Efficient production of citric acid from lignocellulose hydrolysate by metabolically engineered Yarrowia lipolytica†
Minrui
Lu‡
ab,
Yuanyuan
Sha‡
ab,
Yuwei
Zhang
ab,
Mianshen
Ge
ab,
Zhaoxian
Xu
ab and
Mingjie
Jin
 *ab
*ab
aSchool of Environmental and Biological Engineering, Nanjing University of Science & Technology, Nanjing, 210094, China. E-mail: jinmingjie@njust.edu.cn
bBiorefinery Research Institution, Nanjing University of Science and Technology, Nanjing 210094, China
First published on 10th December 2024
Abstract
Lignocellulosic biomass is a reliable renewable feedstock for citric acid fermentation. Low product titer is the bottleneck in the large scale production of cellulosic citric acid by Yarrowia lipolytica. Herein, multiple genetic engineering strategies were explored to construct an engineered Y. lipolytica strain that can efficiently produce citric acid with a high titer and yield. Genes related to TCA cycles were overexpressed to increase citric acid production. Subsequently, genes in the downstream lipid synthesis pathway were deleted to decrease citric acid consumption. The mitochondrial transporter of isocitric acid was also deleted to minimize by-product secretion. Next, six glucose transporter genes, a hexose kinase gene, and a heterologous 6-phosphofructo-1-kinase gene were tested to enhance the efficiency of citric acid production. Consequently, the optimized engineered strain produced 88.2 g L−1 and 73.6 g L−1 citric acid from a pure sugar medium and 30% solid loading hydrolysate, respectively. Finally, in a 3 L bioreactor, 83.6 g L−1 citric acid was produced from 35% solid loading of corn stover hydrolysate via fed-batch fermentation. In this work, an efficient robust yeast cell method was developed for the production of citric acid in a sustainable manner.
Green foundation1. This work advances green chemistry by engineering Yarrowia lipolytica to produce citric acid from lignocellulosic biomass without detoxification, thus promoting sustainable biomanufacturing.2. The DLCA(sa) pretreatment-based cellulosic citric acid biorefinery system does not require detoxification or supplementation of nitrogen sources, simplifying the production process as well as saving the overall cost. This system is more sustainable and eco-friendly than the commercial starch-based citric acid production process. This work sheds light on biosynthesizing organic acid from lignocellulose using a yeast cell factory. 3. Further work will be dedicated to scaling up the biorefinery process for citric acid production, making cellulosic citric acid become commercially viable. |
1. Introduction
Citric acid, a valuable commercial bioproduct, has widespread applications in the food and pharmaceutical industries, polymer production and environmental protection.1,2 In 2022, the global market volume of citric acid reached approximately 2.8 million tons per year, and demand is still increasing at a rate of 5% every year.3 Initially, citric acid was produced commercially from lemon juice; however, many microorganisms have since been identified that can produce significant quantities of citric acid.4 At present, industrial citric acid production mainly occurs through the fermentation of Aspergillus niger, with a production cost approximately 50% lower than that of chemical synthesis.5,6 However, the process of citric acid fermentation by A. niger has many drawbacks. For example, commercial citric acid production by A. niger mainly uses molasses or sucrose as the carbon source, in which excessive trace elements have a great influence on the citric acid production. The elimination of these trace elements generates significant amounts of solid and liquid waste, posing environmental hazards and increasing disposal costs.7 Furthermore, the spores released by A. niger during the fermentation process are potent allergens that can cause pulmonary aspergilloma.8Compared to filamentous fungi, yeasts exhibit greater tolerance to metal ions and high substrate concentrations, are easier to cultivate, have higher fermentation rates, and have therefore been used as an alternative host strain for citric acid fermentation. Yarrowia lipolytica, a generally recognized as safe (GRAS) oleaginous yeast, stands out as a promising option for industrial citric acid production. Y. lipolytica exhibits higher tolerance to metal ions, lower oxygen requirements, a broader substrate spectrum, more genetic modification tools and easier-to-scale-up cultivation processes.9 These advantages make Y. lipolytica an excellent model for obtaining many tricarboxylic acid (TCA) intermediate products such as citric acid, isocitric acid, α-ketoglutarate, and succinic acid.10,11 Many efforts have been devoted to metabolically engineering Y. lipolytica for enhanced production of citric acid. For example, Fu et al. overexpressed a heterologous pyruvate carboxylase-encoding PYC gene in Y. lipolytica, resulting in a recombinant strain that produced 70.2 g L−1 citric acid with a yield of 0.76 g g−1 glucose.12 Yuzbasheva identified the major mitochondrial citrate transporter gene YlYHM2 and co-expressed it with adenosine monophosphate deaminase gene YlAMPD. This co-expression resulted in 49.7 g L−1 and 97.1 g L−1 citric acid in the test tube and fed-batch bioreactor, respectively.13 These results demonstrated the promising application of Y. lipolytica in industrial citric acid production. However, these high yields of citric acid have primarily relied on glucose as a carbon source. In industrial-scale fermentation, the dependence on glucose as a carbon source not only increases production costs but also exacerbates food scarcity issues. Therefore, it is imperative to explore a sustainable citric acid production process using economically viable alternative feedstocks.
Since Y. lipolytica can grow under multiple conditions, many low-cost carbon sources have been employed to decrease the cost of citric acid production. Urak et al. used diluted and fortified carrot juice as a substrate, achieving a citric acid production of 80.5 g L−1.14 Rzechonec et al. cultivated a Y. lipolytica strain with the overexpression of glycerol assimilation genes on crude glycerol, yielding 75.9 g L−1 citric acid.15 The strain Y. lipolytica SWJ-1b achieved 84 g L−1 citric acid from inulin.16 Lignocellulosic biomass, including waste paper, industrial waste, and agricultural and forestry waste, is the most abundant renewable energy on the planet, and it has been an appealing feedstock in the fermentation industry due to its renewable, cheap, and carbon-neutral characteristics. The biorefinery of lignocellulosic biomass by Y. lipolytica has been demonstrated for sustainable production of lipids, limonene, β-farnesene, and α-pinene.17–20 Although efficient production of citric acid has been achieved using the above low-value feedstocks, the potential of lignocellulosic biomass as a carbon source for citric acid production in Y. lipolytica has been less explored. Liu et al. used the hydrolysate of straw cellulose as substrate, achieving a citric acid titer of 42.4 g L−1 in a three-cycle fed-batch cultivation.21 Gao et al. cultivated Y. lipolytica in a corn stover hydrolysate containing glucose and glycerol as substrate, obtaining 63.8 g L−1 citric acid with a yield of 0.66 g g−1.22 However, the low titers of citric acid fall short of the requirements of industrial production, thereby hindering the industrialization process of biorefining lignocellulosic hydrolysate for citric acid production.
