reference, we compared the response of bacteria grown to log phase to the antibiotic across these different culture conditions. In MHB, the bacteria exhibited filamentous elongation, a stress response to antibiotic exposure, with no replication observed after 90 minutes. Several cells stained positive for PI, indicating compromised viability (Fig. 9A). In contrast, in all three urine samples, a substantial portion of cells survived and continued to replicate, with fewer PI-stained cells compared to MHB. The percentage of surviving cells varied among the urine samples, potentially reflecting individual differences in urine composition. This observation was generally consistent with the bulk analysis (see Fig. 1). Fig. 9B illustrates the fold increase in the area of individual cells across different media, highlighting intercellular variability in antibiotic responses depending on the medium. These results further support that the choice of medium, particularly host urine, can influence bacterial susceptibility and contribute to heterogeneity in antibiotic responses. Overall, the microfluidic gel encapsulation platform offers a rapid technique for capturing host-dependent variability in response to antibiotics at the single cell level.Discussion
In this study, we assessed the variability in bacterial killing kinetics at physiologically relevant concentrations of antibiotics in MH2 and urine and developed a fluid-exchange-enabled microfluidic device to track these response kinetics at the single cell level. While previous studies have compared the MIC of bacteria in MH2 and pooled physiological fluid or adapted media,39–43 none have systematically investigated the interindividual heterogeneity in the killing kinetics. Our results highlight the host-dependent heterogeneity of bacterial response to antimicrobials. It is often assumed that if a bacterium can be killed by antibiotics in a broth culture medium, which is favorable for bacterial growth, it can be killed even more effectively by antibiotics in body fluids, which are less ideal for their growth. In contrast, we found that some strains could survive under antibiotic pressure (32× MIC) in urine better than in broth, consistent with previous findings.39–43 Nutrient limitation, immunological stress, or variations in pH might increase the persistence of certain subpopulations under antibiotic pressure or reduce the efficacy of the antibiotic, contributing to the observed effect in urine.44–47 While some susceptible strains were completely killed by the antibiotics within 2 hours, others included subpopulations that survived for a longer time. On the contrary, both resistant strains tested were completely eradicated by the antibiotics in certain urines but not in MH2, corroborating previous findings of bacterial resistance reversal to antibiotics under physiological conditions.39,43 Thus, bacterial responses to the antibiotics varied based on strains, antibiotics, and the individual urine samples. Characterizing this variability in bacterial responses to antibiotics may inform appropriate treatment choice and duration for effective eradication of infection.In addition to resolving interindividual heterogeneity in physiological media, the ability to resolve intercellular time-killing heterogeneity in antimicrobial responses is critical for identifying minority subpopulations of response variants, which may have significant clinical implications. As proof of concept, we demonstrate a microfluidic gel encapsulation platform for analyzing single cell bacterial growth and killing kinetics in response to antimicrobials. The platform enables time-lapse, single cell imaging, resolving heterogeneity within the sample. Compared to the traditional time–kill curve assay with colony counting, the platform eliminates laborious procedures and time-consuming steps, which are critical for future clinical translation. The imaging platform is also compatible with advanced image analysis workflows based on computer vision and supervised learning algorithms. For instance, we have succeeded in predicting the susceptibility and MIC of antibiotic-treated bacteria within an hour by using machine learning analysis of morphological changes in bacterial single cells under the microscope.21
The microfluidic gel encapsulation platform offers several advantages over other microfluidic approaches, such as the microfluidic mother machine, which has been used to study the killing kinetics of bacteria against various antibiotics.24,25 Importantly, the platform enables single cell analysis in the context of intercellular interactions and quorum sensing, which are critical factors in the regulation of tolerance and resistance.48–50 Its imaging capabilities can incorporate viability indicators to assess both bacterial growth and killing. Furthermore, the platform supports medium exchange to evaluate the ability of surviving bacteria to resume growth after antibiotic removal, enabling differentiation between persisters and VBNCs. Unlike other microfluidic systems, the gel encapsulation platform does not require fluid connections or pumps and is compatible with standard liquid handling and imaging equipment, making it suitable for automation. Additionally, it involves simple procedures and can be multiplexed to test multiple conditions and samples in parallel, offering a cost-effective solution with potential for future clinical translation.
