Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Lacticaseibacillus casei HY2782 improves the intestinal barrier and tract environment and ultimately prolongs the lifespan of Caenorhabditis elegans†
Won-Young
Bae‡
a,
Uyen Tran Tu
Nguyen‡
ab,
Tram Anh Ngoc
Le
a,
Son Hung
Tran
ab,
Sohyun
Lee
ab,
Sung-Chul
Hong
ac,
Kwang Hyun
Cha
ab,
Il-Dong
Choi
d and
Kyungsu
Kang
 *ab
*ab
aCenter for Natural Product Systems Biology, Gangneung Institute of Natural Products, Korea Institute of Science and Technology (KIST), Gangneung, Gangwon-do 25451, South Korea. E-mail: kskang@kist.re.kr
bNatural Product Applied Science, KIST School, University of Science and Technology (UST), Gangneung, Gangwon-do 25451, South Korea
cDepartment of Food Science and Biotechnology, Kunsan National University, Gunsan, Jeollabuk-do 54150, South Korea
dR&BD Department, hy Co., Ltd, Yongin, Gyeonggi-do 17086, South Korea
First published on 19th June 2025
Abstract
Caenorhabditis elegans is widely used as a model for investigating longevity owing to its short life cycle and the presence of human orthologs. This study aimed to ultimately prolong the lifespan of C. elegans by evaluating the gastrointestinal tract conditions and intestinal permeability of C. elegans fed Lacticaseibacillus casei HY2782 alone or fermented fecal products. The anti-inflammatory effects of L. casei HY2782 were determined based on interleukin 8 (IL-8) levels and intestinal permeability in human intestinal epithelial cells. In the C. elegans model, intestinal permeability was assessed in the N2 wild-type as well as skn-1, pmk-1, daf-16, and aak-2 mutant worms. During simulated colonic fecal fermentation, changes in short chain fatty acid and microbial composition were investigated. L. casei HY2782 reduced IL-8 production and intestinal permeability from 1646.8 to 1009.1 pg mL−1 and 265.5 to 115.1%, respectively (p < 0.01). Additionally, L. casei HY2872 attenuated intestinal leakage in C. elegans and prolonged its lifespan via the DAF-16/FOXO and SKN-1/NRF2 pathways and gst-4 gene expression. Moreover, L. casei HY2782 inhibited intestinal leakage in aged worms. During fermentation, L. casei HY2782 produced butyrate under both normal and high-protein conditions. Additionally, L. casei HY2782 contributed to butyrate production by genera, such as Faecalibacterium and Lachnospira (p < 0.01), while inhibiting Fusobacterium (p < 0.05). L. casei HY2782 also prolonged lifespan of intestinally damaged worms (p < 0.001). Furthermore, C. elegans fed L. casei HY2782-fermented fecal products lived significantly longer than those fed the vehicle control (p < 0.001). Overall, L. casei HY2782 restored the intestinal tract by mitigating inflammation and microbial metabolic dysbiosis, ultimately extending the lifespan of C. elegans.
1. Introduction
The popularity of high-protein diets has grown steadily owing to their effectiveness in promoting rapid weight loss.1 Not only individuals with obesity, but also weight-class athletes, such as physique and bodybuilding athletes, consume high-protein diets to reduce their body-fat ratio.2 Combined with low-carbohydrate diets, high-protein diets improve glycated hemoglobin levels in patients with type 2 diabetes.3 However, chronic high-protein diets result in intestinal permeability, systemic inflammation, and gut microbiome loss, including Bacteroides and Akkermansia.1Dr Élie Metchnikoff proposed Metchnikoff's hypothesis, which postulated that probiotics contributed to the longevity of Bulgarian peasants. Metchnikoff's hypothesis was based on yogurt containing lactic acid bacteria (LAB) and contributed to shaping the role of probiotics.4 Probiotics are live microorganisms that improve host health when consumed in adequate amounts.5,6 The beneficial effects of probiotics, including regulation of the microbiome and immune system, result from a healthy gut.4,5 However, aging disrupts the immune and digestive systems and the gut microbiome balance, leading to intestinal inflammation.5Caenorhabditis elegans (C. elegans) is a small nematode that has a short life cycle.4,5,7–13 An aged C. elegans shares similarities with the aged intestine in mammals, including poor permeability, small and fewer microvilli, and decreased digestive ability.9 Therefore, C. elegans models have been used to elucidate the correlation between aging and the consumption of functional foods and probiotics.5,6,9,14
Lacticaseibacillus casei (L. casei) is a facultative heterofermentative LAB, typically present in fermented dairy products, vegetables, and the gastrointestinal (GI) tracts of humans and other animals.15 Moreover, L. casei DN-114001, which is used to ferment yogurt, alleviates atopic dermatitis in children.16 Similarly, L. casei HY2782, a probiotic strain isolated from human feces, alleviates depression17 and attenuates cellular and physiological toxicity induced by particulate matter.18 However, the health effects of L. casei HY2782, particularly related to the regulation of aging and longevity, are not fully understood. Moreover, many studies have focused on short-chain fatty acid (SCFA) production by probiotics; however, a comparison of SCFA production under a normal and high-protein diet under simulated colonic conditions has not yet been reported.
Therefore, this study aimed to evaluate the gut conditions and lifespan of C. elegans fed L. casei HY2782 bacteria alone or L. casei HY2782-fermented fecal fermentation products. Moreover, the study compared SCFA production under a normal and high-protein diet under simulated colonic conditions. Ultimately, Metchnikoff's hypothesis was evaluated, thereby revealing the correlation between gut environment and longevity.