In this study, we metabolically engineered Y. lipolytica to improve its citric acid titer and yield and demonstrated its economic viability in producing citric acid from lignocellulosic hydrolysate. Our research employed a push-and-pull strategy to enhance citric acid production. We first evaluated the effects of single and combinatorial overexpression of genes related to the TCA cycle and citric acid transporters on citric acid production and ultimately selected a superior gene combination for improved citric acid production. Next, we weakened the catabolism pathway of citric acid by deleting genes involved in the lipid synthesis pathway. To eliminate the formation of the by-product isocitric acid, we also deleted its mitochondrial transporter. We further overexpressed genes encoding the glucose transporter, hexokinase, and mutant 6-phosphofructo-1-kinase to enhance substrate consumption rates and relieve product inhibition in glycolytic pathways. Through these efforts, we successfully engineered a robust Y. lipolytica capable of producing citric acid with a high yield and reduced by-product formation. In a 3-L bioreactor, this strain achieved a citric acid titer of 83.6 g L−1 in lignocellulosic hydrolysate. This study elucidated the feasibility of utilizing low-cost feedstock lignocellulosic hydrolysate for citric acid production by engineered Y. lipolytica.
2. Materials and methods
2.1 Strains and culture medium
The xylose-utilizing Y. lipolytica BZ1 was used as a host strain and modified to produce citric acid. This strain was constructed by disrupting the KU70 gene in Y. lipolytica po1f and overexpressing Scheffersomyces stipitisD-xylose reductase gene (XR) and D-xylitol dehydrogenase gene (XDH), and Y. lipolytica xylulokinase gene (XK). Escherichia coli DH5α was used for plasmid construction and amplification. Routine cultivation of E. coli DH5α was performed in Luria–Bertani (LB) medium (NaCl 10 g L−1, peptone 10 g L−1, yeast extract 10 g L−1) supplemented with ampicillin or kanamycin at 37 °C and 220 rpm overnight. Y. lipolytica was cultured in a YPD20 medium (glucose 20 g L−1, peptone 10 g L−1, yeast extract 10 g L−1) at 30 °C and 250 rpm overnight. 2*YPD medium (glucose 40 g L−1, peptone 20 g L−1, yeast extract 20 g L−1) was used for the transformation of Y. lipolytica. Plates of the YNB medium without URA (YNB 1.7 g L−1, NH4Cl 5 g L−1, glucose 5 g L−1, 20 g L−1 agar) were used for screening of recombinant strains. Citric acid fermentation medium: glucose 100 g L−1, xylose 50 g L−1, KH2PO4 1.7 g L−1, Na2HPO4 12 g L−1, MgSO4·7H2O 1.25 g L−1, yeast extract 1.26 g L−1, peptone 2.52 g L−1, Vitamin B6 6 mg L−1.2.2 DLCA(sa) hydrolysate preparation
Corn stover was kindly provided by China Oil & Foodstuffs Corporation, and its main components were 31 wt% glucan and 20 wt% xylan. DLCA(sa) pretreatment was performed with sulfuric acid as the pretreatment reagent using the method reported previously.23 Briefly, the corn stover was treated with sulfuric acid at a dose of 0.075 g sulfuric acid per g dry biomass and then pelleted by a pellet machine. The obtained DLC(sa) pellets were then autoclaved at 121 °C for 30 min to promote biomass deconstruction, achieving the DLCA(sa) corn stover. The enzymatic hydrolysis of the DLCA(sa) corn stover was performed with CTec3 HS at a dosage of 20 mg protein per g glucan at solid loadings of 25%, 30% and 35% (w/w). After the hydrolysis process for 72 h, the obtained hydrolysate was centrifuged at 8000 rpm, 10 min, to obtain the supernatant and then supplemented with citric acid fermentation medium components (without peptone and yeast extract) and neutralized with NaOH to pH 6.2.3 Plasmids and recombinant strain construction
All DNA sequencing and oligonucleotide primer syntheses were conducted by Beijing Tsingke Biotech Co., Ltd. Recombinant DNA manipulations and Gibson assembly were used for plasmid construction according to the previously described standard procedures and protocol of the ClonExpress MultiS One-Step Cloning Kit (Vazyme), respectively.24 The heterologous genes (ssXR, ssXDH, pfkA) were codon-optimized and synthesized by Beijing Tsingke Biotech Co., Ltd. The endogenous genes (ICL, YHM2, AMPD, Yht1-6, Hxt1) were amplified from the BZ1 genome by PCR amplification.For the overexpression genes ICL, YHM2 and AMPD, the promoter PTEFin, overexpression genes and the terminator TXPR were PCR-amplified from the genome of strain po1f. These three fragments were then inserted into restriction sites of MssI and EcoRI of plasmid pUC-HUH-rDNA. For the overexpression genes Yht1-6 and Hxt1, the promoter PTEFin, overexpression genes and the terminator Tcyc1 were PCR-amplified from the genome of strain po1f. These three fragments were then inserted into restriction sites of MssI and EcoRI of plasmid pUC-HUH-A2. All the gene deletions were performed by homologous recombination. For the knockout genes like DGA1, the upstream and downstream 1 kb of DGA1 were selected as homologous arms and PCR-amplified from the genome of strain po1f. These two fragments and the origin fragments and ampicillin resistance were overlapped and inserted into restriction sites of NotI and EcoRI of plasmid pUC-HUH-rDNA. The plasmids and engineered Y. lipolytica strains in this study are summarized in Table 1. The primers used for plasmid construction in this study are summarized in Table S1.† All the integration or deletion plasmids are linearized for yeast transformation. The transformation of Y. lipolytica cells was performed according to the protocol of Frozen-EZ Yeast transformation II kit (Zymo Research Corporation), with plating on YNB plates without uracil supplementation and cultivated at 30 °C for 3 days. The colonies picked from the plates were subsequently verified by diagnostic PCR to confirm the integration or deletion of target genes on the Y. lipolytica genome. The recycling of the URA3 selection marker was performed by counter-selecting on a YPG plate containing 1.2 g L−1 5-FOA.25