Time–kill analysis provide valuable information for the management of bacterial infections. For instance, the time–kill curve can suggest the most effective antibiotics (i.e., those that achieve fast killing) for the patient among the multiple agents recommended as effective by AST. K. pneumoniae and E. coli strains susceptible to meropenem, ciprofloxacin, and gentamicin used in this study showed substantial variability in the time–kill kinetics, which depends on bacterial strains, host urines, and antibiotic agents. As demonstrated in our results, time–kill curve can also offer quantitative measures of the synergistic effect between different antimicrobial combinations, enabling clinicians to implement antibiotic combination therapy with rapid killing effects that cannot be informed by MIC alone. Currently, combination therapy is chosen based on disease severity, bacterial species, or the prevalence of antimicrobial resistance in the community to avoid the development of resistance and/or subsequent failure with monotherapy.51
Precision time–kill analysis may also enhance antimicrobial stewardship by optimizing treatment duration. Currently, the duration of antimicrobial therapy is not personalized in clinical practice. Beyond reducing the emergence of antimicrobial resistance, shorter treatment course can reduce adverse effects and cost, improve patient compliance, and minimize collateral damages from gut microbiota dysbiosis. Antimicrobial stewardship has promoted a “less is better” approach to antimicrobial use in hospitals, including daily review of the continued need for intravenous antimicrobials to ensure the shortest effective treatment duration. However, in primary care or outpatient settings, where 90% of fixed-duration oral antimicrobial prescriptions are written for common infections such as uncomplicated UTI, no such ongoing assessment is routinely performed. Treatment durations for common uncomplicated UTI are determined based on comparative studies of treatment success rates over different fixed periods.52–56 While multiple studies report single-dose therapies having similar efficacy to longer-term therapies for uncomplicated UTI, these findings have not been adopted into clinical guidelines.57–61 Guiding antimicrobial treatment based on precision time–kill analysis may open new opportunities to improve antimicrobial stewardship.
Conclusions
We have demonstrated that time–kill curve testing of uropathogens based on single cell analysis performed on individual urine samples can provide additional pharmacodynamic insights beyond conventional static MIC, including better prediction of physiological antimicrobial response in vitro, resolution of interindividual and intercellular response phenotypes, and guidance for single or combinatorial therapy based on effective killing kinetics. With further studies to correlate with clinical outcomes, our precision single cell analysis may offer the opportunity to individualize antimicrobial selection and treatment duration.Data availability
All data generated are included in the manuscript and ESI.† Images generated in this study are available at: https://doi.org/10.5281/zenodo.14171622.Author contributions
Conceptualization: S. Y., P. K. W., R. A., J.-H. L., S. M. C., N. R.-M., which was refined by input from J. C. L. and K. E. M. Methodology: S. Y., P. K. W., R. A., J.-H. L., S. M. C., N. R.-M. Investigation: R. A., J.-H. L., S. M. C. Data analysis: J.-H. L., S. M. C., N. R.-M. Funding acquisition: S. Y., P. K. W., J. C. L. Project administration: S. Y., P. K. W., J. C. L. Supervision: S. Y., P. K. W., J. C. L. Writing – original draft: P. K. W., S. Y., R. A., N. R.-M. All authors discussed the results and contributed to reviewing and editing the manuscript.Conflicts of interest
None to declare.Acknowledgements
This work was partly supported by NIH R01AI153133, R01AI137272, and R21GM147838.References
- D. B. Chastain, S. T. King and K. R. Stover, Rethinking urinary antibiotic breakpoints: analysis of urinary antibiotic concentrations to treat multidrug resistant organisms, BMC Res. Notes, 2018, 11(1), 497 CrossRef PubMed.
- T. A. Stamey, W. R. Fair, M. M. Timothy, M. A. Millar, G. Mihara and Y. C. Lowery, Serum versus urinary antimicrobial concentrations in cure of urinary-tract infections, N. Engl. J. Med., 1974, 291(22), 1159–1163 CrossRef CAS PubMed.
