Open Access Article
Open Access ArticleProteome evaluation of the biological activity of grape seed extract (GSE) on the intestine of rats†
Eduardo
Guisantes-Batán‡
 ab,
Sara
Artigas-Jerónimo‡
ab,
Sara
Artigas-Jerónimo‡
 cd,
Lorena
Mazuecos
cd,
Lorena
Mazuecos
 cd,
Blanca
Rubio
de,
Elena
Bonzón-Kulichenko
cf,
Sergio
Gómez
cd,
Blanca
Rubio
de,
Elena
Bonzón-Kulichenko
cf,
Sergio
Gómez
 ab,
Margarita
Villar
dg and
Nilda
Gallardo
ab,
Margarita
Villar
dg and
Nilda
Gallardo
 *cd
*cd
aRegional Institute for Applied Scientific Research, University of Castilla-La Mancha, Avenida Camilo José Cela 1B, 13071 Ciudad Real, Spain. E-mail: sergio.gomez@uclm.es
bDepartment of Analytical Chemistry and Food Technology, Faculty of Chemical Sciences and Technologies, University of Castilla-La Mancha, Avenida Camilo José Cela 10, 13071 Ciudad Real, Spain
cDOE research group, Institute of Biomedicine of the University of Castilla-La Mancha (IB-UCLM), Spain
dBiochemistry Section, Faculty of Chemical Sciences and Technologies, University of Castilla-La Mancha, Avenida Camilo José Cela 10, 13071 Ciudad Real, Spain
eMolecular Regulation of Heart Failure Research Group, National Cardiovascular Research, Center Carlos III (CNIC), Melchor Fernández Almagro 3, 28029, Madrid, Spain. E-mail: blancamaria.rubio@cnic.es
fBiochemistry Section, Faculty of Environmental Sciences and Biochemistry, University of Castilla-La Mancha, Avda. Carlos III s/n, 45071 Toledo, Spain
gSaBio, Hunting Resources Research Institute IREC-CSIC-UCLM-JCCM, Ronda de Toledo s/n, 13005 Ciudad Real, Spain
First published on 1st May 2025
Abstract
Grape seed extract (GSE) is rich in polyphenols and has garnered attention for its potential health benefits, particularly in the gastrointestinal system. This study explores the principal effects of consuming a low dose of GSE on the intestinal proteome. Using a rat model, we examined the ileum proteomes of rats supplemented with GSE compared with a control group without supplementation. Particularly, this study was conducted using the label-free sequential window acquisition of all theoretical fragment ion spectra mass spectrometry (SWATH-MS) quantitative proteomics to describe ileum proteome changes derived from GSE flavanols consumption, characterizing its possible mechanisms of action. Our results highlight the substantial impact of GSE on the ileum proteome, suggesting potential mechanisms through which GSE exerts its effects. The observed protein regulation patterns indicate significant alterations in metabolic and immune regulatory pathways, highlighting the importance of considering individual variability and physiological context when evaluating the potential effects of GSE on gastrointestinal function.
1. Introduction
Metabolic disorders, such as obesity, type 2 diabetes (T2D), and inflammatory bowel diseases (IBD), are significant global health challenges, often referred to as modern pandemics due to their increasing prevalence and impact on healthcare systems and economies.1,2 Prevention strategies often focus on promoting healthy diets rich in fruits and vegetables, which contain bioactive compounds like flavonoids known to improve metabolic health.3,4 Grape seed extract (GSE), derived from Vitis vinifera seeds, is a particularly rich source of polyphenolic compounds, particularly flavanols, comprising both monomeric catechins and their polymeric counterparts the proanthocyanidins which have been extensively studied for their potent iron chelating capacity, antioxidant and anti-inflammatory properties.5,6Different studies in animals with acute or chronic doses of GSE ranging from 25 to 500 mg per kg per BW per day have observed that the consumption of flavanols improves intestinal barrier function, reduces inflammation and oxidative stress, modulates immune response, and affects the composition of the gut microbiota in both diet-induced obese (DIO) Wistar rats7–9 and rats with chemical-induced intestinal dysfunction.10,11 In our previous study we showed that a low dose of GSE reduces body weight and adiposity, and improves the blood lipid profile in healthy and lean rats.12 In addition, in vivo and in vitro research in animals and cultured human cell lines highlight the potential of flavanols in the prevention of various intestinal diseases, including inflammatory bowel disease (IBD), colitis, and colon cancer.13–17
The ileum, a key segment of the gastrointestinal tract, plays a vital role in nutrient absorption and immune response. It maintains gut homeostasis through complex interactions between its epithelial cells, including intestinal stem cells (ISCs), which exhibit unique metabolic characteristics that are essential for their renewal capacity.18,19 Given the importance of these cells in maintaining intestinal health, studying the effects of dietary compounds like GSE on the ileum is of great interest.
Proteomics offers a powerful approach to exploring how bioactive compounds like GSE influence biological systems at the protein level. By evaluating changes in the ileum proteome, we can gain insights into the molecular mechanisms underlying GSE's effects on intestinal function.
This study aims to evaluate the proteomic changes in the ileum of rats supplemented with GSE, using label-free sequential window acquisition of all theoretical fragment ion spectra mass spectrometry (SWATH-MS). Our goal is to elucidate the biological processes influenced by GSE, particularly its potential to modulate intestinal health and metabolism in young, and healthy rats.
