Open Access Article
Open Access ArticlePA1b-like peptides alleviate mitochondrial dysfunction induced by glucose toxicity through interaction with VDAC1 in β-cells†
Huizhong
Huang
 a,
Xinyu
Zeng
a,
Liying
Zhang
a,
Hongchang
Cheng
a,
Kanghong
Hu
a,
Xiaoke
Shang
*ab and
Chenguang
Yao
*a
a,
Xinyu
Zeng
a,
Liying
Zhang
a,
Hongchang
Cheng
a,
Kanghong
Hu
a,
Xiaoke
Shang
*ab and
Chenguang
Yao
*a
aSino-German Biomedical Center, Hubei Provincial Key Laboratory of Industrial Microbiology, Cooperative Innovation Center of Industrial Fermentation (Ministry of Education & Hubei Province), Key Laboratory of Fermentation Engineering (Ministry of Education), National “111” Center for Cellular Regulation and Molecular Pharmaceutics, Hubei University of Technology, 430068 Wuhan, Hubei, China. E-mail: yaochenguang@hbut.edu.cn; Tel: +027-59750462
bDepartment of Cardiovascular Surgery, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei, China
First published on 27th February 2025
Abstract
PA1b-like peptides, which are extracted from the seeds of members of the Fabaceae family, display remarkable hypoglycemic and β-cell-protective activities when administered orally. However, the direct targets and mechanisms of action of these peptides in islet β-cells remain unclear. In this study, we found that PA1b-like peptides were mainly distributed in the cotyledon of soybean, rather than in the germ and seed coat. We also identified a direct interaction between PA1b-like peptides and voltage-dependent anion channels (VDACs), with binding energies less than −7 kcal mol−1. Molecular dynamics simulations demonstrated that hydrogen bonding, hydrophobic interactions, and van der Waals forces assist these peptides in forming stable and tight complexes with VDAC1. Moreover, as a member of the PA1B-like peptide family, vglycin (VG) protected mitochondrial function by maintaining the ROS level, ATP production, mitochondrial membrane potential (ΔΨm), intracellular Ca2+ inflow and insulin secretion in β-cells under high glucose stimulation. All these effects were reliant on the direct interaction between VG and VDAC1 in β-cells. This study provides a new strategy for the restoration of mitochondrial function in β-cells under glucose toxicity and establishes a theoretical basis for the treatment of type 2 diabetes (T2D) by PA1b-like peptides.
1 Introduction
PA1b-like peptides are a group of polypeptides containing 37 amino acid residues, which have three intra-molecular disulfide bridges at 3–20, 7–22 and 15–32.1 These peptides belong to the cystine knot inhibitor (ICK) family with a consensus sequence CX3–7CX3–8CX0–7CX1–4CX4–13C.2,3 They are resistant to degradation by proteases (such as trypsin, papain, pepsin, Glu-C protease, and proteinase K).4,5 More than 18 kinds of PA1b-like peptides have been identified in the Fabaceae family, and their homology ranges from 61% (between soybean and Alysicarpus ovalifolius) to 86.1% (between aM1 and Glycine max).6,7 PA1b-like peptides are mainly extracted from soaked/germinated pea or soybean seeds.8–10 However, it is not clear whether these peptides are mainly distributed in the seed coat, germ or cotyledon.PA1b-like peptides, such as PA1F, aglycin (AG), vglycin (VG), iglycin (IG), dglycin (DG), Leg-1, Leg-2 and aM1, were reported to have multiple bioactivity properties in both animal and cell models,11–14 including regulating glucose and lipid metabolism, protecting β-cells from apoptosis, alleviating atherosclerosis, alleviating lipopolysaccharide-induced inflammation, promoting intracellular Ca2+ inflow and insulin secretion, inhibiting the growth of colon cancer cells, etc.15–18 Previous studies have shown that cystine-rich microproteins or Pro-rich peptides are cell-penetrating and enter cells by endocytosis.19–21 Although many biological activities of PA1b-like peptides have been discovered, their direct targets and mechanisms of action in β-cells remain to be further elucidated.
Dun et al. identified a protein at a sequence coverage of 60% that interacts with voltage-dependent anion channel 1 (VDAC1) on the mouse pancreatic cell membrane as an interacting protein of both AG and PA1F through peptide mass fingerprinting.5,8 VDAC1, a pore protein located in the outer mitochondrial membrane, can regulate the voltage-gating activity to control OMM permeability and plays an important role as a controlled passage for adenine nucleotides, Ca2+, and other metabolites into and out of mitochondria.22–24 As a gatekeeper, VDAC1 plays a crucial role in maintaining mitochondrial function and cellular viability by regulating its channel conductance and maintaining mitochondrial membrane potential (ΔΨm) homeostasis.25–27 VDAC1 has also been recognized as a key protein in mitochondria-mediated apoptosis, regulating the release of apoptogenic proteins such as cytochrome c (Cyto c) from the mitochondrial inter-membrane space to the cytosol, as well as interacting with anti-apoptotic proteins (Bcl-2, Bax).28,29 Overexpression of VDAC1 induces apoptotic cell death, suggesting that VDAC1 is a conserved mitochondrial element in death pathways operating in both animal and plant systems.30 Zhang et al. demonstrated that VDAC1 was overexpressed and mistargeted to the β-cell plasma membrane in type 2 diabetes (T2D), which impaired β-cell metabolism and secretion with consequent ATP depletion.31 Therefore, we hypothesize that PA1b-like peptides might protect the function of islet β-cells in T2D by interacting with VDAC1. In this study, we set out to explore how PA1b-like peptides interact with VDAC1 and illustrate how the interaction affects the function of islet β-cells in diabetes. This study will provide a new perspective for the development of PA1b-like peptides as potential drugs targeting VDAC1.
2 Materials and methods
2.1 Materials and reagents
The peptide VG was purified from soybeans in our laboratory as previously reported.10 Soybeans were purchased from Bright Biotechnology (Yichang, China) Co., Ltd. The His-VDAC1 protein was expressed and purified in our laboratory. His-VG (HHHHHHVSCNGVCSPFEMPPCGSSACRCIPYGLVVGNCRHPSG) was synthesized by Sangon Biotech Co., Ltd (Shanghai, China). FITC-VG was prepared by connecting FITC to His-VG. Plasmids plko.1-EGFP-puro, pCDH-cmv-mcs-ef1-puro, pCMV-C-EGFP and pCMV-C-EGFP-Linker-VG were stored in our lab. Ni-NTA agarose (L-2008) for His-tag purification was purchased from Shanghai Lingyin Biotechnology Co., Ltd (Shanghai, China). Anti-VDAC1 polyclonal antibody (Cat no.: 10866-1-AP), anti-His monoclonal antibody (Cat no.: 66005-1-lg), anti-GAPDH monoclonal antibody (Cat no.: 60004-1-Ig), anti-β-actin monoclonal antibody (Cat no.: 66009-1-Ig), HRP-conjugated goat anti-mouse IgG(H + L) (Cat no.: SA00001-1), HRP-conjugated goat anti-rabbit IgG(H + L) (Cat no.: SA00001-2) and 4′,6-diamidino-2-phenylindole (DAPI) were purchased from Proteintech (Wuhan, China). Anti-GFP antibody B-2 (Cat no.: sc:9996) was purchased from Santa Cruz Biotechnology Co., Ltd (Shanghai, China).INS-1 cells were purchased from Yuchun Biology Co., Ltd (Shanghai, China). MIN6/HEK293T cells were stored in our lab. The ROS detection kit (S0033S), ATP detection kit (S0027), Fluo-4 calcium assay kit (S1061S), cell mitochondria isolation kit (C3601) and Mito-Tracker Red CMXRos (C1049B) were purchased from Beyotime Co., Ltd (Shanghai, China). The mitochondrial membrane potential assay kit (JC-1) (KTA4001), Annexin V-AbFluor™ 488/PI apoptosis detection kit (KTA0002) and Annexin V-AbFluor™ 647 apoptosis detection kit (KTA0004) were purchased from Abbkine Scientific, Co., Ltd (Wuhan, China). The wide-range insulin immunoassay kit (MS300) was purchased from Ezassay Biotechnology, Co., Ltd (Shenzhen, China). The cell counting kit-8 (CCK-8) and SuperRT III premixed reverse transcription kit (BL1019B) were purchased from Biosharp, Co., Ltd (Hefei, China).
2.2 Extraction and purification of PA1b-like peptides in soybean
VG was isolated from soybean as previously described.10 Briefly, 200 g of soybean cotyledon was separated and dissolved in 1 L of water and extracted with 0.5 mol L−1 acetic acid for 6 h (pH 4.5), and then filtered with a 0.22 μm membrane. The supernatant soybean extract was obtained by freeze-drying. The content of VG in different parts of soybean was analyzed by reverse-phase high-performance liquid chromatography (RP-HPLC) (Ultimate 3000, Thermo, USA) which was conducted using a Syncronis C18 column (250 mm × 4.6 mm, 5 μm particle, Thermo, USA) at a flow rate of 1 mL min−1 with a linear gradient of buffer A [0.1% trifluoroacetic acid (TFA)] and buffer B [0.1% TFA in acetonitrile (ACN)]. The chromatographic procedure was as follows: 10%–50% A for 0–40 min, 50% A for 40–50 min, 50%–10% A for 50–51 min, and 10% A for 51–60 min. The peaks at 34 min were collected with the preparative liquid phase (Waters 2767, USA) and then analyzed by LC-MS.2.3 Identification of PA1b-like peptides using LC-MS
The freeze-dried powder was dissolved in water and filtered through a 0.22 μm membrane for LC-MS analysis. Positive ion mode was used for MS, and the scanning range was 600–4000 m/z. The acquisition mode was data-dependent acquisition (DDA), and the top 5 precursor ions were selected for secondary fragmentation. MS1 resolution was 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and MS2 resolution was 17
000 and MS2 resolution was 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500. The separation was performed with a ZORBAX Eclipse Plus C18 separation column (50 mm × 2.1 mm, 1.8 μm, Agilent, USA) using solvent A (0.1% formic acid) and solvent B (80% ACN). The elution gradient was as follows: 5% B for 0–1 min, 5%–90% B for 1–4 min, 90% B for 4–7 min, 90%–5% B for 7–7.01 min, and 5% B for 7.01–10 min.