- S. C. Ersoy, D. M. Heithoff, L. Barnes, G. K. Tripp, J. K. House and J. D. Marth, et al., Correcting a Fundamental Flaw in the Paradigm for Antimicrobial Susceptibility Testing, EBioMedicine, 2017, 20, 173–181 CrossRef PubMed.
- R. E. W. Hancock, Rethinking the Antibiotic Discovery Paradigm, EBioMedicine, 2015, 2(7), 629–630 CrossRef PubMed.
- J. Z. Kubicek-Sutherland, D. M. Heithoff, S. C. Ersoy, W. R. Shimp, J. K. House and J. D. Marth, et al., Host-dependent Induction of Transient Antibiotic Resistance: A Prelude to Treatment Failure, EBioMedicine, 2015, 2(9), 1169–1178 CrossRef PubMed.
- E. V. K. Ledger, S. Mesnage and A. M. Edwards, Human serum triggers antibiotic tolerance in Staphylococcus aureus, Nat. Commun., 2022, 13(1), 2041 CrossRef CAS PubMed.
- I. Levin-Reisman, A. Brauner, I. Ronin and N. Q. Balaban, Epistasis between antibiotic tolerance, persistence, and resistance mutations, Proc. Natl. Acad. Sci. U. S. A., 2019, 116(29), 14734–14739 CrossRef CAS PubMed.
- C. Michaux, S. Ronneau, R. T. Giorgio and S. Helaine, Antibiotic tolerance and persistence have distinct fitness trade-offs, PLoS Pathog., 2022, 18(11), e1010963 CrossRef CAS PubMed.
- M. Mueller, A. De La Peña and H. Derendorf, Issues in Pharmacokinetics and Pharmacodynamics of Anti-Infective Agents: Kill Curves versus MIC, Antimicrob. Agents Chemother., 2004, 48(2), 369–377 CrossRef CAS PubMed.
- L. Dewachter, M. Fauvart and J. Michiels, Bacterial Heterogeneity and Antibiotic Survival: Understanding and Combatting Persistence and Heteroresistance, Mol. Cell, 2019, 76(2), 255–267 CrossRef CAS PubMed.
- I. Levin-Reisman, I. Ronin, O. Gefen, I. Braniss, N. Shoresh and N. Q. Balaban, Antibiotic tolerance facilitates the evolution of resistance, Science, 2017, 355(6327), 826–830 CrossRef CAS PubMed.
- T. C. Barrett, W. W. K. Mok, A. M. Murawski and M. P. Brynildsen, Enhanced antibiotic resistance development from fluoroquinolone persisters after a single exposure to antibiotic, Nat. Commun., 2019, 10(1), 1177 CrossRef PubMed.
- J. Liu, O. Gefen, I. Ronin, M. Bar-Meir and N. Q. Balaban, Effect of tolerance on the evolution of antibiotic resistance under drug combinations, Science, 2020, 367(6474), 200–204 CrossRef CAS PubMed.
- N. Q. Balaban, S. Helaine, K. Lewis, M. Ackermann, B. Aldridge and D. I. Andersson, et al., Definitions and guidelines for research on antibiotic persistence, Nat. Rev. Microbiol., 2019, 17(7), 441–448 CrossRef CAS PubMed.
- J. Urbaniec, Y. Xu, Y. Hu, S. Hingley-Wilson and J. McFadden, Phenotypic heterogeneity in persisters: a novel ‘hunker’ theory of persistence, FEMS Microbiol. Rev., 2022, 46(1), fuab042 CrossRef CAS PubMed.
- M. Ayrapetyan, T. Williams and J. D. Oliver, Relationship between the Viable but Nonculturable State and Antibiotic Persister Cells, J. Bacteriol., 2018, 200(20), e00249-18 CrossRef PubMed.
- V. I. Band and D. S. Weiss, Heteroresistance: A cause of unexplained antibiotic treatment failure?, PLoS Pathog., 2019, 15(6), e1007726 CrossRef CAS PubMed.
- V. I. Band, S. W. Satola, E. M. Burd, M. M. Farley, J. T. Jacob and D. S. Weiss, Carbapenem-Resistant Klebsiella pneumoniae Exhibiting Clinically Undetected Colistin Heteroresistance Leads to Treatment Failure in a Murine Model of Infection, mBio, 2018, 9(2), e02448-17 CrossRef PubMed.