2. Materials and methods
2.1. Grape seed extract (GSE)
A commercial grape seed extract (GSE) rich in flavanols obtained from white grape seeds was kindly provided by Les Dérives Résiniques et Terpéniques (Dax, France). The compositional characterization of GSE and the analysis of flavanols according to their degree of polymerization (DP) by HPLC-FLD-ESI-QTOF were performed as previously described.12 This comprehensive characterization allowed us to estimate that, as previously reported,12 the rats supplemented with GSE had an average daily intake of 1.59 mg of monomeric flavanols and 2.40 mg of polymeric forms.2.2 Animal model
Three-month-old male Wistar rat purchased from Janvier Laboratories (Pau, France) were randomly housed individually to allow daily control of food intake and body weight, thereby reducing variability in weight, that depend on the amount of feed consumed. All animals were maintained in ventilated-controlled quarters at a temperature of 20–25 °C, 50–55% humidity, and a 12-hour light/dark cycle (8 a.m.–8 p.m.). They were fed ad libitum with a standard chow diet from Harlan Laboratories (2014 Teklad Global 14% Protein Rodent Maintenance Diet, Madison, WI, USA) and had free access to water. Animals were handled according to European Union laws (2010/63/EU) and following Spanish regulations (RD 53/2013) for laboratory animal use. Experimental protocols with animals were approved by the Institutional Committee for Ethical Animal Care CEEA/UCLM (Permit numbers CE/301012 and CE/99-1835-A308, research proposal approved respectively, on October 30, 2012, and May 17, 2019). All efforts were made to reduce the number of animals used and minimize animal suffering. The minimal number of animals required for this study was calculated as previously described.122.3. GSE administration
GSE was administered to young-mature and healthy Wistar rats. Animals were randomly divided into two groups: a Control group (n = 6) and a GSE group (n = 6). The GSE group received supplementation of 25 mg of GSE per kg of body weight per day over a 28-day period, as reported previously.12 After 28 days, rats were 16 hours overnight fasted and anesthetized with CO2 previous sacrifice by decapitation the next day between 9–10 a.m., to avoid variability between different animals. Epididymal adipose tissue (eWAT) was rapidly excised, washed, and frozen in liquid N2 and stored at −80 °C prior analysis. Relative eWAT mass was calculated as a ratio between eWAT mass and body weight mass.2.4. Ileum dissection, protein extraction, quantification, and trypsin digestion procedures
With a pair of scissors, we cut the small intestine about 1 cm away from the stomach and 1 cm immediately before the colon and eliminate any mesenteric connective and/or fat tissue. The entire length was unraveled, washed and rolled following the Swiss roll as described.20 The tissue was frozen in liquid N2 and stored at −80 °C prior analysis. The distal 2 cm of the ileum was dissected from frozen samples using a sterile scalpel. All tissue was ground to a powdery texture in liquid N2 using a mortar and pestle.Total proteins from the ileum samples were isolated using the AllPrep DNA/RNA/Protein Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Isolated proteins were resuspended in 10 mM PBS supplemented with 2% sodium dodecyl sulfate (SDS) and protein concentration was determined using the BCA Protein Assay Kit (Thermo Scientific, Rockford, IL, USA) with bovine serum albumin (BSA) as standard.
Total protein samples (150 μg per sample) were trypsin digested using the FASP Protein Digestion Kit (Expedeon Ltd, UK) and sequencing grade trypsin (Promega, Madison, WI, USA) following manufactureŕs recommendations. The resulting tryptic peptides were desalted onto OMIX Pipette tips C18 (Agilent Technologies, Santa Clara, CA, USA), dried down and stored at −20 °C until mass spectrometry analysis.
2.5. Ileum proteome analysis
The desalted protein digests were resuspended (final concentration of 2 μg μL−1) in 2% acetonitrile and 5% acetic acid in water and analyzed by reverse-phase liquid chromatography coupled online to mass spectrometry (RP-LC-MS/MS) using an Ekspert™ nLC 415 system coupled with a 6600 TripleTOF mass spectrometer (AB Sciex, Framingham, MA, USA) through Information-Dependent Acquisition (IDA) followed by Sequential Windowed data-independent Acquisition of the Total High-Resolution Mass Spectra (SWATH-MS). The peptides were concentrated in a 0.1 × 20 mm C18 RP precolumn (Thermo Scientific, Waltham, MA, USA) with a flow rate of 5 μL min−1 during 10 min in solvent A. Then, peptides were separated in a 0.075 × 250 mm C18 RP column (New Objective, Woburn, MA, USA) with a flow rate of 300 nL min−1. Peptides elution was done in a 120 min gradient from 5% to 30% solvent B followed by a 15 min gradient from 30% to 60% solvent B (solvent A: 0.1% formic acid in water, solvent B: 0.1% formic acid in acetonitrile) and directly injected into the mass spectrometer for analysis.For IDA experiments, 5 μL of each protein digest (6 biological samples per group) were pooled together as a mixed sample for each experimental condition (Control and GSE). Six micrograms of each pooled mixed sample were analyzed. The mass spectrometer was set to IDA scanning full spectra from 350–1400 m/z (250 ms accumulation time) followed by up to 50 MS/MS scans (100–1500 m/z). Candidate ions with a charge state between +2 and +5 and counts per second above a minimum threshold of 100 were isolated for fragmentation. One MS/MS spectrum was collected for 100 ms, before adding those precursor ions to the exclusion list for 15 s (mass spectrometer operated by Analyst R TF 1.6, AB SCIEX). Dynamic background subtraction was turned off. Data were acquired in high sensitivity mode with rolling collision energy on and a collision energy spread of 5.
For SWATH quantitative analysis, 12 independent samples (6 biological replicates from each control and GSE groups) (10 μg each) were subjected to the cyclic Data Independent Acquisition (DIA) of mass spectra using the SWATH variable windows calculator (V 1.0, AB Sciex) and the SWATH acquisition method editor (AB Sciex) similar to previously established methods.21 A set of 50 overlapping windows was constructed (1 m/z for window overlap), covering the precursor mass range of 400–1250 m/z based on data from the IDA runs previously acquired. For these experiments, a 50 ms survey scan (350–1400 m/z) was acquired at the beginning of each cycle, and SWATH MS/MS spectra were collected from 100 to 1500 m/z during 70 ms at high sensitivity mode, resulting in a cycle time of 3.6 s. Collision energy for each window was determined according to the calculation for a charge +2 ion-centered upon the window with a collision energy spread of 15.
2.6 Proteomic data analysis
To create a spectral library of all detectable peptides in the samples, the IDA raw files were combined (2 runs in total) and subjected to database search in unison using ProteinPilot software v. 5.0.1 (AB Sciex) with the Paragon algorithm. Spectra identification was performed by searching against the Uniprot Rattus norvegicus proteome database (79![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 038 entries, in January 2022) with the following parameters: iodoacetamide cysteine alkylation, trypsin digestion, identification focus on biological modification and thorough ID as search effort. The detected protein threshold was set at 0.05. An independent False Discovery Rate (FDR) analysis with the target-decoy approach provided by ProteinPilot, was used to assess the quality of identifications. Positive identifications were considered when identified proteins reached a 1% global FDR. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository,22 with the dataset identifier PXD055819 and https://doi.org/10.6019/PXD055819. For SWATH processing, the spectral alignment and targeted data extraction of DIA samples were performed using the SWATH Acquisition MicroApp2.0 in the PeakView v. 2.2 software (AB Sciex) with the reference spectral library. The parameters used were up to 10 peptides per protein, seven transitions per peptide, 15 ppm ion library tolerance, 5 min XIC extraction window, 0.01 Da XIC width, and considering only peptides with at least 99% confidence and excluding those which were shared or contained modifications. However, to ensure reliable quantitation, only proteins with three or more peptides available for quantitation were selected for XIC peak area extraction and exported for analysis in the MarkerView v. 1.3 software (AB Sciex) (ESI Data 1†).
038 entries, in January 2022) with the following parameters: iodoacetamide cysteine alkylation, trypsin digestion, identification focus on biological modification and thorough ID as search effort. The detected protein threshold was set at 0.05. An independent False Discovery Rate (FDR) analysis with the target-decoy approach provided by ProteinPilot, was used to assess the quality of identifications. Positive identifications were considered when identified proteins reached a 1% global FDR. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository,22 with the dataset identifier PXD055819 and https://doi.org/10.6019/PXD055819. For SWATH processing, the spectral alignment and targeted data extraction of DIA samples were performed using the SWATH Acquisition MicroApp2.0 in the PeakView v. 2.2 software (AB Sciex) with the reference spectral library. The parameters used were up to 10 peptides per protein, seven transitions per peptide, 15 ppm ion library tolerance, 5 min XIC extraction window, 0.01 Da XIC width, and considering only peptides with at least 99% confidence and excluding those which were shared or contained modifications. However, to ensure reliable quantitation, only proteins with three or more peptides available for quantitation were selected for XIC peak area extraction and exported for analysis in the MarkerView v. 1.3 software (AB Sciex) (ESI Data 1†).
The Student's t-test (p < 0.05) was used to perform two-sample comparisons between the averaged area sums of all the transitions derived for each protein across the six biological replicates for each group under comparison, in order to identify proteins that were significantly differentially represented between CONTROL and GSE samples (ESI Data 2†). Data were separately analyzed for underrepresented (ESI Data 2†) and overrepresented (ESI Table 1†) proteins using the Metascape gene annotation and analysis resource (https://metascape.org/gp/index.html#/main/step1), Gene Ontology Resource (https://www.geneontology.org/) and the Reactome Pathway Database (https://reactome.org/).
2.7. Protein identification and quantification by Western Blotting
20 μg of proteins from total ileal protein lysates were separated under reducing conditions on 10% SDS-PAGE. Proteins were transferred to nitrocellulose sheets (0.2 mm) (Bio-Rad, Spain) and incubated overnight at 4 °C with the appropriate primary antibody, followed by incubation at room temperature for 90 min with the corresponding secondary antibody conjugated with horseradish peroxidase. Total OXPHOS rodent WB antibody cocktail (6 μg mL−1, ab110413) containing 5 mouse monoclonal antibodies: anti-CI subunit NDUFB8, anti-CII, anti-CIII-Core protein, anti-CIV subunit I and anti-CV alpha subunit was used from Abcam (Cambridge, UK), Calmodulin polyclonal antibody was used from Thermo Fisher Scientific, MA, USA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, PA5-78373) and anti-β-actin (1
500, PA5-78373) and anti-β-actin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, ab8226) from Abcam, Cambridge, UK. The secondary antibody used was goat anti-mouse conjugated with horseradish peroxidase (1
1000, ab8226) from Abcam, Cambridge, UK. The secondary antibody used was goat anti-mouse conjugated with horseradish peroxidase (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4000, 170-6516) and goat anti-rabbit conjugated with horseradish peroxidase (1
4000, 170-6516) and goat anti-rabbit conjugated with horseradish peroxidase (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, 172-1019) from Bio-Rad, Spain. The immune-protein complexes formed were visualized using the ECL Western-blotting detection kit (Amersham Biosciences, Inc., Piscataway, NJ) and the images were subjected to a densitometric analysis. Equal amounts of proteins from experimental groups were run on the same gel to allow direct comparison between Control and GSE-group. β-Actin was used as control of protein loading.
1000, 172-1019) from Bio-Rad, Spain. The immune-protein complexes formed were visualized using the ECL Western-blotting detection kit (Amersham Biosciences, Inc., Piscataway, NJ) and the images were subjected to a densitometric analysis. Equal amounts of proteins from experimental groups were run on the same gel to allow direct comparison between Control and GSE-group. β-Actin was used as control of protein loading.
2.8. RT-qPCR for gene expression quantification
The cDNA was synthesized from 1 μg of DNase-treated RNA. Relative quantification of target cDNA was performed from 4 ng of cDNA in SYBR-Green One-Step real time PCR Master Mix (Applied Biosystem), with primers supplied by Sigma (Calm3F: 5′GATGGCACCATTACCACCAAG3′ and Calm3R: 5′CGCTGTCTGTATCCTTCATCTTT3′) for Calm3, Mttp, Cd36, Acox1, Atgl and Il-1β expression quantification was performed using TaqMan One-Step real time PCR Master Mix (PE Applied BioSystems, Foster City, CA, USA) and the following probes: Rn01522974_m1 (Mttp), Rn00580728_m1 (Cd36), Rn00569216_m1 (Acox1), Rn01478868_m1 (Atgl) and Rn01514151_m1 (Il-1β) (Applied Biosystem).2.9. Statistical analysis
Data were expressed as mean ± SEM. Statistical analysis was performed using the GraphPad Prism version 8.4 for Windows (GraphPad Software), except proteomic statistical analysis in which the user-friendly web-based platform MetaboAnalyst (https://www.metaboanalyst.ca/) was employed. Differences between the two groups were assessed using the unpaired Student's t-test. All differences were considered significant at p < 0.05.3. Results
3.1. Characterization of grape seed extract (GSE)
In this study we evaluate in vivo the effects of a chronic consumption of GSE on the ileal proteome. Chromatographic analysis of this extract, presented in the Table 1, showed a phenolic composition typical of grape seed,12,23 and revealed that (+)-catechin and (−)-epicatechin were the most common monomers, accounting together 90% of the monomers. Among proanthocyanidins, PB1 and PB2 dimers were the major ones, but some trimers as PC1 were still present. The depolymerisation assay, showed that the mean number of monomeric units per molecule of proanthocyanidin contained in the extract was 4.5, indicating that some tetramers, pentamers and higher proanthocyanidins were also present in the extract as is common in grape seed extracts. This composition is similar to that of the extract used in other intervention studies.24,25| Compounds | Concentration (mg g−1 GSE) |
|---|---|
| Flavan-3-ols monomers | |
| (+)-Catechin | 83.2 ± 1.6 |
| (−)-Epicatechin | 59.5 ± 0.9 |
| (+)-Catechin gallate | 3.9 ± 0.1 |
| (−)-Epicatechin gallate | 12.5 ± 0.2 |
| Flavan-3-ols dimers | |
| Procyanidin B1 | 54.4 ± 1.1 |
| Procyanidin B2 | 39.5 ± 0.8 |
| Procyanidin B3 | 10.8 ± 0.8 |
| Procyanidin B4 | 14.3 ± 1.3 |
| Galloylated dimers | 49.3 ± 0.8 |
| Flavan-3-ols trimers | |
| Procyanidin C1 | 12.3 ± 0.8 |
| Proanthocyanidins | |
| Total proanthocyanidins | 234.5 ± 3.1 |
| Mean degree of polymerization (mDP) | 4.5 ± 0.2 |
| % Galloylation | 7.9 ± 0.2 |
| % Prodelphynidins | 0.5 ± 0.0 |
3.2. Effects of GSE on body weight and visceral adipose tissue
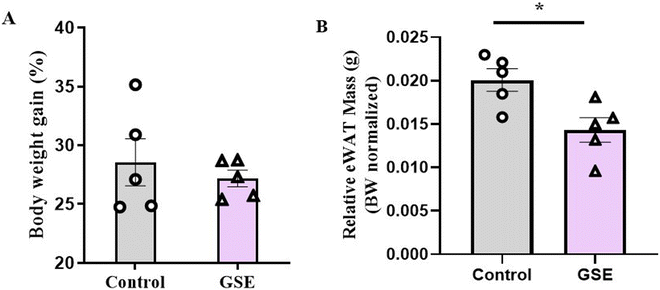
After 28 days of GSE supplementation, no significant changes in body weight were observed between the control and GSE groups (Fig. 1A). However, the GSE-supplemented rats exhibited a significant reduction in epididymal adipose tissue mass (eWAT) (Fig. 1B), suggesting that GSE may influence fat metabolism without altering overall body weight. These findings align with previous reports indicating that GSE can reduce visceral adiposity,12 potentially contributing to improved metabolic homeostasis. In that senses we propose that GSE supplementation could impact intestinal lipid absorption and metabolism, affecting visceral adipose tissue mass accretion.3.3. Ileum proteomics
Proteomic analysis of the ileum using SWATH-MS identified 945 proteins (ESI Data 1†), of which 237 were significantly dysregulated by GSE supplementation (p < 0.05). Among these, 13 proteins were overrepresented, and 224 were underrepresented in the GSE group compared to controls (ESI Data 2†). Principal component analysis (PCA) demonstrated clustering of proteomic profiles between control and GSE-treated rats, highlighting the distinct impact of GSE on the ileum proteome (Fig. 2A).In line with this, a heat map representation was generated to visualize the magnitude of these phenomena. ESI Fig. 1† presents a complete heatmap illustrating the effect of GSE supplementation on the 237 significantly dysregulated proteins in ileum samples, while Fig. 2B displays a focused heatmap depicting the 13 overrepresented proteins and the 24 most differentially underrepresented proteins (log2FC < −2) responsible for the distinct separation between control and GSE-supplemented animals. ESI Table 1† lists the protein names and UniProt accession ID of the proteins shown in Fig. 2. Fig. 6 shows the over- and underrepresented proteins identified in the ileum proteome after GSE supplementation.
For the biological interpretation of these results, we separately analyzed the over- and underrepresented proteins using the Metascape gene annotation analysis resource and the Reactome Pathway Database to gain deeper insights into the cellular processes altered by chronic GSE supplementation at the ileal level. These results underscore the statistically significant impact of GSE supplementation on the ileum proteome.
3.4. Overrepresented proteins: mitochondrial function and smooth muscle contraction
The overrepresented proteins in GSE-supplemented rats were primarily associated with mitochondrial electron transport chain (ETC) activity and smooth muscle contraction pathways (Fig. 3A). Key proteins involved in these pathways included cytochrome c oxidase subunits and apolipoprotein E (ApoE), suggesting enhanced cholesterol export and ATP production through the regulation of mitochondrial function in the ileum. STRING analysis revealed strong interactions between proteins related to the ETC, which may support improved ATP production and antioxidant defense in response to GSE supplementation (Fig. 3B).These results also showed a strong connection between the ETC and calcium signaling (via Calm3). The upregulation of calmodulin (Calm3) and tropomyosin (Tpm4) could indicate potential benefits for smooth muscle function, which could enhance gastrointestinal motility. This observation aligns with previous reports on GSE's ability to modulate gut motility and smooth muscle contractility.26
Moreover, calcium signaling (via Calm3) is crucial for regulating tight junctions, epithelial permeability and then nutrient absorption. Enhanced calcium signaling can also improve the integrity of the intestinal barrier, reducing the likelihood of “leaky gut”, which is often associated with inflammatory and metabolic diseases.27
As shown in Fig. 4, beyond their individual functions, ApoE, Tpm4, and Calm3 may collectively influence nitric oxide (NO) production through the positive regulation of NO synthase activity (GO: 0051000), as suggested our PPI analyses (Fig. 4).
Nitric oxide synthase (NOS) activity plays a significant role in maintaining the integrity of the mucosal wall of the gastrointestinal (GI) tract, largely through its production of nitric oxide (NO). NO is a critical signaling molecule that regulates several physiological processes, including blood flow, nutrient transport, immune response, and smooth muscle contraction, crucial for proper gut motility, nutrient absorption and peristalsis.28
3.5. Underrepresented proteins: RNA processing, protein synthesis, carbon metabolism and immune regulation
Proteins found to be underrepresented in response to GSE supplementation (Fig. 2B), were analyzed to explore their potential functional and/or physical interactions (Fig. 5).The analysis showed a significant networks of protein-protein interactions (PPI) and identified several densely connected groups (MCODE) within the networks. Figure also showed that underrepresented proteins in the GSE group were primarily involved in RNA processing, protein synthesis and fatty acid oxidation, among other processes (Fig. 5). Notably, several ribosomal proteins and components of the translation machinery were also downregulated, suggesting that GSE may reduce protein synthesis and cellular metabolism in the ileum.
Enrichment analysis further indicated significant reductions in pathways related to carbon metabolism (pyruvate metabolism, monocarboxylic acid metabolism), and the innate immune system (Fig. 6).
These metabolic adjustments driven by GSE impact immune function, as evidenced by the downregulation of proteins involved in innate immune responses processes, particularly those related to neutrophil degranulation (ESI Data 3†). This is noteworthy because neutrophil activation has been implicated in the development of various chronic inflammatory disorders, including inflammatory bowel disease (IBD).29 However, further research is needed to determine whether the consumption of GSE components contributes to a beneficial reduction of intestinal inflammation or reflects a broader immune modulation, as suggested by Andersen-Civil et al.30
Overall, these effects of GSE appear to be associated with a reduction in the absorption of nutrients, dietary antigens, and microbiota-derived molecules, such as short-chain fatty acids and other carboxylic acids, which constitute the main carbon flux from the diet through the microbiota to the host. Data presented in Fig. 1 and ESI Fig. 2† confirm that lipid absorption and metabolism are reduced in the ileum of GSE-supplemented rats. These effects resemble those observed during caloric restriction, further supporting the potential role of GSE in promoting longevity.31
Taken together, the antimicrobial properties of GSE, its calorie restriction-like effects, and the pivotal role of the small intestinal microbiota in modulating nutrient absorption32 allow us to suggest that chronic GSE supplementation could, under certain physiological conditions, contribute to poor nutrient uptake, primarily lipids, and potential malnutrition, particularly in individuals with low body mass or higher metabolic demands.
3.6. Western blot and qPCR analysis for proteomic data validation in response to GSE effect
The data presented in ESI Fig. 2,† confirm, in part, our proteomic data, indicating the overrepresentation of proteins and processes primarily associated with mitochondrial electron transport chain (ETC) and calcium signaling via Calm3 in GSE-supplemented rats, as well as the underrepresentation of proteins codified by genes involved in lipid absorption and metabolism, and innate immunity.4. Discussion
This study provides a comprehensive proteomic analysis of the effects of chronic grape seed extract (GSE) supplementation on the ileum of healthy rats. Our results reveal that the ileal proteome is significantly altered in response to GSE affecting key biological processes, including mitochondrial function, energy metabolism and immune regulation. Enrichment analysis indicates that specific molecular pathways are overrepresented among the differentially expressed proteins, reinforcing the notion that GSE exerts coordinated effects on multiple biological processes.The overrepresentation of proteins involved in the ETC, calcium signaling pathway, and nitric oxide (NO) production points to a dynamic mechanism in which calcium binding to calmodulin can influence tight junctions and epithelial permeability, which in turn modulate nutrient absorption, antimicrobial activity, immune response, and smooth muscle contraction.32 This suggests that GSE may play a role in regulating intestinal transport mechanisms in the ileum of healthy rats. Previous literature has shown that, nutrient transport is often accompanied by calcium flux, with calcium absorption being mediated by calcium–calmodulin interaction dependent signaling.33 However, further studies are needed to confirm this mechanism and its physiological relevance.
The upregulation of cytochrome c oxidase and other ETC components is consistent with previous studies demonstrating that GSE polyphenols improved mitochondrial function across different tissues and physiological conditions.13–17 Notably, grape seed proanthocyanidins have been shown to increase the levels and functionality of Complex I, exerting a protective role in irradiated human lung fibroblast where excessive generation of ROS causes cellular damage.34
Additionally, increasing evidence suggests that polyphenol consumption modulates intestinal microbiota and lipid metabolism. These effects have been associated with a reduced load of proinflammatory bacteria in the gut, which may help prevent or alleviate systemic inflammation and metabolic disturbances in individuals at risk of inflammatory diseases or already affected by metabolic disorders.35–37 Hence, the influence of polyphenols on microbiota composition could indirectly contribute to mitochondrial adaptations, reinforcing the potential systemic benefits of GSE supplementation at least in obese individuals or those with comorbidities associated with obesity or dysbiosis. For instance, the downregulation of proteins related to neutrophil degranulation and innate immunity suggests that microbiota changes induced by GSE may protect the ileal mucosa from excessive immune activation and chronic gut inflammation as occurs during autoimmune or inflammatory diseases.29
Although we did not conduct a metagenomic study, our results imply that GSE altered gut mcrobiota composition influencing both nutrient absorption and immune response. For instance, our proteomic analysis suggests a potential reduction in SCFA utilization, which could reflect a decrease in carbon flow from the microbiota to the host. This finding also support the hypothesis that GSE influences microbiota composition and function,7–9 potentially altering SCFA production and subsequent metabolic and immune outcomes in the host, as have been previously proposed.32 However, while reducing inflammation is beneficial in pathological conditions, the suppression of immune-related proteins in healthy animals raises concerns about the potential consequences of prolonged GSE supplementation.
Finally, we propose that the apparent contradictions in our findings could reflect an adaptive response to reduced nutrient absorption, dietary antigens and microbiota-derived metabolites, rather than a dysfunction in metabolic or immune processes induced by GSE. Consequently, GSE supplementation may induce a metabolic reconfiguration in the ileum, adjusting carbon metabolism and immune homeostasis in response to changes in energy availability. Nonetheless, prolonged reduction in nutrient absorption also poses challenges in healthy animals by altering energy homeostasis. Future studies should carefully evaluate the balance between the benefits of GSE and its effects on nutrition, metabolism and immune competence in healthy rats.
5. Conclusions
The observed proteomic changes suggest that GSE supplementation reduced nutrient absorption and metabolism in enterocytes as well as intestinal immune function in healthy animals. Proteomics also indicates the stimulation of calcium signaling pathways and mitochondrial efficiency. We suggest that GSE supplementation activates calcium signaling pathways, potentially enhancing mitochondrial efficiency and nutrient sensing, which could improve nutrient absorption. Additional research focusing on calcium and nutrient transport mechanisms, gut microbiota profiling, and functional assays are needed to confirm the physiological impact of GSE on intestinal health and function in non-obese and healthy rats.Author contributions
Eduardo Guisantes-Batán: methodology, formal analysis, investigation, data curation, writing – review and editing. Sara Artigas-Jerónimo: methodology, formal analysis, data curation, investigation, writing – review and editing. Lorena Mazuecos: methodology, writing – review and editing. Blanca Rubio: methodology, investigation. Elena Bonzón-Kulichenko: formal analysis, data curation. Sergio Gómez: funding acquisition, conceptualization, supervision. Margarita Villar: formal analysis, visualization, supervision, validation, writing – original draft. Nilda Gallardo: funding acquisition, conceptualization, supervision, writing – original draft, review & editing, project administration.Data availability
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the following dataset identifiers: Project Name: Proteomic analysis of the action of Grape Seed Extract (GSE) on the Intestine of rats; Project accession: PXD055819; Project https://doi.org/10.6019/PXD055819. The data supporting this article have been included as part of the manuscript and ESI (Fig. 6, ESI Fig. 1, ESI Data 1, Data 2 and Data 3†).Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This research project was supported by the University of Castilla-La Mancha and the European Regional Development Fund (FEDER) under regional project grants: 2019-GRIN-26992, 2019-GRIN-27164, 2020-GRIN-28803, 2020-GRIN-28744, 2021-GRIN-31106 and 2021-GRIN-30987; and national project grant RTI2018-098643-B-I00 funded by Ministerio de Ciencia, Innovación y Universidades. Eduardo Guisantes-Batan is grateful to the European Social Fund and the University of Castilla-La Mancha for co-funding his predoctoral contract [2018/12504]. Sara Artigas-Jerónimo is supported under the FJC2021-047764-I grant, funded by MCIN/AEI/10.13039/501100011033 and the Europe Union “NextGenerationEU”/PRTR. The authors acknowledge their gratitude to “Les Dérives Résiniques et Terpéniques” for providing the grape seed extract using in this research work and Sergio Moreno for the excellent technical assistance.References
- A. Garus-Pakowska, Int. J. Environ. Res. Public Health, 2023, 20, 6789 CrossRef PubMed.
- T. E. Adolph, M. Meyer, J. Schwärzler, L. Mayr, F. Grabherr and H. Tilg, Nat. Rev. Gastroenterol. Hepatol., 2022, 19, 753–767 CrossRef PubMed.
- R. Jagannathan, S. A. Patel, M. K. Ali and K. M. V. Narayan, Curr. Diabetes Rep., 2019, 19, 44 CrossRef PubMed.
- M. G. Saklayen, Curr. Hypertens. Rep., 2018, 20, 12 CrossRef PubMed.
- A. Scalbert, C. Manach, C. Morand, C. Rémésy and L. Jiménez, Crit. Rev. Food Sci. Nutr., 2005, 45, 287–306 CrossRef PubMed.
- M. Lesjak and S. K. S. Srai, Pharmaceuticals, 2019, 12, 119 CrossRef PubMed.
- K. Gil-Cardoso, I. Ginés, M. Pinent, A. Ardévol, L. Arola, M. Blay and X. Terra, Mol. Nutr. Food Res., 2017, 61, 8 CrossRef PubMed.
- K. Gil-Cardoso, I. Ginés, M. Pinent, A. Ardévol, M. Blay and X. Terra, J. Nutr. Biochem., 2018, 62, 35–42 CrossRef PubMed.
- C. González-Quilen, K. Gil-Cardoso, I. Ginés, R. Beltrán-Debón, M. Pinent, A. Ardévol, X. Terra and M. T. Blay, Nutrients, 2019, 11, 979 CrossRef PubMed.
- K. Gil-Cardoso, R. Comitato, I. Ginés, A. Ardévol, M. Pinent, F. Virgili, X. Terra and M. Blay, Mol. Nutr. Food Res., 2019, 63, e1800720 CrossRef PubMed.
- Y.-H. Wang, B. Ge, X.-L. Yang, J. Zhai, L.-N. Yang, X.-X. Wang, X. Liu, J.-C. Shi and Y.-J. Wu, Int. Immunopharmacol., 2011, 11, 1620–1627 CrossRef PubMed.
- E. Guisantes-Batan, L. Mazuecos, B. Rubio, G. Pereira-Caro, J. M. Moreno-Rojas, A. Andrés, S. Gómez-Alonso and N. Gallardo, Food Funct., 2022, 13, 11353–11368 RSC.
- M. Mabrouk, M. El Ayed, A. Démosthènes, Y. Aissouni, E. Aouani, L. Daulhac-Terrail, M. Mokni and M. Bégou, Front. Immunol., 2022, 13, 960355 CrossRef PubMed.
- J. M. Pérez-Ortiz, L. F. Alguacil, E. Salas, I. Hermosín-Gutiérrez, S. Gómez-Alonso and C. González-Martín, Food Sci. Nutr., 2019, 7, 2948–2957 CrossRef PubMed.
- O. O. Erejuwa, S. A. Sulaiman and M. S. Ab Wahab, Int. J. Mol. Sci., 2014, 15, 4158–4188 CrossRef CAS PubMed.
- H. Li, L. M. Christman, R. Li and L. Gu, Food Funct., 2020, 11, 4878–4891 RSC.
- E. Casanova, L. Baselga-Escudero, A. Ribas-Latre, A. Arola-Arnal, C. Bladé, L. Arola and M. J. Salvadó, BioFactors, 2014, 40, 146–156 CrossRef CAS PubMed.
- C. Stringari, R. A. Edwards, K. T. Pate, M. L. Waterman, P. J. Donovan and E. Gratton, Sci. Rep., 2012, 2, 568 CrossRef PubMed.
- C. Li, Y. Zhou, R. Wei, D. L. Napier, T. Sengoku, M. C. Alstott, J. Liu, C. Wang, Y. Y. Zaytseva, H. L. Weiss, Q. Wang and B. M. Evers, Cell. Mol. Gastroenterol. Hepatol., 2023, 15, 931–947 CrossRef CAS PubMed.
- A. B. Bialkowska, A. M. Ghaleb, M. O. Nandan and V. W. Yang, J. Visualized Exp., 2016, 54161 Search PubMed.
- L. C. Gillet, P. Navarro, S. Tate, H. Röst, N. Selevsek, L. Reiter, R. Bonner and R. Aebersold, Mol. Cell. Proteomics, 2012, 11, O111.016717 CrossRef PubMed.
- Y. Perez-Riverol, J. Bai, C. Bandla, D. García-Seisdedos, S. Hewapathirana, S. Kamatchinathan, D. J. Kundu, A. Prakash, A. Frericks-Zipper, M. Eisenacher, M. Walzer, S. Wang, A. Brazma and J. A. Vizcaíno, Nucleic Acids Res., 2022, 50, D543–D552 CrossRef CAS PubMed.
- J. Pérez-Navarro, P. M. Izquierdo-Cañas, A. Mena-Morales, J. Martínez-Gascueña, J. L. Chacón-Vozmediano, E. García-Romero, I. Hermosín-Gutiérrez and S. Gómez-Alonso, Food Chem., 2019, 295, 350–360 CrossRef PubMed.
- M. Margalef, Z. Pons, F. I. Bravo, B. Muguerza and A. Arola-Arnal, J. Nutr. Biochem., 2015, 26, 987–995 CrossRef CAS PubMed.
- L. E. Downing, R. M. Heidker, G. C. Caiozzi, B. S. Wong, K. Rodriguez, F. Del Rey and M.-L. Ricketts, PLoS One, 2015, 10, e0140267 CrossRef PubMed.
- N. R. Choi, J. N. Kim, M. J. Kwon, J. R. Lee, S. C. Kim, M. J. Lee, W.-G. Choi and B. J. Kim, Int. J. Med. Sci., 2022, 19, 941–951 CrossRef CAS PubMed.
- B. A. Sandoval-Ramírez, Ú. Catalán, A. Pedret, R. M. Valls, M. J. Motilva, L. Rubió and R. Solà, Clin. Nutr., 2021, 40, 1719–1732 CrossRef PubMed.
- J. L. Wallace, Br. J. Pharmacol., 2019, 176, 147–154 CrossRef CAS PubMed.
- A. Rakha, N. Umar, R. Rabail, M. S. Butt, M. Kieliszek, A. Hassoun and R. M. Aadil, Biomed. Pharmacother., 2022, 156, 113945 CrossRef CAS PubMed.
- A. I. S. Andersen-Civil, P. Arora and A. R. Williams, Front. Immunol., 2021, 12, 637603 CrossRef CAS PubMed.
- S. J. Hofer, S. Davinelli, M. Bergmann, G. Scapagnini and F. Madeo, Front. Nutr., 2021, 8, 717343 CrossRef PubMed.
- K. Martinez-Guryn, N. Hubert, K. Frazier, S. Urlass, M. W. Musch, P. Ojeda, J. F. Pierre, J. Miyoshi, T. J. Sontag, C. M. Cham, C. A. Reardon, V. Leone and E. B. Chang, Cell Host Microbe, 2018, 23, 458–469.e5 CrossRef CAS PubMed.
- X. Lu, C. Luo, J. Wu, Y. Deng, X. Mu, T. Zhang, X. Yang, Q. Liu, Z. Li, S. Tang, Y. Hu, Q. Du, J. Xu and R. Xie, J. Cell. Mol. Med., 2023, 27, 2631–2642 CrossRef CAS PubMed.
- X. Yang, T. Liu, B. Chen, F. Wang, Q. Yang and X. Chen, Sci. Rep., 2017, 7, 62 CrossRef PubMed.
- D. Han, Y. Wu, D. Lu, J. Pang, J. Hu, X. Zhang, Z. Wang, G. Zhang and J. Wang, Cell Death Dis., 2023, 14, 1–14 CrossRef PubMed.
- R. Molinari, N. Merendino and L. Costantini, BioFactors, 2022, 48, 255–273 CrossRef CAS PubMed.
- X. Wang, Y. Qi and H. Zheng, Antioxidants, 2022, 11, 1212 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4fo05133e |
| ‡ These authors contributed equally to this study. |
| This journal is © The Royal Society of Chemistry 2025 |