500. The separation was performed with a ZORBAX Eclipse Plus C18 separation column (50 mm × 2.1 mm, 1.8 μm, Agilent, USA) using solvent A (0.1% formic acid) and solvent B (80% ACN). The elution gradient was as follows: 5% B for 0–1 min, 5%–90% B for 1–4 min, 90% B for 4–7 min, 90%–5% B for 7–7.01 min, and 5% B for 7.01–10 min.
2.4 In silico analysis and molecular docking analysis
The Expasy-pI/Mw tool (https://web.expasy.org/computepi/) was utilized to analyze the charge, isoelectric points (pI) and GRAVY (Grand Average of Hydropathy), respectively. The similarity was calculated using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Before the docking analysis (download from https://vina.scripps.edu/downloads/), the peptide ligands and the receptor protein VDAC1 were prepared. hVDAC1 (PDB: 2JK4) was obtained from the RCSB Protein Data Bank (https://www.rcsb.org) as the target protein. The 3D structures of PA1b, AG, VG, IG, DG, aM1, Leg-1 and Leg-2 were constructed using the Swiss-Model website. The protein structure after modeling was scored using the SAVES v6.0 online website, and the PDB file that qualified to illustrate homologous modeling could be used for docking analysis. Before molecular docking, water molecules, native ligands, and other heteroatoms of ligands and receptors were removed using PyMOL. Polar hydrogen was added to the target protein using AutoDock Tools (Version 1.5.7). Subsequently, the receptor and ligand PDB format was converted into PDBQT format with AutoDock Tools (Version 1.5.7). Finally, the active centers were found and set, and 20 models were tested. The parameters were set as follows: center_x = 27.61, center_y = 1.444, and center_z = 5.262. The search space parameters were size_x: 90, size_y: 90, size_z: 90 (the grid spacing was 1.0 Å), and the other parameters were set to the default settings. Hydrogen bonding between ligands and receptors was analyzed using PyMOL software. The ligand–protein interaction diagrams were drawn using LigPlot+ v.2.2.2.5 Molecular dynamics simulation
After the docking study, the AG-VDAC1 and VG-VDAC1 complexes with the best binding energy were used as the initial structures for molecular dynamics (MD) simulations. MD simulations were conducted using GROMACS (version 2020.3-modified).32 The Amber03 force field and the SPC/E water model were used to run MD simulations of the AG/VG-VDAC1 complexes. For preparing MD simulations, the AG-VDAC1 and VG-VDAC1 complexes were placed at the center of a cubic box and dissolved in SPC/E water, and Na+ and Cl− ions were added to neutralize the total charge of the system. Then the systems were energy-minimized using 5000 steps of the steepest descent algorithm. NVT and NPT were used to equilibrate the complex to stabilize the system at a suitable temperature and pressure of 300 K and 1 bar. The total time of MD simulations was 200 ns. After simulation, the root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), radius of gyration (Rg), solvent accessible surface area (SASA), and secondary structure were analyzed using the gmx_rmsd, gmx_rmsf, gmx_gyrate, gmx_sasa, and gmx do_dssp commands, respectively. The RMSD and Rg data were combined and the Gibbs energy landscape was drawn by using the gmx_sham command. Furthermore, the binding free energy between the ligands AG/VG and the receptor VDAC1 was calculated using molecular mechanics Poisson Boltzmann surface area (MMPBSA). The gmx_mmpbsa script was downloaded from https://github.com/Jerkwin/gmxtool/blob/master/gmx_mmpbsa/gmx_mmpbsa.bsh. The specific formulas were as follows:| ΔGbind = ΔEvdw + ΔEelec + ΔGpolar + ΔGnon-polar – TΔS |
2.6 Cell culture
HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM, 12500096, Gibco, USA) with 10% fetal bovine serum (FBS, Biology, Wuhan, China) and 100 U ml−1 penicillin–streptomycin solution (BL505A, Biosharp, Hefei, China) at 37 °C in a humidified incubator containing 5% CO2. INS-1 was maintained in special medium (icell-r036-001b, iCell Bioscience Inc, Shanghai, China) at 37 °C with 5% CO2. The glucose concentration of the basal medium (CK) was 11 mM, and the glucose concentration of the high glucose medium was 20 mM.33 The cell experiments were divided into three groups: CK, H, and H + VG (50 μg mL−1). MIN6 cells were cultured in DMEM supplemented with 10% FBS, 100 U ml−1 penicillin–streptomycin solution and 50 μM β-mercaptoethanol (Aoruisai, Shanghai, China) at 37 °C with 5% CO2. The glucose concentration of the basal medium (CK) was 25 mM, and the glucose concentration of the high glucose medium (H) was 45 mM.34 MIN6 cells were divided into three groups: CK, H, and H + VG (50 μg mL−1). The effects of VG on ATP, reactive oxygen species (ROS), mitochondrial membrane potential (ΔΨm), cell apoptosis, insulin secretion and intracellular Ca2+ content in INS-1 and MIN6 cells under hyperglycemic cultivation were evaluated.2.7 ATP, ROS, ΔΨm and cell apoptosis detection
INS-1 cells and MIN6 cells were seeded in a 6-well plate at a density of 2 × 105 cells in each well. Cells were cultured with or without 50 μg ml−1 VG under high glucose conditions for 24 h, respectively. The content of ATP was measured according to the instructions of the intracellular ATP assay kit (Beyotime, S0026). ROS production was quantified using the ROS detection kit (Beyotime, S0033S) according to the manufacturer's protocol and was detected using a fluorescent enzyme labeling instrument (Thermo, USA). The value of ΔΨm was estimated using the fluorescent probe JC-1 (Abbkine, KTA4001), as described previously.35 Briefly, the treated cells were incubated with 5 μg mL−1 JC-1 for 30 min at 37 °C and detected using a fluorescent enzyme labeling instrument (Thermo, USA). The ratio of red/green fluorescence intensity represents the level of ΔΨm. Cell apoptosis was analyzed using the Annexin V-AbFluor™ 488/PI kit (KTA0002, Abbkine) according to the manufacturer's protocol. Besides, we used the Annexin V-647 and PI kit (KTA0004, Abbkine) to determine the apoptosis of VDAC1 knockdown cells, followed by analysis using a flow cytometer (BD Accuri C6, USA).2.8 Insulin detection assay
The insulin secretion analysis was carried out in accordance with the protocol described by Huang et al.34 INS-1 cells and MIN6 cells were seeded in a 12-well plate with a starting density of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells in each well. The cells were treated with a low glucose medium (5.5 mM in INS-1; 11 mM in MIN6) for 2 h prior to insulin secretion detection, and then treated with VG (50 μg ml−1) for another 2 h. The insulin secretion level was measured using the ELISA kit.
000 cells in each well. The cells were treated with a low glucose medium (5.5 mM in INS-1; 11 mM in MIN6) for 2 h prior to insulin secretion detection, and then treated with VG (50 μg ml−1) for another 2 h. The insulin secretion level was measured using the ELISA kit.
2.9 Ca2+ measurement
Ca2+ measurement was performed as previously described.15 The external Ca2+ solution (NaCl 150 mmol L−1, KCl 2.8 mmol L−1, MgCl2·6H2O2 mmol L−1, CaCl2·2H2O 2.5 mmol L−1, glucose 3 mmol L−1, HEPES 10 mmol L−1) was prepared. The cultured cells were transferred into extracellular fluid containing Ca2+ for 10 min and incubated with the fluorescent Ca2+ indicator Fluo-4AM at 3 μM for 30 min in a CO2 incubator. The cells were washed twice with PBS and the stimulant VG (50 μg ml−1) was added. Images were taken continuously under an SOPTOP ICX41 inverted fluorescence microscope (Sunny Optical, Ningbo, China) for 30 seconds (one picture per second).2.10 Colocalization analysis of FITC-VG and mitochondria
MIN6 cells were pre-grown on a confocal dish (15 mm, Biosharp, Hefei, China) before incubating with 1 μM FITC-VG (1 mg ml−1) at 37 °C for 2 h. The cells were gently washed with PBS and incubated with 200 μM Mito-Tracker Red CMXRos for 30 min. The cells were washed and fixed at room temperature for 10 minutes with 4% paraformaldehyde solution. Nuclei were stained with DAPI (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200) for 5 min. The intracellular distribution of FITC-VG was observed under a confocal laser-scanning microscope (CLSM, Leica, Germany).
200) for 5 min. The intracellular distribution of FITC-VG was observed under a confocal laser-scanning microscope (CLSM, Leica, Germany).
2.11 Immunoprecipitation and immunoblotting
The mitochondria of MIN6 cells were isolated using the cell mitochondria isolation kit (Beyotime, C3601) and cleaved with IP lysate, and then sonicated four times for 4 s at 30 W in an ultrasonic crusher (Ningbo Scientz Biotechnology Co., Ltd). The lysed mitochondria were centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 25 min and the supernatant was collected as mitochondrial protein for the IP assay. 1 mg of synthetic His-VG peptides was dissolved in 200 μl ddH2O and then incubated with Ni-NTA purification agarose beads (200 μl) for 2 h. The purified mitochondrial protein from MIN6 was then added and rocked overnight at 4 °C. After incubation, the samples were centrifuged at 12
000g for 25 min and the supernatant was collected as mitochondrial protein for the IP assay. 1 mg of synthetic His-VG peptides was dissolved in 200 μl ddH2O and then incubated with Ni-NTA purification agarose beads (200 μl) for 2 h. The purified mitochondrial protein from MIN6 was then added and rocked overnight at 4 °C. After incubation, the samples were centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 5 min and the precipitation were washed with TBST for 3 times and then eluted with imidazole (500 mM). The IP samples were subjected to 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE, Epizyme Biotech, Shanghai, China) for immunoblot detection. The antibody information is as follows: rabbit polyclonal anti-VDAC1 antibody (1
000g for 5 min and the precipitation were washed with TBST for 3 times and then eluted with imidazole (500 mM). The IP samples were subjected to 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE, Epizyme Biotech, Shanghai, China) for immunoblot detection. The antibody information is as follows: rabbit polyclonal anti-VDAC1 antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000), anti-His tag antibody (1
2000), anti-His tag antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), HRP-conjugated anti-mouse IgG (1
000), HRP-conjugated anti-mouse IgG (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), and anti-rabbit IgG (1
000), and anti-rabbit IgG (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000). The results were imaged using a ChemiDoc Touch Imaging System (Bio-Rad, 1708370, USA).
000). The results were imaged using a ChemiDoc Touch Imaging System (Bio-Rad, 1708370, USA).
The HEK293T cells were transiently transfected with an empty vector (pCMV-C-EGFP) or plasmid (pCMV-C-EGFP-Linker-VG) for 48 h. The cells were collected, washed with cold PBS and lysed in IP lysis buffer supplemented with protease and phosphatase inhibitors (PMSF). The cell lysates were subjected to the IP assay. The His-VDAC1 protein and Ni-NTA purification agarose beads (200 μl) were mixed for 2 h. The cell lysates were then added and incubated at 4 °C overnight. Rabbit polyclonal anti-VDAC1 antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000, Proteintech) and anti-GFP antibody (1
2000, Proteintech) and anti-GFP antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000, Abcam) were used to detect the endogenous and exogenous VDAC1.
2000, Abcam) were used to detect the endogenous and exogenous VDAC1.
2.12 Surface plasmon resonance (SPR) analysis
The recombinant protein His-VDAC1 (50 μg ml−1) was fixed on the sensor COOH chip by capture-coupling. VG at concentrations of 0, 200, 400, 800, 1600 and 3200 nM were injected into the sample channel at a flow rate of 20 μL min−1 for an association phase of 240 s, followed by 480 s dissociation. The interactions of VG with His-VDAC1 were detected using an OpenSPR™ (Nicoya Lifesciences, Waterloo, Canada) at 25 °C. The wavelength shift corresponded to the change in drug concentration, and a one-to-one diffusion correction model was established. The data were retrieved and analyzed using TraceDrawer software (Ridgeview Instruments AB, Sweden).2.13 Immunofluorescence staining
The immunofluorescence staining experiment for VDAC1 was performed as previously described.35 MIN6 cells were cultured on coverslips and treated with high glucose medium for 48 h. The cells were fixed at room temperature for 10 min with 4% paraformaldehyde solution. Then 0.01% Triton X-100 was used to penetrate the cells for 10 minutes. Next, a primary antibody against VDAC1 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200; Proteintech) was incubated with the cells, and then the Alexa Fluor 488-conjugated goat anti-rabbit IgG (1
200; Proteintech) was incubated with the cells, and then the Alexa Fluor 488-conjugated goat anti-rabbit IgG (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200; Proteintech) was used as the secondary antibody. Finally, 4′,6-diamidino-2-phenylindole (DAPI; 1
200; Proteintech) was used as the secondary antibody. Finally, 4′,6-diamidino-2-phenylindole (DAPI; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200; Proteintech) was added to stain the nuclei. The intracellular expression and sublocalization of VDAC1 were imaged using a confocal laser-scanning microscope (CLSM, Leica, Germany).
200; Proteintech) was added to stain the nuclei. The intracellular expression and sublocalization of VDAC1 were imaged using a confocal laser-scanning microscope (CLSM, Leica, Germany).
2.14 Overexpression of VDAC1 in MIN6 and HEK293T cells
The mouse VDAC1 (mVDAC1) gene was cloned into the pCDH-CMV-MCS-EF1-puro vector. The PCR products of the human VDAC1 (hVDAC1) gene were cloned into the pEGFP vector. For transfection, MIN6 cells were seeded in six-well plates at a density of 2 × 105 cells per well in culture medium and incubated for 12 h until they reached 60–70% confluence. The cells were transfected with 2 μg of plasmids (pCDH or pCDH-VDAC1) using the Neofect™ DNA transfection reagent (Thousand Sunrise, China) following the manufacturer's instructions. Following 48 h of transfection, the cells were collected to detect the expression of VDAC1 through a western blotting experiment.2.15 Lentiviral production and creation of stable cell lines
Lentiviral production was performed as described by Zhou et al..36 The shRNA target design for mouse VDAC1 was GCTACGGCTTTGGCTTAATAA (https://www.sigmaaldrich.cn/). shVDAC1 (short-hairpin) DNA segments were synthesized and then cloned into the pLKO.1 basic vector, according to the pLKO.1 plasmid construction instructions (https://www.addgene.org/protocols/plko/). Specifically, 5 μg of the lentiviral constructs were co-transfected with 5 μg of psPAX2 and 0.5 μg of pMD2.G (viral packaging plasmids) into HEK293T cells in 10 cm dishes to produce lentiviral particles. The viral supernatant was collected after transfection for 48 h and filtered with a 0.22 μm membrane. Subsequently, the viral supernatant was added to MIN6 cells with 10 μg μl−1 of polybrene (Solarbio, H8761) and puromycin resistance screening was performed 48 h after transfection. Then, the clones of puromycin-resistant cells were isolated and expanded for future experiments.2.16 Western blotting
The expression levels of VDAC1 were analyzed in MIN6 and HEK293T cells after plasmid transfection or lentivirus infection. Briefly, the total proteins of the cells were lysed using RIPA buffer (Beyotime, Shanghai, China), and the protein concentration was measured using the Omni-Easy™Instant BCA protein assay kit (Epizyme Biotech, Shanghai, China). 20 μg protein samples were loaded onto 10% SDS-PAGE and then transferred to a PVDF membrane for blotting detection. The antibodies were prepared as follows: VDAC1 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000, Proteintech), GFP (1
2000, Proteintech), GFP (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000, Abcam), GAPDH (1
2000, Abcam), GAPDH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50
50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, Proteintech) and beta-actin (1
000, Proteintech) and beta-actin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, Proteintech). The blots were marked with an enhanced chemiluminescence (ECL) detection kit (Epizyme Biotech, Shanghai, China). The results were imaged using a ChemiDoc Touch Imaging System (Bio-Rad, 1708370, USA).
000, Proteintech). The blots were marked with an enhanced chemiluminescence (ECL) detection kit (Epizyme Biotech, Shanghai, China). The results were imaged using a ChemiDoc Touch Imaging System (Bio-Rad, 1708370, USA).
2.17 Statistical analysis
Statistical analysis and graphing were performed using GraphPad Prism 7.0 (GraphPad Software Inc., San Diego, CA, USA). Differences between two groups were analyzed using Student's t-test and those among multiple groups were analyzed using one-way analysis of variance (ANOVA) followed by Bonferroni's test. Data are shown as the mean ± standard deviation (SD) and a significance level of p < 0.05 was considered statistically significant.3 Results
3.1 Identification of PA1b-like peptides in soybean
As multifunctional bioactive polypeptides, PA1b-like peptide family members were previously extracted from germinated pea or soybean. However, it was not clear where these peptides were located in the seeds of members of the Fabaceae family. Here, the seed coat, germ and cotyledon of germinated soybean were separated, and the content of PA1b-like polypeptides was detected, respectively. We found that the retention time of PA1b-like peptides was around 33 minutes (Fig. 1A), as is the location of the standard VG, one of the PA1b-like peptide family members (red arrow, Fig. 1A). Most PA1b-like peptides were identified in the cotyledon, less in the germ, and almost none in the seed coat (Fig. 1A), which indicates that soybean cotyledon may be the best raw material for preparing PA1b-like peptides.Subsequently, the peptides extracted from soybean cotyledon by RP-HPLC were collected and analyzed by LC-MS. The prominent peaks eluted at 4.14 min were identified to be PA1b-like peptides using typical base peak ion chromatograms (Fig. 1B). More than 10 variants were identified in soybean cotyledon (Fig. 1C). The molecular weights of the top four abundant PA1b-like peptides were 3740.61 Da (m/z = 1247.88), 3902.66 Da (m/z = 1301.90), 3918.65 Da (m/z = 1307.23) and 3950.65 Da (m/z = 1317.89) (Fig. 1D). The peptide residues corresponding to these molecular weights (MW) were matched to the reported sequences AS![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) NGV
NGV![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) SPFEMPP
SPFEMPP![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) GTSA
GTSA![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) R
R![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) IPVGLVIGY
IPVGLVIGY![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) RNPSG (MW = 3742.3 Da),8 AD
RNPSG (MW = 3742.3 Da),8 AD![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) NGA
NGA![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) SPFEVPP
SPFEVPP![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) RSRD
RSRD![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) R
R![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) VPIGLFVGF
VPIGLFVGF![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) IHPTG (MW = 3917.4 Da),11 and VS
IHPTG (MW = 3917.4 Da),11 and VS![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) NGV
NGV![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) SPFEIPP
SPFEIPP![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) GTPL
GTPL![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) R
R![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) IPYGLFVGN
IPYGLFVGN![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) RHPYG (MW = 3958.2 Da).8
RHPYG (MW = 3958.2 Da).8
3.2 Structural analysis of PA1b-like peptides
We summarized several reported PA1b-like peptides and their biological activities (Table 1). PA1b (AS![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) NGV
NGV![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) SPFEMPP
SPFEMPP![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) GTSA
GTSA![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) R
R![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) IPVGLVIGY
IPVGLVIGY![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) RNPSG), an anti-insecticidal peptide from pea seed, had a MW of 3742 Da.1,2 PA1F (AS
RNPSG), an anti-insecticidal peptide from pea seed, had a MW of 3742 Da.1,2 PA1F (AS![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) NGV
NGV![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) SPFEMPP
SPFEMPP![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) GTSA
GTSA![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) R
R![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) IPVGLVIGY
IPVGLVIGY![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) RNPSG), isolated from pea seeds, could dramatically increase blood glucose concentration by subcutaneous injection with a dosage of 5 or 10 μg g−1 (body weight) in normal and T2D mice.8 AG (AS
RNPSG), isolated from pea seeds, could dramatically increase blood glucose concentration by subcutaneous injection with a dosage of 5 or 10 μg g−1 (body weight) in normal and T2D mice.8 AG (AS![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) NGV
NGV![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) SPFEMPP
SPFEMPP![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) GSSA
GSSA![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) R
R![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) IPVGLVVGY
IPVGLVVGY![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) RHPSG) regulated glucose homeostasis in T2D mice by activating the IR/IRS1 pathway.9 VG (VS
RHPSG) regulated glucose homeostasis in T2D mice by activating the IR/IRS1 pathway.9 VG (VS![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) NGV
NGV![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) SPFEMPP
SPFEMPP![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) GSSA
GSSA![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) R
R![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) IPYGLVVGN
IPYGLVVGN![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) RHPSG) could be useful in T2D for restoring impaired insulin signaling, improving glucose tolerance, directly promoting the proliferation of β-cells via the IR/Akt/Erk pathway,10,37 and enhancing fatty acid metabolism in mice by activating the AMPK pathway.35 IG (IS
RHPSG) could be useful in T2D for restoring impaired insulin signaling, improving glucose tolerance, directly promoting the proliferation of β-cells via the IR/Akt/Erk pathway,10,37 and enhancing fatty acid metabolism in mice by activating the AMPK pathway.35 IG (IS![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) NGV
NGV![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) SPFDIPP
SPFDIPP![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) GTPL
GTPL![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) R
R![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) IPAGLFVGK
IPAGLFVGK![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) RHPYG) ameliorated insulin resistance in high-fat diet fed C57BL/6J mice and differentiated 3T3L1 adipocytes with improvement in insulin signaling and mitochondrial function.14 DG (VS
RHPYG) ameliorated insulin resistance in high-fat diet fed C57BL/6J mice and differentiated 3T3L1 adipocytes with improvement in insulin signaling and mitochondrial function.14 DG (VS![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) NGV
NGV![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) SPFDIPP
SPFDIPP![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) GTPL
GTPL![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) R
R![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) IPYGLFVGN
IPYGLFVGN![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) RHPYG) alleviated atherosclerosis in apolipoprotein E-deficient mice.13 Leg-1 (AD
RHPYG) alleviated atherosclerosis in apolipoprotein E-deficient mice.13 Leg-1 (AD![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) NGA
NGA![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) SPFEVPP
SPFEVPP![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) RSRD
RSRD![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) R
R![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) VPIGLFVGF
VPIGLFVGF![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) IHPTG) and Leg-2 (AD
IHPTG) and Leg-2 (AD![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) NGA
NGA![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) SPFEMPP
SPFEMPP![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) RSRD
RSRD![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) R
R![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) VPIGLVAGF
VPIGLVAGF![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) IHPTG) were also identified in soybean and significantly induced 2DG uptake in mouse C2C12 cells.11 aM1 (VD
IHPTG) were also identified in soybean and significantly induced 2DG uptake in mouse C2C12 cells.11 aM1 (VD![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) SGA
SGA![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) SPFEVPP
SPFEVPP![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) GSRD
GSRD![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) R
R![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) IPIGLVVGF
IPIGLVVGF![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) IYPTG) targeted Akt signaling and alleviated insulin resistance.20
IYPTG) targeted Akt signaling and alleviated insulin resistance.20
| PA1b-like peptides | Species | Amino acid sequence | Mass (Da)a | Chargeb | PIc | GRAVYd | Similarity %e | Ref. |
|---|---|---|---|---|---|---|---|---|
| a Mass (Da) = reported mass. b Charge: the total charge was the sum of positive (lysine, arginine, and histidine residues) and negative (glutamate and aspartate residues) charges present in the sequence. c PI was calculated using ProtParam (https://web.expasy.org/protparam/). d GRAVY (Grand Average of Hydropathy) was calculated using ProtParam. e Similarity was calculated online using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). | ||||||||
| PA1F | Pea |
AS![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) NGV NGV![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) SPFEMPP SPFEMPP![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) GTSA GTSA![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) R R![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) IPVGLVIGY IPVGLVIGY![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) RNPSG RNPSG
|
3742.3 | +1 | 7.81 | 0.38 | 100.00 | 8 |
| Aglycin (AG) | Soybean |
AS![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) NGV NGV![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) SPFEMPP SPFEMPP![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) GSSA GSSA![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) R R![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) IPVGLVVGY IPVGLVVGY![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) RHPSG RHPSG
|
3742.3 | +1 | 7.82 | 0.38 | 91.89 | 9 |
| Vglycin (VG) | Soybean |
VS![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) NGV NGV![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) SPFEMPP SPFEMPP![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) GSSA GSSA![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) R R![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) IPYGLVVGN IPYGLVVGN![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) RHPSG RHPSG
|
3786.4 | +1 | 7.77 | 0.23 | 83.78 | 10 |
| Iglycin (IG) | Soybean seed |
IS![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) NGV NGV![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) SPFDIPP SPFDIPP![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) GTPL GTPL![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) R R![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) IPAGLFVGK IPAGLFVGK![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) RHPYG RHPYG
|
3880.0 | +2 | 8.34 | 0.37 | 70.27 | 14 |
| Dglycin (DG) | Soybean |
VS![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) NGV NGV![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) SPFDIPP SPFDIPP![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) GTPL GTPL![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) R R![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) IPYGLFVGN IPYGLFVGN![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) RHPYG RHPYG
|
3940.0 | +1 | 7.76 | 0.29 | 70.27 | 13 |
| Leg-1 | Green soybean |
AD![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) NGA NGA![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) SPFEVPP SPFEVPP![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) RSRD RSRD![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) R R![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) VPIGLFVGF VPIGLFVGF![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) IHPTG IHPTG
|
3917.4 | 0 | 6.74 | 0.27 | 59.46 | 11 |
| Leg-2 | Green soybean |
AD![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) NGA NGA![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) SPFEMPP SPFEMPP![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) RSRD RSRD![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) R R![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) VPIGLVAGF VPIGLVAGF![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) IHPTG IHPTG
|
3873.4 | 0 | 6.74 | 0.18 | 64.86 | 11 |
| α-Astratide (aM1) | Huang qi |
VD![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) SGA SGA![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) SPFEVPP SPFEVPP![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) GSRD GSRD![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) R R![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) IPIGLVVGF IPIGLVVGF![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) IYPTG IYPTG
|
3811.8 | −1 | 4.56 | 0.61 | 62.16 | 20 |
We found that these peptides had 37 amino acid residues and contained highly conserved six cysteine residues with the homology ranging from 62% to 92%, forming three pairs of disulfide bonds C3–C20, C7–C22, and C15–C32 (Table 1). The isoelectric point (pI) of PA1b-like peptides ranged from 4.56 (aM1) to 8.34 (IG). PA1b, PA1F, AG, VG, IG, and DG were positively charged. The two peptides Leg-1 and Leg-2 were neutral, whereas aM1 was negatively charged. Besides, these PA1b-like peptides had low GRAVY values, indicating that they had strong hydrophilicity (Table 1). Based on the cysteine-spacing pattern, the peptides formed five conserved inter-cysteinyl loops. A sequence logo of PA1b-like peptides was generated to illustrate the similarity and occurrence of the amino acid positions (Fig. 2A). In general, in addition to the six conserved cysteine residues, five prolines (Pro9, Pro13, Pro14, Pro24, and Pro35), four glycines (Gly5, Gly26, Gly30 and Gly37), Ser8, Phe10, Arg21, and Leu27 were absolutely conserved in all PA1b-like peptides (Fig. 2A). Secondary structure sequence alignment of PA1b-like peptides was modeled using ESPript3.0, which revealed that these peptides had three β-fold regions (Fig. 2B), presenting lots of potential for interaction with target proteins. PA1b-like peptides had a T-knot scaffold containing 3 β-strands (βA: Ala6-Ser8; βB: Cys20-Pro24; βC: Gly30-His34), with two adjacent β-strands, βB and βC, connected by a distorted type-I β-turn around Gly26-Val29. The β-strands made up a two-stranded antiparallel β-sheet which is stacked by a long N-terminal loop containing βA, 2 type-I β-turns around Ser8–Glu11 and Ser17–Cys20, and a cis Pro at position 13 (Fig. 2B).38,39
 | ||
| Fig. 2 Advanced structural analysis of PA1b-like peptides. (A) Conservation analysis of amino acid residues in PA1b-like peptides using WebLogo – Create Sequence Logos (berkeley.edu). Conservation is indicated by the overall height of the stack, whereas the frequency of amino acids at specific positions is revealed by the height of the symbols within the stack. The colors red, blue, pink, and green indicate cysteine, negatively charged residues (D and E), positively charged residues (H, K, R), and aromatic amino acid residues (F, W, Y), respectively. (B) Secondary structure sequence alignment of PA1b-like peptides using ESPript3.0 (https://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). η, β-turn; β, β-fold; TT, β-sheet angle; green numbers represent the order of the three disulfide bonds. (C) 3D structural modeling of PA1b-like peptides. PA1F (PDB: 1P8B) and Leg-1 (PBD:1JU8) were downloaded from the PDB database. The 3D structures of the peptides were generated using Swiss-Model online (https://swissmodel.expasy.org/) and visualized using PyMOL (download from https://www.pymol.org/). (D) Structural similarity of the PA1b-like peptides was evaluated using RMSD. | ||
Three-dimensional (3D) structures of AG, VG, IG, DG, aM1 and Leg-2 were built using Swiss-Model online with the solution structure of PA1B (PDB: 1P8b) as a template, and then analyzed using PyMOL software (Fig. 2C). The sequence of Leg-1 was the same as the solution structure of leginsulin (PDB: 1JU8). RMSD values (ranging from 0.073 to 1.799) showed that the 3D structures of these PA1b-like peptides were highly similar (Fig. 2D). It is speculated that they may have similar biological functions or similar targets.
3.3 Molecular docking analysis of PA1b-like peptides and VDAC1
Previous studies showed that PA1b-like peptides AG and PA1F may have an affinity for VDAC1 in pancreatic membrane protein extracts from porcine and mouse pancreases.5,8 In the present study, the interactions of PA1b-like peptides and VDAC1 (PDB: 2jk4) were further investigated by molecular docking. The ligands (AG, VG, IG, DG, aM1, Leg-1, Leg-2 and PA1F) were prepared using Swiss-Model online as mentioned in Fig. 2C. The binding energies of the interaction complexes were found to range from −7.324 (PA1F/VDAC1) to −9.256 (AG/VDAC1) kcal mol−1 (Table 2). AG has the lowest binding energy (−9.256 kcal mol−1), while VG shows the second lowest binding energy (−9.254 kcal mol−1) (Table 2). Human VDAC1 (hVDAC1) showed an uneven number of 19 strands and 18 loop-like connections with a less distinct distribution of shortened loop structures to one side of the membrane (Fig. 3A). The horizontal dimensions of hVDAC1 are 3.5 × 3.1 nm (Fig. 3A).40 We illustrated the theoretical binding pattern of the peptide AG/VG to the VDAC1 binding sites. The docking models for AG/VG demonstrated the presence of hydrophobic interactions and hydrogen bonding (Fig. 3A and C). The amino acid residues Ala1, Ser2, Ser18, and Gly37 in AG formed hydrogen bonds with the VDAC1 active sites Glu169, Asn171, Asp133, Thr9, and Asn127 (Fig. 3B). The amino acid residues Ser2, Asn4, Val6, Asn31, Tyr25 and Gly37 in VG formed hydrogen bonds with the VDAC1 active sites Lys122, Lys118, Glu87, Arg142, and Asn15 (Fig. 3D). The docking diagram of other ligands (IG, DG, aM1, Leg-1, Leg-2 and PA1F) and VDAC1 is shown in Fig. S1.† Structurally, the pore on VDAC1 for the exchange of metabolites blocked by PA1b-like peptides may affect the mitochondrial function in cells.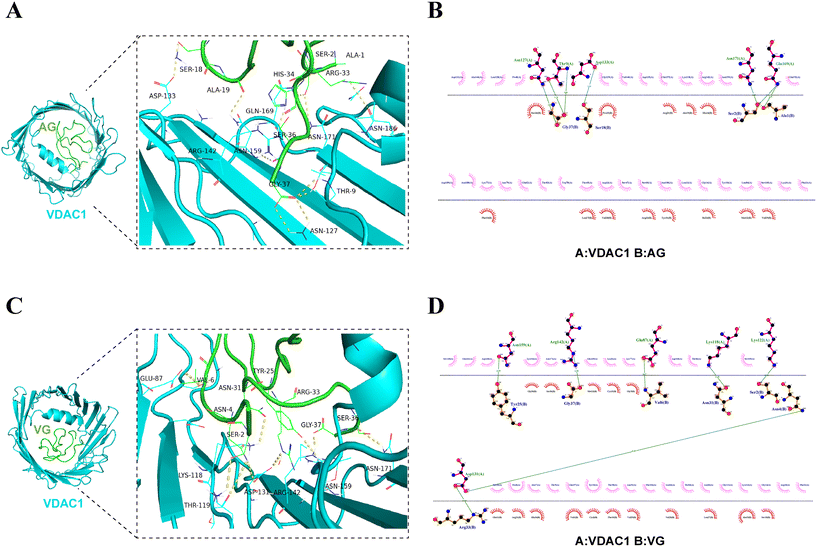 | ||
| Fig. 3 Molecular docking results of peptides AG/VG and VDAC1 (PDB: 2JK4). (A and C) Three-dimensional models of the interaction between AG/VG and VDAC1. (B and D) The binding geometry analysis of AG/VG and VDAC1. Three-dimensional models of the interactions between AG/VG and VDAC1 are drawn using PyMOL, and 2D analyses were conducted using LigPlot+. Green dashed lines represent H-bonds. Red-dashed lines represent hydrophobic interactions. Full side chains are shown for residues involved in H-bond formation. | ||
| Ligands | Receptor | Binding free energya (kcal mol−1) |
|---|---|---|
| a Binding free energy <−7.0 kcal mol−1 indicated a strong interaction between the receptors and the ligands. | ||
| AG | VDAC1 (2JK4) | −9.256 |
| VG | VDAC1 | −9.254 |
| IG | VDAC1 | −8.497 |
| Leg-2 | VDAC1 | −8.239 |
| DG | VDAC1 | −9.108 |
| aM1 | VDAC1 | −7.765 |
| Leg-1 (1JU8) | VDAC1 | −7.408 |
| PA1F (1P8B) | VDAC1 | −7.324 |
3.4 Molecular dynamics simulation analysis
To investigate the binding behavior of PA1b-like peptides and VDAC1 at the atomic level, we conducted a 200 ns molecular dynamics simulation. The RMSD, RMSF, Rg, SASA, secondary structure and binding free energy of the complexes were calculated (Fig. 4).3.5 VG directly interacted with VDAC1 in mitochondria of β-cells
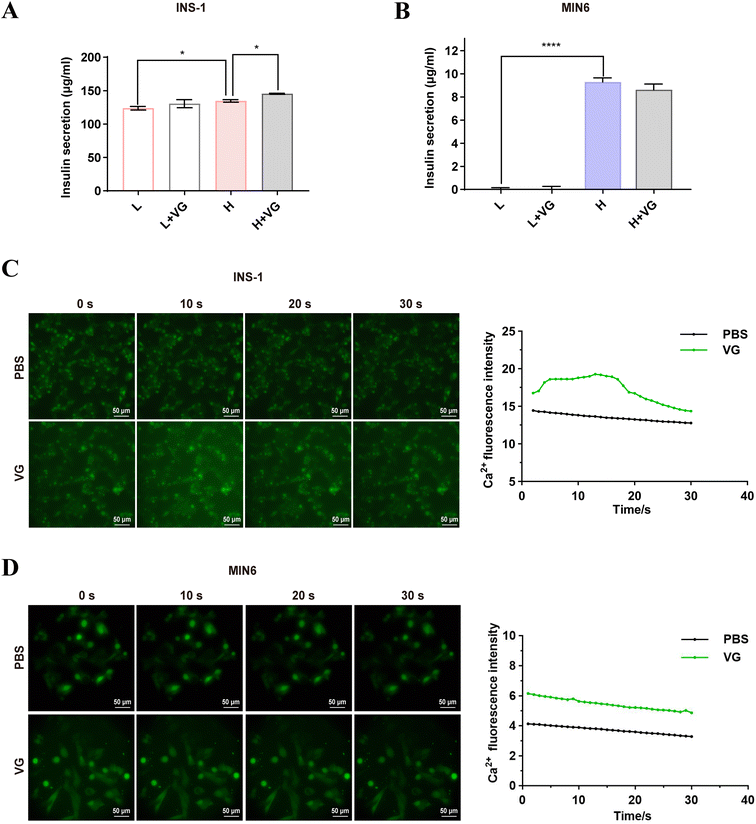
The molecular docking and molecular dynamics simulations showed a strong affinity of PA1b-like peptides and VDAC1. Next, we wanted to determine whether these peptides can interact with VDAC1 protein in vivo and in vitro by colocalization, immunoprecipitation and SPR analysis. To validate this prediction, we synthesized a fusion peptide (His-VG) that contained a 6× His tag at the N terminal. The synthesized His-VG had a purity >95%, and a molecular weight of 4608.21 Da (Fig. S3A and B†). We also prepared a FITC-labeled peptide VG (FITC-VG), which was confirmed by MALDI-TOF MS (Fig. S3C†). Previous studies showed that cystine-rich microproteins or proline-rich peptides are cell-penetrating.20 Live-cell imaging of MIN6 showed that FITC-VG was internalized and distributed throughout the cytoplasm, and co-localized with mitochondria (Fig. 5A). The co-IP results showed that His-VG can pull down the VDAC1 protein from the mitochondrial lysate in MIN6 cells (Fig. 5B). To further demonstrate the universality of this interaction, we overexpressed VG by transfecting plasmid pEGFP-VG into HEK293T cells. The size of GFP-VG was about 30.7 kDa, which was in line with the expected size. We further overexpressed VG in HEK293T cells and found that VG can be pulled down by His-VDAC1 (Fig. 5C). SPR results showed that VDAC1 can bind VG with an affinity constant of 653 nM (Fig. 5D). These results strongly indicate a direct interaction between the peptide VG and VDAC1 in β-cells.3.6 VG ameliorated the high glucose-induced mitochondrial dysfunction in β-cells
Long-term high glucose conditions can lead to elevation of ROS, mitochondrial dysfunction, and eventually induce the deregulation of metabolism and gradual apoptosis in β-cells.31,44 In our study, high glucose cultivation mildly promoted apoptosis in MIN6 and INS-1 cells, which was in line with the previous reports.45 Comparatively, VG treatment reduced the percentages of apoptotic cells in both cell lines (Fig. 6A and B). Similarly, high glucose cultivation increased the ROS level, a trigger of mitochondrial dysfunction and cell apoptosis in INS-1 and MIN6 cells, while VG suppressed ROS production (Fig. 6C and D). We also found that high glucose cultivation damaged the mitochondrial function, as evidenced by the decreased levels of ATP and ΔΨm. However, VG treatment reversed the two events in INS-1 and MIN6 cells (Fig. 6E–H). The results suggested that VG ameliorated high-glucose induced mitochondrial dysfunction and apoptosis. VDAC1 is a key structural component of the mitochondrial permeability transition pore.30 The mitochondrial permeability transition pore opening directly impairs mitochondrial cellular integrity, mitochondrial membrane potential and ATP stores.46 Our IP experiment demonstrated that VDAC1 was the interacting protein of the peptide VG. Here, VG improves mitochondrial function, which may be related to VDAC1.3.7 VG promoted insulin secretion and intracellular Ca2+ in β-cells
Stimulation by glucose and nutrients (amino acids, free fatty acids) in plasma modulates the entry of Ca2+ through the opening of L-type voltage-dependent calcium channels, which leads to insulin secretion.47,48 High glucose (20 mM) stimulated an increase in insulin secretion in INS-1 cells compared to low glucose (5.5 mM). VG treatment promoted insulin secretion under high glucose cultivation, rather than under low glucose cultivation (Fig. 7A). However, in MIN6 cells, we did not detect an obvious influence of VG on the release of insulin both under high glucose and low glucose cultivation (Fig. 7B). These results indicate that VG can restore the function of β-cells under high glucose while avoiding hypoglycemia. We found that the stimulation with the peptides significantly increased intracellular green fluorescence, that is, the concentration of Ca2+ instantaneously increased in INS-1 cells (Fig. 7C), rather than in MIN6 cells (Fig. 7D). The results were consistent with what has been reported before. PA1b can open β-cell voltage-dependent L-type Ca2+ channels, and stimulate cell secretion.153.8 VG protected mitochondrial function through VDAC1 in β-cells
It has been reported that VDAC1 is upregulated in islets from T2D and non-diabetic organ donors under glucotoxicity conditions.31 Given that we demonstrated VDAC1 is a direct target of VG, we then investigated whether VG restores the mitochondrial function through targeting VDAC1. Immunofluorescence results showed that the expression of VDAC1 was increased under high glucose cultivation (Fig. 8A). Long-term high glucose cultivation induced an increase in caspase 3, indicating apoptosis in MIN6 cells (Fig. 8B). Overexpression of VDAC1 elevated the mitochondrial ΔΨm in MIN6 cells, and VG treatment further increased ΔΨm (Fig. 8C and D). We also observed the same phenomenon in HEK293T cells (Fig. 8E–G). Moreover, we knocked down VDAC1 in MIN6 cells and found that ΔΨm decreased significantly (Fig. 8H and I). VG treatment could further reduce ΔΨm in MIN6 cells with knockdown of VDAC1 (Fig. 8I). These results provided evidence that VG may modulate ΔΨm by targeting VDAC1. Knockdown of VDAC1 decreased cell viability under cultivation with 25 mM glucose. VG did not affect cell viability under cultivation with 45 mM glucose (Fig. 8J). Compared with the control group, the ΔΨm of MIN6 cells with VDAC1 knockdown did not decrease significantly under high glucose cultivation (Fig. 8K). Moreover, VG treatment could not improve ΔΨm (Fig. 8K). High glucose significantly increased ROS in wild-type MIN6 cells, and VG treatment reversed this. However, under high glucose cultivation, knockdown of VDAC1 led to a significant decrease in ROS levels, while VG treatment elevated them (Fig. 8L). Similarly, under high glucose cultivation, the percentage of apoptotic cells in VDAC1-knockdown MIN6 cells decreased, and VG could not exhibit a protective effect (Fig. 8M). These results indicated that VG protected the mitochondrial function through VDAC1 in β-cells.4 Discussion
Currently, approximately 45% of all clinically approved drugs are plant-derived, with a molecular weight <1 kDa.49 37% of synthetic drugs are inspired by small molecule metabolites.50 In contrast, peptide drugs are preferred by the public due to their higher specificity and fewer toxic side effects. Previous studies showed that PA1b-like peptides, also called 37-amino-acid hormone-like ICK paradigm peptides, have high stability and resist protease hydrolysis.8,17,51 C3–C20 and C7–C22 of the three disulfide bridge bonds of PA1b-like peptides formed a skeletal structure, and C15–C32 penetrated the skeleton.1,2 We analyzed the structure of some PA1b-like peptides (PA1F, AG, VG, IG, DG, Leg-1, Leg-2 and aM1) and found that in addition to the six conserved cysteine residues, five prolines (Pro9, Pro13, Pro14, Pro24, and Pro35), four glycines (Gly5, Gly26, Gly30 and Gly37), Ser8, Phe10, Arg21, and Leu27 were absolutely conserved.VDAC1, an outer mitochondrial membrane protein, is involved in energy homeostasis by transporting metabolites.52 The connection from the α-helix to the first β-strand is a flexible and highly conserved sequence (Gly21-Tyr22-Gly23-Phe24-Gly25), which increased mobility in this region.53 Specifically, rotation at this hinge region to move the helix away from the wall of the pore would place the N-terminal domain into a position of greater obstruction within the cavity where it would block the transport of metabolites while still permitting ion flux.54 High blood glucose levels can lead to increased expression of VDAC1, which damages mitochondrial function by elevating oxidative stress in β-cells, and increases the risk of diabetic complications.31,55 VDAC1 is considered as a promising therapeutic target to regulate vital metabolic processes and apoptotic cell death.56,57 In this work, we found that nearly all the PA1b-like peptides had a strong interaction with VDAC1 by blocking the ion channel, which may inhibit the expression or oligomerization of VDAC1 in β-cells, thereby protecting the mitochondria from damage under high glucose conditions. Our results indicated that VG improved mitochondrial function via rebuilding mitochondrial membrane potential in both MIN6 and INS-1 cells, especially under a stimulation of high glucose.
We also found that the mitochondrial membrane potential was not affected by high glucose in VDAC1 knockdown cells. We speculated that the significant decrease in VDAC1 contributes to the normalization of mitochondrial function under high glucose stimulation. This result is consistent with the previous research that silencing VDAC1 led to a less pronounced manifestation of all the signs of damage to mitochondria in human skin fibroblasts exposed to high glucose.58 It suggests that reduced expression of VDAC1 in the cell contributes to the activation of compensatory mechanisms for increased glucose utilization, which may include, for example, an increase in mitochondrial mass. The PA1b-like peptide VG not only restored mitochondrial function, but also inhibited apoptosis in β-cells, indicating that VG protects islet β-cells as a VDAC1 inhibitor.
Author contributions
Huizhong Huang: methodology, conceptualization, investigation, formal analysis, and writing – original draft. Xinyu Zeng, Liying Zhang and Hongchang Cheng: methodology and data curation. Kanghong Hu: comments and suggestion. Xiaoke Shang and Chenguang Yao: writing – review and editing. All authors approved the final version for submission.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
The authors report no conflicts of interest.Acknowledgements
This study is supported by Open Project Funding of the Key Laboratory of Fermentation Engineering (Ministry of Education, Grant No. 202409FE19), and the Collaborative Grant-in-Aid of the HBUT National “111” Center for Cellular Regulation to C. Yao.References
- T. J. Higgins, P. M. Chandler, P. J. Randall, D. Spencer, L. R. Beach, R. J. Blagrove, A. A. Kortt and A. S. Inglis, Gene structure, protein structure, and regulation of the synthesis of a sulfur-rich protein in pea seeds, J. Biol. Chem., 1986, 261, 11124–11130 CrossRef CAS PubMed.
- L. Jouvensal, L. Quillien, E. Ferrasson, Y. Rahbé, J. Guéguen and F. Vovelle, PA1b, an insecticidal protein extracted from pea seeds (Pisum sativum): 1H-2-D NMR study and molecular modeling, Biochemistry, 2003, 42, 11915–11923 CrossRef CAS PubMed.
- D. J. Craik, N. L. Daly and C. Waine, The cystine knot motif in toxins and implications for drug design, Toxicon, 2001, 39, 43–60 Search PubMed.
- F. Gressent, P. Da Silva, V. Eyraud, L. Karaki and C. Royer, Pea Albumin 1 subunit b (PA1b), a promising bioinsecticide of plant origin, Toxins, 2011, 3, 1502–1517 Search PubMed.
- X. P. Dun, J. H. Wang, L. Chen, J. Lu, F. F. Li, Y. Y. Zhao, E. Cederlund, G. Bryzgalova, S. Efendic, H. Jörnvall, Z. W. Chen and T. Bergman, Activity of the plant peptide aglycin in mammalian systems, FEBS J., 2007, 274, 751–759 CrossRef CAS PubMed.
- S. Louis, B. Delobel, F. Gressent, G. Duport, O. Diol, I. Rahioui, H. Charles and Y. Rahbé, Broad screening of the legume family for variability in seed insecticidal activities and for the occurrence of the A1b-like knottin peptide entomotoxins, Phytochemistry, 2007, 68, 521–535 CrossRef CAS PubMed.
- J. Huang, K. H. Wong, S. V. Tay, A. Serra, S. K. Sze and J. P. Tam, Astratides: Insulin-Modulating, Insecticidal, and Antifungal Cysteine-Rich Peptides from Astragalus membranaceus, J. Nat. Prod., 2019, 82, 194–204 Search PubMed.
- X. P. Dun, F. F. Li, J. H. Wang and Z. W. Chen, The effect of pea albumin 1F on glucose metabolism in mice, Peptides, 2008, 29, 891–897 Search PubMed.
- J. Lu, Y. Zeng, W. Hou, S. Zhang, L. Li, X. Luo, W. Xi, Z. Chen and M. Xiang, The soybean peptide aglycin regulates glucose homeostasis in type 2 diabetic mice via IR/IRS1 pathway, J. Nutr. Biochem., 2012, 23, 1449–1457 CrossRef CAS PubMed.
- H. Jiang, J. Feng, Z. Du, H. Zhen, M. Lin, S. Jia, T. Li, X. Huang, C. G. Ostenson and Z. Chen, Oral administration of soybean peptide Vglycin normalizes fasting glucose and restores impaired pancreatic function in Type 2 diabetic Wistar rats, J. Nutr. Biochem., 2014, 25, 954–963 CrossRef CAS PubMed.
- T. Hashidume, T. Sakano, A. Mochizuki, K. Ito, S. Ito, Y. Kawarasaki and N. Miyoshi, Identification of soybean peptide leginsulin variants in different cultivars and their insulin-like activities, Sci. Rep., 2018, 8, 16847 CrossRef PubMed.
- C. Yao, G. Ye, Q. Yang, Z. Chen and M. Yang, The Disulfide Bond-Mediated Cyclization of Oral Peptides, Curr. Protein Pept. Sci., 2024, 25, 438–442 CrossRef CAS PubMed.
- H. Zhao, P. Dan, J. Xi, Z. Chen, P. Zhang, W. Wei and Y. Zhao, Novel soybean polypeptide dglycin alleviates atherosclerosis in apolipoprotein E-deficient mice, Int. J. Biol. Macromol., 2023, 251, 126347 CrossRef CAS PubMed.
- Y. Wu, R. Zhao, M. Li, H. Li, Z. Chen and Y. Zhao, Novel soybean peptide iglycin ameliorates insulin resistance of high-fat diet fed C57BL/6J mice and differentiated 3T3L1 adipocytes with improvement of insulin signaling and mitochondrial function, 2022 Search PubMed.
- Z. Hu, X. Dun, M. Zhang, H. Zhu, L. Xie, Z. Wu, Z. Chen and T. Xu, PA1b, a plant peptide, induces intracellular [Ca2+] increase via Ca2+ influx through the L-type Ca2+ channel and triggers secretion in pancreatic beta cells, Sci. China, Ser. C:Life Sci., 2007, 50, 285–291 CrossRef CAS PubMed.
- C. Gao, R. Sun, Y. R. Xie, A. L. Jiang, M. Lin, M. Li, Z. W. Chen, P. Zhang, H. Jin and J. P. Feng, The soy-derived peptide Vglycin inhibits the growth of colon cancer cells in vitro and in vivo, Exp. Biol. Med., 2017, 242, 1034–1043 Search PubMed.
- K. Hu, H. Huang, H. Li, Y. Wei and C. Yao, Legume-Derived Bioactive Peptides in Type 2 Diabetes: Opportunities and Challenges, Nutrients, 2023, 15, 1096 CrossRef CAS PubMed.
- P. Dan, H. Zhao, J. Xi and Y. Zhao, Bioinsecticide PA1b alleviates lipopolysaccharide-induced inflammation and impairment of bovine mammary epithelial cells, Pestic. Biochem. Physiol., 2024, 201, 105866 CrossRef CAS PubMed.
- A. Kam, S. Loo, B. Dutta, S. K. Sze and J. P. Tam, Plant-derived mitochondria-targeting cysteine-rich peptide modulates cellular bioenergetics, J. Biol. Chem., 2019, 294, 4000–4011 CrossRef CAS PubMed.
- B. Dutta, S. Loo, A. Kam and J. P. Tam, Plant-derived cell-penetrating microprotein α-astratide aM1 targets Akt signaling and alleviates insulin resistance, Cell. Mol. Life Sci., 2023, 80, 293 CrossRef CAS PubMed.
- B. Dutta, S. Loo, A. Kam, S. K. Sze and J. P. Tam, Ginsentide TP1 Protects Hypoxia-Induced Dysfunction and ER Stress-Linked Apoptosis, Cells, 2023, 12, 1401 CrossRef CAS PubMed.
- T. K. Rostovtseva, K. L. Sheldon, E. Hassanzadeh, C. Monge, V. Saks, S. M. Bezrukov and D. L. Sackett, Tubulin binding blocks mitochondrial voltage-dependent anion channel and regulates respiration, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 18746–18751 CrossRef CAS PubMed.
- D. Ben-Hail, R. Palty and V. Shoshan-Barmatz, Measurement of mitochondrial Ca2+ transport mediated by three transport proteins: VDAC1, the Na+/Ca2+ exchanger, and the Ca2+ uniporter, Cold Spring Harb. Protoc., 2014, 2014, 161–166 Search PubMed.
- P. Sander, T. Gudermann and J. Schredelseker, A Calcium Guard in the Outer Membrane: Is VDAC a Regulated Gatekeeper of Mitochondrial Calcium Uptake?, Int. J. Mol. Sci., 2021, 22, 946 CrossRef CAS PubMed.
- F. Tomasello, A. Messina, L. Lartigue, L. Schembri, C. Medina, S. Reina, D. Thoraval, M. Crouzet, F. Ichas, V. De Pinto and F. De Giorgi, Outer membrane VDAC1 controls permeability transition of the inner mitochondrial membrane in cellulo during stress-induced apoptosis, Cell Res., 2009, 19, 1363–1376 CrossRef CAS PubMed.
- J. Kim, R. Gupta, L. P. Blanco, S. Yang, A. Shteinfer-Kuzmine, K. Wang, J. Zhu, H. E. Yoon, X. Wang, M. Kerkhofs, H. Kang, A. L. Brown, S. J. Park, X. Xu, E. Zandee van Rilland, M. K. Kim, J. I. Cohen, M. J. Kaplan, V. Shoshan-Barmatz and J. H. Chung, VDAC oligomers form mitochondrial pores to release mtDNA fragments and promote lupus-like disease, Science, 2019, 366, 1531–1536 CrossRef CAS PubMed.
- C. H. Lipper, J. T. Stofleth, F. Bai, Y. S. Sohn, S. Roy, R. Mittler, R. Nechushtai, J. N. Onuchic and P. A. Jennings, Redox-dependent gating of VDAC by mitoNEET, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 19924–19929 CrossRef CAS PubMed.
- D. Ben-Hail, R. Begas-Shvartz, M. Shalev, A. Shteinfer-Kuzmine, A. Gruzman, S. Reina, V. De Pinto and V. Shoshan-Barmatz, Novel Compounds Targeting the Mitochondrial Protein VDAC1 Inhibit Apoptosis and Protect against Mitochondrial Dysfunction, J. Biol. Chem., 2016, 291, 24986–25003 CrossRef CAS PubMed.
- Y. Ma, X. Sun and X. Yao, The role and mechanism of VDAC1 in type 2 diabetes: An underestimated target of environmental pollutants, Mitochondrion, 2024, 78, 101929 CrossRef CAS PubMed.
- V. Shoshan-Barmatz, V. De Pinto, M. Zweckstetter, Z. Raviv, N. Keinan and N. Arbel, VDAC, a multi-functional mitochondrial protein regulating cell life and death, Mol. Aspects Med., 2010, 31, 227–285 CrossRef CAS PubMed.
- E. Zhang, I. Mohammed Al-Amily, S. Mohammed, C. Luan, O. Asplund, M. Ahmed, Y. Ye, D. Ben-Hail, A. Soni, N. Vishnu, P. Bompada, Y. De Marinis, L. Groop, V. Shoshan-Barmatz, E. Renström, C. B. Wollheim and A. Salehi, Preserving Insulin Secretion in Diabetes by Inhibiting VDAC1 Overexpression and Surface Translocation in β Cells, Cell Metab., 2019, 29, 64–77 CrossRef CAS PubMed.
- H. Rakhshani, E. Dehghanian and A. Rahati, Enhanced GROMACS: toward a better numerical simulation framework, J. Mol. Model., 2019, 25, 355 CrossRef PubMed.
- M. Masini, M. Anello, M. Bugliani, L. Marselli, F. Filipponi, U. Boggi, F. Purrello, M. Occhipinti, L. Martino, P. Marchetti and V. De Tata, Prevention by metformin of alterations induced by chronic exposure to high glucose in human islet beta cells is associated with preserved ATP/ADP ratio, Diabetes Res. Clin. Pract., 2014, 104, 163–170 CrossRef CAS PubMed.
- Q. Huang, W. You, Y. Li, Y. Sun, Y. Zhou, Y. Zhang, D. Liu, S. Zhan, Y. Zhu and X. Han, Glucolipotoxicity-Inhibited miR-299–5p Regulates Pancreatic β-Cell Function and Survival, Diabetes, 2018, 67, 2280–2292 CrossRef CAS PubMed.
- C. C. Yao, Y. X. Tong, H. Jiang, D. R. Yang and Z. W. Chen, Native polypeptide vglycin prevents nonalcoholic fatty liver disease in mice by activating the AMPK pathway, J. Funct. Foods, 2020, 73, 104110 CrossRef CAS.
- B. Zhou, J. Kreuzer, C. Kumsta, L. Wu, K. J. Kamer, L. Cedillo, Y. Zhang, S. Li, M. C. Kacergis, C. M. Webster, G. Fejes-Toth, A. Naray-Fejes-Toth, S. Das, M. Hansen, W. Haas and A. A. Soukas, Mitochondrial Permeability Uncouples Elevated Autophagy and Lifespan Extension, Cell, 2019, 177, 299–314 CrossRef CAS PubMed.
- H. Jiang, Y. Tong, D. Yan, S. Jia, C. G. Ostenson and Z. Chen, The Soybean Peptide Vglycin Preserves the Diabetic β-cells through Improvement of Proliferation and Inhibition of Apoptosis, Sci. Rep., 2015, 5, 15599 CrossRef CAS PubMed.
- A. Heitz, L. Chiche, D. Le-Nguyen and B. Castro, 1H 2D NMR and distance geometry study of the folding of Ecballium elaterium trypsin inhibitor, a member of the squash inhibitors family, Biochemistry, 1989, 28, 2392–2398 CrossRef PubMed.
- T. Yamazaki, M. Takaoka, E. Katoh, K. Hanada, M. Sakita, K. Sakata, Y. Nishiuchi and H. Hirano, A possible physiological function and the tertiary structure of a 4 kDa peptide in legumes, Eur. J. Biochem., 2003, 270, 1269–1276 CrossRef CAS PubMed.
- M. Bayrhuber, T. Meins, M. Habeck, S. Becker, K. Giller, S. Villinger, C. Vonrhein, C. Griesinger, M. Zweckstetter and K. Zeth, Structure of the human voltage-dependent anion channel, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 15370–15375 CrossRef CAS PubMed.
- T. Wang, N. Bo, G. Sha, Y. Guan, D. Yang, X. Shan, Z. Lv, Q. Chen, G. Yang, S. Gong, Y. Ma and M. Zhao, Identification and molecular mechanism of novel hypoglycemic peptide in ripened pu-erh tea: Molecular docking, dynamic simulation, and cell experiments, Food Res. Int., 2024, 194, 114930 CrossRef CAS PubMed.
- M. T. Khan, A. Ali, Q. Wang, M. Irfan, A. Khan, M. T. Zeb, Y. J. Zhang, S. Chinnasamy and D. Q. Wei, Marine natural compounds as potents inhibitors against the main protease of SARS-CoV-2-a molecular dynamic study, J. Biomol. Struct. Dyn., 2021, 39, 3627–3637 CrossRef CAS PubMed.
- S. Gorelov, A. Titov, O. Tolicheva, A. Konevega and A. Shvetsov, DSSP in GROMACS: Tool for Defining Secondary Structures of Proteins in Trajectories, J. Chem. Inf. Model., 2024, 64, 3593–3598 CrossRef CAS PubMed.
- E. Haythorne, M. Rohm, M. van de Bunt, M. F. Brereton, A. I. Tarasov, T. S. Blacker, G. Sachse, M. Silva Dos Santos, R. Terron Exposito, S. Davis, O. Baba, R. Fischer, M. R. Duchen, P. Rorsman, J. I. MacRae and F. M. Ashcroft, Diabetes causes marked inhibition of mitochondrial metabolism in pancreatic β-cells, Nat. Commun., 2019, 10, 2474 CrossRef PubMed.
- M. Federici, M. Hribal, L. Perego, M. Ranalli, Z. Caradonna, C. Perego, L. Usellini, R. Nano, P. Bonini, F. Bertuzzi, L. N. Marlier, A. M. Davalli, O. Carandente, A. E. Pontiroli, G. Melino, P. Marchetti, R. Lauro, G. Sesti and F. Folli, High glucose causes apoptosis in cultured human pancreatic islets of Langerhans: a potential role for regulation of specific Bcl family genes toward an apoptotic cell death program, Diabetes, 2001, 50, 1290–1301 CrossRef CAS PubMed.
- V. Shoshan-Barmatz, E. Nahon-Crystal, A. Shteinfer-Kuzmine and R. Gupta, VDAC1, mitochondrial dysfunction, and Alzheimer's disease, Pharmacol. Res., 2018, 131, 87–101 CrossRef CAS PubMed.
- M. Vakilian, Y. Tahamtani and K. Ghaedi, A review on insulin trafficking and exocytosis, Gene, 2019, 706, 52–61 CrossRef CAS PubMed.
- S. Y. Park, J. F. Gautier and S. Chon, Assessment of Insulin Secretion and Insulin Resistance in Human, Diabetes Metab. J., 2021, 45, 641–654 CrossRef PubMed.
- J. Huang, K. H. Wong, S. V. Tay, A. How and J. P. Tam, Cysteine-Rich Peptide Fingerprinting as a General Method for Herbal Analysis to Differentiate Radix Astragali and Radix Hedysarum, Front. Plant Sci., 2019, 10, 973 CrossRef PubMed.
- B. M. Schmidt, D. M. Ribnicky, P. E. Lipsky and I. Raskin, Revisiting the ancient concept of botanical therapeutics, Nat. Chem. Biol., 2007, 3, 360–366 Search PubMed.
- D. Spencer, T. J. Higgins, M. Freer, H. Dove and J. B. Coombe, Monitoring the fate of dietary proteins in rumen fluid using gel electrophoresis, Br. J. Nutr., 1988, 60, 241–247 CrossRef CAS PubMed.
- E. Blachly-Dyson, E. B. Zambronicz, W. H. Yu, V. Adams, E. R. McCabe, J. Adelman, M. Colombini and M. Forte, Cloning and functional expression in yeast of two human isoforms of the outer mitochondrial membrane channel, the voltage-dependent anion channel, J. Biol. Chem., 1993, 268, 1835–1841 CrossRef CAS PubMed.
- V. Shoshan-Barmatz, A. Shteinfer-Kuzmine and A. Verma, VDAC1 at the Intersection of Cell Metabolism, Apoptosis, and Diseases, Biomolecules, 2020, 10, 1485 CrossRef CAS PubMed.
- R. Ujwal, D. Cascio, J. P. Colletier, S. Faham, J. Zhang, L. Toro, P. Ping and J. Abramson, The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 17742–17747 CrossRef CAS PubMed.
- X. Li, H. Wang, B. Yao, W. Xu, J. Chen and X. Zhou, lncRNA H19/miR-675 axis regulates cardiomyocyte apoptosis by targeting VDAC1 in diabetic cardiomyopathy, Sci. Rep., 2016, 6, 36340 CrossRef CAS PubMed.
- J. T. Varughese, S. K. Buchanan and A. S. Pitt, The Role of Voltage-Dependent Anion Channel in Mitochondrial Dysfunction and Human Disease, Cells, 2021, 10, 1737 CrossRef CAS PubMed.
- A. Atlante, D. Valenti, V. Latina and G. Amadoro, Dysfunction of Mitochondria in Alzheimer’s Disease: ANT and VDAC Interact with Toxic Proteins and Aid to Determine the Fate of Brain Cells, Int. J. Mol. Sci., 2022, 23, 7722 CrossRef CAS PubMed.
- K. N. Belosludtsev, D. A. Serov, A. I. Ilzorkina, V. S. Starinets, M. V. Dubinin, E. Y. Talanov, M. N. Karagyaur, A. L. Primak and N. V. Belosludtseva, Pharmacological and Genetic Suppression of VDAC1 Alleviates the Development of Mitochondrial Dysfunction in Endothelial and Fibroblast Cell Cultures upon Hyperglycemic Conditions, Antioxidants, 2023, 12, 1459 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5fo00054h |
| This journal is © The Royal Society of Chemistry 2025 |