| Strains or plasmids | Characteristics | Ref. |
|---|---|---|
| Strain | ||
| Y. lipolytica | ||
| Po1f | matA, ura3-302, leu2-270, xpr2-322, axp2-delta NU49, XPR2::SUC2 | |
| Po1fΔKu70 | Po1f, ΔKu70 | This study |
| BZ | Po1fΔKu70, integration of ssXR-ssXDH-ylXK cassette | This study |
| BZ-I | BZ, integration of ICL1 cassette | This study |
| BZ-A | BZ, integration of AMPD cassette | This study |
| BZ-Y | BZ, integration of YHMP cassette | This study |
| BZ-C1 | BZ, integration of CIT1 cassette | This study |
| BZ-C2 | BZ, integration of CIT2 cassette | This study |
| BZ-AY | BZ, integration of AMPD-YHMP cassette | This study |
| BZ-IY | BZ, integration of ICL-YHMP cassette | This study |
| BZ-IAY | BZ, integration of ICL-AMPD-YHMP cassette | This study |
| BZ-ΔD1 | BZ, ΔDGA1 | This study |
| BZ-ΔD2 | BZ, ΔDGA2 | This study |
| BZ-ΔA | BZ, ΔACL | This study |
| BZ-Δ12 | BZ-Δ1, ΔDGA2 | This study |
| BZ-Δ1A | BZ-Δ1, ΔACL | This study |
| BZ-Δ2A | BZ-Δ2, ΔACL | This study |
| BZ-IYΔ1 | BZ-Δ1, integration of ICL-YHMP cassette | This study |
| BZ-IYΔ2 | BZ-Δ2, integration of ICL-YHMP cassette | This study |
| BZ-IYΔA | BZ-ΔA, integration of ICL-YHMP cassette | This study |
| BZ-IYΔ12 | BZ-Δ12, integration of ICL-YHMP cassette | This study |
| BZ-IYΔ1A | BZ-Δ1A, integration of ICL-YHMP cassette | This study |
| BZ-IYΔ2A | BZ-Δ2A, integration of ICL-YHMP cassette | This study |
| BZ-IYΔ12S | BZ-IYΔ12, ΔylSCF | This study |
| BZ-Δ12Y1–6 | BZ-Δ12, integration of Yht1-6 cassettes, respectively | This study |
| BZ-Δ12Hxk1 | BZ-Δ12, integration of Hxk1 cassette | This study |
| BZ-IYΔ12SY3 | BZ-IYΔ12S, integration of Yht3 cassette | This study |
| Plasmid | ||
| pUC-E3-HUH | Cloning vector | |
| pUC-rDNA-HUH | Cloning vector | This study |
| pUC-A2-HUH | Cloning vector | This study |
| pUC-X | pUC-rDNA-HUH derivative expressing ssXR, ssXDH and ylXK | This study |
| pUC-I | pUC-rDNA-HUH derivative expressing ICL | This study |
| pUC-A | pUC-rDNA-HUH derivative expressing AMPD | This study |
| pUC-Y | pUC-rDNA-HUH derivative expressing YHMP | This study |
| pUC-C | pUC-rDNA-HUH derivative expressing CIT1 | This study |
| pUC-IY | pUC-rDNA-HUH derivative expressing ICL, YHMP | This study |
| pUC-AY | pUC-rDNA-HUH derivative expressing AMPD, YHMP | This study |
| pUC-IAY | pUC-rDNA-HUH derivative expressing synthetic ICL, AMPD, YHMP | This study |
| pUC-ΔDGA1-HUH | pUC-HUH insert in the homologous arm of DGA1 for gene knockout | This study |
| pUC-ΔDGA2-HUH | pUC-HUH insert in the homologous arm of DGA2 for gene knockout | This study |
| pUC-ΔACL-HUH | pUC-HUH insert in the homologous arm of ACL for gene knockout | This study |
| pUC-ΔylSCF-HUH | pUC-HUH insert in the homologous arm of ylSCF for gene knockout | This study |
| pUC-yht1–6 | pUC-rDNA-HUH derivative expressing yht1-6 genes | This study |
| pUC-Hxk1 | pUC-rDNA-HUH derivative expressing Hxk1 genes | This study |
| pUC-PFK | pUC-rDNA-HUH derivative expressing pfkA genes | This study |
2.4 Culture conditions in a shake flask
A single colony was selected from a plating medium and cultivated in a test tube with 5 mL of YPD medium at 30 °C for 12 h. Then, a portion of the microbial medium in the test tube was transferred to a flask with 25 mL YPD medium as seed culture. The broth in the flask was cultivated at 30 °C for 12 h and centrifuged, and the obtained pellet was inoculated in a fermentation medium with an initial OD600 of 2.0. The culture pH was controlled at 6.0 by adding 40 mM NaOH every 12 h. Fermentation was performed at 30 °C and 200 rpm for 168 h. Samples were taken every 24 h for the measurement of cell density, sugar concentration and citric acid concentration.2.5 Citric acid fermentation in 3 L bioreactors
Strain BZ-IY3Δ12S was selected as the fermentation strain, and the solid loading of DLCA(sa)-pretreated corn stover was 30%. The medium components were the same as those in the shake flask fermentation, without the addition of peptone and yeast extract. The medium pH was monitored by an automatic system and controlled at 6 by automatically feeding 1 M NaOH. A stirring speed of 800 rpm, airflow rate of 1 vvm, temperature of 30 °C and dissolved oxygen at 50% were maintained. In the batch fermentation, the fermentation volume was 1.2 L. In the fed-batch fermentation, the initial fermentation volume was 1.2 L, then 30 mL concentrated hydrolysate of 30% solid loading (405.2 g L−1 glucose and 239.2 g L−1 xylose) was added when the glucose was used up. Samples were withdrawn every 12 h for the analysis of residual glucose, xylose and citric acid.2.6 Analytical methods
The concentrations of sugar and citric acid in the fermentation broth were quantified by HPLC equipped with a Bio-rad Aminex HPX-87H column and a refractive index detector. A 15.0 μL injection volume was used in a mobile phase composed of 0.005 M sulfuric acid with a flow rate of 0.6 mL min−1. The column temperature was maintained at 30 °C. Cell growth of Y. lipolytica was determined by measuring the OD600 value of fermentation broth.Lipid production was measured using a previously described procedure. Briefly, 50 μL culture broth was centrifuged and the supernatant was discarded. The pellet was washed and resuspended by ddH2O (double distilled water) and then mixed with 1 mL sulfuric acid. The mixture was heated at 100 °C for 10 min and then cooled for 10 min. Then, 2.5 mL vanillin-phosphoric acid was added to react at 37 °C for 15 min and cooled for 10 min. The absorbance was determined at 530 nm against the reference samples prepared with 50 μL deionized water.
3. Results and discussion
3.1 Rewiring the carbon flux of the TCA cycle toward citric acid
As an oleaginous microorganism, Y. lipolytica possesses a vigorous TCA cycle flux, which confers it an ideal chassis strain for citric acid production. Under nitrogen depletion conditions, adenosine monophosphate deaminase (AMPD) inhibits the activity of isocitrate dehydrogenase (ICDH), leading to the accumulation of citric acid and isocitric acid.26 Then, the accumulated citric acid was transported from the mitochondria to the cytoplasm by the mitochondrial citrate carrier (YlYhm2). In addition, iso-citrate lyase (ICL) catalyzes isocitric acid into acetyl-CoA and oxaloacetate, which then react to form citric acid.27 Approaches of overexpressing AMPD, YHM2, and ICL genes have been confirmed to be effective in enhancing citric acid accumulation.13,16 Therefore, the endogenous genes AMPD, YHM2 and ICL were individually and/or combinatorial overexpressed in a xylose-utilizing strain BZ to rewire the carbon flux of the TCA cycle towards citric acid synthesis and decrease the formation of isocitric acid (Fig. 1A).The AMPD, YHM2, and ICL genes were first individually integrated into the 16s rDNA locus of Y. lipolytica in multiple copies. Since the overexpressed genes were randomly inserted into the chromosome with various copy numbers, transformants may exhibit distinct metabolic characteristics. Thus, 8 transformants were randomly selected for each overexpressed gene to evaluate their fermentation profiles, focusing on those with enhanced sugar consumption rate and citric acid titer. As shown in Fig. 1B, all the overexpression strains showed higher citric acid titer and sugar conversion yield than the original strain BZ, as expected. Overexpression of YHM2 (BZ-Y) resulted in the highest citric acid accumulation of 62.4 g L−1, with a yield of 0.49 g g−1. AMPD overexpression strain BZ-A produced lower citric acid of 58.6 g L−1 but a similar yield (0.50 g g−1). The ICL overexpression strain BZ-I produced 48.8 g L−1 of citric acid, and the yield was 0.48 g g−1. In addition, the sugar consumption and OD600 were not significantly affected by overexpressing these genes (Fig. 1C). Strains BZ-Y exhibited superior sugar consumption, consuming 100 g L−1 glucose and 21.5 g L−1 xylose at 168 h. Strain BZ-A and strain BZ also consumed glucose, but the xylose consumption was relatively lower than that in BZ-Y. All the strains showed a similar OD600 in the range of 38.2 to 43.7. The concentration of isocitric acid was also tested (Fig. S1†). Strain BZ-I showed a significantly decreased isocitric acid titer up to 62.8% compared to the control strain, followed by BZ-Y with a 33.1% decrease in isocitric acid titer. The BZ-Y only showed a slight decrease in isocitric acid production.
Given the positive effect of individual overexpression of three genes on citric acid production, the combinational overexpression of two or three genes was also tested. Based on the highest citric acid titer achieved in strain BZ-Y, the gene AMPD and ICL were subsequently overexpressed in BZ-Y, resulting in strain BZ-AY and BZ-IY, respectively. Still, 6 transformants were randomly chosen for each overexpressed gene, and transformants with improved sugar consumption rates and citric acid titers were selected. As shown in Fig. 1B, strain BZ-IY obtained a higher citric acid titer than strain BZ-AY (70.6 g L−1 and 65.3 g L−1, respectively). The yields of citric acid for the two strains were comparable (0.58 g g−1 for BZ-IY and 0.56 g g−1 for BZ-AY). Both strain BZ-AY and strain BZ-IY consumed up to 100 g L−1 glucose within 168 h, while strain BZ-IY consumed more xylose (Fig. 1C). Then, we tried to overexpress the AMPD gene in strain BZ-IY. However, the attempt to further raise the production of citric acid failed, yielding only 65.8 g L−1 citric acid. This could be attributed to the excessive interference with the TCA cycle. Therefore, the strain BZ-IY was selected for the subsequent genetic modification.
3.2 Reducing fatty acid synthesis to improve citric acid production
As an oleaginous yeast, Y. lipolytica naturally synthesizes lipids as energy storage. However, the lipid synthesis pathway is the downstream catabolism pathway of citric acid and disperses carbon resources away from citric acid synthesis. The accumulated citric acid is cleaved into acetyl-CoA and oxaloacetic acid by ACL, which is then catalyzed to form acyl-CoA and stored as triglycerides (TAG) under the catalysis of acyl-CoA diacylglycerol acyltransferase 1 and 2 (Dga1, Dga2).28–30 Therefore, these three genes were knocked out to decrease lipid accumulation and reduce citric acid consumption (Fig. 2A).The lipid content in strain BZ-IY was 10.76% (wt/wt). We first tested the effect of single-gene knockout on lipid content and citric acid production. As shown in Fig. 2B, each single-gene disruption showed a reduced accumulation of intracellular lipids. Among these, the effect of disrupting DGA1 and ACL were significant. The lipid content of BZ-IYΔ1 decreased to 4.8% (wt/wt), and the lipid content of BZ-IYΔA reduced to 3.2% (wt/wt), both resulting in more than 60% reduction in the total lipid content. In contrast, deletion of DGA2 showed a slight decline to 8.6% (wt/wt) in lipid content. Notably, only strain BZ-IYΔ1 showed an improved citric acid yield of 0.75 g g−1, while BZ-IYΔ2 and BZ-IYΔA strains exhibited decreased titers, with yields remaining relatively unchanged. This may be because the deletion of ACL reduced the supply of acetyl-CoA, thereby affecting cell growth, which was indicated by the weakened sugar consumption rate (Fig. S2†). In addition, DGA1 may appear to be the primary TAG-synthesizing gene, thus leading to a lesser impact on lipid synthesis when only DGA2 is deleted.31,32
Considering the above results, we iteratively disrupted DGA2 and ACL1 genes in the strain BZ-IYΔ1, generating the double-mutation strains BZ-IYΔ12 and BZ-IYΔ1A, respectively. We also disrupted the DGA2 gene in strain BZ-IYΔA to generate strain BZ-IYΔ2A. All generated strains resulted in a reduced lipid content (Fig. 2B). The most significant reduction in lipid content was achieved by the strain BZ-IYΔ1A, which obtained a lipid content of 3.54% (wt/wt). However, this engineered strain showed a significantly decreased citric acid titer of 20.5 g L−1 (Fig. 2B), as well as a slow sugar consumption (Fig. S2†), suggesting that simultaneous disruption of DGA1 and ACL1 may affect cell growth. As reported, the deletion of DGA1 or ACL1 affected the formation of TAG, which may result in the shortage of fatty acid for membrane biogenesis, the disruption of cellular TAG homeostasis and the inhibitory feedback on fatty acids biosynthesis.28,31,33 Strains BZ-IYΔ12 and BZ-IYΔ2A also exhibited reduced lipid contents of 3.98% (wt/wt) and 4.55% (wt/wt), respectively, along with lower sugar consumption compared to the control strain BZ-IY (Fig. S4†). This led to a final citric acid titer of 70.5 g L−1 in BZ-IYΔ12, similar to the 70.6 g L−1 observed in strain BZ-IY. However, the citric acid yield for BZ-IYΔ12 increased to 0.83 g g−1, representing a 37.9% enhancement over the 0.58 g g−1 yield of strain BZ-IY (Fig. 2B). These results proved that iteratively disrupting DGA1 and DGA2 to downregulate lipid synthesis is effective in mitigating citric acid consumption, thus increasing the yield from the substrate to citric acid.