- F. Zhang, M. Ding, X. Yan, J. Bai, Q. Li and B. Zhang, et al., Carbapenem-resistant K. pneumoniae exhibiting clinically undetected amikacin and meropenem heteroresistance leads to treatment failure in a murine model of infection, Microb. Pathog., 2021, 160, 105162 CrossRef CAS PubMed.
- C. Pereira, J. Larsson, K. Hjort, J. Elf and D. I. Andersson, The highly dynamic nature of bacterial heteroresistance impairs its clinical detection, Commun. Biol., 2021, 4(1), 521 CrossRef CAS PubMed.
- K. C. Tjandra, N. Ram-Mohan, M. Roshardt, E. J. Zudock, Z. Qu and K. E. Mach, et al., Growth independent morphometric machine learning workflow for single-cell antimicrobial susceptibility testing of Klebsiella pneumoniae to meropenem, Front. Imaging, 2024, 3, 1418669 CrossRef.
- K. C. Tjandra, N. Ram-Mohan, R. Abe, M. M. Hashemi, J. H. Lee and S. M. Chin, et al., Diagnosis of Bloodstream Infections: An Evolution of Technologies towards Accurate and Rapid Identification and Antibiotic Susceptibility Testing, Antibiotics, 2022, 11(4), 511 CrossRef CAS PubMed.
- H. Li, K. Hsieh, P. K. Wong, K. E. Mach, J. C. Liao and T. H. Wang, Single-cell pathogen diagnostics for combating antibiotic resistance, Nat. Rev. Methods Primers, 2023, 3(1), 6 CrossRef CAS.
- Y. Zhang, I. Kepiro, M. G. Ryadnov and S. Pagliara, Single Cell Killing Kinetics Differentiate Phenotypic Bacterial Responses to Different Antibacterial Classes, Microbiol. Spectrum, 2023, 11(1), e0366722 CrossRef PubMed.
- W. Postek, N. Pacocha and P. Garstecki, Microfluidics for antibiotic susceptibility testing, Lab Chip, 2022, 22(19), 3637–3662 RSC.
- J. Q. Boedicker, L. Li, T. R. Kline and R. F. Ismagilov, Detecting bacteria and determining their susceptibility to antibiotics by stochastic confinement in nanoliter droplets using plug-based microfluidics, Lab Chip, 2008, 8(8), 1265–1272 RSC.
- H. Li, P. Torab, K. E. Mach, C. Surrette, M. R. England and D. W. Craft, et al., Adaptable microfluidic system for single-cell pathogen classification and antimicrobial susceptibility testing, Proc. Natl. Acad. Sci. U. S. A., 2019, 116(21), 10270–10279 CrossRef CAS PubMed.
- C. H. Chen, Y. Lu, M. L. Y. Sin, K. E. Mach, D. D. Zhang and V. Gau, et al., Antimicrobial Susceptibility Testing Using High Surface-to-Volume Ratio Microchannels, Anal. Chem., 2010, 82(3), 1012–1019 CrossRef CAS PubMed.
- Y. Lu, J. Gao, D. D. Zhang, V. Gau, J. C. Liao and P. K. Wong, Single Cell Antimicrobial Susceptibility Testing by Confined Microchannels and Electrokinetic Loading, Anal. Chem., 2013, 85(8), 3971–3976 CrossRef CAS PubMed.
- D. J. Farrell, I. Morrissey, D. De Rubeis, M. Robbins and D. Felmingham, A UK multicentre study of the antimicrobial susceptibility of bacterial pathogens causing urinary tract infection, J. Infect., 2003, 46(2), 94–100 CrossRef CAS PubMed.
- Clinical and Laboratory Standards Institute, M100 Performance Standards for 551 Antimicrobial Susceptibility Testing, 28th edn, 552, http://file.qums.ac.ir/repository/mmrc/CLSI-2018-M100-S28.pdf Search PubMed.
- R. P. Bax, W. Bastain, A. Featherstone, D. M. Wilkinson, M. Hutchison and S. J. Haworth, The pharmacokinetics of meropenem in volunteers, J. Antimicrob. Chemother., 1989, 24(Suppl A), 311–320 CrossRef CAS PubMed.
- F. M. E. Wagenlehner, M. Kinzig-Schippers, F. Sörgel, W. Weidner and K. G. Naber, Concentrations in plasma, urinary excretion and bactericidal activity of levofloxacin (500 mg) versus ciprofloxacin (500 mg) in healthy volunteers receiving a single oral dose, Int. J. Antimicrob. Agents, 2006, 28(6), 551–559 CrossRef CAS PubMed.
- E. Labovitz, M. E. Levison and D. Kaye, Single-dose daily gentamicin therapy in urinary tract infection, Antimicrob. Agents Chemother., 1974, 6(4), 465–470 CrossRef CAS PubMed.
- H. Wolf, P. O. Madsen and P. Rhodes, The ampicillin concentration in prostatic tissue and prostatic fluid, Urol. Int., 1967, 22(5), 453–460 CrossRef CAS PubMed.
- G. Butler, J. Bos, R. H. Austin, S. R. Amend and K. J. Pienta, Escherichia coli survival in response to ciprofloxacin antibiotic stress correlates with increased nucleoid length and effective misfolded protein management, R. Soc. Open Sci., 2023, 10(8), 230338 CrossRef CAS PubMed.
- F. Silva, O. Lourenço, J. A. Queiroz and F. C. Domingues, Bacteriostatic versus bactericidal activity of ciprofloxacin in Escherichia coli assessed by flow cytometry using a novel far-red dye, J. Antibiot., 2011, 64(4), 321–325 CrossRef CAS PubMed.
- J. M. Diver and R. Wise, Morphological and biochemical changes in Escherichia coli after exposure to ciprofloxacin, J. Antimicrob. Chemother., 1986, 18(Suppl D), 31–41 CrossRef CAS PubMed.
- E. Thulin, M. Thulin and D. I. Andersson, Reversion of High-level Mecillinam Resistance to Susceptibility in Escherichia coli During Growth in Urine, EBioMedicine, 2017, 23, 111–118 CrossRef PubMed.
- H. Laue, T. Valensise, A. Seguin, S. Hawser, S. Lociuro and K. Islam, Effect of human plasma on the antimicrobial activity of iclaprim in vitro, J. Antimicrob. Chemother., 2007, 60(6), 1388–1390 CrossRef CAS PubMed.
- M. Castanheira, L. R. Duncan, P. R. Rhomberg and H. S. Sader, Enhanced activity of cefepime-tazobactam (WCK 4282) against KPC-producing Enterobacteriaceae when tested in media supplemented with human serum or sodium chloride, Diagn. Microbiol. Infect. Dis., 2017, 89(4), 305–309 CrossRef CAS PubMed.
- K. D. Leuthner, C. M. Cheung and M. J. Rybak, Comparative activity of the new lipoglycopeptide telavancin in the presence and absence of serum against 50 glycopeptide non-susceptible staphylococci and three vancomycin-resistant Staphylococcus aureus, J. Antimicrob. Chemother., 2006, 58(2), 338–343 CrossRef CAS PubMed.
- D. M. Heithoff, V. L. Barnes, S. P. Mahan, J. C. Fried, L. N. Fitzgibbons and J. K. House, et al., Re-evaluation of FDA-approved antibiotics with increased diagnostic accuracy for assessment of antimicrobial resistance, Cell Rep. Med., 2023, 4(5), 101023 CrossRef CAS PubMed.
- R. A. Fisher, B. Gollan and S. Helaine, Persistent bacterial infections and persister cells, Nat. Rev. Microbiol., 2017, 15(8), 453–464 CrossRef CAS PubMed.
- L. Dewachter, M. Fauvart and J. Michiels, Bacterial Heterogeneity and Antibiotic Survival: Understanding and Combatting Persistence and Heteroresistance, Mol. Cell, 2019, 76(2), 255–267 CrossRef CAS PubMed.
- S. M. Amato, C. H. Fazen, T. C. Henry, W. W. K. Mok, M. A. Orman and E. L. Sandvik, et al., The role of metabolism in bacterial persistence, Front. Microbiol., 2014, 5, 70 Search PubMed.
- J. M. Woodcock, J. M. Andrews, N. P. Brenwald, J. P. Ashby and R. Wise, The in-vitro activity of faropenem, a novel oral penem, J. Antimicrob. Chemother., 1997, 39(1), 35–43 CrossRef CAS PubMed.
- R. Sikdar and M. H. Elias, Evidence for Complex Interplay between Quorum Sensing and Antibiotic Resistance in Pseudomonas aeruginosa. Rampioni G, editor, Microbiol. Spectrum, 2022, 10(6), e0126922 CrossRef PubMed.
- D. V. Raju, A. Nagarajan, S. Pandit, M. Nag, D. Lahiri and V. Upadhye, Effect of bacterial quorum sensing and mechanism of antimicrobial resistance, Biocatal. Agric. Biotechnol., 2022, 43, 102409 CrossRef CAS.
- X. Zhao, Z. Yu and T. Ding, Quorum-Sensing Regulation of Antimicrobial Resistance in Bacteria, Microorganisms, 2020, 8(3), 425 CrossRef CAS PubMed.
- M. J. Rybak and B. J. McGrath, Combination antimicrobial therapy for bacterial infections, Guidelines for the clinician, Drugs, 1996, 52(3), 390–405 CrossRef CAS PubMed.
- T. M. Hooton, K. Running and W. E. Stamm, Single-dose therapy for cystitis in women. A comparison of trimethoprim-sulfamethoxazole, amoxicillin, and cyclacillin, JAMA, J. Am. Med. Assoc., 1985, 253(3), 387–390 CrossRef CAS.
- T. Vogel, R. Verreault, M. Gourdeau, M. Morin, L. Grenier-Gosselin and L. Rochette, Optimal duration of antibiotic therapy for uncomplicated urinary tract infection in older women: a double-blind randomized controlled trial, CMAJ, 2004, 170(4), 469–473 Search PubMed.
- W. E. Stamm and T. M. Hooton, Management of urinary tract infections in adults, N. Engl. J. Med., 1993, 329(18), 1328–1334 CrossRef CAS PubMed.
- I. E. Nygaard and J. M. Johnson, Urinary tract infections in elderly women, Am. Fam. Physician, 1996, 53(1), 175–182 Search PubMed.
- S. R. Norrby, Short-term treatment of uncomplicated lower urinary tract infections in women, Rev. Infect. Dis., 1990, 12(3), 458–467 CrossRef CAS PubMed.
- H. W. Asbach, Single Dose Oral Administration of Cefixime 400mg in the Treatment of Acute Uncomplicated Cystitis and Gonorrhoea, Drugs, 1991, 42(Supplement 4), 10–13 CrossRef PubMed.
- F. J. Buckwold, P. Ludwig, G. K. Harding, L. Thompson, M. Slutchuk and J. Shaw, et al., Therapy for acute cystitis in adult women. Randomized comparison of single-dose sulfisoxazole vs trimethoprim-sulfamethoxazole, JAMA, J. Am. Med. Assoc., 1982, 247(13), 1839–1842 CrossRef CAS.
- A. Huttner, A. Kowalczyk, A. Turjeman, T. Babich, C. Brossier and N. Eliakim-Raz, et al., Effect of 5-Day Nitrofurantoin vs Single-Dose Fosfomycin on Clinical Resolution of Uncomplicated Lower Urinary Tract Infection in Women: A Randomized Clinical Trial, JAMA, J. Am. Med. Assoc., 2018, 319(17), 1781 CrossRef CAS PubMed.
- G. A. Richard, C. P. Mathew, J. M. Kirstein, D. Orchard and J. Y. Yang, Single-dose fluoroquinolone therapy of acute uncomplicated urinary tract infection in women: results from a randomized, double-blind, multicenter trial comparing single-dose to 3-day fluoroquinolone regimens, Urology, 2002, 59(3), 334–339 CrossRef PubMed.
- F. A. van Balen, F. W. Touw-Otten and R. A. de Melker, Single-dose pefloxacin versus five-days treatment with norfloxacin in uncomplicated cystitis in women, J. Antimicrob. Chemother., 1990, 26(Suppl B), 153–160 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4lc00765d |
| ‡ Equal contributions. |
| This journal is © The Royal Society of Chemistry 2025 |