Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Enzymatic synthesis of β-galactosylated xylitol derivatives modulates gut microbiota and improves obesity-related metabolic parameters in mice†
Carlos
Sabater
 abc,
Martina
Buonaccorsi
d,
Paloma
Delgado-Fernández
e,
Nieves
Corzo
e,
Blanca
de las Rivas
f,
Rosario
Muñoz
f,
Alice
Alba
d,
Pilar
Utrilla
d and
F. Javier
Moreno
abc,
Martina
Buonaccorsi
d,
Paloma
Delgado-Fernández
e,
Nieves
Corzo
e,
Blanca
de las Rivas
f,
Rosario
Muñoz
f,
Alice
Alba
d,
Pilar
Utrilla
d and
F. Javier
Moreno
 *e
*e
aRowett Institute, University of Aberdeen, Foresterhill, Aberdeen, UK AB25 2ZD
bGroup of Functionality and Ecology of Beneficial Microorganisms (MicroHealth), Dairy Research Institute of Asturias (IPLA-CSIC), Francisco Pintado Fe nº 26, 33011, Oviedo, Asturias, Spain
cHealth Research Institute of Asturias (ISPA), Avenida Hospital Universitario s/n, 33011, Oviedo, Asturias, Spain
dDepartamento de Farmacología, Centro de Investigaciones Biomédicas en Red –Enfermedades Hepáticas y Digestivas (CIBER-EHD), Centro de Investigación Biomédica, Universidad de Granada, Granada, Spain
eInstituto de Investigación en Ciencias de la Alimentación (CIAL) (CSIC-UAM) CEI (CSIC+UAM), Nicolas Cabrera 9, 28049, Madrid, Spain. E-mail: javier.moreno@csic.es; Tel: (+34) 91 0017948
fInstituto de Ciencia y Tecnología de Alimentos y Nutrición, ICTAN (CSIC), José Antonio Novais, 6 28040, Madrid, Spain
First published on 9th June 2025
Abstract
Obesity and its associated metabolic disorders are major global health concerns, highlighting the need for novel dietary interventions. Xylitol, a polyol widely used as a sugar substitute, has shown metabolic benefits beyond its sweetening properties. However, the potential physiological effects of its enzymatically modified derivatives, particularly β-galactosylated xylitol (XylGal), remain largely unexplored. In this study, we evaluated the impact of XylGal and unmodified xylitol (Xyl) on metabolic health and gut microbiota composition in a murine model of diet-induced obesity. Lean and obese C57BL/6J mice received daily doses of Xyl (50 mg kg−1) or XylGal (50 and 100 mg kg−1) for seven weeks. Our findings indicate that both Xyl and XylGal significantly reduced body weight gain, adipose tissue accumulation, and liver weight in obese mice, without affecting food intake. Additionally, Xyl and XylGal modulated glucose homeostasis, with Xyl-treated mice exhibiting improved glucose tolerance. A significant reduction in inflammatory cytokine expression (TNF-α, IL-1β) in abdominal fat was observed, suggesting decreased macrophage infiltration and attenuation of obesity-induced inflammation. High-throughput sequencing of 16S rRNA revealed that both compounds promoted beneficial bacterial genera, including Lachnospiraceae NK4A136 and Eubacterium xylanophilum, while reducing potentially obesity-associated taxa such as Blautia and Colidextribacter. These results suggest that XylGal and Xyl exert prebiotic effects that contribute to their metabolic benefits. Our study provides new insights into the potential of these compounds as functional ingredients for obesity management and metabolic health improvement.
Introduction
Obesity and its associated metabolic disorders represent a major global health concern, driving the need for innovative dietary strategies to mitigate their impact.1,2 Obesity is a multifactorial disease characterized by excessive fat accumulation and associated with numerous metabolic disorders, including type II diabetes, cardiovascular diseases, and non-alcoholic fatty liver disease. The prevalence of obesity has risen dramatically worldwide, in large part due to sedentary lifestyles and significant changes in dietary patterns over recent decades.3 A key dietary factor implicated in this trend is the excessive consumption of sugars. High sugar intake, commonly from sugar-sweetened beverages and processed foods, has been linked to rapid weight gain, insulin resistance, and metabolic syndrome,4,5 contributing to the onset and progression of obesity and its related complications. Recent epidemiological studies have demonstrated a clear association between dietary sugar load and increased risk of metabolic syndrome, highlighting the urgent need to explore alternative dietary interventions that could counteract these adverse effects.4,6,7In this context, the exploration of non-digestible and modified sugar analogues, such as enzymatically-produced oligosaccharides, represents a promising strategy. Unlike common sugars, these modified compounds may provide a lower caloric load while exerting additional metabolic benefits, such as modulation of gut microbiota. This dual function, reducing caloric intake and acting on the gut ecosystem, could be critical for developing new functional foods aimed at the prevention or management of obesity-related disorders.8–10 In recent years, polyols such as xylitol have gained significant attention in the food industry as sugar substitutes due to their lower caloric content and beneficial health properties11,12 Xylitol, a naturally occurring sugar alcohol, is widely used for its sweetening power and non-cariogenic properties.13 In addition, it has been shown to exert favorable metabolic effects, such as attenuating postprandial glycemic responses and improving glucose tolerance.14–16 Additionally, previous research has indicated that xylitol can influence lipid metabolism and immune function, suggesting a broader spectrum of metabolic benefits.17,18
Beyond its use as a sugar alternative, xylitol's potential biological effects may be enhanced when enzymatically modified to generate functional derivatives with prebiotic or metabolic modulatory properties19–23 Thus, a recent study has explored the in vitro prebiotic and metabolic properties of glycosylated polyols, particularly β-galactosylated xylitol derivatives. These compounds have demonstrated the ability to modulate gut microbiota composition, promoting beneficial bacterial populations such as Bifidobacterium and Lactobacillus, among other genera, and enhancing short-chain fatty acid (SCFA) production.24 Despite these promising findings, studies investigating the in vivo physiological effects of β-galactosylated xylitol derivatives and their comparison to xylitol are still scarce. While previous research has explored their impact on cecal fermentation and serum parameters in rats,19 their role in obesity prevention and metabolic health improvement remains largely unexplored. To address this gap, we hypothesized that β-galactosylated xylitol derivatives exert beneficial metabolic effects by modulating gut microbiota composition and improving metabolic parameters in an in vivo model. In this study, we administered these compounds at doses of 50 and 100 mg kg−1 to lean and obese five-week-old male C57BL/6J mice and evaluated their metabolic and gut microbiota modulatory effects. Our results indicate that these derivatives contribute to a beneficial modulation of gut microbiota, which may be a key mechanism underlying their metabolic benefits. Given the recognized role of gut microbiota in energy homeostasis, adipogenesis, and inflammation,25–28 our findings highlight the potential of microbiome-targeted interventions in obesity management.
This study provides novel insights into the potential of β-galactosylated xylitol derivatives and xylitol itself as functional ingredients with anti-obesogenic properties. By demonstrating their efficacy in vivo, we highlight their relevance not only as sugar substitutes but also as promising dietary interventions for metabolic health improvement.
Experimental
Enzymatic synthesis, purification and characterization of β-galactosylated xylitol derivatives
β-Galactosylated xylitol derivatives were biosynthesized through a transglycosylation reaction catalyzed by the recombinant LacA β-galactosidase from Lactiplantibacillus plantarum WCFS1 (1.16 U mL−1). The reaction took place at 37 °C in 50 mM sodium phosphate buffer (pH 6.5) supplemented with 2 mM MgCl2, using lactose (VWR, Barcelona, Spain) as the galactosyl donor and xylitol (Sigma Aldrich, Stenheim, Germany) as the acceptor. The initial substrate ratio was set at 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20% (w/w) for xylitol and lactose, following the methodology described by Delgado-Fernandez et al.29 Before assessing their in vivo effects, the synthesized carbohydrates were purified from the reaction mixtures. The purification process consisted of two key steps as described by Calvete-Torre et al.:24 (i) the selective hydrolysis of residual lactose, used as the donor substrate, was carried out using a β-galactosidase from Kluyveromyces lactis (Lactozym® Pure 6500 L) (Novozyme, Bagsværd, Denmark), ensuring the preservation of β-galactosylated xylitol structures under optimized conditions described by Julio-Gonzalez et al.;30 (ii) free monosaccharides and unreacted xylitol were removed via activated charcoal treatment. A solution containing 1 g of oligosaccharides and 6 g of activated charcoal (Darco G60, 100 mesh, Sigma–Aldrich) was prepared in 200 mL of water and stirred for 60 minutes. The mixture was then vacuum-filtered using Whatman no. 1 paper (Whatman International Ltd, Maidstone, UK), and the charcoal was washed three times with 50 mL of water. Oligosaccharides were eluted from the charcoal using 100 mL of 50% (v/v) ethanol, stirred for 30 minutes, and filtered again.31 Finally, the purified oligosaccharides were concentrated using a Büchi R-200 rotary evaporator (Büchi Labortechnik AG, Flawil, Switzerland) at temperatures below 25 °C and subsequently freeze-dried.
20% (w/w) for xylitol and lactose, following the methodology described by Delgado-Fernandez et al.29 Before assessing their in vivo effects, the synthesized carbohydrates were purified from the reaction mixtures. The purification process consisted of two key steps as described by Calvete-Torre et al.:24 (i) the selective hydrolysis of residual lactose, used as the donor substrate, was carried out using a β-galactosidase from Kluyveromyces lactis (Lactozym® Pure 6500 L) (Novozyme, Bagsværd, Denmark), ensuring the preservation of β-galactosylated xylitol structures under optimized conditions described by Julio-Gonzalez et al.;30 (ii) free monosaccharides and unreacted xylitol were removed via activated charcoal treatment. A solution containing 1 g of oligosaccharides and 6 g of activated charcoal (Darco G60, 100 mesh, Sigma–Aldrich) was prepared in 200 mL of water and stirred for 60 minutes. The mixture was then vacuum-filtered using Whatman no. 1 paper (Whatman International Ltd, Maidstone, UK), and the charcoal was washed three times with 50 mL of water. Oligosaccharides were eluted from the charcoal using 100 mL of 50% (v/v) ethanol, stirred for 30 minutes, and filtered again.31 Finally, the purified oligosaccharides were concentrated using a Büchi R-200 rotary evaporator (Büchi Labortechnik AG, Flawil, Switzerland) at temperatures below 25 °C and subsequently freeze-dried.
The composition of the tested β-galactosylated xylitol derivatives was analyzed in terms of carbohydrate content, expressed as a percentage of total carbohydrates as shown in Table 1. The structural characterization of these compounds was performed using NMR, as described by Delgado-Fernandez et al.29
| Component | Percentage (%) |
|---|---|
| β-D-Galactopyranosyl-(1 → 1)-xylitol | 46.3 |
| β-D-Galactopyranosyl-(1 → 3)-xylitol | 11.4 |
| Xylitol | 14.7 |
| Galactooligosaccharides (disaccharides) | 5.2 |
| Galactooligosaccharides (trisaccharides) | 10.3 |
| Lactose | 2.9 |
| Glucose | 6.3 |
| Galactose | 2.7 |
Animals
Male C57BL/6J mice were obtained from Janvier (St Berthevin Cedex, France). The animals were housed in T3 type polycarbonate plastic boxes, which were located in the facilities of the Animal Experimentation Center of the University of Granada (Campus of Cartuja). Water was supplied in 500 ml polycarbonate bottles, with a stainless-steel pipette, while pelleted food was placed in the hopper of the box for ad libitum availability. All mice were kept at a constant temperature of 24 ± 1 °C, with a 12 hour light-dark cycle. The animals’ body weight and food intake were monitored regularly. All experiments were performed following the Guide for the Care and Use of Laboratory Animals (National Institute of Health (NIH) publication no. 85-23, revised 1996) and were approved by our Institutional Commission for the ethical care of animals (Consejería de Agricultura, Ganadería, Pesca y Desarrollo Sostenible. Dirección General de la Producción Agrícola y Ganadera. Protocol no. 18/05/2022/069). At the end of the experiment, the mice were sacrificed to collect tissue samples for further processing.Experimental groups
Five-week-old C57BL/6J mice were randomly assigned (n = 6–8 per group) into seven different groups defined by both diet and compound administration:Standard diet (lean) groups:
(a) Lean control (LC)
(b) Lean treated with xylitol (Xyl) 50 mg kg−1 (LXyl)
(c) Lean treated with β-galactosylated xylitol derivatives (XylGal) 100 mg kg−1 (LXylGal)
High-fat diet (obese) groups:
(d) Obese controls (OC)
(e) Obese treated with Xyl 50 mg kg−1 (OXyl)
(f) Obese treated with XylGal 50 mg kg−1 (OXylGal50)
(g) Obese treated with XylGal 100 mg kg−1 (OXylGal100)
The treated mice received the Xyl or XylGal compounds dissolved in water by esophageal gavage daily, and the untreated (control) mice only received the vehicle. Treatment with the compounds continued for 7 weeks. Food intake and body weight was monitored every 3 days, throughout the feeding period. Additionally, stool samples were taken at day 0 and at the end of treatment for analysis. LC, LXyl and LXylGal mice were maintained on a standard maintenance diet (13% calories from fat, 20% calories from protein, 67% calories from carbohydrates; 3.8 kcal g−1) (Global diet 2014, Harlan Laboratories, Barcelona, Spain), whereas OC, OXyl, OxylGal50, and OXylGal100 mice were fed a Western-type High-Fat Diet (HFD) in which 60% of its caloric content came from fat (60.3% of calories from fat (lard), 18.4% calories from protein, 21.3% calories from carbohydrates; 5.4 kcal g−1) (Purified diet 230 HF, Scientific Animal Food & Engineering, Augy, France). This standard–diet control provides a physiologically relevant baseline; future studies including a calorie-matched low-fat diet may further refine our understanding of how fat content versus total energy intake shape metabolic and microbiota outcomes. At the end of treatment, mice were sacrificed under anesthesia (Ketamine 50 mg ml−1 – Atropine 1 mg ml−1 – Diazepam 5 mg ml−1). Liver, abdominal fat, and visceral fat were collected, weighed, and stored at −80 °C until analysis.
The terms ‘lean’ and ‘obese’ refer to mice fed a standard diet or a high-fat diet, respectively. Including both dietary states allows us to assess the effects of Xyl and XylGal under normal and obesity-related metabolic conditions. Lean mice served as baseline controls to delineate the metabolic and microbiota changes attributable to each compound in the absence of diet-induced dysregulation, thereby strengthening the evaluation of their efficacy across distinct metabolic states.
To justify the doses used in this study, we first examined the typical xylitol content in common food sources. For example, sugar-free chewing gum generally contains approximately 1 g of xylitol per piece. Considering a typical serving (1 piece) for an average adult weighing 60–70 kg, this corresponds to roughly 14–17 mg kg−1. Using the standard body surface area conversion factor (mouse Km = 3; human (adult) Km = 37) as described in Reagan-Shaw et al.,32 the 50 mg kg−1 dose administered to mice translates to a human equivalent dose (HED) of approximately 4 mg kg−1, which equals about 240 mg for a 60 kg human. This indicates that the doses employed in our study are physiologically relevant and can be achieved through the consumption of a realistic number of servings of xylitol-containing foods. Additionally, the safety profile of xylitol is well supported by its regulatory status. Specifically, EFSA has not defined a specific numerical ADI for xylitol, reflecting its low toxicity and safe use at customary dietary levels. Likewise, the FDA's designation of xylitol as GRAS further confirms its safety for use in food products. Thus, the doses applied in our experiments are not only physiologically relevant but also fall within a range that is considered safe for human consumption.
Glucose tolerance test
A glucose tolerance test (GTT) was performed according to the method described by Benedé-Ubieto et al.33 GTT was assessed in mice fasting for 12 hours. Briefly, animals received a 20% glucose solution in water at a dose of 1.5 g kg−1 body weight by intraperitoneal injection, and blood was collected from the tail vein after 0, 15, 45, and 120 minutes. Glucose levels were measured using an Accu-Chek blood glucose meter (Roche Diagnostics, Mainz-Hechtsheim, Germany).RNA isolation and quantitative RT-PCR
Total RNA was isolated from frozen samples of liver and abdominal and visceral adipose tissue using the RNeasy kit (QIAGEN, Tokyo, Japan) following the manufacturer's protocol. Briefly, tissues were homogenized using a Next Advance Bullet Blender device, then transferred to the supplied columns where the RNA was retained; it was washed repeatedly and then eluted and stored until use at −80 °C. It was quantified using a NanoDrop 1000 Spectrophotometer device (Thermo Scientific). cDNA was synthesized using reverse transcriptase (M-MLV) and oligo (dt)15 Primer from Promega. Real-time PCR was performed with a Techne thermal cycler (Techne, Cambridge, UK) using the primers described in ESI Table S1.† Quantification was performed using the ΔΔCt method. Internal normalization was performed with Glyceraldehyde-3-Phosphate dehydrogenase (GAPDH).Statistical analysis of in vivo study endpoints
Based on our extensive prior experience with similar datasets and through careful visual inspection of the data distribution (e.g., histograms and Q–Q plots), normality and homogeneity of variances were assumed. Accordingly, the mean and standard deviation were calculated for quantitative variables. For the comparison of means among multiple groups, data were analyzed using one-way Analysis of Variance (ANOVA) followed by Tukey's post hoc test for multiple comparisons. When comparing two independent groups, the Student's t-test was applied with a 95% confidence level. Statistically significant differences were considered to exist when p < 0.05.High-throughput sequencing of 16S rRNA
High-throughput sequencing of 16S rRNA was performed to characterise the taxonomic profiles of a total 35 fecal samples corresponding to seven different groups of mice under study as previously described. Amplicon sequencing of the V3–V4 region was carried out in an Illumina MiSeq instrument at the Sequencing Facilities of “Fundación Parque Científico de Madrid”. For this purpose, the following primers were used: CCTACGGGNGGCWGCAG (forward) and GACTACHVGGGTATCTAATCC (reverse). Then, raw sequencing reads were processed using an in-house script of QIIME2 v.2021.8 software,34 developed in previous works.35,36 This script included quality control and filtering of paired-end reads as well as taxonomic identification of Amplicon Sequence Variants (ASVs) at genus level using reference database SILVA 138 release. Raw reads generated have been deposited in the Short Reads Archive (SRA) under BioProject accession number PRJNA1223685.Bioinformatic analysis
Further bioinformatic analyses were performed on R (v.4.3.2.). Microbial diversity estimators including alpha (Chao1, Shannon, Simpson, Inverse Simpson) and beta (Bray Curtis dissimilarity distance) diversity indices were first calculated using Microbiome (v.1.24.0)37 and Phyloseq (v.1.46.0)38 R packages. To get a graphical overview of microbiota differences among groups of mice, cluster analysis based on Bray Curtis dissimilarity distance, principal coordinate analysis (PCoA) and microbiota composition bar plots were computed using Microbiome (v.1.24.0) R package.37To better characterise the modulatory effect of Xyl and XylGal compounds on specific microbial genera, statistically significant (p < 0.05 and padj < 0.25) differences in the microbiota composition of groups under study were calculated using advanced statistical methods (ANCOM, ANCOMBC, LEfSe and metagenomeSeq) implemented in microbiomeMarker (v.1.8.0) R package.39–43
Finally, statistically significant (p < 0.05) correlations between microbial genera and mice metadata (final weight, food consumption, abdominal and visceral fat, liver weight, triglyceride levels and glucose area under the curve, AUC) and cytokine levels (abdominal and visceral increments of IL-1β and IL-1β) were computed and expressed as Pearson correlation coefficients using base R (v.4.3.2.) functions.
Results
Impact on obesity-related physiological and anthropometric parameters
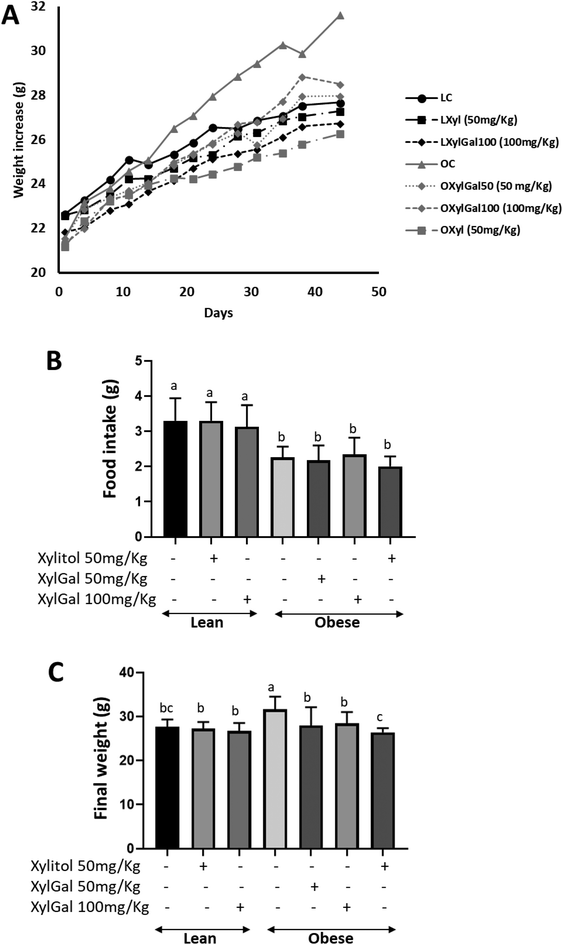
In the present study, the dietary intervention in obese mice led to a significant weight increase in untreated animals (OC) compared to those fed a standard diet (Fig. 1A). Treatment with the XylGal compound (50 and 100 mg kg−1) resulted in a significant weight reduction at both doses in obese mice (OxylGal50 and OXylGal100), while in lean mice, the 100 mg kg−1 dose (LXylGal 100) also induced a significant decrease (p < 0.05). Similarly, xylitol (50 mg kg−1) also promoted weight loss in both lean (LXyl) and obese (OXyl) mice. A comparison of food intake between treated and untreated animals revealed no significant differences within either the standard diet or HFD groups (Fig. 1B).The reduced body weight observed in XylGal-treated mice (at both doses) and xylitol-treated mice (Fig. 1C) correlated with lower abdominal and visceral fat accumulation at the end of the study. As shown in Fig. 2A and B, mice treated with xylitol exhibited the lowest fat accumulation.
No significant difference in liver weight was observed between LC and OC (p > 0.05; Fig. 2C) however, both Xyl- and XylGal-treated groups (LXyl, LXylGal100, OXyl, OXylGal50, OXylGal100) exhibited significantly lower liver weights compared to their respective diet-matched controls (p < 0.05).
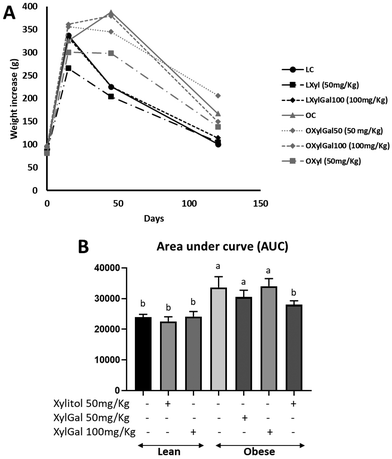
Glucose tolerance test (GTT) and glycemic response
In the glucose tolerance test (GTT), both Xyl- and XylGal-treated animals exhibited a reduction in blood glucose levels following glucose overload (Fig. 3A). Given the carbohydrate nature of the tested compounds, their potential impact on glycemia must be considered. Previous studies have shown that xylitol does not alter glycemic profiles post-ingestion and may even improve postprandial glucose levels in rats.15 Additionally, the digestibility of the XylGal compounds, as analyzed by Rosado et al.23 using an in vitro digestion model, suggests that these compounds are poorly absorbed in the small intestine, minimizing their direct impact on blood glucose levels.A clear difference in glycemic response following glucose overload was observed between lean and obese animals. Xyl-treated animals displayed a significant reduction in blood glucose levels. However, XylGal-treated animals did not show a decrease in the area under the curve (AUC), possibly due to the presence of galactose, despite its predicted limited intestinal absorption (Fig. 3B).
Inflammatory cytokine expression in adipose tissues
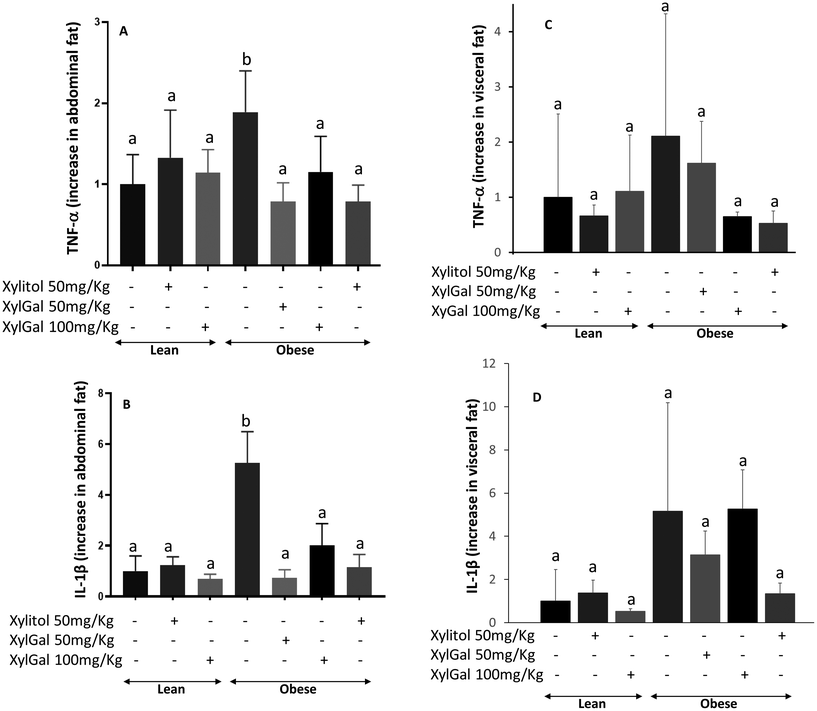
Following RNA extraction from abdominal and visceral adipose tissue, qPCR analysis was performed to assess macrophage and monocyte infiltration in obese animals. As shown in Fig. 4, untreated obese mice exhibited significantly higher expression levels of inflammatory cytokines, such as TNF-α and IL-1β, in abdominal fat compared to untreated lean mice. Treatment with XylGal (both doses) and Xyl significantly reduced the expression of pro-inflammatory cytokines in adipose tissue, indicating a local reduction in inflammatory signalling that may reflect changes in immune cell activation or presence.TNF-α production by macrophages infiltrating adipose tissue plays a key role in insulin resistance and the development of type II diabetes in overweight and obese individuals. Consistent with this, the reduced TNF-α expression observed in Xyl-treated obese mice (Fig. 4A) correlates with improved performance in the glucose overload test. Similarly, IL-1β is known to contribute to obesity-induced metabolic dysfunction, as it plays a key role in promoting systemic inflammation and pancreatic β-cell impairment, ultimately affecting insulin secretion and glucose homeostasis.44,45 For XylGal-treated mice, the observed reduction in inflammatory cytokine expression aligns with the decrease in body weight and fat accumulation (Fig. 4B). However, in the glucose overload test, the presence of galactose appears to interfere with the test outcome.
qPCR analysis of visceral fat gene expression yielded results that, while not statistically significant, followed a trend similar to that observed in abdominal fat. High intra-group variability and the low quantity of visceral fat, particularly in lean and treated obese animals, likely contributed to the lack of statistical significance. Despite this limitation, the observed reduction in proinflammatory cytokines in visceral fat suggests a potential association with lower cardiovascular risk, which could be further explored in longer-term studies (Fig. 4C and D).
Xylitol (Xyl) and β-galactosylated xylitol derivatives (XylGal) promote beneficial genera in lean (normal) and obese mice
The microbiota modulatory effect of Xyl and XylGal on lean and obese mice has been assessed. In addition, potential dose-dependent (50 and 100 mg kg−1) effects of XylGal in obese mice have been investigated.Several alpha diversity estimators (Chao1, Shannon, Simpson, Inverse Simpson) measuring microbial diversity within each group of mice were first calculated (Fig. 5). As can be seen, no significant (p > 0.05) differences were observed in the Chao1 index measuring the total number of genera determined. In contrast, LXyl showed significantly (p < 0.05) lower Simpson and Inverse Simpson indices than obese mice following the same intervention (OXyl). Similarly, LXylGal100 showed significantly (p < 0.05) lower Shannon index than obese mice treated with XylGal at two different doses (OXylGal50 and OXylGal100). These results suggest characteristic differences in the microbial diversity of obese and lean mice, defined as significant differences in microbial diversity values (p < 0.05) between LXyl and OXyl, and LXylGal100, OXylGal50 and OXylGal100. These differences in microbial diversity suggest a different modulatory effect of Xyl and XylGal. While a lower Shannon index typically indicates reduced microbial diversity, this finding may reflect a selective enrichment of certain taxa in response to the high-dose XylGal treatment, rather than a global loss of microbiota richness.
Then, beta diversity measuring microbial diversity between groups was calculated based on Bray Curtis dissimilarity distances (Fig. 6). No significant (p > 0.05) differences among intervention groups were observed. Bray Curtis dissimilarity distances were also used to cluster taxonomic profiles of samples (Fig. 7). As can be seen, cluster analysis completely discriminated lean and obese groups. In addition, samples corresponding to the same substrate (Xyl or XylGal) administered at the same dose (50 or 100 mg kg−1) were clustered together in the same branches. Cluster analysis highlights different microbiota modulatory properties of Xyl and XylGal in obese and lean mice as well as potential dose-dependent effects. Similarly, PCoA discriminated obese and lean groups and samples corresponding to different doses of substrates were differentiated (Fig. 8). However, PCoA classification was less accurate than the one observed for cluster analysis.
Once differences in the total microbiota profiles were determined, differences in the abundance of microbial genera composition of intervention groups were investigated (Fig. 9). With regard to lean groups, an unidentified Muribaculaceae genus and Lachnospiraceae NK4A136 group were the most abundant genera. Major genera found in obese groups include Blautia, Eubacterium fissicatena group and Colidextribacter.
Statistical analysis of the microbiota revealed higher (p < 0.05 and padj < 0.25) abundances of Bacteroides, Clostridia UCG-014 and Desulfovibrio in non-treated mice compared to prebiotic-administered groups (Table 2). These differences were specially accentuated in lean mice. In addition, Blautia, Lachnospiraceae GCA-900066575, Lachnoclostridium and Eubacterium nodatum group showed higher (p < 0.05 and padj < 0.25) abundances in non-treated obese mice compared to the rest of intervention groups (Table 2).
| Statistical differences: control and obesity groups | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LC | LXyl | LXylGal100 | OC | OXyl | OXylGal50 | OXylGal100 | |||||||||
| Genera | Higher in: | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD |
| Bacteroides | LC | 4.30 | 1.98 | 3.93 | 3.86 | 3.34 | 2.43 | 1.71 | 1.05 | 1.14 | 0.92 | 1.12 | 0.74 | 0.77 | 0.42 |
| Clostridia UCG-014 | LC | 4.08 | 3.33 | 2.00 | 1.79 | 2.41 | 1.90 | 0.05 | 0.03 | 0.04 | 0.04 | 0.03 | 0.03 | 0.01 | 0.01 |
| Desulfovibrio | LC | 1.65 | 0.67 | 1.52 | 0.91 | 0.97 | 0.50 | 0.06 | 0.12 | 0.00 | 0.00 | 0.02 | 0.01 | 0.02 | 0.01 |
| RF39 | LC/LXylGal100 | 0.87 | 0.67 | 0.62 | 0.64 | 0.86 | 0.56 | 0.42 | 0.59 | 0.39 | 0.17 | 0.11 | 0.07 | 0.73 | 1.17 |
| Unidentified Muribaculaceae | LXyl | 32.44 | 13.17 | 41.44 | 21.44 | 34.61 | 13.95 | 2.64 | 1.56 | 5.54 | 2.93 | 8.33 | 2.61 | 3.78 | 3.27 |
| Rikenellaceae RC9 gut group | LXyl | 0.13 | 0.09 | 1.93 | 1.36 | 1.33 | 0.77 | 1.15 | 0.47 | 1.46 | 0.74 | 0.21 | 0.18 | 0.83 | 1.07 |
| Alloprevotella | LXyl | 1.89 | 0.83 | 2.35 | 2.71 | 0.19 | 0.28 | 0.75 | 0.32 | 1.03 | 0.89 | 0.10 | 0.07 | 0.19 | 0.09 |
| Lachnospiraceae NK4A136 group | LXylGal100 | 24.38 | 14.52 | 18.96 | 20.08 | 30.51 | 13.93 | 2.32 | 2.00 | 0.65 | 0.11 | 0.56 | 0.38 | 1.99 | 0.92 |
| Alistipes | LXylGal100 | 2.06 | 0.80 | 2.69 | 0.64 | 3.19 | 2.41 | 2.23 | 1.08 | 1.97 | 1.04 | 0.27 | 0.12 | 0.93 | 1.07 |
| [Eubacterium] xylanophilum group | LXylGal100 | 2.37 | 1.30 | 3.08 | 4.13 | 3.12 | 1.51 | 0.14 | 0.18 | 0.01 | 0.02 | 0.04 | 0.02 | 0.07 | 0.14 |
| Blautia | OC | 0.52 | 0.22 | 0.93 | 1.06 | 1.76 | 1.07 | 25.43 | 13.47 | 19.09 | 3.67 | 12.48 | 4.31 | 16.64 | 6.91 |
| GCA-900066575 | OC | 0.78 | 0.54 | 0.33 | 0.39 | 0.90 | 0.45 | 1.87 | 0.88 | 1.76 | 0.26 | 0.99 | 0.25 | 1.38 | 0.53 |
| Lachnoclostridium | OC | 0.55 | 0.50 | 0.39 | 0.39 | 0.43 | 0.29 | 1.52 | 0.39 | 0.93 | 0.44 | 0.29 | 0.18 | 1.15 | 0.98 |
| [Eubacterium] nodatum group | OC | 0.04 | 0.02 | 0.01 | 0.01 | 0.05 | 0.04 | 1.17 | 0.58 | 1.11 | 0.33 | 0.92 | 0.44 | 0.42 | 0.42 |
| [Eubacterium] fissicatena group | OXylGal50 | 0.29 | 0.32 | 0.66 | 1.03 | 0.00 | 0.01 | 14.61 | 4.22 | 19.27 | 9.70 | 21.99 | 7.35 | 6.92 | 4.28 |
| Akkermansia | OXylGal50 | 10.80 | 6.55 | 6.98 | 5.34 | 1.20 | 2.64 | 2.66 | 3.43 | 4.78 | 4.99 | 18.28 | 12.28 | 1.37 | 1.31 |
| Colidextribacter | OXylGal100 | 1.54 | 0.94 | 1.44 | 0.99 | 1.84 | 0.84 | 11.62 | 2.07 | 12.07 | 2.95 | 8.14 | 3.08 | 18.09 | 6.05 |
| Coriobacteriaceae UCG-002 | OXylGal100 | 0.54 | 0.65 | 0.62 | 0.39 | 0.11 | 0.13 | 8.61 | 3.80 | 3.70 | 4.09 | 6.11 | 6.50 | 9.75 | 12.21 |
| Uncultured genus | OXylGal100 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.01 | 7.77 | 3.48 | 10.42 | 2.13 | 11.06 | 5.09 |
| Oscillibacter | OXylGal100 | 0.97 | 0.73 | 0.87 | 0.73 | 1.14 | 0.80 | 1.98 | 0.91 | 1.94 | 0.28 | 0.97 | 0.41 | 2.60 | 0.74 |
| Parabacteroides | OXylGal100 | 1.06 | 0.99 | 0.19 | 0.15 | 0.08 | 0.09 | 1.64 | 0.87 | 1.69 | 0.80 | 1.88 | 0.97 | 2.33 | 2.56 |
| Faecalibaculum | OXylGal100 | 0.11 | 0.11 | 0.01 | 0.01 | 0.04 | 0.07 | 3.32 | 1.35 | 0.93 | 0.70 | 0.67 | 0.40 | 3.49 | 2.44 |
| Ileibacterium | OXylGal100 | 0.12 | 0.08 | 0.02 | 0.01 | 0.05 | 0.08 | 2.31 | 0.92 | 0.28 | 0.23 | 0.31 | 0.19 | 4.73 | 4.23 |
| Tuzzerella | OXylGal100 | 0.14 | 0.12 | 0.08 | 0.10 | 0.18 | 0.14 | 1.15 | 0.30 | 1.11 | 0.73 | 0.54 | 0.51 | 1.37 | 0.63 |
Differences in the modulatory effect of substrates under study were observed in agreement with cluster analysis (Fig. 7). In this sense, Xyl administration led to an increment (p < 0.05 and padj < 0.25) in an unidentified Muribaculaceae genus, Rikenellaceae RC9 gut group and Alloprevotella in lean mice (Table 2). Similarly, XylGal administration at 100 mg kg−1 stimulated (p < 0.05 and padj < 0.25) the growth of health-promoting Lachnospiraceae NK4A136 group, Alistipes and Eubacterium xylanophilum group in lean mice (Table 2). It should be noted that the majority of these genera were not promoted by Xyl and XylGal in obese mice. In contrast, XylGal administration at 50 mg kg−1 resulted in an increment (p < 0.05 and padj < 0.25) in Eubacterium fissicatena group and Akkermansia in obese mice while high doses (100 mg kg−1) of XylGal promoted a wider variety of genera in these mice including Parabacteroides and Faecalibaculum.
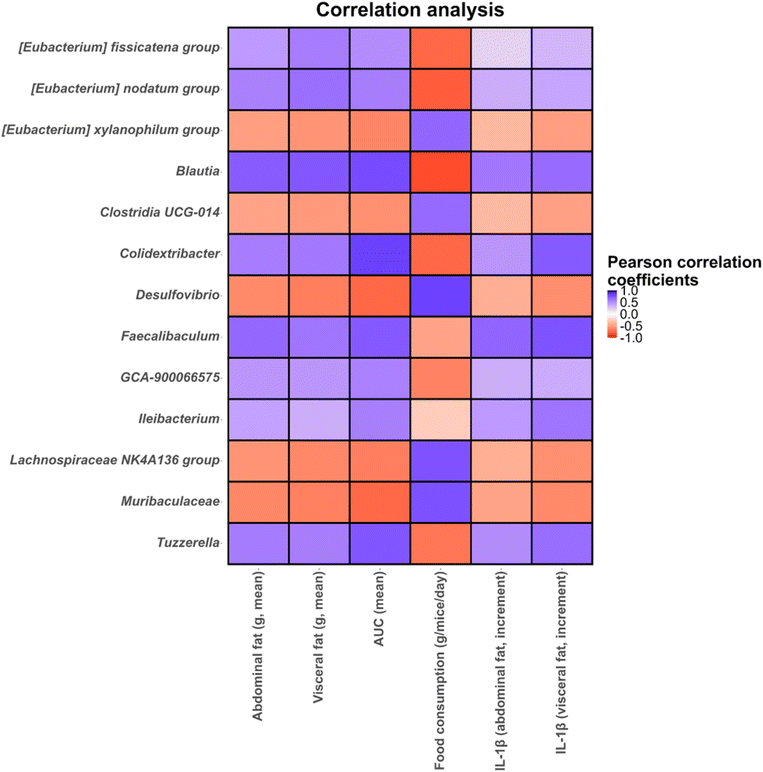
Finally, statistical correlations (p < 0.05) between microbial genera found in the microbiota of different intervention groups and mice metadata and biochemical parameters were determined (Fig. 10). In this regard, several genera including Blautia, Colidextribacter, Eubacterium fissicatena and Eubacterium nodatum groups, Faecalibaculum, Ileibacterium, Lachnospiraceae GCA-900066575 and Tuzzerella showed positive correlations with abdominal and visceral fat and IL-1β increments and glucose AUC, and negative correlations with food consumption (Fig. 10). These taxa showed high abundances in both treated and untreated obese mice highlighting the higher levels of abdominal and visceral fat, IL-1β and glucose AUC compared to lean mice. In contrast, Desulfovibrio, Eubacterium xylanophilum group, Lachnospiraceae NK4A136 group and an unidentified Muribaculaceae genus showed positive correlations with food consumption and negative correlations with abdominal and visceral fat and glucose AUC (Fig. 10). Most of these taxa showed the highest increments in lean mice treated with Xyl or XylGal highlighting the positive effect of these substrates on biochemical parameters and inflammatory markers associated with obesity.
Discussion
The present study demonstrates that Xyl and XylGal supplementation mitigated obesity-related metabolic disturbances in mice, with significant reductions in body weight and fat accumulation, particularly in abdominal and visceral depots. The inclusion of lean mice provided a necessary control, revealing that while the metabolic impact of Xyl and XylGal is evident in both lean and obese states, the magnitude and mechanisms differ. Future studies may focus on delineating these pathways further. The weight loss observed in treated animals has previously been attributed to the satiating properties of xylitol, as described by Gasmi Benahmed et al.12 However, in our study, no clear evidence of satiety was observed at the doses used, as food intake did not significantly differ between treated and control animals within each dietary group. Interestingly, obese mice exhibited lower overall food consumption than their lean counterparts, yet still gained significantly more weight. This observation is not entirely uncommon, as high-fat diets are more energy-dense and often result in reduced food volume intake despite increased caloric consumption.46,47 However, in previous experiments conducted in our laboratory using the same HFD formulation, we observed higher overall food intake in obese mice, suggesting possible variability due to strain, experimental conditions, or gut microbial status.Despite these nuances, reductions in adipose tissue and liver weight were observed in both Xyl- and XylGal-treated animals. Given that fatty liver is a frequent complication of obesity and liver weight serves as a proxy for hepatic lipid accumulation, these findings suggest a potential role for the compounds in attenuating hepatic steatosis. Similar observations were reported in an 8 week study by Amo et al.15 in rats, where xylitol reduced visceral fat, upregulated genes involved in hepatic fatty acid oxidation, and enhanced lipid degradation in adipose tissue. Conversely, other reports, including those reviewed by Gasmi Benahmed et al.,12 have shown less consistent outcomes, underscoring the complexity of xylitol's metabolic effects and the need for additional mechanistic studies.
In line with our findings, a recent study in HFD-fed rats treated with β-galactooligosaccharides (GOS) demonstrated reductions in body weight and fat accumulation, which were linked to alterations in gut microbiota composition.48 Although our study used β-galactosylated xylitol derivatives rather than GOS, the presence of galactose in XylGal could similarly contribute to prebiotic-like effects. Moreover, glucose homeostasis was significantly improved in Xyl-treated animals, as evidenced by a reduced area under the curve (AUC) in the glucose tolerance test (GTT). However, XylGal did not significantly lower AUC values, possibly due to the presence of galactose, which may have interfered with the test outcome. Additionally, both Xyl and XylGal effectively reduced the expression of inflammatory cytokines in abdominal adipose tissue, demonstrating a significant reduction in the expression of macrophage-associated markers, which may reflect decreased immune cell presence in adipose tissue and suggesting an attenuation of obesity-related low-grade inflammation. While qPCR does not directly quantify immune cell infiltration, the expression levels of macrophage and monocyte markers have been widely used as molecular proxies for tissue inflammation and immune cell recruitment in adipose tissue. Although the visceral fat results did not reach statistical significance, they followed a similar trend, indicating a potential systemic effect of these compounds on inflammation and metabolic health.
These metabolic and inflammatory changes observed in vivo correlate with the modulatory effects of Xyl and XylGal on gut microbiota composition, which may underlie some of the observed health benefits. The microbiota modulatory effect of Xyl and XylGal compounds at different doses in obese and lean mice has been investigated. Previous studies suggest that Xyl helps in glycemic and obesity control,12 in agreement with our results where several genera promoted by Xyl and XylGal where negatively correlated with glucose AUC and obesity-related inflammatory markers (Fig. 10). To our knowledge, this is the first study to examine the effect of Xyl on gut microbiota in an obesity context. Xyl administration led to an increment in Rikenellaceae RC9 gut group and Alloprevotella in lean mice, while XylGal stimulated the growth of Lachnospiraceae NK4A136 group, Alistipes and Eubacterium xylanophilum group. In obese mice, high doses of XylGal promoted the enrichment of Parabacteroides and Faecalibaculum. These taxa are considered beneficial gut commensals with established or emerging links to host metabolic health. Among them, members of Lachnospiraceae, including the NK4A136 group, have been associated with improved gut barrier integrity and reduced systemic inflammation, particularly in the context of obesity in both humans and mice.49,50 They are also considered promising targets for personalized therapeutic interventions to manage cardiometabolic risk.51Eubacterium species, similarly promoted by XylGal, are characteristic of a healthy gut microbiota compared to taxononomic profiles associated with inflammatory diseases like chronic spontaneous urticaria.52Eubacterium species may improve intestinal dysbiosis in high-fat diet-fed mice,53 and play a major role in immunomodulation and suppression of inflammation in the gut,54 in agreement with our results, where Eubacterium showed negative correlations with obesity-related inflammatory markers. Other taxa modulated by treatment, including Rikenellaceae, Alistipes and Parabacteroides have been positively associated with circulating lipid metabolites and inversely associated with body mass index. These correlations suggest a potential role for these taxa in lipid metabolism and energy homeostasis. Moreover, these microbial genera have also been involved in the anti-obesity mechanisms following interventions such as sleeve gastrectomy.55 This aligns with our observation that Xyl and XylGal supplementation significantly reduced body weight and fat accumulation, potentially mediated by shifts in gut microbiota composition.
In particular, Parabacteroides is an emerging probiotic that shows a wide range of metabolic activities, including xylanases and xylosidases.56 Previous works involving in vitro fermentation experiments of beer bagasse fractions suggest that Parabacteroides may ameliorate gut dysbiosis associated with inflammatory diseases.36 Moreover, this genus is inversely associated with obesity among females and participants aged 40–69 years.57 Therefore, Parabacteroides species have been proposed as next-generation probiotics as potential preventive and therapeutic agents against obesity.58 Given the observed metabolic improvements in our study, it is plausible that the increase in Parabacteroides following XylGal supplementation contributed to the reduction in obesity-related inflammation and metabolic dysfunction.
Faecalibaculum, likewise enriched in XylGal-treated obese mice, has been negatively associated with body-weight gain in high fat-induced obesity mice supplemented with apple pectin, contributing to obesity control.59 The presence of this genus in XylGal-treated obese mice further supports its potential role in modulating body composition and metabolic health.
Interestingly, despite these microbial shifts, the LXylGal100 group showed a significantly lower Shannon diversity index compared to obese controls (Fig. 5). While this reduction may appear unexpected, it likely reflects selective enrichment of health-promoting taxa rather than a loss of microbial richness. Indeed, not all decreases in alpha diversity are indicative of dysbiosis. For example, Zarrinpar et al.60 demonstrated that time-restricted feeding reduced diversity in mice while improving metabolic outcomes, and Thaiss et al.61 showed that circadian control of the microbiota can lead to transient reductions in diversity with functional gains. Moreover, Thomas et al.62 reported higher microbial richness in colorectal cancer patients than in healthy individuals, largely due to the presence of ectopic taxa from the oral cavity. Similarly, significantly lower Shannon diversity values have been reported after functional polysaccharide and xylanase supplementation in African catfish63 and mice64 with chronic pancreatitis, respectively, suggesting that these interventions selectively promoted certain microbial populations. These examples reinforce the idea that microbial diversity alone does not fully define gut health, and that composition and function may be more relevant indicators in certain contexts. Thus, the microbial shifts observed in our study may contribute to the metabolic improvements seen in XylGal-treated mice, even in the presence of reduced overall diversity.
Finally, while our study used a standard maintenance diet for the control groups, we acknowledge that a calorie-matched low-fat diet could help more clearly differentiate between effects driven by dietary composition and total energy intake. We have highlighted this point as a limitation and an opportunity for refinement in future study designs.
In conclusion, our study demonstrates that supplementation with Xyl and XylGal improves obesity-related metabolic disturbances, reduces inflammatory markers, and modulates gut microbiota composition in a diet- and dose-dependent manner. Our data suggest that the beneficial effects of XylGal, particularly in the modulation of key microbial genera, could play a pivotal role in reducing metabolic inflammation and adiposity in obese subjects. These findings support further exploration of these compounds as functional ingredients for managing obesity and metabolic dysfunction. Future research should explore the long-term clinical applicability of these compounds and further elucidate the mechanistic links between gut microbiota modulation and improved metabolic outcomes.
Conclusions
Taken together, these findings support a connection between Xyl and XylGal supplementation, metabolic improvements, and gut microbiota modulation. Reductions in body weight, fat accumulation, and inflammatory cytokines observed in vivo were accompanied by specific gut microbiota shifts, including an increase in potentially beneficial bacterial genera such as Lachnospiraceae NK4A136, Eubacterium xylanophilum, and Parabacteroides, which have been previously associated with improved gut barrier function, reduced inflammation, and enhanced lipid metabolism. Simultaneously, a decrease in obesity-related taxa such as Blautia, Colidextribacter, and Eubacterium fissicatena was observed, suggesting a potential role in mitigating gut dysbiosis linked to metabolic disorders. While our study was not designed to establish causality, these microbiota modifications may contribute to the observed metabolic benefits, including improved glucose tolerance and reduced obesity-associated inflammation, and suggest a possible prebiotic role for β-galactosylated xylitol derivatives. Future research should investigate the mechanistic pathways linking gut microbial changes to host metabolism outcomes and explore the long-term clinical relevance of these compounds in human studies. These insights may support their development as functional ingredients for metabolic health management.Author contributions
CS: formal analysis, data curation, writing – original draft, writing – review & editing, methodology; MB: formal analysis, methodology; PDF: formal analysis, methodology; NC: supervision, funding acquisition, conceptualization; BdlR: methodology, validation; RM: methodology, validation; AA: formal analysis, methodology; PU: supervision, writing – original draft, writing – review & editing, resources, methodology, validation; FJM: supervision, funding acquisition, conceptualization, writing – original draft, writing – review & editing.Data availability
The sequencing data supporting the findings of this study have been deposited in the Short Reads Archive (SRA) under BioProject accession number PRJNA1223685 and can be accessed at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1223685. Additional data, including experimental procedures and analysis details, are provided in the ESI† of this article.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Authors acknowledge PID2021-123862OB-100 funded by MCIN/AEI/10.13039/501100011033 and 2021AEP102 funded by MCIN/AEI within the State Plan for Scientific and Technical Research and Innovation (PEICTI).References
- P. González-Muniesa, M. A. Mártinez-González, F. B. Hu, J. P. Després, Y. Matsuzawa, R. J. F. Loos, L. A. Moreno, G. A. Bray and J. A. Martinez, Obesity, Nat. Rev. Dis. Primers., 2017, 3, 17034 CrossRef PubMed.
- X. Wen, B. Zhang, B. Wu, H. Xiao, Z. Li, R. Li, X. Xu and T. Li, Signaling pathways in obesity: mechanisms and therapeutic interventions, Signal Transduct. Target. Ther., 2022, 7, 298 CrossRef PubMed.
- M. Ng, T. Fleming, M. Robinson, B. Thomson, N. Graetz, C. Margono, E. C. Mullany, S. Biryukov, C. Abbafati, S. F. Abera, J. P. Abraham, N. M. Abu-Rmeileh, T. Achoki, F. S. AlBuhairan, Z. A. Alemu, R. Alfonso, M. K. Ali, R. Ali, N. A. Guzman, W. Ammar, P. Anwari, A. Banerjee, S. Barquera, S. Basu, D. A. Bennett, Z. Bhutta, J. Blore, N. Cabral, I. C. Nonato, J. C. Chang, R. Chowdhury, K. J. Courville, M. H. Criqui, D. K. Cundiff, K. C. Dabhadkar, L. Dandona, A. Davis, A. Dayama, S. D. Dharmaratne, E. L. Ding, A. M. Durrani, A. Esteghamati, F. Farzadfar, D. F. Fay, V. L. Feigin, A. Flaxman, M. H. Forouzanfar, A. Goto, M. A. Green, R. Gupta, N. Hafezi-Nejad, G. J. Hankey, H. C. Harewood, R. Havmoeller, S. Hay, L. Hernandez, A. Husseini, B. T. Idrisov, N. Ikeda, F. Islami, E. Jahangir, S. K. Jassal, S. H. Jee, M. Jeffreys, J. B. Jonas, E. K. Kabagambe, S. E. Khalifa, A. P. Kengne, Y. S. Khader, Y. H. Khang, D. Kim, R. W. Kimokoti, J. M. Kinge, Y. Kokubo, S. Kosen, G. Kwan, T. Lai, M. Leinsalu, Y. Li, X. Liang, S. Liu, G. Logroscino, P. A. Lotufo, Y. Lu, J. Ma, N. K. Mainoo, G. A. Mensah, T. R. Merriman, A. H. Mokdad, J. Moschandreas, M. Naghavi, A. Naheed, D. Nand, K. M. Narayan, E. L. Nelson, M. L. Neuhouser, M. I. Nisar, T. Ohkubo, S. Oti, A. Pedroza, D. Prabhakaran, N. Roy, U. Sampson, H. Seo, S. G. Sepanlou, K. Shibuya, R. Shiri, I. Shiue, G. M. Singh, J. A. Singh, V. Skirbekk, N. J. Stapelberg, L. Sturua, B. L. Sykes, M. Tobias, B. X. Tran, L. Trasande, H. Toyoshima, S. van de Vijver, T. J. Vasankari, J. L. Veerman, V. V. Vlassov, S. E. Vollset, T. Vos, C. Wang, X. Wang, E. Weiderpass, A. Werdecker, J. L. Wright, Y. C. Yang, H. Yatsuya, J. Yoon, S. J. Yoon, Y. Zhao, M. Zhou, S. Zhu, A. D. Lopez, C. D. Murray, E. Gakidou and J. G. Caporaso, Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: a systematic analysis for the Global Burden of Disease Study 2013, Lancet, 2014, 384, 766–781 CrossRef PubMed.
- V. S. Malik, B. M. Popkin, G. A. Bray, J. P. Després, W. C. Willett and F. B. Hu, Sugar-sweetened beverages and risk of metabolic syndrome and type 2 diabetes: a meta-analysis, Diabetes Care, 2010, 33, 2477–2483 CrossRef PubMed.
- A. Muñoz-Labrador, O. Hernandez-Hernandez and F. J. Moreno, A review of the state of sweeteners science: the natural versus artificial non-caloric sweeteners debate. Stevia rebaudiana and Siraitia grosvenorii into the spotlight, Crit. Rev. Biotechnol., 2024, 44, 1080–1102 CrossRef PubMed.
- R. K. Johnson, L. J. Appel, M. Brands, B. V. Howard, M. Lefevre, R. H. Lustig, F. Sacks, L. M. Steffen and J. Wylie-Rosett, Dietary sugars intake and cardiovascular health: a scientific statement from the American Heart Association, Circulation, 2009, 120, 1011–1020 CrossRef CAS PubMed.
- World Health Organization, Guideline: Sugars Intake for Adults and Children, WHO Press, Geneva, 2015, vol. 57, pp. 1716–1722 Search PubMed.
- G. R. Gibson, R. Hutkins, M. E. Sanders, S. L. Prescott, R. A. Reimer, S. J. Salminen, K. Scott, C. Stanton, K. Swanson, P. D. Cani, K. Verbeke and G. Reid, Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics, Nat. Rev. Gastroenterol. Hepatol., 2017, 14, 491–502 CrossRef PubMed.
- R. A. Rastall, M. Diez-Municio, S. D. Forssten, B. Hamaker, A. Meynier, F. J. Moreno, F. Respondek, B. Stahl, K. Venema and M. Wiese, Structure and function of non-digestible carbohydrates in the gut microbiome, Benefic. Microbes, 2022, 13, 95–168 CAS.
- S. Yang, C. Wu, Q. Yan, X. Li and Z. Jiang, Nondigestible Functional Oligosaccharides: Enzymatic Production and Food Applications for Intestinal Health, Annu. Rev. Food Sci. Technol., 2023, 14, 297–322 CrossRef CAS PubMed.
- G. Livesey, Health potential of polyols as sugar replacers, with emphasis on low glycaemic properties, Nutr. Res. Rev., 2003, 16, 163–191 CrossRef CAS PubMed.
- A. Gasmi Benahmed, A. Gasmi, M. Arshad, M. Shanaida, R. Lysiuk, M. Peana, I. Pshyk-Titko, S. Adamiv, Y. Shanaida and G. Bjørklund, Health benefits of xylitol, Appl. Microbiol. Biotechnol., 2020, 104, 7225–7237 CrossRef CAS PubMed.
- B. N. Paulino, G. Molina, G. M. Pastore and J. L. Bicas, Current perspectives in the biotechnological production of sweetening syrups and polyols, Curr. Opin. Food Sci., 2021, 41, 36–43 CrossRef CAS.
- S. S. Natah, K. R. Hussien, J. A. Tuominen and V. A. Koivisto, Metabolic response to lactitol and xylitol in healthy men, Am. J. Clin. Nutr., 1997, 65, 947–950 CrossRef CAS PubMed.
- K. Amo, H. Arai, T. Uebanso, M. Fukaya, M. Koganei, H. Sasaki, H. Yamamoto, Y. Taketani and E. Takeda, Effects of xylitol on metabolic parameters and visceral fat accumulation, J. Clin. Biochem. Nutr., 2011, 49, 1–7 CrossRef CAS PubMed.
- E. Flad, A. Altstädt, C. Beglinger, J. F. Rehfeld, L. Van Oudenhove, B. K. Wölnerhanssen and A. C. Meyer-Gerspach, Effects of Oral Xylitol, Sucrose, and Acesulfame Potassium on Total Energy Intake During a Subsequent ad libitum Test Meal: A Randomized, Controlled, Crossover Trial in Healthy Humans, Nutrients, 2025, 17, 484 CrossRef CAS PubMed.
- K. Salli, M. J. Lehtinen, K. Tiihonen and A. C. Ouwehand, Xylitol's health benefits beyond dental health: a comprehensive review, Nutrients, 2019, 11, 1813 CrossRef CAS PubMed.
- B. K. Wölnerhanssen, A. C. Meyer-Gerspach, C. Beglinger and M. S. Islam, Metabolic effects of the natural sweeteners xylitol and erythritol: a comprehensive review, Crit. Rev. Food Sci. Nutr., 2020, 60, 1986–1998 CrossRef.
- J. Juśkiewicz, R. Klewicki and Z. Zduńczyk, Consumption of galactosyl derivatives of polyols beneficially affects cecal fermentation and serum parameters in rats, Nutr. Res., 2006, 26, 531–536 CrossRef.
- R. Klewicki, Effect of selected parameters of lactose hydrolysis in the polyol derivatives, Eng. Life Sci., 2007, 7, 268–274 CrossRef CAS.
- E. Klewicka and R. Klewicki, In vitro fermentation of galactosyl derivatives of polyols by Lactobacillus strains, Czech J. Food Sci., 2009, 27, 65–70 CrossRef CAS.
- L. Lipińska, R. Klewicki, M. Sójka, R. Bonikowski, D. Żyżelewicz, K. Kołodziejczyk and E. Klewicka, Antifungal activity of Lactobacillus pentosus ŁOCK 0979 in the presence of polyols and galactosyl-polyols, Probiotics Antimicrob. Proteins, 2018, 10, 186–200 CrossRef PubMed.
- E. Rosado, P. Delgado-Fernández, B. de Las Rivas, R. Muñoz, F. J. Moreno, N. Corzo and C. Mateo, Production and digestibility studies of β-galactosyl xylitol derivatives using heterogeneous catalysts of LacA β-galactosidase from Lactobacillus plantarum WCFS1, Molecules, 2022, 27, 1235 CrossRef CAS PubMed.
- I. Calvete-Torre, C. Sabater, P. Delgado-Fernández, A. Muñoz-Labrador, B. de las Rivas, R. Muñoz, N. Corzo, F. J. Moreno, A. Margolles and L. Ruiz, Microbiota modulatory properties of novel non-digestible xylitol-derived galacto-oligosaccharides and non-digestible lactulose derived carbohydrate mixtures, LWT-Food Sci. Technol., 2024, 206, 116580 CrossRef CAS.
- H. Tilg, N. Zmora, T. E. Adolph and E. Elinav, The intestinal microbiota fuelling metabolic inflammation, Nat. Rev. Immunol., 2020, 20, 40–54 CrossRef CAS PubMed.
- X. Li, H. C. H. Lau and J. Yu, Microbiota-mediated phytate metabolism activates HDAC3 to contribute intestinal homeostasis, Signal Transduction Targeted Ther., 2020, 5, 211 CrossRef CAS PubMed.
- K. D. Corbin, E. A. Carnero, B. Dirks, D. Igudesman, F. Yi, A. Marcus, T. L. Davis, R. E. Pratley, B. E. Rittmann, R. Krajmalnik-Brown and S. R. Smith, Host-diet-gut microbiome interactions influence human energy balance: a randomized clinical trial, Nat. Commun., 2023, 14, 3161 CrossRef CAS PubMed.
- M. Xiao, C. Zhang, H. Duan, A. Narbad, J. Zhao, W. Chen, Q. Zhai, L. Yu and F. Tian, Cross-feeding of bifidobacteria promotes intestinal homeostasis: a lifelong perspective on the host health, npj Biofilms Microbiomes, 2024, 10, 47 CrossRef CAS PubMed.
- P. Delgado-Fernandez, L. Plaza-Vinuesa, S. Lizasoain-Sánchez, B. de Las Rivas, R. Muñoz, M. L. Jimeno, E. García-Doyagüez, F. J. Moreno and N. Corzo, Hydrolysis of lactose and transglycosylation of selected sugar alcohols by LacA β-galactosidase from Lactobacillus plantarum WCFS1, J. Agric. Food Chem., 2020, 68, 7040–7050 CrossRef CAS PubMed.
- L. C. Julio-Gonzalez, O. Hernández-Hernández, F. J. Moreno, A. Olano and N. Corzo, High-yield purification of commercial lactulose syrup, Sep. Purif. Technol., 2019, 224, 475–480 CrossRef CAS.
- O. Hernández, A. I. Ruiz-Matute, A. Olano, F. J. Moreno and M. L. Sanz, Comparison of fractionation techniques to obtain prebiotic oligosaccharides, Int. Dairy J., 2009, 19, 531–536 CrossRef.
- S. Reagan-Shaw, M. Nihal and N. Ahmad, Dose translation from animal to human studies revisited, FASEB J., 2008, 22, 659–661 CrossRef CAS PubMed.
- R. Benedé-Ubieto, O. Estévez-Vázquez, P. Ramadori, F. J. Cubero and Y. A. Nevzorova, Guidelines and considerations for metabolic tolerance tests in mice, Diabetes, Metab. Syndr. Obes.: Targets Ther., 2020, 13, 439–450 CrossRef PubMed.
- E. Bolyen, J. R. Rideout, M. R. Dillon, N. A. Bokulich, C. C. Abnet, G. A. Al-Ghalith, H. Alexander, E. J. Alm, M. Arumugam, F. Asnicar, Y. Bai, J. E. Bisanz, K. Bittinger, A. Brejnrod, C. J. Brislawn, C. T. Brown, B. J. Callahan, A. M. Caraballo-Rodríguez, J. Chase, E. K. Cope, R. Da Silva, C. Diener, P. C. Dorrestein, G. M. Douglas, D. M. Durall, C. Duvallet, C. F. Edwardson, M. Ernst, M. Estaki, J. Fouquier, J. M. Gauglitz, S. M. Gibbons, D. L. Gibson, A. Gonzalez, K. Gorlick, J. Guo, B. Hillmann, S. Holmes, H. Holste, C. Huttenhower, G. A. Huttley, S. Janssen, A. K. Jarmusch, L. Jiang, B. D. Kaehler, K. B. Kang, C. R. Keefe, P. Keim, S. T. Kelley, D. Knights, I. Koester, T. Kosciolek, J. Kreps, M. G. I. Langille, J. Lee, R. Ley, Y.-X. Liu, E. Loftfield, C. Lozupone, M. Maher, C. Marotz, B. D. Martin, D. McDonald, L. J. McIver, A. V. Melnik, J. L. Metcalf, S. C. Morgan, J. T. Morton, A. T. Naimey, J. A. Navas-Molina, L. F. Nothias, S. B. Orchanian, T. Pearson, S. L. Peoples, D. Petras, M. L. Preuss, E. Pruesse, L. B. Rasmussen, A. Rivers, M. S. Robeson II, P. Rosenthal, N. Segata, M. Shaffer, A. Shiffer, R. Sinha, S. J. Song, J. R. Spear, A. D. Swafford, L. R. Thompson, P. J. Torres, P. Trinh, A. Tripathi, P. J. Turnbaugh, S. Ul-Hasan, J. J. J. van der Hooft, F. Vargas, Y. Vázquez-Baeza, E. Vogtmann, M. von Hippel, W. Walters, Y. Wan, M. Wang, J. Warren, K. C. Weber, C. H. D. Williamson, A. D. Willis, Z. Z. Xu, J. R. Zaneveld, Y. Zhang, Q. Zhu, R. Knight and J. G. Caporaso, Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2, Nat. Biotechnol., 2019, 37, 852–857 CrossRef CAS PubMed.
- I. Calvete-Torre, C. Sabater, M. J. Antón, F. J. Moreno, S. Riestra, A. Margolles and L. Ruiz, Prebiotic potential of apple pomace and pectins from different apple varieties: modulatory effects on key target commensal microbial populations, Food Hydrocolloids, 2022, 133, 107958 CrossRef CAS.
- I. Calvete-Torre, C. Sabater, A. Montilla, F. J. Moreno, S. Riestra, A. Margolles and L. Ruiz, Physichochemical characterization and microbiota modulatory potential of brewer's spent grain and arabinoxylan-derived fractions: a valorization study, LWT-Food Sci. Technol., 2023, 185, 115107 CrossRef CAS.
- L. Lahti and S. Shetty, Tools for microbiome analysis in R. Version 2.1.24, 2017, https://microbiome.github.com/microbiome. last accessed 22/02/2024 Search PubMed.
- P. J. McMurdie and S. Holmes, phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data, PLoS One, 2013, 8, e61217 CrossRef CAS PubMed.
- N. Segata, J. Izard, L. Waldron, D. Gevers, L. Miropolsky, W. S. Garrett and C. Huttenhower, Metagenomic biomarker discovery and explanation, Genome Biol., 2011, 12, 1–18 CrossRef.
- J. N. Paulson, O. C. Stine, H. C. Bravo and M. Pop, Differential abundance analysis for microbial marker-gene surveys, Nat. Methods, 2013, 10, 1200–1202 CrossRef CAS PubMed.
- S. Mandal, W. van Treuren, R. A. White, M. Eggesbø, R. Knight and S. D. Peddada, Analysis of composition of microbiomes: a novel method for studying microbial composition, Microb. Ecol. Health Dis., 2015, 26, 27663 Search PubMed.
- H. Lin and S. D. Peddada, Analysis of compositions of microbiomes with bias correction, Nat. Commun., 2020, 11, 1–11 CrossRef PubMed.
- Y. Cao, Q. Dong, D. Wang, P. Zhang, Y. Liu and C. Niu, microbiomeMarker: an R/Bioconductor package for microbiome marker identification and visualization, Bioinformatics, 2022, 38, 4027–4029 CrossRef CAS PubMed.
- R. Stienstra, L. A. Joosten, T. Koenen, B. van Tits, J. A. van Diepen, S. A. van den Berg, P. C. Rensen, P. J. Voshol, G. Fantuzzi, A. Hijmans, S. Kersten, M. Müller, W. B. van den Berg, N. van Rooijen, M. Wabitsch, B. J. Kullberg, J. W. van der Meer, T. Kanneganti, C. J. Tack and M. G. Netea, The inflammasome-mediated caspase-1 activation controls adipocyte differentiation and insulin sensitivity, Cell Metab., 2010, 12, 593–605 CrossRef CAS PubMed.
- M. Donath and S. Shoelson, Type 2 diabetes as an inflammatory disease, Nat. Rev. Immunol., 2011, 11, 98–107 CrossRef CAS PubMed.
- N. Hariri and L. Thibault, High-fat diet-induced obesity in animal models, Nutr. Res. Rev., 2010, 23, 270–299 CrossRef CAS PubMed.
- S. Hu, L. Wang, D. Yang, L. Li, J. Togo, Y. Wu, Q. Liu, B. Li, M. Li, G. Wang, X. Zhang, C. Niu, J. Li, Y. Xu, E. Couper, A. Whittington-Davies, M. Mazidi, L. Luo, S. Wang, A. Douglas and J. R. Speakman, Dietary fat, but not protein or carbohydrate, regulates energy intake and causes adiposity in mice, Cell Metab., 2018, 28, 415–431 CrossRef CAS PubMed.
- S. Kong, X. Huang, H. Cao, Y. Bai, Q. Che, H. Nie and Z. Su, Anti-obesity effects of galacto-oligosaccharides in obese rats, Eur. J. Pharmacol., 2022, 917, 174728 CrossRef CAS PubMed.
- L. Ma, Y. Ni, Z. Wang, W. Tu, L. Ni, F. Zhuge, A. Zheng, L. Hu, Y. Zhao, L. Zheng and Z. Fu, Spermidine improves gut barrier integrity and gut microbiota function in diet-induced obese mice, Gut Microbes, 2020, 12, 1832857 CrossRef PubMed.
- H. Li, F. Liu, J. Lu, J. Shi, J. Guan, F. Yan, B. Li and G. Huo, Probiotic mixture of Lactobacillus plantarum strains improves lipid metabolism and gut microbiota structure in high fat diet-fed mice, Front. Microbiol., 2020, 11, 512 CrossRef PubMed.
- Z. Li, E. Zhou, C. Liu, H. Wicks, S. Yildiz, F. Razack, Z. Ying, S. Kooijman, D. P. Y. Koonen, M. Heijink, S. Kostidis, M. Giera, I. M. J. G. Sanders, E. J. Kuijper, W. K. Smits, K. W. van Dijk, P. C. N. Rensen and Y. Wang, Dietary butyrate ameliorates metabolic health associated with selective proliferation of gut Lachnospiraceae, bacterium 28–4, JCI Insight, 2023, 8, e166655 CrossRef PubMed.
- D. Ćesić, L. Lugović Mihić, P. Ozretić, I. Lojkić, M. Buljan, M. Šitum, M. Zovak, D. Vidović, A. Mijić, N. Galić and A. Tambić Andrašević, Association of Gut Lachnospiraceae and Chronic Spontaneous Urticaria, Life, 2023, 13, 1280 CrossRef PubMed.
- J. Wei, Y. Zhao, C. Zhou, Q. Zhao, H. Zhong, X. Zhu, T. Fu, L. Pan, Q. Shang and G. Yu, Dietary polysaccharide from Enteromorpha clathrata attenuates obesity and increases the intestinal abundance of butyrate-producing bacterium, Eubacterium xylanophilum, in mice fed a high-fat diet, Polymers, 2021, 13, 3286 CrossRef CAS PubMed.
- A. Mukherjee, C. Lordan, R. P. Ross and P. D. Cotter, Gut microbes from the phylogenetically diverse genus Eubacterium, and their various contributions to gut health, Gut Microbes, 2020, 12, 1802866 CrossRef PubMed.
- C. Liu, Q. Xu, S. Dong, H. Ding, M. Zhong and G. Zhang, New mechanistic insights of anti-obesity by sleeve gastrectomy-altered gut microbiota and lipid metabolism, Front. Endocrinol., 2024, 15, 1338147 CrossRef PubMed.
- C. Sabater, I. Calvete-Torre, M. Villamiel, F. J. Moreno, A. Margolles and L. Ruiz, Vegetable waste and by-products to feed a healthy gut microbiota: current evidence, machine learning and computational tools to design novel microbiome-targeted foods, Trends Food Sci. Technol., 2021, 118, 399–417 CrossRef CAS.
- F. Zhang, X. Zhang, J. Fu, Z. Duan, W. Qiu, Y. Cai, W. Ma, H. Zhou, Y. Chen, J. Zheng and Y. He, Sex- and age-dependent associations between parabacteroides and obesity: evidence from two population cohort, Microorganisms, 2023, 11, 2087 CrossRef CAS PubMed.
- N. G. Vallianou, D. Kounatidis, D. Tsilingiris, F. Panagopoulos and G. S. Christodoulatos, A: Evangelopoulos, I. Karampela and M. Dalamaga, The Role of next-generation probiotics in obesity and obesity-associated disorders: current knowledge and future perspectives, Int. J. Mol. Sci., 2023, 24, 6755 CrossRef CAS PubMed.
- Y. Zhao, J. Bi, J. Yi, J. Peng and Q. Ma, Dose-dependent effects of apple pectin on alleviating high fat-induced obesity modulated by gut microbiota and SCFAs, Food Sci. Hum. Wellness, 2022, 11, 143–154 CrossRef CAS.
- A. Zarrinpar, A. Chaix, S. Yooseph and S. Panda, Diet and feeding pattern affect the diurnal dynamics of the gut microbiome, Cell Metab., 2014, 20, 1006–1017 CrossRef CAS PubMed.
- C. A. Thaiss, D. Zeevi, M. Levy, G. Zilberman-Schapira, J. Suez, A. C. Tengeler, L. Abramson, M. N. Katz, T. Korem, N. Zmora, Y. Kuperman, I. Biton, S. Gilad, A. Harmelin, H. Shapiro, Z. Halpern, E. Segal and E. Elinav, Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis, Cell, 2014, 159, 514–529 CrossRef CAS PubMed.
- A. M. Thomas, P. Manghi, F. Asnicar, E. Pasolli, F. Armanini, M. Zolfo, F. Beghini, S. Manara, N. Karcher, C. Pozzi, S. Gandini, D. Serrano, S. Tarallo, A. Francavilla, G. Gallo, M. Trompetto, G. Ferrero, S. Mizutani, H. Shiroma, S. Shiba, T. Shibata, S. Yachida, T. Yamada, J. Wirbel, P. Schrotz-King, C. M. Ulrich, H. Brenner, M. Arumugam, P. Bork, G. Zeller, F. Cordero, E. Dias-Neto, J. C. Setubal, A. Tett, B. Pardini, M. Rescigno, L. Waldron, A. Naccarati and N. Segata, Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation, Nat. Med., 2019, 25, 667–678 CrossRef CAS PubMed.
- S. J. Gericke, K. Salie, L. de Wet and N. J. Goosen, Effects of dietary supplementation of endo-(1, 4)-β-xylanase in plant-based diets on growth performance, hindgut microbial diversity, and blood chemistry in large on-growing African catfish (Clarias gariepinus), J. Appl. Aquac., 2023, 35, 561–584 CrossRef.
- Y. Hu, C. Teng, S. Yu, X. Wang, J. Liang, X. Bai, L. Dong, T. Song, M. Yu and J. Qu, Inonotus obliquus polysaccharide regulates gut microbiota of chronic pancreatitis in mice, Amb Express, 2017, 7, 1–11 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Table S1. See DOI: https://doi.org/10.1039/d5fo00978b |
| This journal is © The Royal Society of Chemistry 2025 |