3.3 Abolishing the isocitric acid transporter to reduce by-products
As the intermediates of the TCA cycle, the synthesis of citric acid is often accompanied by the formation of isocitric acid. The inhibited activity of isocitrate dehydrogenase (IDH) under nitrogen depletion conditions disrupted the conversion of isocitric acid in the cycle. Both citric acid and isocitric acid accumulate in the mitochondria and diffuse into the cytoplasm and culture medium, which improves the difficulty of downstream separation and purification.34 Since the inner membrane of mitochondria is impermeable to citric acid and isocitric acid, a transporter is required for isocitric acid secretion. A native isocitric acid mitochondrial carrier YLSfc1 in Y. lipolytica was demonstrated to be the main factor in isocitric acid secretion.35 Inactivation of ylSFC1 gene may lead to a decreased isocitric acid production and isocitric acid/citric acid ratio.To reduce the secretion of the by-product isocitric acid, the ylSFC1 gene was deleted in the strain BZ-IYΔ12, resulting in the strain BZ-IYΔ12S (Fig. 3A). As shown in Fig. 3B, the titer of by-product isocitric acid was decreased from 5.5 g L−1 in strain BZ-IYΔ12 to 2.6 g L−1 in strain BZ-IYΔ12S as expected, indicating the validity of knocking out ylSFC1 gene as reported.35 However, the citric acid production decreased from 70.4 g L−1 in strain BZ-IYΔ12 to 64.8 g L−1 in strain BZ-IYΔ12S. The citric acid yield of BZ-IYΔ12S (0.81 g g−1) remained similar to that of BZ-IYΔ12 (0.83 g g−1). The reduced titer of citric acid contributed to lower sugar consumption compared to the parent strain BZ-IYΔ12. According to Fig. 3C and D, slightly lower glucose consumption was observed in strain BZ-IYΔ12S (80 g L−1) than that of strain BZ-IYΔ12 (85 g L−1) within 168 h. The highest value of OD600 reached 33.1 and 28.1 in strains BZ-IYΔ12 and BZ-IYΔ12S, respectively. These results indicated the deletion of ylSFC1 had a negative effect on cell growth.35 Xylose concentration remained unchanged due to the carbon catabolite repression. Despite the deletion of the isocitric acid transporter, which had no contribution to the increment of citric acid production, the reduced concentration of the by-product was beneficial for the separation and purification process of citric acid from the fermentation broth.
3.4 Boosting citric acid productivity by enhancing substrate utilization efficiency
The above-mentioned metabolic engineering strategies successfully improved the sugar conversion rate from 0.34 g g−1 in the original strain to 0.83 g g−1 in the BZ-IYΔ12S strain, whereas it comes at the cost of decreased glucose utilization rate. This is unfavorable in the industrial application with hydrolysate as a carbon source because Y. lipolytica preferentially uptakes glucose over xylose in mixed sugar.36 A low glucose utilization rate impedes xylose fermentation, resulting in higher costs due to the extended fermentation period. To promote glucose consumption, six native hexose transporter genes, Yht1-6, and a native hexose kinase gene, Hxt1, were overexpressed in strain BZ-Δ12S, respectively, resulting in strain BZ-Y1-6Δ12S and strain BZ-HΔ12S (Fig. 4A).The sugar utilization rate was recovered by strain overexpressing Yht1, Yht3, Yht5 and Yht6. The Yht1, Yht5 and Yht6 overexpressing strains exhibited higher sugar consumption rates but with lower citric acid titer than the original strain BZ-Δ12S (Fig. S3†). As stated, this reduction may be due to the redirection of carbon flux towards the pentose phosphate pathway, resulting in mannitol and erythritol secretion.37,38 Only overexpressing Yht3 resulted in an enhanced sugar uptake rate and higher citric acid titer. Consequently, gene Yht3 was overexpressed in strain BZ-IYΔ12S, obtaining strain BZ-IY3Δ12S. As shown in Fig. 4B, glucose was almost consumed (up to 100 g L−1) within 120 h and only 12 g L−1 xylose was left after 168 h fermentation of strain BZ-IY3Δ12S. A high citric acid titer of 88.2 g L−1 was obtained, while the citric acid yield was reduced to 0.68 g g−1.
6-Phosphofructo-1-kinase (PFK1) is one of the key regulatory enzymes in eukaryotic microorganisms, catalyzing the first irreversible reaction of glycolysis by phosphorylating fructose-6-phosphate. PFK1 is tightly regulated within the glycolytic pathway and has six allosteric ligands. Citrate is one of the allosteric inhibitors. An intracellular overflow of citrate can lead to the inhibition of PFK1 activity and cause deregulated glycolysis. By constructing a modified pfkA gene in A. niger, a highly active and citrate-resistant PFK1 enzyme was obtained, increasing the citric acid production by up to 70% compared to the parental strain.39 To further accelerate the glucose consumption rate and improve citric acid production, we overexpressed the codon-optimized modified pfkA gene in Y. lipolytica, obtaining the strain BZ-IY3FΔ12S (Fig. 4C). However, the sugar consumption rate and titer of citric acid was not further improved (Fig. 4D). This may suggest that PFK1 is not the rate-limiting enzyme in citric acid synthesis in Y. lipolytica. More research studies, including metabolome and proteome analysis, are needed to clarify the complex metabolic pathways. Therefore, we choose the BZ-IY3Δ12S strain for the subsequent experiment.
3.5 Citric acid fermentation from corn stover hydrolysate in the flask and 3 L bioreactor
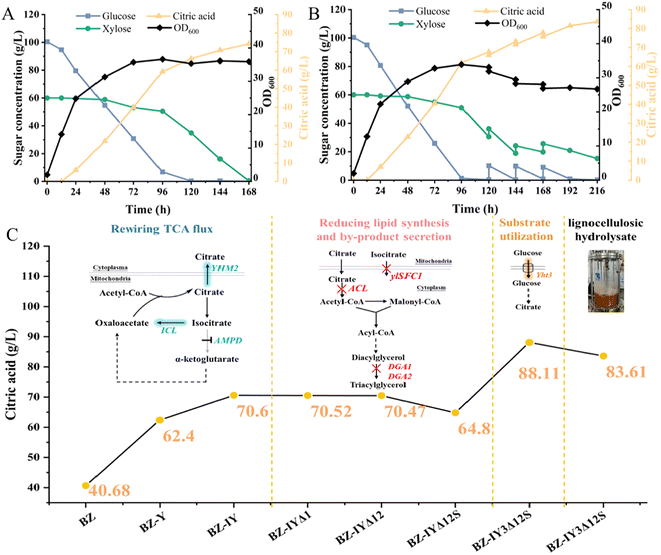
Given the optimized citric acid production of the engineered strain BZ-IY3Δ12S in pure sugar medium, we then tested its performance in the corn stove hydrolysate in the shake flask. Lignocellulose hydrolysate is the most abundant renewable resource worldwide, and it has been utilized as a low-value feedstock for the production of various biofuels and chemicals. Numerous studies have been conducted to produce citric acid from hydrolysates; however, these often necessitate detoxification due to the toxic compounds generated during the pretreatment process. Additionally, the titers achieved are relatively low because of the diminished sugar concentration at low solid loading, which ultimately hinders the process from reaching a commercial scale.21,40,41 Recently, a promising pretreatment method known as “densifying lignocellulosic biomass with chemicals (DLC)” has been developed, leading to low inhibition and high enzymatic hydrolysis, and has been successfully applied for high production of ethanol, lipids and lactic acid.42–44 It was hence selected as feedstock for the investigation of citric acid production by the engineered strain BZ-IY3Δ12S.The engineered strain BZ-IY3Δ12S was tested for fermenting corn stover hydrolysate at high solid loadings of 25%, 30%, and 35%. The BZ-IY3Δ12S strain survived well in 25% and 30% solid loading corn stover hydrolysate and efficiently converted sugars into citric acid. In the hydrolysate with 25% solid loading, 68.1 g L−1 citric acid was produced with a yield of 0.50 g g−1 (Fig. 5A). In contrast, a higher titer of 73.6 g L−1 citric acid was achieved in 30% solid loading with a citric acid yield of 0.49 g g−1, suggesting the robustness of the engineered strain and the high fermentability of DLCA(sa)-pretreated corn stover hydrolysate (Fig. 5B). However, when the solid loading increased to 35%, the strain could not fully consume glucose in 144 h and only 48.6 g L−1 citric acid was achieved (Fig. 5C). This may be attributed to the inhibitory effects from the high initial sugar concentration and high inhibitor contents (Table S2†). Therefore, DLCA(sa)-pretreated corn stover hydrolysate with 30% solid loading was selected for the following experiments.
To further improve citric acid production by the strain BZ-IY3Δ12S using undetoxified hydrolysates, batch and fed-batch fermentation using hydrolysate at 30% solid loading was implemented in the 3-L bioreactor. 100.4 g L−1 glucose and 60.8 g L−1 xylose were detected in the DLCA(sa)-pretreated corn stover hydrolysate at 30% solid loading. In the batch fermentation (Fig. 6A), with a controlled temperature of 30 °C and pH at 6, strain BZ-IY3Δ12S consumed all the glucose within 120 h and xylose within 168 h. The highest titer of citric acid reached 74.2 g L−1 at 168 h with a yield of 0.46 g g−1 of fermentable sugar. A fed-batch fermentation was subsequently conducted to further enhance the production of citric acid. The feeding started at 120 h and was performed three times with a final solid loading of 35% (Fig. 6B). Along with the progressed feeding strategy, the increased inhibitor content led to a decrease in xylose utilization, which was consistent with previous studies.45 Thus, the fermentation was terminated at 216 h. Finally, 129.6 g L−1 glucose and 61.5 g L−1 xylose were consumed, resulting in a higher citric acid titer of 83.6 g L−1. The yield and productivity of citric acid were 0.44 g g−1 and 0.39 g L−1 h−1, respectively. There have been barely any studies reporting high citric acid titers of more than 80 g L−1 from Lignocellulosic biomass (as detailed in Table 2). To our knowledge, this is the highest citric acid production achieved from the non-detoxified lignocellulosic hydrolysate by Y. lipolytica. Additionally, it is noteworthy that no supplementary nitrogen source was added to the hydrolysate during the fermentation process. This absence of added nitrogen not only simplifies the production protocol but also contributes to overall cost savings, making the process more economically viable for large-scale applications.