2. Materials and methods
2.1 Reagents, microorganisms, and cell culture
Recombinant human interleukin (IL)-1β was purchased from PeproTech (Rocky Hill, NJ, USA). Fluorescein isothiocyanate–dextran (FITC-dextran, average molecular weight = 4 kDa), 3,3′-diindolylmethane (DIM), 2-methylpentanoic acid, and volatile-free acid mix were obtained from Sigma-Aldrich (St. Louis, MO, USA). Brilliant Blue FCF dye was purchased from Tokyo Chemical Industry (Tokyo, Japan).The L. casei HY2782 was provided by hy Co. Ltd (Yongin, Korea) and was cultured in de Man-Rogosa-Sharpe medium (BD Biosciences, Franklin Lakes, NJ, USA) at 37 °C for 24 h. Pseudomonas aeruginosa ATCC 15692 (PAO1) (P. aeruginosa PAO1) was obtained from the American Type Culture Collection (Manassas, VA, USA), and Escherichia coli (E. coli) OP50 was obtained from Caenorhabditis Genetics Center (Minneapolis, MN, USA). P. aeruginosa PAO1 and E. coli OP50 were cultured in Luria Bertani broth at 37 °C for 15 h at an agitation speed of 150 rpm.
Human intestinal epithelial cells (HT-29 and Caco-2) were purchased from the Korean Cell Line Bank (Seoul, Korea). The cells were maintained in Dulbecco's modified Eagle's medium (Cytiva, Marlborough, MA, USA) supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin solution at 37 °C in a humidified atmosphere containing 5% CO2. During incubation, the medium was replaced every 2–3 d, and the cells were grown until they reached 80% confluence. For differentiation, Caco-2 cells (4 × 105 cells) were seeded in a 60 mm culture dish, and the medium that contained 1% (v/v) non-essential amino acid solution was changed every two days as described previously.19,20
2.2 Evaluation of cell viability and IL-8 production
HT-29 cells were seeded in 96-well microplates (1 × 104 cells per well) and incubated for 24 h. After incubation, the HT-29 cells were treated with DIM (10 μM) or 5 × 107 colony-forming unit (CFU) mL−1 of L. casei HY2782 for 24 h. An inflammatory reaction was induced by adding IL-1β (10 ng mL−1) to DIM- or L. casei HY2782-treated cells for 5 h. DIM was used as a positive control because it exerts anti-inflammatory activity in intestinal epithelial cells.19 Following reaction completion, the supernatants were harvested and centrifuged at 2000g for 20 min at 4 °C. IL-8 production in the supernatant was analyzed using ELISA MAX™ Deluxe Set Human IL-8 (BioLegend, San Diego, CA, USA) according to the manufacturer's guidelines. The viability of DIM- or L. casei-treated cells was measured using an EZ-Cytox kit (DoGenBio, Seoul, Korea). The absorbance of water-soluble tetrazolium salts was measured at 450 nm using a microplate reader (Synergy HTX Multi-Mode Reader; BioTek, Winooski, VT, USA).19,202.3 Assessment of cellular permeability in human intestinal epithelial cells
Intestinal permeability was investigated by measuring the transepithelial electrical resistance (TEER) and FITC-dextran efflux in Caco-2 cells.18–20 Briefly, 3 × 105 Caco-2 cells per well were seeded on the apical side of a 12-well Transwell® microplate. The cells were cultured for 12–15 days to allow differentiation into a polarized monolayer. Following polarization, the cells were treated with IL-1β (50 ng mL−1) and DIM (10 μM) or 5 × 107 CFU mL−1 of L. casei HY2782 for 72 h. DIM was used as a positive control as it recovered the intestinal barrier function in cultured Caco-2 cell monolayers.19 The change in TEER values was measured using a Millicell® ERS-2 Electrical Resistance System (MilliporeSigma, Burlington, MA, USA). After TEER measurements, the medium on the apical side was removed and replaced with fresh medium containing 1 mg mL−1 FITC-dextran. FITC-dextran efflux was then performed for 2 h, and the fluorescence intensity of the basolateral side was evaluated using a microplate reader (Infinite® M1000; Tecan, Männedorf, Switzerland) at 485 nm excitation and 535 nm emission wavelengths.2.4 C. elegans maintenance
The C. elegans wild-type N2 Bristol variety was used with the following mutant strains: EU31, skn-1(zu135) IV/nT1 [unc-?(n754) let-?] (IV;V); KU25, pmk-1(km25) IV; CF1038, daf-16(mu86) I; RB754, aak-2(ok524) X; and CL2166, dvIs19 [(pAF15)gst-4p::GFP::NLS] III. All strains were maintained on nematode growth media (NGM) at 20 °C, seeded with E. coli OP50 as a food source. Synchronized L4 worms were examined for intestinal permeability and lifespan.10,19,202.5 Intestinal permeability in L. casei HY2782-treated C. elegans
The P. aeruginosa PAO1 cell-free supernatant (CFS) was prepared using a 0.45 μm membrane filter. Larvae at the synchronized L4 stage were transferred onto NGM agar and incubated with 795 μL of E. coli OP50 or L. casei HY2782 (∼1 × 108 CFU mL−1) for 48 h. During incubation, C. elegans was treated with 5 μL of CFS to induce intestinal permeability dysfunction. After incubation, all worms were washed with S-buffer and further incubated with FITC-dextran (20 μg mL−1) and E. coli OP50 for 15 h. The FITC-dextran-treated worms were washed with S-buffer and fixed with 4% formaldehyde. All fixed worms were mounted using mounting media in a 96-well microplate and observed using a high content analysis system (Operetta® CLS™; PerkinElmer, Waltham, MA, USA) with an enhanced green fluorescent protein (EGFP) filter.10,19,202.6 Lifespan measurement of intestinal-damaged C. elegans
Synchronized worms (L4 stage) were transferred onto NGM agar (60 mm) coated with 20 μL of P. aeruginosa CFS mixed in 380 μL of E. coli OP50 or L. casei HY2782 culture.21 After incubation, the plates were observed under a stereomicroscope (SMZ800N; Nikon, Tokyo, Japan). Worms that did not respond to gentle poking with a platinum wire were considered dead.7,9,192.7 Detection of Smurf phenotype in aged worms
The physiological condition of aged worms was observed using the Smurf assay with slight modifications.22 Briefly, age-synchronized L4-stage worms were transferred onto NGM agar (60 mm) supplemented with E. coli OP50 or L. casei HY2782. On day 10 of adulthood, worms were collected and washed with S-buffer. The washed worms were incubated in Brilliant Blue FCF dye 5% (w/v) for 3 h at 20 °C except for the vehicle control (non-dyed worms). Worms on day 4 of adulthood that were fed E. coli OP50 were used as non-Smurf worms. Representative images were taken with a stereomicroscope (SMZ800N; Nikon). Worms exhibiting blue dye in the germline only were not classified as Smurf worms unless the dye also leaked into the body cavity.2.8 Glutathione S-transferase 4 gene (gst-4) expression in transgenic worms
The CL2166 worm strain, which contains the gst-4p::GFP reporter construct, was synchronized at the L4 stage. After synchronization, the worms were transferred onto NGM agar coated with E. coli OP50 alone, L. casei HY2782 alone, E. coli OP50 mixed with P. aeruginosa CFS, or L. casei HY2782 mixed with P. aeruginosa CFS. The GFP expression was evaluated using a High Content Analysis System with an EGFP filter at 24, 48, and 72 h.112.9 Simulated colonic fecal fermentation with L. casei HY2782
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min. The supernatants were further extracted to obtain fatty acids by adding 500 μL of anhydrous ethyl ether and shaking for 30 s. Following centrifugation at 14
000g for 10 min. The supernatants were further extracted to obtain fatty acids by adding 500 μL of anhydrous ethyl ether and shaking for 30 s. Following centrifugation at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min, the fatty acids were collected in the upper ether layer, and 2-methylpentanoic acid (1%) was used as an internal standard.
000g for 10 min, the fatty acids were collected in the upper ether layer, and 2-methylpentanoic acid (1%) was used as an internal standard.
The GC-FID system comprised a Bruker Varian 450-GC instrument (Bruker Corporation, Billerica, MA, USA). A Nukol™ Capillary GC Column (30 m × 0.25 mm, 0.25 μm film thickness; Supelco, Burlington, MA, USA), which is an acid-modified polyethylene glycol phase column, was employed. The injection port and flame ionization detector temperature were maintained at 225 °C. The sample injection volume was 2 μL in splitless mode. Nitrogen was applied as the carrier gas, and oven temperature was maintained at 170 °C. A 10 mM volatile-free acid mix was used as a certified reference material for quantification.
2.10 Statistical analysis
Statistical analyses were performed using JMP version 13 (JMP Statistical Discovery, Cary, NC, USA). Mean values were analyzed using student's t-test and one-way ANOVA followed by Tukey's range, Duncan's multiple range, and Games-Howell at p < 0.05. To analyze the lifespan of C. elegans, the two groups were compared using the log-rank test, with significance set at p < 0.05. To analyze Smurf phenotype in aged worms, the two groups were compared using a binomial test at p < 0.05.3. Results
3.1 Anti-inflammatory effects of L. casei HY2782 in human intestinal epithelial cells
The IL-8 production and intestinal permeability of human intestinal cells treated with L. casei HY2782 are shown in Fig. 1. L. casei HY2782 did not exhibit cytotoxicity in HT-29 cells (Fig. 1A). Moreover, L. casei HY2782 significantly decreased IL-8 levels from 1646.8 to 1009.1 pg mL−1 (p < 0.001) in HT-29 cells under IL-1β-induced inflammation (Fig. 1B). Furthermore, IL-1β significantly decreased the TEER value from 478 to 329 Ω cm2 (p < 0.01), whereas that in L. casei HY2782-treated cells slightly decreased from 478 to 439 Ω cm2 (Fig. 1C). In addition, L. casei HY2782 significantly decreased FITC-dextran permeability from 265.5 to 115.1% (p < 0.01, Fig. 1D). Taken together, L. casei HY2782 exerted anti-inflammatory effects and improved intestinal barrier function in the cultured human intestinal cells.3.2 Inhibition of intestinal permeability in C. elegans by L. casei HY2782
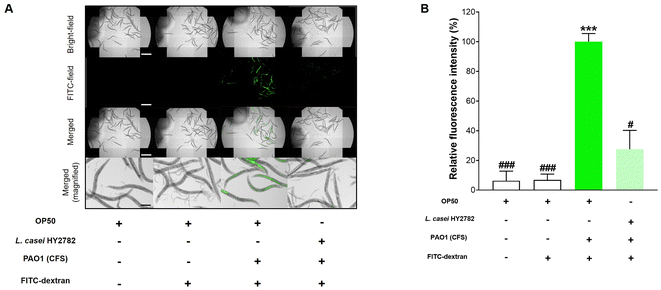
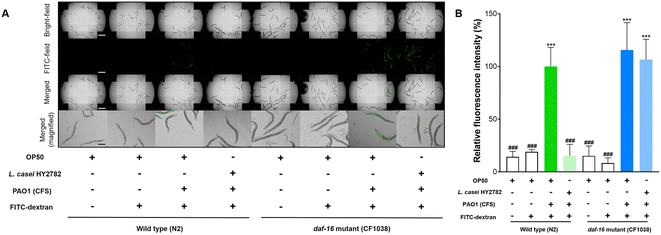
To evaluate the effect of L. casei HY2782 on the intestinal barrier function of C. elegans, the intestinal FITC-dextran permeability was determined using fluorescence microscopy. As shown in Fig. 2, P. aeruginosa PAO1 CFS induced significant gut leakage and permeability of FITC-dextran in wild-type C. elegans (p < 0.001, Fig. 2). However, L. casei HY2782 significantly decreased FITC-dextran permeability in C. elegans administered P. aeruginosa CFS (p < 0.05) (Fig. 2B).Mutant worms that were deficient in skn-1, pmk-1, daf-16, or aak-2 were used to identify the genes responsible for the beneficial effects of L. casei HY2782. In contrast to the wild-type, skn-1, daf-16, and aak-2 mutant worms did not show L. casei HY2782-induced recovery from gut leakage (Fig. 3, 5 and 6). However, the pmk-1 mutant (KU25) showed a significant decrease in fluorescence intensity from 175.1% to 54.6% (p < 0.001) (Fig. 4). These results indicate that skn-1, daf-16, and aak-2 are involved in the L. casei HY2782-facilitated recovery of intestinal permeability in C. elegans.
3.3 Effect of L. casei HY2782 consumption on lifespan of C. elegans damaged by P. aeruginosa
To further verify whether the improvement in intestinal permeability by L. casei HY2782 was associated with lifespan extension, the effect of L. casei HY2782 consumption on lifespan extension was examined in P. aeruginosa CFS-treated C. elegans. As shown in Table 1 and Fig. 7, the control group (E. coli OP50 with P. aeruginosa CFS) exhibited a shorter mean lifespan of 8.5 ± 0.3 days and a maximum lifespan of 21 days than those of L. casei HY2782 feeding group (L. casei HY2782 with P. aeruginosa CFS). Conversely, L. casei HY2782 extended the mean lifespan to 12.1 ± 0.4 days and maximum lifespan to 25 days (p < 0.001).| Food source | Lifespan (days) | p-Values | ||
|---|---|---|---|---|
| Minimum | Maximum | Mean | ||
| Data are shown as mean ± standard deviation of each group. Significant difference was determined using the log-rank test. | ||||
| Escherichia coli OP50 | 4 | 21 | 8.5 ± 0.3 | <0.001 |
| Lacticaseibacillus casei HY2782 | 4 | 25 | 12.1 ± 0.4 | |
3.4 Effect of L. casei HY2782 consumption on intestinal permeability in aged C. elegans
To further investigate the improvement of intestinal permeability induced by L. casei HY2782 consumption, the incidence of Smurf phenotype was examined in L. casei HY2782-fed aged C. elegans (Day 10 of adulthood). The Smurf phenotype appeared in 34.8% (8/24 worms) of E. coli OP50-fed aged worms, which exhibited aging-induced intestinal permeability. In contrast, only three Smurf worms (total of 23 worms) were found in the L. casei HY2782-fed aged worms (p < 0.001) (Fig. 8).3.5 L. casei HY2782 induced gst-4 expression in C. elegans
To assess whether gst-4 gene expression contributed to intestinal damage prevention in C. elegans, GFP fluorescence intensity was measured in CL2166 worms, which contained the gst-4p::GFP reporter construct. Upon exposure to L. casei HY2782 (without P. aeruginosa CFS), the CL2166 worms exhibited a significant increase in fluorescence intensity from 100.0% to 127.9% (p < 0.05), 161.8% (p < 0.01), and 168.1% (p < 0.05) at 24, 48, and 72 h, respectively. Worms exposed to the P. aeruginosa CFS showed increased green fluorescence intensity (140.4%) at 24 h. However, the worms did not maintain GFP expression at 48 h (92.0%) or 72 h (107.5%) (p > 0.05), whereas GFP expression in worms exposed to CFS and L. casei HY2782 increased to 238.6%, 222.6% (p < 0.001), and 227.7% (p < 0.001) at 24, 48, and 72 h, respectively. In addition, significant differences were observed between worms exposed to CFS alone and those exposed to L. casei HY2782 and CFS at 24, 48, and 72 h (p < 0.001) (Fig. 9). These data imply that L. casei HY2782 consumption increased gst-4 expression, which plays an important role in the removal of oxidative stress during P. aeruginosa infection.293.6 SCFA production under simulated colonic fecal fermentation with L. casei HY2782
SCFAs are metabolized end-products of fermentation by the gut microbiota and are closely associated with human health benefits.30–33 SCFA production in the GI tract is affected by the microbiome niche; therefore, the SCFA concentration was measured under simulated colonic fecal fermentation conditions. In normal colon condition, the vehicle group increased acetate concentration (up to 88.2 mM) and propionate and butyrate was rarely measured during fermentation (Fig. 10A). However, L. casei HY2782 consistently increased butyrate concentration (from 21.0 mM to 93.4 mM) and inhibited acetate production during fermentation in normal colon condition (Fig. 10B). In vehicle group of high-protein diet colon condition, acetate was the most abundant SCFA whereas that of propionate and butyrate production was inhibited during fermentation (Fig. 10C). Compared with those in the vehicle group, the L. casei HY2782 increased butyrate concentration from 78.0 mM to 114,78 mM whereas acetate and propionate slightly increased from 22.2 mM to 48.9 mM and 13.2 mM to 25.2 mM, respectively (Fig. 10D).3.7 Microbiome diversity under simulated colonic fecal fermentation
Microbial diversity in the simulated colon was measured to analyze changes in the microbiome niche of the GI tract. After fermentation, the OTU levels differed between the control and L. casei HY2782-fortified groups. At the phylum level, Firmicutes (containing order Lactobacillales) increased, whereas Bacteroidetes decreased in the L. casei HY2782-fermented group (Fig. 11A). Considering alpha diversity (Shannon index), L. casei HY2782 showed the potential to proliferate in the GI tract and inhibit other opportunistic pathogens, such as Bacteroidetes (Fig. 11B). During fermentation, L. casei HY2782 assisted in the growth of Faecalibacterium and Lachnospira, which belong to a butyrate-producing family (p < 0.01), whereas Fusobacterium growth was inhibited (p < 0.05) (Fig. 11D and E).3.8 Lifespan of fermented product-fed C. elegans
Finally, C. elegans worms were fed fermented product samples. The survival curves are presented in Fig. 12 and the mean lifespans are summarized in Table 2. Briefly, worms fed with normal colonic ferments had a mean lifespan of 10.3 ± 0.2 days. Feeding with L. casei HY2782-incoulated fecal ferments from the normal colon increased the mean lifespan to 14.5 ± 0.3 days (p < 0.001, Table 2). Feeding with high-protein diet colon ferments shortened the mean lifespan to 7.9 ± 0.1 days, whereas feeding with the high-protein diet colon ferments containing L. casei HY2782 resulted in the mean lifespan of 13.7 ± 0.2 (p < 0.001). Altogether, the lifespan of C. elegans significantly increased when fed L. casei HY2782-fermented product.| Artificial colonic conditions | Lifespan (days) | p-Values | |||
|---|---|---|---|---|---|
| Minimum | Maximum | Mean | |||
| Data are shown as mean ± standard deviation of each group. Significant difference was determined using the log-rank test. | |||||
| Normal | Control | 3 | 17 | 10.3 ± 0.2 | <0.001 |
| Lacticaseibacillus casei HY2782 | 4 | 20 | 14.5 ± 0.3 | ||
| High-protein diet | Control | 3 | 16 | 7.9 ± 0.1 | <0.001 |
| Lacticaseibacillus casei HY2782 | 4 | 19 | 13.7 ± 0.2 | ||
4. Discussion
The intestinal microbiome plays a critical role in protein metabolism.34 The proteolytic activity of the intestinal microbiome contributes to the development of a metabolite pool and regulates amino acids in the intestine.31 Acetate and propionate mainly accumulate in the intestine during dietary protein fermentation in the GI tract,35 whereas butyrate accumulates via a restricted pathway—the acetate-CoA-transferase pathway—in Firmicutes.36 In addition, butyrate is synthesized from amino acids, such as lysine and glutamine,37 and organic acids, such as succinate and lactate.32 Furthermore, butyrate contributes to healthy GI tract conditions by regulating the microbial niche and inflammation, reinforcing the mucosal barrier, and promoting epithelial growth.38 In a high-protein diet, proteolytic fermentation decreases butyrate levels and induces inflammatory reactions and cardiovascular diseases.39 Therefore, several studies have investigated butyrate production by LAB to improve GI tract conditions. For example, Moens et al.32 investigated four LAB species (Lactobacillus acidophilus, Lactiplantibacillus plantarum, Lacticaseibacillus rhamnosus, and Enterococcus faecium) that promote the production of butyrate and IL-10, an anti-inflammatory cytokine produced in human intestinal models. In a clinical trial, Ford et al.39 reported that a high-protein diet suppressed butyrate production, and probiotic supplements containing Bifidobacterium longum, Bifidobacterium breve, L. acidophilus, and L. plantarum relieved indigestion in high-protein diet consumers. In this study, L. casei HY2782 supplementation increased butyrate production during the simulated colon fermentation, which aligns well with previous studies.The human body comprises the following bacterial phyla: Firmicutes, Bacteroidetes, Proteobacteria, Fusobacteria, Actinobacteria, and Verrucomicrobia.33 These bacteria interact with the host by secreting metabolites, such as SCFAs, ρ-cresol, ρ-cresyl-glucuronide, indole-3 acetic acid, indoxyl sulfate, and trimethylamine N-oxide.40 The Firmicutes/Bacteroidetes (F/B) ratio is a necessary index for maintaining homeostasis.33 Stojanov et al.33 reported that an imbalance when the F/B ratio is decreased causes inflammatory bowel disease. Moreover, an imbalance in the F/B ratio has also been observed in an intestinal bowel disease mouse model.41Faecalibacterium36,37 and Lachnospira32,37 are representative genera of butyrate producers, which are members of the Firmicutes. Notably, L. casei HY2782 enhanced the Faecalibacterium and Lachnospira ratio during the simulated colonic fecal fermentation, which increased the F/B ratio, thereby resulting in butyrate production.
Nematodes and humans share conserved aging mechanisms. For example, DAF-16 and SKN-1, orthologs of human forkhead box protein O (FOXO) and nuclear factor erythroid 2-related factor 2 (Nrf2), respectively, play critical roles in regulating aging, stress responses, metabolism, and homeostasis in C. elegans.11 Lee and Lee12 reported that insulin/insulin-like growth factor-1 (IGF-1) signaling (IIS), regulated by the DAF-2/IGF-1 receptor (IGFR), affects aging in diverse species, ranging from C. elegans to mammals. The IIS cascade inhibits the phosphorylation of DAF-2/IGFR, thus resulting in its cytoplasmic retention.11,12 The anti-aging effects of IIS inhibition and translocation of DAF-16/FOXO and SKN-1/Nrf2 have been investigated.11 Adenosine monophosphate-activated protein kinase-2 (AAK-2/AMPK), encoded by aak-2 in C. elegans, is also crucial in aging.42 AAK-2/AMPK mediates the phosphorylation of DAF-16/FOXO42,43 and the DAF-16/FOXO-dependent transcription of superoxide dismutase to extend the lifespan of C. elegans.43 Onken and Driscoll44 suggested that AAK-2/AMPK activates SKN-1/Nrf2 translocation to regulate reactive oxygen species. Additionally, Peng et al.45 showed that the AAK-2/AMPK pathway is involved in the antioxidant mechanism of chicoric acid in aak-2 mutant worm strains. Similarly, this study suggests that AAK-2/AMPK, DAF-16/FOXO, and SKN-1/Nrf2 play essential roles in the L. casei HY2782-induced longevity of C. elegans, as evidenced by the experiments in which L. casei HY2782 was consumed by aak-2, daf-16, and skn-1 mutant worms, respectively.
Glutathione (GSH) is another critical factor affecting the lifespan of C. elegans.11,46 Similar to IIS, GSH biosynthesis in C. elegans resembles that of mammals. In C. elegans, the SKN-1/Nrf-2 transcription factor promotes gamma-glutamylcysteine synthetase and GST-4, which are phase II detoxification enzymes that resist oxidative or xenobiotic stress.13 Ferguson and Bridge46 suggested that C. elegans has 56 GST genes that encode specific GSTs in response to certain xenobiotic compounds. Thus, gst-4 expression in C. elegans damaged by P. aeruginosa is related to a specific reaction, and L. casei HY2782 probably promotes gst-4 expression to attenuate this damage.
5. Conclusions
This study demonstrates improvements in intestinal permeability and lifespan extension in C. elegans as a result of consumption of the probiotic bacteria L. casei HY2782. Moreover, this study is the first to mimic the intestinal system by combining aerobic fecal fermentation and the model nematode C. elegans. L. casei HY2782 consumption prevented intestinal permeability dysfunction and prolonged the lifespan of C. elegans through the DAF-16/FOXO and SKN-1/NRF2 molecular pathways. Furthermore, gst-4 expression was upregulated by L. casei HY2782. Additionally, fermented media from the simulated colon fecal fermentation, which reconstructed the microbiome niche from human fecal samples, further supported lifespan extension in C. elegans. Overall, these findings suggest that L. casei HY2782 is a promising probiotic candidate for improving intestinal health as well as longevity.Conflicts of interest
The authors declare no competing interests.Data availability
The 16S rRNA amplicon was deposited in the Sequence Read Archive under accession number PRJNA1071338 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1071338). The datasets analyzed in this study can be made available from the corresponding author upon reasonable request.Acknowledgements
This work was supported by the Bio-Industry Technology Development Program (20019431, Development of postbiotics based on microbiome for treatment resistant depression and anxiety disorders), which is funded by the Korea Evaluation Institute of Industrial Technology (KEIT) and the Ministry of Trade Industry & Energy (MOTIE), and the Korea Institute of Science and Technology Intramural Grant (2E33521, for the Development of data utilization technology for natural product research, a project that creates the KIST Dashboard).We thank Ms Ngoc Minh Ha for contributing to the C. elegans experiments.
References
- M. Snelson, R. E. Clarke, T. V. Nguyen, S. A. Penfold, J. M. Forbes, S. M. Tan and M. T. Coughlan, Long term high protein diet feeding alters the microbiome and increases intestinal permeability, systemic inflammation and kidney injury in mice, Mol. Nutr. Food Res., 2021, 65, e2000851 CrossRef PubMed.
- J. Roberts, A. Zinchenko, K. T. Mahbubani, J. Johnstone, L. Smith, V. Merzbach, M. Blacutt, O. Banderas, L. Villasenor, F. T. Vårvik and M. Hanselmans, Satiating effect of high protein diets on resistance-trained in energy deficit, Nutrients, 2018, 11, 56 CrossRef PubMed.
- M. J. Skytte, A. Samkani, A. D. Petersen, M. N. Thomsen, A. Astrup, E. Chabanova, J. Frystyk, J. J. Holst, H. S. Thomsen, S. Madsbad, T. M. Larsen, S. B. Haugaard and T. Krarup, A carbohydrate-reduced high-protein diet improves HbA1c and liver fat content in weight stable participants with type 2 diabetes: A randomised controlled trial, Diabetologia, 2019, 62, 2066–2078 CrossRef CAS PubMed.
- A. Kumar, T. Joishy, S. Das, M. C. Kalita, A. K. Mukherjee and M. R. Khan, A potential probiotic Lactobacillus plantarum JBC5 improves longevity and healthy aging by modulating antioxidative, innate immunity and serotonin-signaling pathways in Caenorhabditis elegans, Antioxidants, 2022, 11, 268 CrossRef CAS PubMed.
- M. Roselli, E. Schifano, B. Guantario, P. Zinno, D. Uccelletti and C. Devirgiliis, Caenorhabditis elegans and probiotics interactions from a prolongevity perspective, Int. J. Mol. Sci., 2019, 20, 5020 CrossRef CAS PubMed.
- S. Kishimoto, M. Nono, Y. Makizaki, Y. Tanaka, H. Ohno, E. Nishida and M. Uno, Lactobacillus paracasei subsp. paracasei, 2004 improves health and lifespan in Caenorhabditis elegans, Sci. Rep., 2024, 14, 10453 CrossRef CAS PubMed.
- U. T. T. Nguyen, E. Youn, T. A. N. Le, N. M. Ha, S. H. Tran, S. Lee, J. W. Cha, J. S. Park, H. C. Kwon and K. Kang, Photodynamic treatment increases the lifespan and oxidative stress resistance of Caenorhabditis elegans, Free Radicals Biol. Med., 2024, 221, 98–110 CrossRef CAS PubMed.
- N. M. Ha, S. H. Tran, Y. H. Shim and K. Kang, Caenorhabditis elegans as a powerful tool in natural product bioactivity research, Appl. Biol. Chem., 2022, 65, 18 CrossRef.
- A. N. Kang, D. Mun, S. Ryu, J. J. Lee, S. Oh, M. K. Kim, M. Song, S. Oh and Y. Kim, Culturomic-, metagenomic-, and transcriptomic-based characterization of commensal lactic acid bacteria isolated from domestic dogs using Caenorhabditis elegans as a model for aging, J. Anim. Sci., 2022, 100, 1–16 Search PubMed.
- T. A. N. Le, B. Selvaraj, J. W. Lee and K. Kang, Measuring the effects of bacteria and chemicals on the intestinal permeability of Caenorhabditis elegans, J. Visualized Exp., 2019, 154, e60419 Search PubMed.
- R. Li, Q. Yi, J. Wang, Y. Miao, Q. Chen, Y. Xu and M. Tao, Paeonol promotes longevity and fitness in Caenorhabditis elegans through activating the DAF-16/FOXO and SKN-1/Nrf2 transcription factors, Biomed. Pharmacother., 2024, 173, 116368 CrossRef CAS PubMed.
- H. Lee and S. V. Lee, Recent progress in regulation of aging by insulin/IGF-1 signaling in Caenorhabditis elegans, Mol. Cells, 2022, 45, 763–770 CrossRef CAS PubMed.
- A. Zhu, F. Zheng, W. Zhang, L. Li, Y. Li, H. Hu, Y. Wu, W. Bao, G. Li, Q. Wang and H. Li, Oxidation and antioxidation of natural products in the model organism Caenorhabditis elegans, Antioxidants, 2022, 11, 705 CrossRef CAS PubMed.
- C. T. S. Ana, J. Ju, F. A. R. de Barros and K. H. Kim, Macauba (Acrocomia aculeata) pulp oil reduces fat accumulation and enhances the lifespan of Caenorhabditis elegans at low temperatures via fat-1- and fat-7-dependent pathway, J. Food Sci., 2024, 89, 5101–5112 CrossRef CAS PubMed.
- Y. Cui and X. Qu, Genetic mechanisms of prebiotic carbohydrate metabolism in lactic acid bacteria: Emphasis on Lacticaseibacillus casei and Lacticaseibacillus paracasei as flexible, diverse and outstanding prebiotic carbohydrate starters, Trends Food Sci. Technol., 2021, 115, 486–499 CrossRef CAS.
- D. W. Olson and K. J. Aryana, Probiotic incorporation into yogurt and various novel yogurt-based products, Appl. Sci., 2022, 12, 12607 CrossRef CAS.
- X. Ma, Y. J. Shin, H. S. Park, J. W. Jeong, J. Y. Kim, J. J. Shim, J. L. Lee and D. H. Kim, Lactobacillus casei and its supplement alleviate stress-induced depression and anxiety in mice by the regulation of BDNF expression and NF-κB activation, Nutrients, 2023, 15, 2488 CrossRef CAS PubMed.
- J. Y. Kim, S. Y. Lee, S. H. Jung, M. R. Kim, I. D. Choi, J. L. Lee, J. H. Sim, C. H. Pan and K. Kang, Protective effect of Lactobacillus casei HY2782 against particulate matter toxicity in human intestinal CCD-18Co cells and Caenorhabditis elegans, Biotechnol. Lett., 2020, 42, 519–528 CrossRef CAS PubMed.
- J. Y. Kim, T. A. N. Le, S. Y. Lee, D. G. Song, S. C. Hong, K. H. Cha, J. W. Lee, C. H. Pan and K. Kang, 3,3′-Diindolylmethane improves intestinal permeability dysfunction in cultured human intestinal cells and the model animal Caenorhabditis elegans, J. Agric. Food Chem., 2019, 67, 9277–9285 CrossRef CAS PubMed.
- M. R. Kim, S. Y. Cho, H. H. Lee, J. Y. Kim, U. T. T. Nguyen, N. M. Ha, K. Y. Choi, K. H. Cha, J. H. Kim, W. K. Kim and K. Kang, Schisandrin C improves leaky gut conditions in intestinal cell monolayer, organoid, and nematode models by increasing tight junction protein expression, Phytomedicine, 2022, 103, 154209 CrossRef CAS PubMed.
- A. C. Walker, R. Bhargava, M. J. Bucher, Y. M. Argote, A. S. Brust and D. M. Czyż, Identification of proteotoxic and proteoprotective bacteria that non-specifically affect proteins associated with neurodegenerative diseases, iScience, 2024, 27, 110828 CrossRef CAS PubMed.
- S. Gelino, J. T. Chang, C. Kumsta, X. She, A. Davis, C. Nguyen, S. Panowski and M. Hansen, Intestinal autophagy improves healthspan and longevity in C. elegans during dietary restriction, PLoS Genet., 2016, 12, e1006135 CrossRef PubMed.
- J. B. Jin, J. W. Cha, I. S. Shin, J. Y. Jeon, K. H. Cha and C. H. Pan, Supplementation with Chlorella vulgaris, Chlorella protothecoides, and Schizochytrium sp. increases propionate-producing bacteria in in vitro human gut fermentation, J. Sci. Food Agric., 2020, 100, 2938–2945 CrossRef CAS PubMed.
- K. H. Cha, E. H. Lee, H. S. Yoon, J. H. Lee, J. Y. Kim, K. Kang, J. S. Park, J. B. Jin, G. Ko and C. H. Pan, Effects of fermented milk treatment on microbial population and metabolomic outcomes in a three-stage semi-continuous culture system, Food Chem., 2018, 263, 216–224 CrossRef CAS PubMed.
- S. Scortichini, M. C. Boarelli, S. Silvi and D. Fiorini, Development and validation of a GC-FID method for the analysis of short chain fatty acids in rat and human faeces and in fermentation fluids, J. Chromatogr. B:Anal. Technol. Biomed. Life Sci., 2020, 1143, 121972 CrossRef CAS PubMed.
- D. W. Fadrosh, B. Ma, P. Gajer, N. Sengamalay, S. Ott, R. M. Brotman and J. Ravel, An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform, Microbiome, 2014, 2, 6 CrossRef PubMed.
- E. Bolyen, J. R. Rideout, M. R. Dillon, N. A. Bokulich, C. C. Abnet, G. A. Al-Ghalith, H. Alexander, E. J. Alm, M. Arumugam, F. Asnicar, Y. Bai, J. E. Bisanz, K. Bittinger, A. Brejnrod, C. J. Brislawn, C. T. Brown, B. J. Callahan, A. M. Caraballo-Rodríguez, J. Chase, E. K. Cope, R. Da Silva, C. Diener, P. C. Dorrestein, G. M. Douglas, D. M. Durall, C. Duvallet, C. F. Edwardson, M. Ernst, M. Estaki, J. Fouquier, J. M. Gauglitz, S. M. Gibbons, D. L. Gibson, A. Gonzalez, K. Gorlick, J. Guo, B. Hillmann, S. Holmes, H. Holste, C. Huttenhower, G. A. Huttley, S. Janssen, A. K. Jarmusch, L. Jiang, B. D. Kaehler, K. B. Kang, C. R. Keefe, P. Keim, S. T. Kelley, D. Knights, I. Koester, T. Kosciolek, J. Kreps, M. G. I. Langille, J. Lee, R. Ley, Y. X. Liu, E. Loftfield, C. Lozupone, M. Maher, C. Marotz, B. D. Martin, D. McDonald, L. J. Mclver, A. V. Melnik, J. L. Metcalf, S. C. Morgan, J. T. Morton, A. T. Naimey, J. A. Navas-Molina, L. F. Nothias, S. B. Orchanian, T. Pearson, S. L. Peoples, D. Petras, M. L. Preuss, E. Pruesse, L. B. Rasmussen, A. Rivers, M. S. Robeson, P. Rosenthal, N. Segata, M. Shaffer, A. Shiffer, R. Sinha, S. J. Song, J. R. Spear, A. D. Swafford, L. R. Thompson, P. J. Torres, P. Trinh, A. Tripathi, P. J. Turnbaugh, S. Ul-Hasan, J. J. J. van der Hooft, F. Vargas, Y. Vázquez-Baeza, E. Vogtmann, M. von Hippel, W. Walters, Y. Wan, M. Wang, J. Warren, K. C. Weber, C. H. D. Williamson, A. D. Willis, Z. Z. Xu, J. R. Zaneveld, Y. Zhang, Q. Zhu, R. Knight and J. G. Caporaso, Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2, Nat. Biotechnol., 2019, 37, 852–857 CrossRef CAS PubMed.
- Y. Lu, G. Zhou, J. Ewald, Z. Pang, T. Shiri and J. Xia, MicrobiomeAnalyst 2.0: Comprehensive statistical, functional and integrative analysis of microbiome data, Nucleic Acids Res., 2023, 51, W310–W318 CrossRef CAS PubMed.
- R. van der Hoeven, K. C. McCallum, M. R. Cruz and D. A. Garsin, Ce-Duox1/BLI-3 generated reactive oxygen species trigger protective SKN-1 activity via p38 MAPK signaling during infection in C. elegans, PLoS Pathog., 2011, 7, e1002453 CrossRef CAS PubMed.
- A. Oliver, Z. Alkan, C. B. Stephensen, J. W. Newman, M. E. Kable and D. G. Lemay, Diet, microbiome, and inflammation predictors of fecal and plasma short-chain fatty acids in humans, J. Nutr., 2024, 11, 3298–3311 CrossRef PubMed.
- R. Jäger, J. Zaragoza, M. Purpura, S. Iametti, M. Marengo, G. M. Tinsley, A. J. Anzalone, J. M. Oliver, W. Fiore, A. Biffi, S. Urbina and L. Taylor, Probiotic administration increases amino acid absorption from plant protein: A placebo-controlled, randomized, double-blind, multicenter, crossover study, Probiotics Antimicrob. Proteins, 2020, 12, 1330–1339 CrossRef PubMed.
- F. Moens, P. Van den Abbeele, A. W. Basit, C. Dodoo, R. Chatterjee, B. Smith and S. Gaisford, A four-strain probiotic exerts positive immunomodulatory effects by enhancing colonic butyrate production in vitro, Int. J. Pharm., 2019, 555, 1–10 CrossRef CAS PubMed.
- S. Stojanov, A. Berlec and B. Štrukelj, The influence of probiotics on the Firmicutes/Bacteroidetes ratio in the treatment of obesity and inflammatory bowel disease, Microorganisms, 2020, 8, 1715 CrossRef CAS PubMed.
- F. Mei, Z. Duan, M. Chen, J. Lu, M. Zhao, L. Li, X. Shen, G. Xia and S. Chen, Effect of a high-collagen peptide diet on the gut microbiota and short-chain fatty acid metabolism, J. Funct. Foods, 2020, 75, 104278 CrossRef CAS.
- P. Markowiak-Kopeć and K. Śliżewska, The effect of probiotics on the production of short-chain fatty acids by human intestinal microbiome, Nutrients, 2020, 12, 1107 CrossRef PubMed.
- S. Kim, S. Park, T. G. Choi and S. S. Kim, Role of short chain fatty acids in epilepsy and potential benefits of probiotics and prebiotics: Targeting “health” of epileptic patients, Nutrients, 2022, 14, 2982 CrossRef CAS PubMed.
- S. Zhao, R. Lau, Y. Zhong and M. H. Chen, Lactate cross-feeding between Bifidobacterium species and Megasphaera indica contributes to butyrate formation in the human colonic environment, Appl. Environ. Microbiol., 2024, 90, e0101923 CrossRef PubMed.
- J. Zhang, L. Zhong, S. Chi, W. Chu, Y. Liu and Y. Hu, Sodium butyrate supplementation in high-soybean meal diets for juvenile rice field eel (Monopterus albus): Effects on growth, immune response and intestinal health, Aquaculture, 2020, 520, 734952 CrossRef CAS.
- A. L. Ford, V. Nagulesapillai, A. Piano, J. Auger, S. A. Girard, M. Christman, T. A. Tompkins and W. J. Dahl, Microbiota stability and gastrointestinal tolerance in response to a high-protein diet with and without a prebiotic, probiotic, and synbiotic: A randomized, double-blind, placebo-controlled trial in older women, J. Acad. Nutr. Diet., 2020, 120, 500–516 CrossRef PubMed.
- H. Usuda, T. Okamoto and K. Wada, Leaky gut: Effect of dietary fiber and fats on microbiome and intestinal barrier, Int. J. Mol. Sci., 2021, 22, 7613 CrossRef CAS PubMed.
- F. Wan, H. Han, R. Zhong, M. Wang, S. Tang, S. Zhang, F. Hou, B. Yi and H. Zhang, Dihydroquercetin supplement alleviates colonic inflammation potentially through improved gut microbiota community in mice, Food Funct., 2021, 12, 11420–11434 RSC.
- A. Mallick, A. Ranawade, W. van den Berg and B. P. Gupta, Axin-mediated regulation of lifespan and muscle health in C. elegans requires AMPK-FOXO signaling, iScience, 2020, 23, 101843 CrossRef CAS PubMed.
- M. Hao, Z. Zhang, Y. Guo, H. Zhou, Q. Gu and J. Xu, Rubidium chloride increases life span through an AMPK/FOXO-dependent pathway in Caenorhabditis elegans, J. Gerontol., Ser. A, 2022, 77, 1517–1524 CrossRef CAS PubMed.
- B. Onken and M. Driscoll, Metformin induces a dietary restriction–like state and the oxidative stress response to extend C. elegans healthspan via AMPK, LKB1, and SKN-1, PLoS One, 2010, 5, e8758 CrossRef PubMed.
- Y. Peng, Q. Sun, R. Gao and Y. Park, AAK-2 and SKN-1 are involved in chicoric-acid-induced lifespan extension in Caenorhabditis elegans, J. Agric. Food Chem., 2019, 67, 9178–9186 CrossRef CAS PubMed.
- G. D. Ferguson and W. J. Bridge, The glutathione system and the related thiol network in Caenorhabditis elegans, Redox Biol., 2019, 24, 101171 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5fo01239b |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |