Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Suppression of fecal phenol production by oral supplementation of sesamol: inhibition of tyrosine phenol-lyase by sesamol†
Daiki
Oikawa
 ab,
Zion
Byun
ab,
Zion
Byun
 b,
Bunzo
Mikami
b,
Bunzo
Mikami
 cd,
Aina
Gotoh
cd,
Aina
Gotoh
 b,
Toshihiko
Katoh
b,
Toshihiko
Katoh
 b,
Ryo
Ueno
b,
Aruto
Nakajima
b,
Ryo
Ueno
b,
Aruto
Nakajima
 b,
Satoshi
Yamashita
b,
Satoshi
Yamashita
 e,
Wakako
Ikeda-Ohtsubo
e,
Wakako
Ikeda-Ohtsubo
 f,
Seiji
Takahashi
f,
Seiji
Takahashi
 a,
Toshiyuki
Waki
a,
Toshiyuki
Waki
 a,
Koichi
Kikuchi
g,
Takaaki
Abe
a,
Koichi
Kikuchi
g,
Takaaki
Abe
 g,
Takane
Katayama
g,
Takane
Katayama
 *b and
Toru
Nakayama
*b and
Toru
Nakayama
 *a
*a
aDepartment of Biomolecular Engineering, Graduate School of Engineering, Tohoku University, 6-6-11, Aza Aoba, Aramaki, Aoba-ku, Sendai, Miyagi 980-8579, Japan. E-mail: toru.nakayama.e5@tohoku.ac.jp
bDivision of Integrated Life Science, Graduate School of Biostudies, Kyoto University, Oiwake-cho, Kitashirakawa, Sakyo-ku, Kyoto 606-8502, Japan. E-mail: katayama.takane.6s@kyoto-u.ac.jp
cResearch Institute for Sustainable Humanosphere, Kyoto University, Uji, Kyoto 611-011, Japan
dInstitute of Advanced Energy, Kyoto University, Uji, Kyoto 611-011, Japan
eDepartment of Material Chemistry, Graduate School of Natural Science and Technology, Kanazawa University, Kakuma, Kanazawa, Ishikawa 902-1192, Japan
fFood and Feed Immunology Group, Laboratory of Animal Products Chemistry, Graduate School of Agricultural Science, Tohoku University, 468-1 Aoba, Aramaki, Aoba-ku, Sendai, Miyagi 980-8572, Japan
gDivision of Nephrology, Endocrinology and Vascular Medicine, Tohoku University Graduate School of Medicine, Sendai, Miyagi 980-8575, Japan
First published on 7th April 2025
Abstract
Phenol is produced from dietary L-tyrosine by the action of tyrosine phenol-lyase (TPL) of gut bacteria and contributes to various physiological disorders, including skin diseases, certain cancers, and kidney dysfunction. We found that oral supplementation of sesamol (36 or 180 μg mL−1) ad libitum for 14 days in mice significantly suppressed fecal phenol production. Fecal microbiota structure analysis in sesamol-supplemented and control groups revealed that their overall microbiota structures were indistinguishable. To explain the sesamol-induced suppression of fecal phenol production, we characterized inhibition of bacterial TPL by sesamol in vitro. Sesamol specifically inhibited bacterial TPL in a mixed-type fashion (Ki, 135 μM), which was rationalized by computational docking studies using the crystal structure of Pantoea agglomerans TPL that was determined at 1.3 Å resolution. Sesamol was detected at 0–0.295 μmol g−1 feces in the sesamol-supplemented group. Given the Ki value of sesamol for TPL inhibition, these levels may not have been sufficient to fully inhibit TPL and suppress fecal phenol production. Therefore, the observed suppression of fecal phenol production upon oral sesamol supplementation arose not solely from the inhibition of TPL by sesamol, but also potentially from the effects of metabolites derived from sesamol and the antioxidant activities of sesamol and related metabolites. Nevertheless, these findings highlight the potential for using sesamol to prevent physiological disorders associated with phenol production by the gut microbiota.
1. Introduction
Phenol is one of the microbial products metabolized from dietary L-tyrosine by gut bacteria.1 Phenol produced by gut bacteria is related to some physiological disorders in humans. As produced phenol is absorbed in the gut and delivered via the bloodstream to the skin where it accumulates, it makes the skin dull and incompletely cornified in mouse models,2 or it inhibits the differentiation of keratinocytes in female human skin.3 Phenol was also found to promote the exacerbation of skin cancer in a mouse model.4 In addition, in vitro studies showed that phenol decreases the viability of colonic epithelial cells in humans.5 Moreover, phenol produced by gut bacteria has been implicated in the onset of leukemia.6 In terms of other pathogenic effects, phenol also induces thrombocytopathy and defective platelet aggregation in patients with kidney dysfunction.7 In the liver, phenol absorbed from the gut is converted to phenyl sulfate, and its level was found to increase with the progression of diabetes in rats. Furthermore, albuminuria and podocyte damage in kidney cells were induced by its administration in experiments on diabetes models, which exacerbated kidney dysfunction.8Phenol is produced from L-tyrosine by a gut bacterial enzyme, tyrosine phenol-lyase (TPL, eqn (1)).
| L-Tyrosine + H2O → phenol + pyruvate + ammonia | (1) |
The TPL-catalyzed reaction requires pyridoxal 5′-phosphate (PLP) as a coenzyme, which is widely used in many enzymatic reactions in the amino acid metabolic pathways, including transamination and α,β- and α,γ-eliminations.9 Inhibition of bacterial TPL would be a promising strategy to prevent and/or treat physiological disorders caused by phenol microbially produced in the gut. In fact, oral administration of 2-aza-L-tyrosine, a competitive inhibitor of TPL with a Ki value of 135 μM,10 to diabetic mice not only decreased the level of phenyl sulfate in plasma and tissues but also suppressed albuminuria in the model.8
Sesame (Sesamum indicum L.) seed oil exerts nephroprotective effects and contributes to the prevention of diabetes-induced disease.11,12 However, its molecular mechanism remains to be clarified. Sesame seed oil contains lignan-related compounds, such as sesamolin, sesamin, and sesamol (3,4-methylenedioxyphenol, Fig. 1), which reportedly benefit human health in a wide variety of ways.13 Among these compounds, sesamol, which is formed by the thermal hydrolysis of sesamolin,14 is the only molecule with a phenolic group in its molecular structure and potentially mimics phenol and the phenolic moiety of L-tyrosine. Consistent with this, our preliminary results showed that sesamol indeed inhibits bacterial TPL,15 implying that it might contribute to the prevention and/or treatment of physiological disorders caused by phenol produced by gut microbiota.
In this study, to further address this issue, sesamol was orally supplemented to mice to assess its effects on fecal phenol production and microbiota structure. Detailed kinetic studies were also carried out to investigate the mode of TPL inhibition by sesamol. The X-ray crystal structure of TPL from Pantoea agglomerans (PaTPL) was then determined at 1.3 Å resolution, and using this structure a molecular docking study was performed to structurally rationalize the results of enzyme inhibition kinetics. The findings obtained in this study highlight the potential for using sesamol to prevent various physiological disorders caused by phenol production by the gut microbiota, including skin diseases, certain cancers, and kidney dysfunction.
2. Materials and methods
2.1. Chemicals
(+)-Sesamin, sesaminol, and sesamol of ≥99% purity were purchased from Nagara Science (Gifu, Japan). 2-O-(β-D-Glucopyranosyl)-β-D-glucopyranosylsesaminol [SDG(β1-2)] and 6-O-(β-D-glucopyranosyl)-β-D-glucopyranosylsesaminol [SDG(β1-6)] were prepared as described previously.16 Rabbit muscle L-lactate dehydrogenase (LDH) was obtained from Sigma-Aldrich (St Louis, MO, USA), while pig heart malate dehydrogenase (MDH) was obtained from Wako (Sendai, Japan). All other chemicals were of analytical grade.2.2. Bacterial strains
The following bacterial strains were obtained from Riken BioResource Research Center (Tsukuba, Japan): Clostridium cochlearium JCM 1396T, Citrobacter freundii JCM 1657T, Citrobacter koseri JCM 1658T, Kluyvera intermedia JCM 1238T, Lacrimispora sphenoides JCM 1415T, and Morganella morganii JCM 1672T.2.3. Animal experiments
C57BL/6N male mice of 8 weeks of age were obtained from SHIMIZU Laboratory Supplies Co., Ltd (Kyoto, Japan). The mice were co-housed in four polycarbonate cages under standard conditions [22 °C, 12 h light/dark cycle (lights on from 8 a.m. to 8 p.m.)], and randomly exchanged between the cages every day during a 7-day acclimation period. Food (MF; Oriental Yeast Co., Ltd, Tokyo, Japan) and water were available ad libitum. The mice were randomly assigned to three groups: C, L, and H. Group C served as the control, while Group L received a low dose of sesamol (36 μg mL−1 in water) and Group H received a high dose of sesamol (180 μg mL−1 in water). Each group consisted of seven mice (n = 7), which were housed individually. Body weight was measured as an indicator of food intake and health. Fecal pellets were collected from each mouse on Days 0, 7, and 14 within 6 h of defecation and stored at −30 °C until use. The fecal samples were weighed and transferred to a 2 mL plastic tube containing stainless-steel beads (φ = 5.0 mm), zirconia beads (φ = 0.1 mm, equivalent to 100 μL volume), and 5 volumes of 50% ethanol. The tubes were shaken at 1500 rpm for 10 min using a Shake Master Neo (Bio Medical Science, Tokyo, Japan), and the supernatant was collected after centrifugation. The pellets were suspended again with 50% ethanol and similarly shaken. The supernatants were combined, filtered through a 0.45 μm filter (Millipore), and subjected to the quantification of phenol and sesamol using a liquid chromatography–mass spectrometry (LC-MS) system. Animal experiments were approved by Kyoto University Animal Experimentation Committee (Lif-K24009) and performed in December 2024.2.4. Microbiota analysis
Extraction of mouse fecal DNA and subsequent 16S rRNA gene-based microbiota analysis were conducted as described previously with slight modifications.17 In brief, lyophilized fecal pellets were suspended in 300 μL of InhibitEX buffer appended to a QIAmpFast DNA Stool Mini Kit (Qiagen, Hilden, Germany), disrupted by bead-beating with a Shake Master NEO (Bio Medical Science, Tokyo, Japan), and the supernatants were collected by centrifugation. The following DNA extraction procedures were performed in accordance with the protocol provided with the kit. The V3–V4 region was amplified using a two-step PCR approach, in which KOD FX Neo (Toyobo, Osaka, Japan) was used for the first and second PCR. The sequencing was performed using a MiSeq instrument with MiSeq Reagent kit v3 (Illumina, San Diego, CA, USA). The primer sequences were trimmed with FastX_trimmer (FASTX-Toolkit, version 0.0.14), and low-quality reads (quality value <20, length <130 bp) were removed by Sickle (version 1.33). The paired-end sequences were assembled using FLASH (version 1.2.11). The reads were analyzed using the QIIME2 software package (version 2024.10). Potential chimeric sequences were removed using DADA2.18 Taxonomic classification was performed using a Naive Bayes classifier trained on the Greengenes 13.8 database clustered at the 97% identity threshold. Linear discriminant analysis (LDA) effect size (LEfSe) was used to determine the taxa that most strongly discriminated between different groups.19 Principal coordinate analysis (PCoA) was conducted based on Bray–Curtis distances, for which the vegan package in R (version 4.1.0) was used. The 16S rRNA amplicon sequencing data have been deposited in the DNA Data Bank of Japan under BioProject ID number PRJDB16730.2.5. Enzyme preparation
Human alanine aminotransferase (ALT) and aspartate aminotransferase (AST) were heterologously expressed in Escherichia coli cells, as described previously,15 and the cell-free extracts were used for enzyme assays. PaTPL20 was heterologously expressed in E. coli cells and purified as described previously.152.6. Inhibition of phenol production of gut bacteria encoding TPL-homolog by sesamol
Overnight cultures of the strains described above in Gifu Anaerobic Medium (GAM; Nissui Pharmaceutical, Tokyo, Japan) were separately transferred to fresh EC-GY medium [GAM without added sugars (Nissui Pharmaceutical) supplemented with 1% (w/v) glycerol and 0.1% (w/v) L-tyrosine] at 1/100 dilution, and the cultivation was continued anoxically at 37 °C for 24 h. The cells were harvested and disrupted by ultrasonication, followed by centrifugation. The reaction mixture (final volume, 100 μL) contained 100 mM potassium phosphate buffer (pH 8.0), 100 μM PLP, 2 mM 2-mercaptoethanol (2-ME), 1 mM L-tyrosine, 1–25 pkat of enzyme, and 500 μM sesamol. The mixture was pre-incubated at 37 °C for 10 min in the absence of the enzyme, and the reaction was initiated by enzyme addition. After incubation at 37 °C for 30 min, the reaction was stopped by adding 100 μL of 4% (v/v) trifluoroacetic acid in a 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 (v/v) mixture of acetonitrile and water. After centrifugation, the amount of phenol in the supernatant was quantified by high-performance liquid chromatography (HPLC).
6 (v/v) mixture of acetonitrile and water. After centrifugation, the amount of phenol in the supernatant was quantified by high-performance liquid chromatography (HPLC).
2.7. Effects of sesamol on activities of mammalian PLP- and NADH-dependent enzymes
ALT, AST, LDH, and MDH activities were assayed as described previously.21,22 Enzyme activities were measured in the presence of different concentrations (i.e., 0, 20, 50, or 100 μM) of sesamol.2.8. Effects of sesamol on viability of some gut bacteria
Overnight culture of bacteria (C. koseri, L. sphenoides, and M. morganii) in GAM medium was separately transferred to fresh EC-GY medium at 1/100 dilution, and the cultivation was continued anoxically at 37 °C for 24 h. The cells were harvested and suspended in an appropriate volume of a mixture containing 100 mM potassium phosphate buffer (pH 8.0), 200 μM PLP, and 1 mM 2-ME, to which 5 mM sesamol was added when necessary. The resulting cell suspensions, equivalent to OD600 of 2.0, were anaerobically incubated in the presence or absence of 0.1% L-tyrosine at 37 °C for 24 h. The cells were harvested and suspended in 0.85% NaCl at serial dilutions, 100 μL of which was plated on GAM agar plates to estimate colony-forming units (CFU) per mL of the cell suspension.2.9. Enzyme kinetics
The reaction mixture (final volume, 100 μL) contained 100 mM potassium phosphate buffer (pH 8.0), 100 μM PLP, 2 mM 2-ME, varying concentrations of L-tyrosine, 1–25 pkat mL−1PaTPL, and 15 μM sesamol or other sesame-derived compounds. The mixture was pre-incubated at 37 °C for 10 min in the absence of the enzyme, and the reaction was initiated by adding PaTPL. After incubation at 37 °C for 10 min, the reaction was stopped by adding 100 μL of 4% (v/v) trifluoroacetic acid in a 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 (v/v) mixture of acetonitrile and water. After centrifugation, the amount of phenol in the supernatant was quantified by HPLC, as described below. The inhibition mode was determined using double-reciprocal plots. Inhibition constants were determined by fitting the data to the Michaelis–Menten equation with competitive or mixed-type inhibition mode (eqn (2) or (3), respectively) using SigmaPlot (HULINKS, Tokyo, Japan).
6 (v/v) mixture of acetonitrile and water. After centrifugation, the amount of phenol in the supernatant was quantified by HPLC, as described below. The inhibition mode was determined using double-reciprocal plots. Inhibition constants were determined by fitting the data to the Michaelis–Menten equation with competitive or mixed-type inhibition mode (eqn (2) or (3), respectively) using SigmaPlot (HULINKS, Tokyo, Japan). | (2) |
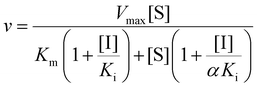 | (3) |
Here, v and Vmax denote initial and maximum velocities of enzymatic reaction, [S] and [I] denote concentrations of substrate (S) and inhibitor (I), and Km and Ki denote the Michaelis constant for S and the inhibition constant for I, respectively. α refers to the degree to which the binding of inhibitor changes the affinity of the enzyme for the substrate.
2.10. X-ray crystallographic studies of PaTPL
A tryptophanase-deficient E. coli JM101 strain transformed with a pUC-derived plasmid carrying the PaTPL gene20 was grown at 30 °C for 24 h in 500 mL of LB medium supplemented with 0.1% (w/v) L-tyrosine as an inducer. After cultivation, cells were harvested and disrupted by sonication. The cell-free extracts were subjected to 30% saturation of ammonium sulfate. Ammonium sulfate was added to the supernatant to 70% saturation, and the resulting precipitate was recovered by centrifugation. The pellet was dissolved in 3 mL of 20 mM potassium phosphate buffer (pH 8.0) containing 200 μM PLP, 2 mM 2-ME, and 1 mM EDTA (pH 8.0), and the protein was concentrated using an Amicon Ultra-4 (10 K) centrifugal filter device (Merck Millipore, Burlington, MA, USA). The protein was then loaded onto a MonoQ 5/50 GL column (GE HealthCare, Tokyo, Japan) pre-equilibrated with the same buffer and eluted by a linear gradient of 0–1 M NaCl. The enzyme was further purified by SOURCE 15PHE column chromatography (GE Healthcare), in which the elution was carried out by a linear gradient of 1–0 M ammonium sulfate. The protein fractions with high purity were collected, concentrated by ammonium sulfate precipitation, and dialyzed against a crystallization solution consisting of 10 mM KCl, 200 μM PLP, 4 mM 2-ME, and 1 mM EDTA using Slide-A-Lyzer G2 dialysis cassettes (10 K; Thermo Fisher Scientific, Tokyo, Japan). The purified enzyme was stored at −80 °C.Crystals of PaTPL were grown at 20 °C using the hanging-drop vapor-diffusion method, by mixing 1 μL of the protein solution with an equal volume of reservoir solution consisting of 0.2 M ammonium acetate (pH 7.5), 0.1 M sodium dihydrogen citrate (pH 4.2), 0.12 M sodium hydroxide, and 21% (v/v) PEG 4000. The crystals were flash-cooled in the cold nitrogen gas stream. Diffraction data were collected at 100 K at beamline BL26B1 of SPring-8 (Harima, Japan). The data sets were processed using X-ray Detector Software.23 The structure was refined with PHENIX·refine24 using the TPL model (PDB code, 1C7G) as an initial model. Model building was performed using COOT.25 Molecular graphic images were prepared using PyMOL (version 2.5.4) (Schrödinger, NY, USA) or UCSF Chimera version 1.17.1 (Resource for Biocomputing, Visualization and Informatics, CA, USA). DALI server (https://ekhidna.biocenter.helsinki.fi/dali) was used for structural comparison between PaTPL and CfTPL (PDB code, 6DUR).26
2.11. Molecular docking analysis
Computational docking simulations were performed through the SwissDock server program (https://www.swissdock.ch/)27 using the crystal structures of a holo-enzyme of PaTPL (this study; PDB code, 7FJK) and a phenylalanine-bound form of CfTPL (PDB code, 6DUR). A search for binding sites of sesamol was performed in the 15 × 15 × 15 Å cube around the PLP-binding (Lys257) residue of TPL. Based on the obtained results, representative models with estimated ΔG values more negative than −5.2 kcal mol−1 were selected. Models in which the ligand clashed with the protein surface of PaTPL and CfTPL were excluded.2.12. HPLC and LC-MS
Phenol and sesamol were quantified by either HPLC or LC-MS. For HPLC, a Shimadzu Prominence system (Shimadzu, Kyoto, Japan) equipped with a photodiode array detector (model SPD M-20A; Shimadzu) and a J′sphere ODS-H80 column (4.6 × 150 mm; YMC, Kyoto, Japan) was used. For the detection of phenol, elution was carried out at a flow rate of 0.7 mL min−1 with a linear gradient between solvent A [0.1% (v/v) trifluoroacetic acid in water] and solvent B [0.1% trifluoroacetic acid (v/v) in a 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) mixture of acetonitrile and water]. The gradient system consisted of 20% B for 0–3 min, 20%–80% B for 3–11 min, 100% B for 11–14 min, and 20% B for 14–19 min. Column temperature was maintained at 25 °C, and the run was monitored by the absorbance at 280 nm. For the detection of sesamol, elution was started with 40% B for 3 min, followed by a linear gradient of 40%–65% B for 8 min, 100% B for 3 min, and 40% B for 5 min. The other conditions were the same as used for the phenol detection.
1 (v/v) mixture of acetonitrile and water]. The gradient system consisted of 20% B for 0–3 min, 20%–80% B for 3–11 min, 100% B for 11–14 min, and 20% B for 14–19 min. Column temperature was maintained at 25 °C, and the run was monitored by the absorbance at 280 nm. For the detection of sesamol, elution was started with 40% B for 3 min, followed by a linear gradient of 40%–65% B for 8 min, 100% B for 3 min, and 40% B for 5 min. The other conditions were the same as used for the phenol detection.
LC-MS was also used for the detection of phenol and sesamol. For the detection of phenol, a Shimadzu LC-MS8030 system with an Agilent 1260 Infinity II LC equipped with an InertSustain C18 HP column (3 μm, 2.1 × 100 mm; GL Sciences, Tokyo, Japan) was used. Elution was carried out at a flow rate of 0.25 mL min−1 with a linear gradient of 40%–90% (v/v) methanol in water in 8 min, and the eluate was ionized by atmospheric-pressure chemical ionization with negative mode. Data acquisition for phenol was performed in the Q1 selected ion monitoring mode at m/z 93. For the detection of sesamol, the same LC-MS system was used. The elution was carried out at a flow rate of 0.3 mL min−1 with a linear gradient of 20%–30% (v/v) methanol in water in 9 min. An electrospray interface was used in negative mode. Data acquisition was performed in the Q1 selected ion monitoring mode at m/z 137.12. For both HPLC and LC-MS, the known concentrations of phenol and sesamol were used to produce standard curves.
2.13. Statistics
Data are shown as mean ± standard deviation (SD) or median with interquartile range. Statistical analysis was performed using GraphPad Prism version 10.4.1 (GraphPad Software Inc., San Diego, CA, USA). P values of less than 0.05 were considered statistically significant.3. Results
3.1. Effect of sesamol supplementation on fecal phenol production in mice
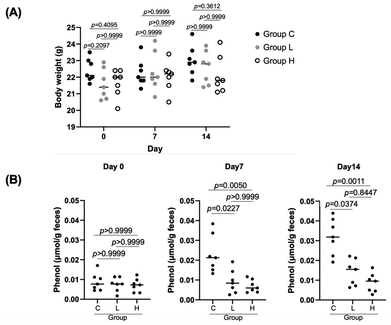
Acclimated 9-week-old C57BL/6N male mice were randomly assigned to three groups (Groups C, L, and H; n = 7 per group) and housed in parallel for 14 days with ad libitum access to food and water. The low-dose sesamol-supplemented group (Group L) received water containing 36 μg mL−1 sesamol, while the high-dose group (Group H) received water with a sesamol concentration five times higher (180 μg mL−1). The control group (Group C) received water without sesamol. Note that the dose of sesamol for Group L was determined on the basis of our previous study,8 in which 2-aza-L-tyrosine, a competitive inhibitor of TPL, was orally administrated to a mouse at 0.3 mg a day. During the course of the experiment, there were no sudden deaths, loss of appetite, or immobility among the mice used, and thus no mice were excluded. No significant differences were found among the three groups in terms of body weight on Days 0, 7, and 14 (Fig. 2A). Fecal samples were collected from the seven individuals in each of Groups C, L, and H on Days 0, 7, and 14 after supplementation. This resulted in seven fecal samples for each of nine conditions: C0, C7, C14, L0, L7, L14, H0, H7, and H14 (where, for example, C0 represents Group C on Day 0). The phenol content of these samples was measured using LC-MS. The phenol concentrations in fecal samples were 0.0230 ± 0.0095, 0.0092 ± 0.0060, and 0.0069 ± 0.0032 μmol g−1 feces for Groups C7, L7, and H7, and 0.0321 ± 0.0092, 0.0140 ± 0.0066, and 0.0087 ± 0.0049 μmol g−1 feces for Groups C14, L14, and H14, respectively (Fig. 2B). The results indicated that the fecal phenol content in Group C mice increased during the intervention, whereas those in Groups L and H remained significantly lower than in Group C.3.2. Effect of oral sesamol supplementation on fecal microbiota
Then, 16S rRNA gene-based fecal microbiota analysis was carried out with fecal samples of C0, C14, L0, L14, H0, and H14. We first analyzed the species richness (α-diversity) in the microbial communities of the fecal samples from these six conditions by comparing the number of amplicon sequence variants (ASVs), which reflects taxonomic diversity. No significant difference in the number of ASVs was detected among these six conditions (Fig. 3A). We then analyzed the phylogenetic diversity (i.e., the β-diversity) among the fecal microbiota structures using PCoA based on Bray–Curtis dissimilarities. The PCoA matrices contained the dissimilarity in species composition between all combinations of subjects, which quantifies the phylogenetic β-diversity between groups; a group with a smaller Bray–Curtis dissimilarity value has lower phylogenetic variation (i.e., smaller β-diversity) in its microbiota structure. In the results, the distributions of the plots of the seven fecal samples of the six conditions appeared to be essentially indistinguishable from each other (Fig. 3B), suggesting that the oral supplementation of sesamol did not cause a remarkable difference or shift in the microbiota structure of feces among Groups C, L, and H or within a single group over time. We also analyzed the fecal microbiota phylogenies of C0, C14, L0, L14, H0, and H14 at the phylum and genus levels (Fig. 3C). Because the relative abundances of certain phyla and genera appeared to vary slightly among the groups (Fig. 3C), we conducted LEfSe analysis to statistically examine their differences among the samples in greater detail. The results showed no statistically significant variation in the relative abundance of any bacterial taxa among C0, L0, H0, C14, L14, and H14. When performing analysis using C14, L14, and H14 to assess the effects of sesamol supplementation, we found that Staphylococcus was the only genus that showed a significant decrease in abundance in L14 and H14 compared with that at C14 (Fig. 3D). However, because the fecal abundance of this taxon was originally very low, the size of this change was negligible. Overall, these findings suggest that the gut microbiota structures of mice of Groups C, L, and H remained largely unchanged after the 14-day intervention, indicating that sesamol supplementation had minimal impact on the structure of the gut microbiota in mice.3.3. Enzymatic characterization of TPL inhibition by sesamol
In vitro studies were then conducted to assess the specificity of TPL inhibition by sesamol. Sesamol was found to inhibit the TPL activity of a wide range of gut bacteria, which had previously been shown to either accumulate high concentrations of phenol in culture supernatants or encode a TPL homolog28,29 (Fig. 4A; see also ESI Table S1†). However, it did not inhibit the activity of the following enzymes: ALT (EC 2.6.1.2) and AST (EC 2.6.1.1), which are PLP-dependent enzymes in humans, or LDH (EC 1.1.1.27, from rabbit muscle) and MDH (EC 1.1.1.37, from pig heart), which are NADH-dependent enzymes (Fig. 4B). Thus, sesamol exhibited high specificity toward bacterial TPL. Furthermore, sesamol did not affect bacterial cell viability (Fig. 4C), highlighting that its effects are limited to inhibiting gut bacterial metabolism involved in phenol production.TPL inhibition by sesamol was then kinetically analyzed using PaTPL. Double-reciprocal plots of the activity data in the presence and absence of sesamol (15 μM) intersect to the left of the 1/v-axis and above the 1/[S]-axis (Fig. 5A). These results indicate that the data exhibit the best fit to a mixed-type inhibition model (Fig. 5B), in which sesamol binds to E and ES to form catalytically inactive EI and ESI complexes, with a Ki value of 135 μM and α value of 1.57 (Fig. 5A, Table 1; see the legend to Fig. 5 for the definition of the α value). For comparison, other lignans that are structurally related to sesamol [(+)-sesamin, sesaminol, SDG(β1-2), and SDG(β1-6)] were examined for their ability to inhibit PaTPL [Table 1; see ESI Fig. S1† for chemical structures of (+)-sesamin, sesaminol, SDG(β1-2), and SDG(β1-6)]. (+)-Sesamin, sesaminol, SDG(β1-2), and SDG(β1-6) also inhibited PaTPL with similar Ki values (Table 1).
| Inhibitor | K i (μM) | α value for mixed-type inhibition | Inhibition type |
|---|---|---|---|
| a The values are presented as the average of three independent determinations with ±SDs. | |||
| 2-Aza-L-tyrosine | 71.5 ± 7.6 | — | Competitive |
| (+)-Sesamin | 77.9 ± 2.9 | 0.995 | Mixed-type |
| Sesaminol | 74.1 ± 2.3 | 6.89 | Mixed-type |
| Sesamol | 135 ± 67 | 1.57 | Mixed-type |
| SDG(β1-2) | 264 ± 77 | 0.586 | Mixed-type |
| SDG(β1-6) | 182 ± 54 | 0.208 | Mixed-type |
To structurally rationalize the results of kinetic studies of TPL inhibition by sesamol (see above), we performed X-ray crystallographic studies of PaTPL (the enzyme used for enzyme assays in this study). The crystal structure of the PLP-bound holo-enzyme of PaTPL was solved at 1.30 Å resolution (PDB code, 7FJK; Fig. 6A, see also Table 2 for data collection statistics). The crystal structure of PaTPL, which is 91% identical in amino acid sequence to CfTPL, exhibited significant structural similarity to that of CfTPL (PDB code, 6DUR),26 with root mean square deviation of 0.4 Å for 455 Cα atoms (z-score = 67.0), as revealed by a Dali search (https://ekhidna.biocenter.helsinki.fi/dali) (see ESI Fig. S2† for further details of a comparison of the crystal structures of PaTPL and CfTPL). We also attempted to obtain crystal structures of PaTPL in complex with sesamol by co-crystallization and immersion experiments, but all attempts failed. Therefore, we performed a computational docking simulation by using SwissDock (https://www.swissdock.ch/). When sesamol was docked with the crystal structure of PLP-bound PaTPL (PDB code, 7FJK), the sesamol molecule was predicted to bind to the catalytic center of the enzyme (Fig. 6B, left). In the modeled structure, the methylenedioxy ring of the bound sesamol is located near Tyr71 of PaTPL (Fig. 6B, middle), which corresponds to the residue from the neighboring subunit in the catalytic dimer.30 Moreover, a comparison of this structural model with the crystal structure (PDB code, 6DUR) of CfTPL complexed with L-phenylalanine, a competitive inhibitor of TPL, suggested that the bound sesamol is located where the pheny moiety of the bound phenylalanine is accommodated in CfTPL (Fig. 6B, right). These docking results were consistent with the proposed inhibition model, where sesamol competes with L-tyrosine for the active site of TPL (Fig. 5B, step i-1). We also examined the possible formation of an ESI complex (Fig. 5B, step i-2) by computational docking of sesamol with the L-phenylalanine-bound CfTPL structure (PDB code, 6DUR), which mimics the ES complex. It was predicted that sesamol binds to the entrance of the active-site cavity (Fig. 6C) of the binary complex to form ESI and inhibit product formation (Fig. 5B, step i-2). These docking results are consistent with the proposed model of TPL inhibition by sesamol based on our kinetic analysis (Fig. 5).
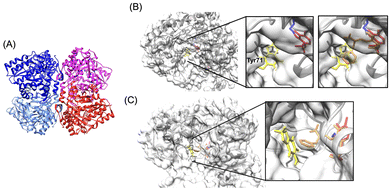 | ||
| Fig. 6 The crystal structure of PaTPL and the predicted structures of sesamol–TPL complexes. (A) The crystal structure of PaTPL determined at 1.3 Å resolution (PDB code, 7FJK). The overall structure consists of four identical subunits, each of which is differently colored, and can be regarded as a “dimer of a dimer”. The subunits of one of these dimers (left) are shown in blue and sky blue, and those of the other dimer (right) are shown in magenta and red. Each of these dimers produces the active site in its monomer–monomer interface and conjugates at the N-terminus, resulting in the formation of a tetrameric structure. For further details of the characterization of the crystal structure of PaTPL, see ESI Fig. S2.† (B) Predicted structure of a binary PaTPL–sesamol complex. One of the results of docking simulations with the estimated ΔG values more negative than −5.2 kcal mol−1 is shown. Left, The structure of a dimer portion of PaTPL is shown using an overlaid ribbon and surface diagram, with catalytic Lys257 (blue), Tyr71 (dark gray), bound sesamol (yellow), and PLP (red) shown in a stick model. Middle, Magnified images showing the predicted location of the bound sesamol molecule in the catalytic site. Right, Superposition of the sesamol-bound model of PaTPL and L-phenylalanine molecule bound in the CfTPL structure (PDB code, 6DUR), in which overlapping monomer structures of PaTPL (PDB code, 7FJK) and CfTPL (PDB code, 6DUR) are shown using a ribbon model (PaTPL, dark gray; CfTPL, light gray). The positions of sesamol and the phenyl moiety of L-phenylalanine partially overlap. (C) Predicted structure of a ternary CfTPL–sesamol–L-phenylalanine complex. One of the representative models with estimated ΔG values more negative than −5.2 kcal mol−1 is shown. One of the top results of docking simulations is shown. Close-up image around the active site of PaTPL and CfTPL is also presented next to the overall structure with a stick model of catalytic residue Lys257 (PaTPL, blue; CfTPL, sky blue), Tyr71 in PaTPL (black), cofactor PLP (red), L-phenylalanine quinonoid complex binding to CfTPL (orange), and sesamol (yellow). | ||
| TPL/citrate | |
|---|---|
| A. Diffraction data | |
| X-ray source | SPring-8/BL26B1 |
| Detector | Eiger4M |
| Wavelength (Å) | 1.0 |
| Resolution range (Å) | 50.00–1.30 (1.37–1.30) |
| Space group | P21212 |
| Unit cell parameters | |
| a, b, c (A) | 111.62, 161.08, 100.15 |
| Unique reflections | 442![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 369 (70 369 (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 556) 556) |
| Multiplicity | 4.17 (4.11) |
| Completeness (%) | 99.6 (98.9) |
| Mean I/σ(I) | 12.0 (2.6) |
| Wilson B-factor (Å2) | 19.1 |
| R merge (%) | 6.3 (44.6) |
| R meas (%) | 7.3 (51.1) |
| CC1/2 (%) | 99.7 (85.6) |
| B. Refinement statistics | |
| Refinement program | Phenix 1.18.2 |
| Resolution range used refinement | 45.68–1.30 (1.31–1.30) |
| Number of reflections | 442![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 342 (13 342 (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 991) 991) |
| R work (%) | 12.9 (24.7) |
| R free (%) | 15.5 (28.6) |
| Number of protein residues | 456 (1–456) × 4 (chain A, B, C, D) |
| PLP/CIT/EOD/PEG/PGE/water | 4/4/3/65/2/1/2040 |
| R.m.s.d., bond lengths (Å) | 0.005 |
| R.m.s.d., bond angles (°) | 0.86 |
| Ramachandran favored (%) | 98.1 |
| Rarnachandran outliers (%) | 0 |
| Rotamer outliers (%) | 1.2 |
| Clash score | 5.3 |
| PDB ID | 7FJK |
3.4. Determination of fecal sesamol content
Fecal sesamol contents of the mice in Groups L and H were measured on Days 0, 7, and 14. A low level of sesamol was found in almost all of the mice in both groups on Day 7 and Day 14, whereas no detectable level of sesamol could be found in each group on Day 0 (Table 3).| Group | ||||
|---|---|---|---|---|
| L | H | |||
| Day | Mouse no. | Sesamol content (μmol g−1 feces) | Mouse no. | Sesamol content (μmol g−1 feces) |
| a Below detection limit (0.005 μmol g−1 feces). | ||||
| 0 | 1 | 1 | ||
| 2 | 2 | |||
| 3 | 3 | |||
| 4 | 4 | |||
| 5 | 5 | |||
| 6 | 6 | |||
| 7 | 7 | |||
| 7 | 1 | 1 | 0.025 | |
| 2 | 0.006 | 2 | 0.016 | |
| 3 | 0.014 | 3 | 0.027 | |
| 4 | 0.007 | 4 | 0.020 | |
| 5 | 0.011 | 5 | 0.034 | |
| 6 | 0.014 | 6 | 0.015 | |
| 7 | 7 | 0.014 | ||
| 14 | 1 | 0.042 | 1 | 0.234 |
| 2 | 0.051 | 2 | 0.129 | |
| 3 | 0.041 | 3 | 0.023 | |
| 4 | 0.048 | 4 | 0.045 | |
| 5 | 0.026 | 5 | 0.074 | |
| 6 | 0.005 | 6 | 0.033 | |
| 7 | 0.026 | 7 | 0.295 | |
4. Discussion
A wide variety of human health benefits conferred by the bioactivities of sesamol have been reported,13 which include antioxidant,31 neuroprotective,32 cardioprotective,33 anticancer,34,35 anti-inflammatory,36 radioprotective,37 and skin-protective effects.38 In this study, we demonstrated in mice that oral supplementation of sesamol significantly suppressed phenol production in feces (Fig. 2B), while not causing significant changes in the gut microbiota structure (Fig. 3). These results provide evidence supporting an additional potential role of sesamol as a nutraceutical (a food chemical that provides health benefits in addition to its nutritional content), namely, for the prevention and treatment of physiological disorders caused by phenol produced by gut microbiota, including skin diseases, certain cancers, and kidney dysfunction.A previous study using a mouse model showed that the oral administration of 2-aza-L-tyrosine also decreased phenol levels in feces and achieved a nephroprotective effect (i.e., suppressing albuminuria).8 2-Aza-L-tyrosine is a competitive inhibitor of TPL,10 and these observations have been ascribed to the fact that this amino acid derivative inhibits TPL of gut bacteria to suppress phenol production.8 Therefore, we further investigated the mechanistic aspects of the observed suppression of fecal phenol production by sesamol. Results from the present TPL inhibition studies revealed that sesamol acts as an inhibitor of TPL irrespective of the microbial origin of the enzyme, without killing TPL-producing microbes (Fig. 4A and C). Our previous study showed that sesamol also acted as an inhibitor of bacterial tryptophan indole-lyase, which is responsible for the formation of indoxyl sulfate, a uremic toxin.15 For comparison, however, sesamol was shown not to inhibit human ALT, human AST, rabbit muscle LDH, and pig heart MDH (Fig. 4B). Therefore, enzyme inhibition by sesamol appears to be specific to PLP-dependent enzymes catalyzing the α,β-elimination of aromatic amino acids. Kinetic analysis indicated that sesamol acts as a mixed-type inhibitor of TPL, with a Ki value of 135 μM, which is comparable to the Ki of 2-aza-L-tyrosine [135 μM,10 72 μM (this study; see Table 1)] known for its ability to decrease fecal phenol levels.8 The specific inhibition of TPL by sesamol was further supported by structural models of TPL–sesamol complexes. It was predicted that sesamol binds to the active site of TPL via its interaction with Tyr71 (Fig. 6B), which is functionally important for TPL catalysis.30 It was also suggested that sesamol possibly binds near the active site of the TPL enzyme–substrate complex to inhibit catalytic reactions (Fig. 6C).
Sesamol was detected in the feces of nearly all mice in Groups L and H on Days 7 and 14. However, its concentrations were low. Given the Ki value of sesamol for TPL inhibition, these levels may not have been sufficient to fully inhibit TPL and suppress fecal phenol production. The low recovery of orally administered sesamol in mouse feces is likely due to xenobiotic metabolism. In rats, sesamol undergoes hepatic metabolism, converting it into 2-methoxybenzene-1,4-diol, benzene-1,2,4-triol, and glucuronide or sulfate conjugates.39,40 Although sesamol metabolism in mice has not been extensively studied, similar metabolic pathways are likely present, reducing the proportion of orally supplemented sesamol that reaches the colon. Additionally, sesamol conjugates formed in the liver may be excreted into bile, transported to the colon, and subsequently deconjugated by microbial activity.41 Fecal sesamol concentrations may also be affected by water absorption in the colon. Variability in these processes and water intake from ad libitum administration could account for the sporadic detection of sesamol in certain mice (e.g., mouse No. 1 and 7 in Group H). Thus, the observed reduction in fecal phenol production following oral sesamol supplementation was likely due not only to the direct inhibition of TPL by sesamol but also, at least in part, to unidentified effects of its metabolites generated through hepatic xenobiotic metabolism. Further research is needed to identify these metabolites and clarify their role in phenol suppression. Moreover, recent studies have suggested that the antioxidant properties of certain phenolic compounds can influence metabolic pathways independently of their effects on gut microbiota.42 Sesamol is known for its strong antioxidant activity,43 which may modulate amino acid metabolism, particularly that of L-tyrosine. Additionally, oral administration of a diet containing sesaminol, a sesame-derived lignan with antioxidant properties, has been shown to increase tocopherol levels in the plasma and tissues of rats.44 These findings suggest that the suppression of phenol production by sesamol in mice may involve similar indirect mechanisms.
A key challenge in the effective use of nutraceuticals for disease prevention and therapy is the limited understanding of their mechanisms of action, which underpins their efficacy.45,46 The identification of sesamol as a bacterial TPL inhibitor strongly suggests its potential as a mechanism-based agent46 for both preventive and therapeutic strategies in addressing physiological disorders caused by phenol production from gut microbiota, including skin diseases, certain cancers, and kidney dysfunction. However, the bioavailability of sesamol—specifically, the proportion of ingested sesamol that reaches the target site (the gut) in its active form—must be taken into account. Only sesamol that reaches and is distributed to the gut can exert its beneficial health effects via TPL inhibition. To achieve this, several strategies could be explored. For example, the encapsulation of sesamol in enteric-coated capsules and its administration to deliver concentrations equivalent to its Ki in the colon should be investigated in future studies.
5. Conclusions
Oral supplementation of sesamol in mice significantly suppressed fecal phenol production with only a minimal effect on the gut microbiota structure. Although sesamol specifically inhibited bacterial TPL activity, the suppression of fecal phenol production could be only partly attributed to TPL inhibition. Nevertheless, the findings from this study underscore the potential of using sesamol to prevent physiological disorders linked to phenol production by the gut microbiota, including skin diseases, certain cancers, and kidney dysfunction.Abbreviations
| 2-ME | 2-Mercaptoethanol |
| ALT | Alanine aminotransferase |
| AST | Aspartate transaminase |
| ASV | Amplicon sequence variant |
| HPLC | High-performance liquid chromatography |
| LC-MS | Liquid chromatography-mass spectrometry |
| LDA | Linear discriminant analysis |
| LDH | Lactate dehydrogenase |
| LEfSe | Linear discriminant analysis effect size |
| MDH | Malate dehydrogenase |
| NADH | Nicotinamide adenine dinucleotide reduced form |
| PLP | Pyridoxal 5′-phosphate |
| TPL | Tyrosine phenol-lyase |
Author contributions
Daiki Oikawa: conceptualization, funding acquisition, investigation, data curation, validation, writing – original draft preparation. Zion Byun: investigation, data curation. Bunzo Mikami: conceptualization, investigation, data curation, validation, writing – reviewing and editing. Aina Gotoh: investigation, data curation. Toshihiko Katoh: investigation, data curation. Ryo Ueno: investigation. Aruto Nakajima: investigation. Satoshi Yamashita: methodology, data curation. Wakako Ikeda-Ohtsubo: investigation, data curation. Seiji Takahashi: investigation, data curation. Toshiyuki Waki: investigation, data curation. Koichi Kikuchi: investigation, data curation. Takaaki Abe: conceptualization, funding acquisition, supervision, project administration. Takane Katayama: conceptualization, funding acquisition, supervision, investigation, data curation, validation, project administration, writing – reviewing and editing. Toru Nakayama: conceptualization, funding acquisition, supervision, investigation, data curation, validation, project administration, writing – reviewing and editing.Data availability
Coordinates and structure factors of the crystal structure of the PLP-bound holo-enzyme of PaTPL have been deposited in the Protein Data Bank with accession code 7FJK. The 16S rRNA amplicon sequencing data have been deposited in the DNA Data Bank of Japan under BioProject ID number PRJDB16730. Data will be made available on request.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
Diffraction data were collected at the BL26B1 beamline of SPring-8, Sayo, Hyogo, Japan, with the approval of JASRI (proposal no. 2020A2744). This work was supported in part by a National Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (20K20604 and 21H02932 to TA, JP22J00724 to DO, and 23H05470 to TN), grants from the Japan Agency for Medical Research and Development (AMED) (22ek0120133, JP22ak010127, JP23fk0108655, and JP23zf127001 to TA), and a grant from Noda Institute for Scientific Research (to DO). We thank Edanz Group (https://jp.edanz.com) for editing a draft of this manuscript.References
- E. A. Smith and G. T. Macfarlane, Formation of phenolic and indolic compounds by anaerobic bacteria in the human large intestine, Microb. Ecol., 1997, 33, 180–188 CrossRef CAS PubMed.
- R. Iizuka, K. Kawakami, N. Izawa and K. Chiba, Phenols produced by gut bacteria affect the skin in hairless mice, Microb. Ecol. Health Dis., 2009, 21, 50–56 CrossRef CAS.
- R. Iizuka, K. Kawakami and K. Chiba, Gut bacteria producing phenols disturb keratinocyte differentiation in human skin, Microb. Ecol. Health Dis., 2009, 21, 221–227 CrossRef CAS.
- R. K. Boutwell and D. K. Bosch, The tumor-promoting action of phenol and related compounds for mouse skin, Cancer Res., 1959, 19, 413–424 CAS.
- K. Windey, V. De Preter and K. Verbeke, Relevance of protein fermentation to gut health, Mol. Nutr. Food Res., 2012, 56, 184–196 CrossRef CAS PubMed.
- T. A. McDonald, N. T. Holland, C. Skibola, P. Duramad and M. T. Smith, Hypothesis: phenol and hydroquinone derived mainly from diet and gastrointestinal flora activity are causal factors in leukemia, Leukemia, 2001, 15, 10–20 CrossRef CAS PubMed.
- S. F. Rabiner and F. Molinas, The role of phenol and phenolic acids on the thrombocytopathy and defective platelet aggregation of patients with renal failure, Am. J. Med., 1970, 49, 346–351 CrossRef CAS PubMed.
- K. Kikuchi, D. Saigusa, Y. Kanemitsu, Y. Matsumoto, P. Thanai, N. Suzuki and T. Abe, Gut microbiome-derived phenyl sulfate contributes to albuminuria in diabetic kidney disease, Nat. Commun., 2019, 10, 1835 CrossRef PubMed.
- R. S. Phillips, Chemistry and diversity of pyridoxal-5′-phosphate dependent enzymes, Biochim. Biophys. Acta, Proteins Proteomics, 2015, 1854, 1167–1174 CrossRef CAS PubMed.
- E. B. Watkins and R. S. Phillips, Inhibition of tyrosine phenol-lyase from Citrobacter freundii by 2-azatyrosine and 3-azatyrosine, Biochemistry, 2001, 40, 14862–14868 CrossRef CAS PubMed.
- D. Z. Hsu, Y. H. Li, P. Y. Chu, S. Periasamy and M. Y. Liu, Sesame oil prevents acute kidney injury induced by the synergistic action of aminoglycoside and iodinated contrast in rats, Antimicrob. Agents Chemother., 2011, 55, 2532–2536 CrossRef CAS PubMed.
- D. Sankar, A. Ali, G. Sambandam and R. Rao, Sesame oil exhibits synergistic effect with anti-diabetic medication in patients with type 2 diabetes mellitus, Clin. Nutr., 2011, 30, 351–358 CrossRef CAS PubMed.
- A. Shah, R. Lobo, N. Krishnadas and R. Surubhotla, Sesamol and health—a comprehensive review, Indian J. Pharm. Educ. Res., 2019, 53, S28–S42 CrossRef CAS.
- H. Yoshida and S. Takagi, Effects of seed roasting temperature and time on the quality characteristics of sesame (Sesamum indicum) oil, J. Sci. Food Agric., 1997, 75, 19–26 CrossRef CAS.
- D. Oikawa, S. Yamashita, S. Takahashi, T. Waki, K. Kikuchi, T. Abe and T. Nakayama, (+)-Sesamin, a sesame lignan, is a potent inhibitor of gut bacterial tryptophan indole-lyase that is a key enzyme in chronic kidney disease pathogenesis, Biochem. Biophys. Res. Commun., 2022, 590, 158–162 CrossRef CAS PubMed.
- A. Nair, A. Kuwahara, A. Nagase, H. Yamaguchi, T. Yamazaki, M. Hosoya, A. Omura, K. Kiyomoto, M. Yamaguchi, T. Shimoyama, S. Takahashi and T. Nakayama, Purification, gene cloning, and biochemical characterization of a β-glucosidase capable of hydrolyzing sesaminol triglucoside from Paenibacillus sp. KB0549, PLoS One, 2013, 8, e60538 CrossRef CAS PubMed.
- T. Katoh, C. Yamada, M. D. Wallace, A. Yoshida, A. Gotoh, M. Arai, T. Maeshibu, T. Kashima, A. Hagenbeek, M. N. Ojima, H. Takada, M. Sakanaka, H. Shimizu, K. Nishiyama, H. Ashida, J. Hirose, M. Suarez-Diez, M. Nishiyama, I. Kimura, K. A. Stubbs, S. Fushinobu and T. Katayama, A bacterial sulfoglycosidase highlights mucin O-glycan breakdown in the gut ecosystem, Nat. Chem. Biol., 2023, 19, 778–789 CrossRef CAS PubMed.
- B. J. Callahan, P. J. McMurdie, M. J. Rosen, A. W. Han, A. J. A. Johnson and S. P. Holmes, DADA2: high-resolution sample inference from Illumina amplicon data, Nat. Methods, 2016, 13, 581–583 CrossRef CAS PubMed.
- N. Segata, J. Izard, L. Waldron, D. Gevers, L. Miropolsky, W. S. Garrett and C. Huttenhower, Metagenomic biomarker discovery and explanation, Genome Biol., 2011, 12, R60 CrossRef PubMed.
- T. Koyanagi, T. Katayama, A. Hirao, H. Suzuki and H. Kumagai, Construction of an effective protein expression system using the TPL promoter in Escherichia coli, Biotechnol. Lett., 2005, 27, 1267–1271 CrossRef CAS PubMed.
- H. U. Bergmeyer and E. Berndt, in Methods of Enzymatic Analysis, ed. H. U. Bergmeyer, Academic Press, New York, 2nd edn, 1974, pp. 574–579 Search PubMed.
- R. Rej, Aspartate aminotransferase activity and isoenzyme proportions in human liver tissues, Clin. Chem., 1978, 24, 1971–1979 CrossRef CAS.
- W. Kabsch, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 125–132 CrossRef CAS PubMed.
- D. Liebschner, P. V. Afonine, M. L. Baker, G. Bunkoczi, V. B. Chen, T. I. Croll, B. Hintze, L.-W. Hung, S. Jain, A. J. McCoy, N. W. Moriarty, R. D. Oeffner, B. K. Poon, M. G. Prisant, R. J. Read, J. S. Richardson, D. C. Richardson, M. D. Sammito, O. V. Sobolev, D. H. Stockwell, T. C. Terwilliger, A. G. Urzhumtsev, L. L. Videau, C. J. Williams and P. D. Adams, Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix, Acta Crystallogr., Sect. D: Struct. Biol., 2019, 75, 861–877 CrossRef CAS PubMed.
- P. Emsley, B. Lohkamp, W. G. Scott and K. Cowtan, Features and development of Coot, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 486–501 CrossRef CAS PubMed.
- R. S. Phillips and S. Craig, Crystal structures of wild-type and F448A mutant Citrobacter freundii, tyrosine phenol-lyase complexed with a substrate and inhibitors: implications for the reaction mechanism, Biochemistry, 2018, 57, 6166–6179 CrossRef CAS PubMed.
- A. Grosdidier, V. Zoete and O. Michielin, SwissDock, a protein-small molecule docking web service based on EADock DSS, Nucleic Acids Res., 2011, 39, W270–W277 CrossRef CAS PubMed.
- Y. Saito, T. Sato, K. Nomoto and H. Tsuji, Identification of phenol- and p-cresol-producing intestinal bacteria by using media supplemented with tyrosine and its metabolites, FEMS Microbiol. Ecol., 2018, 94, fiy125 CrossRef CAS PubMed.
- W. Yuan, S. Zhong, Y. Xiao, Z. Wang and J. Sun, Efficient biocatalyst of L-DOPA with Escherichia coli expressing a tyrosine phenol-lyase mutant from Kluyvera intermedia, Appl. Biochem. Biotechnol., 2020, 190, 1187–1200 CrossRef CAS PubMed.
- T. Katayama and H. Kumagai, Tyrosine Phenol–Lyase, in Encyclopedia of Industrial Biotechnology, ed. and M. C. Flickinger, John Wiley & Sons, 2010, pp. 4752–4757.
- J. Lee and E. Choe, Extraction of lignan compounds from roasted sesame oil and their effects on the autoxidation of methyl linoleate, J. Food Sci., 2006, 71, C430–C436 CAS.
- A. K. Sachdeva, S. Misra, I. P. Kaur and K. Chopra, Neuroprotective potential of sesamol and its loaded solid lipid nanoparticles in ICV-STZ-induced cognitive deficits: behavioral and biochemical evidence, Eur. J. Pharmacol., 2015, 747, 132–140 CrossRef CAS PubMed.
- P. Jayaraj, C. A. Narasimhulu, S. Rajagopalan, S. Parthasarathy and R. Desikan, Sesamol: a powerful functional food ingredient from sesame oil for cardioprotection, Food Funct., 2020, 11, 1198–1210 RSC.
- T. Geetha, P. K. Deol and I. P. Kaur, Role of sesamol-loaded floating beads in gastric cancers: a pharmacokinetic and biochemical evidence, J. Microencapsulation, 2015, 32, 478–487 CAS.
- G. J. Kapadia, M. A. Azuine, H. Tokuda, M. Takasaki, T. Mukainaka, T. Konoshima and H. Nishino, Chemopreventive effect of resveratrol, sesamol, sesame oil and sunflower oil in the Epstein-Barr virus early antigen activation assay and the mouse skin two-stage carcinogenesis, Pharmacol. Res., 2002, 45, 499–505 CrossRef CAS PubMed.
- P. Y. Chu, D. Z. Hsu, P. Y. Hsu and M. Y. Liu, Sesamol down-regulates the lipopolysaccharide-induced inflammatory response by inhibiting nuclear factor-kappa B activation, Innate Immun., 2010, 16, 333–339 CrossRef CAS PubMed.
- V. K. Parihar, K. R. Prabhakar, V. P. Veerapur, M. S. Kumar, Y. R. Reddy, R. Joshi, M. K. Unnikrishnan and C. M. Rao, Effect of sesamol on radiation-induced cytotoxicity in Swiss albino mice, Mutat. Res., Genet. Toxicol. Environ. Mutagen., 2006, 611, 9–16 CrossRef CAS PubMed.
- M. Srisayam, N. Weerapreeyakul, S. Barusrux and K. Kanokmedhakul, Antioxidant, antimelanogenic, and skin-protective effect of sesamol, J. Cosmet. Sci., 2014, 65, 69–79 CAS.
- K. C. Jan, C. T. Ho and L. S. Hwang, Bioavailability and tissue distribution of sesamol in rat, J. Agric. Food Chem., 2008, 56, 7032–7037 CrossRef CAS PubMed.
- K. C. Jan, C. T. Ho and L. S. Hwang, Elimination and metabolism of sesamol, a bioactive compound in sesame oil, in rats, Mol. Nutr. Food Res., 2009, 53, 36–43 Search PubMed.
- C. D. Klaassen and J. Y. Cui, Mechanisms of how the intestinal microbiota alters the effects of drugs and bile acids, Drug Metab. Dispos., 2015, 43, 1505–1521 CrossRef CAS PubMed.
- X. Yang, R. Hu, L. Yao, W. Zhang, M. Shi, J. Gong, X. Yuan, Y. Li, J. Yan, Y. Wang, Q. Zhang, Z. He, D.-X. Hou, Z. Fan, H. Zhang, L. Chen, X. He, J. He and S. Wu, The role of uterus mitochondrial function in high-fat diet-related adverse pregnancy outcomes and protection by resveratrol, Food Funct., 2024, 15, 4852–4861 RSC.
- M. Andargie, M. Vinas, A. Rathgeb, E. Möller and P. Karlovsky, Lignans of sesame (Sesamum indicum L.): A comprehensive review, Molecules, 2021, 26, 883 CrossRef CAS PubMed.
- K. Yamashita, Y. Yamada, S. Kitou, S. Ikeda, C. Abe, N. M. Saarinen and R. Santti, Hydroxymatairesinol and sesaminol act differently on tocopherol concentrations in rats, J. Nutr. Sci. Vitaminol., 2007, 53, 393–399 CrossRef CAS PubMed.
- A. Santini, G. C. Tenore and E. Novellino, Nutraceuticals: a paradigm of proactive medicine, Eur. J. Pharm. Sci., 2017, 96, 53–61 CrossRef CAS PubMed.
- B. Wilffert, J. Swen, H. Mulder, D. Touw, A.-H. M. Van der Zee and V. Deneer, From evidence based medicine to mechanism based medicine. Reviewing the role of pharmacogenetics, Int. J. Clin. Pharm., 2013, 35, 369–375 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4fo04839c |
| This journal is © The Royal Society of Chemistry 2025 |