| Strains | Carbon source | Fermentation strategy | Substrate consumption | Citric acid (g L−1) | Yield (g g−1) | Productivity (g L−1 h−1) | Ref. |
|---|---|---|---|---|---|---|---|
| a N. A = data not available. b Olive mill wastewater. c The conversion yield was based on the reducing sugars (OMWs contained some quantities of reducing sugars) consumed by the strains. d Estimates based on graphical data from a related paper. | |||||||
| Y. lipolytica SWJ-1b | Straw | Batch | 33.8 g L−1 glucose | 26.7 | 0.79 | 0.23 | 21 |
| Fed-batch | N. Aa | 42.4 | 0.43 | 0.18 | |||
| Y. lipolytica LGAM S (7) | OMWb | Batch in shake flask, OMW and glycerol blend | 50 g L−1 glycerol and reducing sugars in OMW | 30.3c | 0.62 | 0.11d | 46 |
| Y. lipolytica ACA-DC 50109 | OMWb | Batch in shake flask, mixture of OMW with synthetic medium containing commercial glucose | 65 g L−1 glucose | 28.9c | 0.53 | 0.08d | 47 |
| Y. lipolytica W29 | OMWb | Shake flask, OMW and glucose blend | 34 g L−1 glucose | 15.8c | 0.46 | 0.11 | 48 |
| Y. lipolytica ACA-YC 5033 | OMWb | Shake flask, OMW and glucose blend | N.Aa | 52c | 0.64 | 0.12 | 49 |
| Y. lipolytica CGMCC 2.1506 | Corn stover | Batch in 5-L bioreactor, hydrolysate with 0.25% TritonX-100 addition | 50 g L−1 glucose, 46.3 g L−1 glycerol and 10–12b g L−1 xylose | 63.8 | 0.60d | 0.33 | 22 |
| Y. lipolytica BZ-IY3Δ12S | Corn stover | Batch in 3-L bioreactor, no additional nitrogen source | 100.4 g L−1 glucose and 60.8 g L−1 xylose | 74.2 | 0.46 | 0.44 | This study |
| Y. lipolytica BZ-IY3Δ12S | Corn stover | Fed-batch in 3-L bioreactor, no additional nitrogen source | 129.6 g L−1 glucose and 61.5 g L−1 xylose | 83.6 | 0.44 | 0.39 | This study |
Overall, the engineered Y. lipolytica strain BZ-IY3Δ12S achieved a high citric acid titer of 83.6 g L−1 in lignocellulosic hydrolysate without further detoxification. However, the citric acid titer still fell behind the value in the synthetic medium (Fig. 6C). This may be attributed to the inhibition effects from high inhibitor contents. Further studies should focus on improving the robustness of the engineered strain.
4. Conclusion
In the present study, we constructed a Y. lipolytica platform for sustainable production of citric acid by overexpressing TCA related genes to enhance citric acid production and reduce isocitric acid formation, deleting DGA1, DGA2, and ACL genes to reduce the consumption of citric acid in the downstream lipid synthesis pathway, knocking out the isocitric acid transporter gene to further reduce by-products in the culture medium to improve product purity, and overexpressing glucose transportation and glycolysis pathway genes to improve the substrate utilization rate. Finally, 83.6 g L−1 citric acid was achieved from DLCA(sa)-pretreated corn stover in a 3-L bioreactor by fed-batch fermentation. This work showed the potential of using lignocellulose hydrolysate as a low-cost carbon source to enable sustainable and efficient citric acid production with a yeast platform.Author contributions
Minrui Lu: conceptualization, methodology, investigation, formal analysis, roles/writing – original draft; Yuanyuan Sha: conceptualization, methodology, investigation, formal analysis, data curation, formal analysis, roles/writing – original draft, validation; Yuwei Zhang: investigation, writing – review & editing; Mianshen Ge: formal analysis; Zhaoxian Xu: formal analysis; Mingjie Jin: conceptualization, data curation, project administration, supervision, resources, writing – review & editing, validation, funding acquisition.Data availability
The data that support the findings of this study are available from the corresponding author, Mingjie Jin, upon reasonable request.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This work was supported by the “Natural Science Foundation of Jiangsu Province” (No. BK20241485).References
- Z. Li, X. Wen and H. Liu, Green Chem., 2022, 24, 1650–1658 RSC.
- J. Li, L. Rong, Y. Zhao, S. Li, C. Zhang, D. Xiao, J. L. Foo and A. Yu, Biotechnol. Adv., 2020, 43, 107605 CrossRef PubMed.
- E. Książek, Molecules, 2023, 29, 22 CrossRef.
- S. Mores, L. P. S. Vandenberghe, A. I. M. Junior, J. C. de Carvalho, A. F. M. de Mello, A. Pandey and C. R. Soccol, Bioresour. Technol., 2021, 320, 124426 CrossRef.
- M. Laltha, Y. Sewsynker-Sukai and E. B. G. Kana, Bioresour. Technol., 2022, 361, 127700 CrossRef PubMed.
- S. Zhou, N. Ding, R. Han and Y. Deng, Bioresour. Technol., 2023, 379, 128986 CrossRef.
- B. Igliński, U. Kiełkowska and G. Piechota, Clean Technol. Environ. Policy, 2022, 24, 2061–2079 CrossRef.
- S. V. Kamzolova, Processes, 2023, 11, 3435 CrossRef.
- Y. K. Park and R. Ledesma-Amaro, Trends Biotechnol., 2023, 41, 242–254 CrossRef.
- N. Poorinmohammad and E. J. Kerkhoven, Biotechnol. Bioeng., 2021, 118, 3640–3654 CrossRef.
- S. Zhang, F. Guo, Q. Yang, Y. Jiang, S. Yang, J. Ma, F. Xin, T. Hasunuma, A. Kondo, W. Zhang and M. Jiang, Green Chem., 2023, 25, 183–195 RSC.
- G. Y. Fu, Y. Lu, Z. Chi, G. L. Liu, S. F. Zhao, H. Jiang and Z. M. Chi, Mar. Biotechnol., 2016, 18, 1–14 CrossRef PubMed.
- E. Y. Yuzbasheva, G. Agrimi, T. V. Yuzbashev, P. Scarcia, E. B. Vinogradova, L. Palmieri, A. V. Shutov, I. M. Kosikhina, F. Palmieri and S. P. Sineoky, Metab. Eng., 2019, 54, 264–274 CrossRef.
- S. Urak, O. Yeniay and S. Karasu-Yalcin, Ann. Microbiol., 2014, 65, 639–649 CrossRef.
- D. A. Rzechonek, A. Dobrowolski, W. Rymowicz and A. M. Mironczuk, Bioresour. Technol., 2019, 271, 340–344 CrossRef CAS PubMed.
- X. Y. Liu, Z. Chi, G. L. Liu, C. Madzak and Z. M. Chi, Mar. Biotechnol., 2013, 15, 26–36 CrossRef PubMed.
- T. Sun, Y. Yu, L. Wang, Y. Qi, T. Xu, Z. Wang, L. Lin, R. Ledesma-Amaro and X. J. Ji, ACS Synth. Biol., 2023, 12, 761–767 CrossRef.
- F. Yao, S. C. Liu, D. N. Wang, Z. J. Liu, Q. Hua and L. J. Wei, FEMS Yeast Res., 2020, 20, foaa046 CrossRef PubMed.
- H. Bi, C. Xv, C. Su, P. Feng, C. Zhang, M. Wang, Y. Fang and T. Tan, Fermentation, 2022, 8, 532 CrossRef.
- L. J. Wei, Y. T. Zhong, M. Y. Nie, S. C. Liu and Q. Hua, J. Agric. Food Chem., 2021, 69, 275–285 CrossRef PubMed.
- X. Liu, J. Lv, T. Zhang and Y. Deng, Prep. Biochem. Biotechnol., 2015, 45, 825–835 CrossRef.
- Y. Gao, F. Wang, X. Li, G. Mao, H. Xie, A. Song, J. C. D. Santos and Z. Zhang, Ind. Crops Prod., 2022, 189, 115820 CrossRef.
- X. Yuan, X. Chen, G. Shen, S. Chen, J. Yu, R. Zhai, Z. Xu and M. Jin, Renewable Energy, 2022, 182, 377–389 CrossRef.
- Z. D. Wang, L. L. Zhou, M. R. Lu, Y. W. Zhang, S. Perveen, H. R. Zhou, Z. Q. Wen, Z. X. Xu and M. J. Jin, Appl. Microbiol. Biotechnol., 2021, 105, 1745–1758 CrossRef.
- Q. Guo, T. Q. Shi, Q. Q. Peng, X. M. Sun, X. J. Ji and H. Huang, J. Agric. Food Chem., 2021, 69, 13831–13837 CrossRef PubMed.
- L. Zhang, M. Y. Nie, F. Liu, J. Chen, L. J. Wei and Q. Hua, Biotechnol. Lett., 2021, 43, 1277–1287 CrossRef PubMed.
- A. Forster, K. Jacobs, T. Juretzek, S. Mauersberger and G. Barth, Appl. Microbiol. Biotechnol., 2007, 77, 861–869 CrossRef PubMed.
- T. Dulermo, Z. Lazar, R. Dulermo, M. Rakicka, R. Haddouche and J. M. Nicaud, Biochim. Biophys. Acta, 2015, 1851, 1107–1117 CrossRef.
- K. Athenstaedt, Biochim. Biophys. Acta, 2011, 1811, 587–596 CrossRef.
- A. Beopoulos, R. Haddouche, P. Kabran, T. Dulermo, T. Chardot and J. M. Nicaud, Appl. Microbiol. Biotechnol., 2012, 93, 1523–1537 CrossRef PubMed.
- H. Zhang, H. G. Damude and N. S. Yadav, Yeast, 2012, 29, 25–38 CrossRef PubMed.
- J. Blazeck, A. Hill, L. Liu, R. Knight, J. Miller, A. Pan, P. Otoupal and H. S. Alper, Nat. Commun., 2014, 5, 3131 CrossRef PubMed.
- G. Zhang, C. Zhang, Z. Wang, Q. Wang, J. Nielsen and Z. Dai, Metab. Eng., 2022, 73, 225–234 CrossRef PubMed.
- S. V. Kamzolova and I. G. Morgunov, Appl. Microbiol. Biotechnol., 2019, 103, 9321–9333 CrossRef PubMed.
- E. Y. Yuzbasheva, P. Scarcia, T. V. Yuzbashev, E. Messina, I. M. Kosikhina, L. Palmieri, A. V. Shutov, M. O. Taratynova, R. L. Amaro, F. Palmieri, S. P. Sineoky and G. Agrimi, Metab. Eng., 2021, 65, 156–166 CrossRef PubMed.
- G. M. Rodriguez, M. S. Hussain, L. Gambill, D. F. Gao, A. Yaguchi and M. Blenner, Biotechnol. Biofuels, 2016, 9, 1–15 CrossRef PubMed.
- S. Xu, X. Zhang, Y. Zhang, Q. Li, L. Ji and H. Cheng, J. Agric. Food Chem., 2023, 71, 11567–11578 CrossRef PubMed.
- P. Hapeta, P. Szczepanska, T. Witkowski, J. M. Nicaud, A. M. Crutz-Le Coq and Z. Lazar, Int. J. Mol. Sci., 2021, 22, 9282 CrossRef PubMed.
- M. Capuder, T. Solar, M. Bencina and M. Legisa, J. Biotechnol., 2009, 144, 51–57 CrossRef.
- P. P. Zhou, J. Meng and J. Bao, Bioresour. Technol., 2017, 224, 563–572 CrossRef PubMed.
- W. Hou and J. Bao, Bioresour. Technol., 2018, 253, 72–78 CrossRef.
- Y. Zhang, Z. Xu, M. Lu, X. Ma, S. Chen, Y. Wang, W. Shen, P. Li and M. Jin, Bioresour. Technol., 2023, 388, 129729 CrossRef.
- X. Chen, X. Yuan, S. Chen, J. Yu, R. Zhai, Z. Xu and M. Jin, Green Chem., 2021, 23, 4828–4839 RSC.
- G. Shen, X. Yuan, Y. Cheng, S. Chen, Z. Xu and M. Jin, Green Chem., 2023, 25, 8026–8039 RSC.
- S. Chen, Z. Xu, B. Ding, Y. Zhang, S. Liu, C. Cai, M. Li, B. E. Dale and M. Jin, Sci. Adv., 2023, 9, eadd8835 CrossRef PubMed.
- M. Dourou, A. Kancelista, P. Juszczyk, D. Sarris, S. Bellou, I.-E. Triantaphyllidou, A. Rywinska, S. Papanikolaou and G. Aggelis, J. Cleaner Prod., 2016, 139, 957–969 CrossRef.
- S. Papanikolaou, M. Galiotou-Panayotou, S. Fakas, M. Komaitis and G. Aggelis, Bioresour. Technol., 2008, 99, 2419–2428 CrossRef.
- D. Sarris, M. Galiotou-Panayotou, A. A. Koutinas, M. Komaitis and S. Papanikolaou, J. Chem. Technol. Biotechnol., 2011, 86, 1439–1448 CrossRef.
- D. Sarris, N. G. Stoforos, A. Mallouchos, I. K. Kookos, A. A. Koutinas, G. Aggelis and S. Papanikolaou, Eng. Life Sci., 2017, 17, 695–709 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4gc05521g |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |