 Open Access Article
Open Access ArticleEffect of protein texturization on amino acids and protein in vitro bioaccessibility of pea and rice protein†
Irene
Peñaranda
 a,
María Dolores
Garrido
a,
Purificación
García-Segovia
a,
María Dolores
Garrido
a,
Purificación
García-Segovia
 b,
Javier
Martínez-Monzó
b,
Javier
Martínez-Monzó
 b and
Marta
Igual
b and
Marta
Igual
 *b
*b
aDepartment of Food Science and Technology, Veterinary Faculty, University of Murcia, Espinardo, 30100, Murcia, Spain
bUniversitat Politècnica de València, i-Food, Instituto de Ingeniería de Alimentos (FoodUPV), Camino de Vera s/n, 46022 València, Spain. E-mail: marigra@upvnet.upv.es; Tel: +34963877361
First published on 4th February 2025
Abstract
The growing demand for vegetable proteins, driven by population increase and interest in sustainable protein-rich diets, has generated a focus on the use of legumes and cereals as protein sources for the production of meat analogues due to their complementary nutritional profile and lower environmental impact. However, the analogues should not only imitate the nutritional profile of meat, but their texture is also important for good commercialization. One of the processes used to improve these properties is protein texturization by thermoextrusion, which could modify the availability and digestibility of amino acids. Thus, the objective of this work is to evaluate the in vitro bioaccessibility of proteins and amino acids from pea and rice protein isolates (I) and texturized proteins (T). Proteins from pea (PP), rice (RP) and a 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 mixture (PRP) were processed by extrusion, evaluating the effects on digestibility and bioaccessibility of essential (EAA) and non-essential (NEAA) amino acids. The results showed higher in vitro digestibility and bioaccessibility of NEAA of texturized proteins compared to isolated proteins, although this effect is not significant for PP. However, texturization significantly reduced the bioaccessibility of some EAA such as lysine. Rice protein showed greater stability during the extrusion process, maintaining a more balanced amino acid profile. Texturization can therefore be a useful tool to improve the functionality of vegetable proteins, but it is necessary to optimize the process to minimize nutritional losses.
50 mixture (PRP) were processed by extrusion, evaluating the effects on digestibility and bioaccessibility of essential (EAA) and non-essential (NEAA) amino acids. The results showed higher in vitro digestibility and bioaccessibility of NEAA of texturized proteins compared to isolated proteins, although this effect is not significant for PP. However, texturization significantly reduced the bioaccessibility of some EAA such as lysine. Rice protein showed greater stability during the extrusion process, maintaining a more balanced amino acid profile. Texturization can therefore be a useful tool to improve the functionality of vegetable proteins, but it is necessary to optimize the process to minimize nutritional losses.
Introduction
In the world today, where the pursuit of a healthy lifestyle is an undisputed priority, the importance of a balanced, protein-rich diet has become paramount. Protein, the macronutrient essential for body growth and tissue repair, as well as for producing metabolic and digestive enzymes, is therefore a crucial nutritional element in the daily diet.1 However, while traditionally the predominant source of protein has been of animal origin, an emerging trend is changing the nutrition landscape due to the growing interest in and consumption of plant-based proteins.2 The 2030 Agenda for Sustainable Development and the United Nations Sustainable Development Goals (SDGs)3 point towards a necessary change in the way food is produced and consumed, not only to contribute to improving human health but also to increasing productivity and sustainability, there is a clear commitment to diets based fundamentally on plant-based foods, with a variable composition depending on the ethical considerations of each region, which must be produced using environmentally friendly techniques and obtained from fair sources for all members of the food chain.This growing demand for plant protein production is in line with the population growth expected in the coming years, which seems, among other things, to make massive consumption of animal protein unsustainable, as the growth in global meat protein consumption during this decade is expected to increase by 14% by 2030.4,5
Among the most commercially used plant proteins for the production of meat substitutes, soybean stands out for its numerous economic and functional benefits, as it supports well the secondary structures of soybean proteins in the extrusion process and for its high nutritional quality.6,7 However, due to its allergenic power other proteins from the legume family are gaining importance as a protein source for meat analogues such as peas, beans and chickpeas,8,9 although its use is not gaining traction because it has a less complete amino acid profile than soy, lacks the essential amino acids necessary for human function and its iron and manganese content is lower.10 Among these legumes, pea stands out as it is hypoallergenic protein rich in the essential amino acids lysine (7 g g−1 protein) and branched-chain amino acids, but still deficient in methionine (1 g g−1 protein) and vitamins.11,12 Hence, other alternatives are being sought, such as combining legumes with cereals to achieve a more balanced nutrient composition and a high biological value protein profile,13 especially rice due to its lower allergenic potential than wheat, mild taste, and better processing characteristics.14,15 In particular, rice is rich in methionine and cysteine (3.5 g g−1 protein) and deficient in lysine.15 Therefore, the combination of pulses with rice balances the essential amino acid profile, achieving a protein value closer to the WHO/FAO ideal standard (2.5 g g−1 protein of methionine and cysteine per g protein and 4.5 g lysine per g protein).1
Analogues must not only mimic the nutritional profile of meat, their appearance, texture and mouthfeel are especially important for good marketability.16 When considering vegetable proteins, a question arises: protein isolate or textured protein? Protein isolate refers to a purified, concentrated form of protein, while textured protein is characterised by its more fibrous, meat-like structure.17 While both options have their advantages, textured protein is gaining ground due to its ability to mimic the texture and taste of meat,9,18 making it particularly attractive to those looking to reduce their consumption of animal products without compromising their nutritional and organoleptic quality.
The texturization process of vegetable proteins involves extrusion and heat treatment, which transforms the proteins into a fibrous meat-like structure. During extrusion, plant proteins are modified by pressure, thermal and mechanical forces, which can lead to protein denaturation and solubility, as well as changes in molecular structure, which can lead to the formation of protein aggregates.19 Other factors such as the availability and digestibility of amino acids may be modified by extrusion due to the decrease of anti-nutrients in many vegetables such as phytates or tannins.20 This process may offer consumers meat analogues with an improved nutritional profile and potentially improved functionality.
However, when considering the nutritional value of plant proteins, it is essential to take into account their bioaccessibility, i.e. the amount of nutrients that the body can absorb and utilise after digestion.21 Protein digestibility is affected by factors such as protein structure, food source (animal or plant), anti-nutritional compounds, and human digestive physiology. Animal proteins are highly digestible, with rates exceeding 95%, while plant proteins range from 65–90% due to structural differences and anti-nutritional factors that hinder digestion and absorption.22 In this sense, the use of standardised bioaccessibility assessment techniques, such as the INFOGEST model, is crucial to determine their physiological application.23
Ultimately, the growing interest in plant proteins, especially from pea and rice for their hypoallergenic, amino acid profile and optimal processing characteristics,12,13,15 is transforming the landscape of modern nutrition. The choice between isolated and textured proteins, together with the assessment of their bioaccessibility, are key aspects to consider when integrating these proteins into a balanced and healthy diet. Therefore, the aim of this study was to investigate the bioaccessibility of amino acids from isolates and texturized pea and/or rice protein using the in vitro gastrointestinal digestion model.
Materials and methods
Raw materials
Pea protein isolate powder (PP) (Nutralys S85F) and rice isolate protein powder (RP) (Nutralys rice I800XF) were supplied by Roquette Laisa España S.A. (Benifaió, Valencia, Spain).Mixtures and preparation of texturized protein
Three types of samples were processed: PP, RP and a 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 mixture (PRP). All three were hydrated till 30% of water content by continuously mixing at medium speed in the same mixer, previously to extrusion. Then, samples were introduced in a single-screw Kompaktextruder KE 19/25 extruder (Brabender, Duisburg, Germany). The conditions used for extrusion were: 3
50 mixture (PRP). All three were hydrated till 30% of water content by continuously mixing at medium speed in the same mixer, previously to extrusion. Then, samples were introduced in a single-screw Kompaktextruder KE 19/25 extruder (Brabender, Duisburg, Germany). The conditions used for extrusion were: 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 compression ratio, dosing speed of 18 rpm (feed rate range, 1.34 kg h−1), a nozzle of 3 mm of diameter, 150 rpm of screw rotation, and 40, 80, 120, and 120 °C of temperature section barrel. These conditions have been tested in other works obtaining satisfactory texturized products.9
1 compression ratio, dosing speed of 18 rpm (feed rate range, 1.34 kg h−1), a nozzle of 3 mm of diameter, 150 rpm of screw rotation, and 40, 80, 120, and 120 °C of temperature section barrel. These conditions have been tested in other works obtaining satisfactory texturized products.9
Barrel temperatures (T1 and T2), melted pressure, screw speed and motor torque were registered using Extruder Winext software (Brabender). Extruded products were immediately dried at 25 °C for 18 h. Dried samples were stored in polyethylene bags at room temperature (25 °C) and used for further analysis.
Specific mechanical energy (SME) was calculated according eqn (1):
 | (1) |
In vitro digestion
Sample in vitro digestibility was evaluated following the standardized static in vitro digestion protocol designed for food (COST INFOGEST network) as outlined by Minekus et al.24 and Baugreet et al.25 The process consisted of four sequential phases: oral phase, gastric phase, intestinal phase and filtration. Oral phase consisted on mixing samples with simulate salivary fluid (SSF) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and amylase (75 U mL−1) for 2 min at pH 7. In gastric phase, oral bolus is mixed with simulate gastric fluid (SGF) (1
1) and amylase (75 U mL−1) for 2 min at pH 7. In gastric phase, oral bolus is mixed with simulate gastric fluid (SGF) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), pepsin (2000 U mL−1) and gastric lipase (60 U mL−1) for 2 h at pH 3. For intestinal phase, gastric chime was mixed with simulate intestinal fluid (SIF) (1
1), pepsin (2000 U mL−1) and gastric lipase (60 U mL−1) for 2 h at pH 3. For intestinal phase, gastric chime was mixed with simulate intestinal fluid (SIF) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and pancreatin (trypsin activity 100 U mL−1) for 2 h at pH 7. Finally, mixture was centrifuged at 2600g for 30 min and then filtering through a 1 μm glass-fibre membrane.26 Enzymes concentration used was estimated according to the activity certificated by the analysis from the manufacturer (Sigma-Aldrich). Simulates fluids was prepared according Minekus et al.24 The in vitro digestibility (IVD) (%) was computed according to the method described by Batista et al.27 All analyses were conducted in triplicate. Post-digestion samples were collected following the procedure detailed by Minekus et al.24 and subsequently freeze-dried with the addition of a protease inhibitor (Pefabloc SC, Sigma-Aldrich).
1) and pancreatin (trypsin activity 100 U mL−1) for 2 h at pH 7. Finally, mixture was centrifuged at 2600g for 30 min and then filtering through a 1 μm glass-fibre membrane.26 Enzymes concentration used was estimated according to the activity certificated by the analysis from the manufacturer (Sigma-Aldrich). Simulates fluids was prepared according Minekus et al.24 The in vitro digestibility (IVD) (%) was computed according to the method described by Batista et al.27 All analyses were conducted in triplicate. Post-digestion samples were collected following the procedure detailed by Minekus et al.24 and subsequently freeze-dried with the addition of a protease inhibitor (Pefabloc SC, Sigma-Aldrich).
Crude protein (CP)
The nitrogen content was determined by the Kjeldahl procedure on a SpeedDigester K-436 and distiller K-350 (Büchi, Labortechnik AG, Flawil, Switzerland) according to the official method 955.04 of the AOAC International.28 The crude protein (CP) was calculated as the nitrogen content multiplied by the nitrogen factor according to each formulation. The nitrogen-protein factor used for pea and rice flour was 5.34 and 5.95, respectively.29 With CP values calculated, the bio-accessibility was determined using eqn (2) proposed by Khouzam et al.30 and Sahuquillo et al.31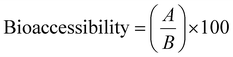 | (2) |
Amino acids (AA)
Amino acids (AA) were analysed by prior digestion with HCl 6 N of the sample, according to the procedure described by Utrera et al.32 The extract was neutralized and filtered by Nylon 0.45 μm and 1 μL was injected in LCMS system. Analysis were carried out in a Liquid Chromatography (1290 Infinity II, Agilent Technologies, Santa Clara, CA, USA) and coupled to an Agilent 6470 QqQ Mass Spectrometer (Agilent Technologies, Santa Clara, CA, USA) using an Agilent Jet Stream Dual electrospray (AJS-Dual ESI, Agilent Technologies, Santa Clara, CA, USA) interface. The mass spectrometer was operated in positive mode and was run in MS/MS mode to each amino acid. The control of the HPLC and QqQ detector were made by the MassHunter Workstation Data Acquisition software (Rev. B.08.00, Agilent Technologies, Santa Clara, CA, USA). The results are expressed in g amino acid per 100 g sample. All analyses were conducted in triplicate.Statistical analysis
Analysis of variance (ANOVA), with a confidence level of 95% (p < 0.05), using Statgraphics Centurion XVII Software (version 17.2.04, Statgraphics Technologies, Inc., The Plains, VA, USA) was applied to evaluate the differences among samples. The method used to discriminate between means is Fisher's least significant difference procedure. Principal Component Analysis (PCA) was applied to explore the relationships among AA studied.Results and discussion
Extrusion parameters
Table 1 shows the control parameters during extrusion to obtain the texturized samples. The extrusion temperatures did not show significant (p > 0.05) differences between the samples studied. In terms of water content, RP showed significantly (p < 0.05) higher values than PP but both showed no significant (p > 0.05) differences with PRP. RP reached significantly (p < 0.05) higher pressure during extrusion than the rest of the samples. The opposite trend was observed for PP. The specific mechanical energy values (SME) were significantly (p < 0.05) higher in PP than in the other samples, being the values significantly (p < 0.05) lower for RP. This tendency was observed by other works in high-moisture extrusion when incorporating RP in PP.33 The insolubility of RP reduced viscosity, led to shorter retention times in the extruder barrel, thus required less mechanical and thermal energy during the extrusion process.34 For both, pressure (P) and SME, PRP reached intermediate values because its composition is a mixture of 50% of each of the above.| Sample | T 1 (°C) | T 2 (°C) | P (Pa) | SME (J kg−1) | X w (gw per 100 g) |
|---|---|---|---|---|---|
| The same letter in superscript within the column indicates homogeneous groups established by ANOVA (p < 0.05). PP: pea protein; RP: rice protein; PRP: pea-rice protein. | |||||
| PP | 123.1 (1.3)a | 127 (2)a | 19 (5)c | 1.413 (0.009)a | 20.6 (0.5)b |
| RP | 123.7 (0.5)a | 124 (2)a | 56 (3)a | 1.035 (0.008)c | 22.8 (0.6)a |
| PRP | 123.4 (0.6)a | 124 (2)a | 39 (4)b | 1.255 (0.005)b | 21.9 (0.6)ab |
Protein and amino acid (AA) content of samples
Comparative analysis of the amino acid content in different plant protein samples provides an in-depth insight into their nutritional composition and potential value for human health. In Table 2, the amino acid profile of pea (PP) and rice (RP) proteins, along with a combination of both (PRP), in both isolated (I) and texturized (T) forms, is presented.| Amino acids | IPP | IRP | IPRP | TPP | TRP | TPRP |
|---|---|---|---|---|---|---|
| IPP: isolate pea protein; IRP: isolate rice protein; IPRP: isolate pea-rice protein; TPP: texturized pea protein; TRP: texturized rice protein; TPRP: texturized pea-rice protein. The same letter in superscript within row indicates homogeneous groups established by ANOVA (p < 0.05). | ||||||
| Essential amino acids (EAA) | ||||||
| Histidine | 2.1 (0.3)a | 2.0 (0.5)ab | 1.6 (0.3)ab | 1.3 (0.2)ab | 1.4 (0.2)ab | 1.3 (0.3)b |
| Arginine | 6.97 (0.13)ab | 7.8 (0.5)a | 6.82 (0.14)bc | 6.0 (0.4)cd | 5.4 (0.5)d | 5.72 (0.12)d |
| Threonine | 3.2 (0.3)a | 2.85 (0.02)ab | 2.8 (0.2)ab | 2.7 (0.4)ab | 2.4 (0.3)b | 2.7 (0.2)ab |
| Valine | 4.75 (0.11)a | 4.7 (0.7)a | 4.3 (0.2)a | 3.5 (0.2)b | 4.16 (0.07)ab | 4.04 (0.13)ab |
| Methionine | 0.93 (0.02)c | 1.82 (0.06)a | 1.00 (0.15)bc | 0.73 (0.04)d | 1.67 (0.02)a | 1.10 (0.02)b |
| Tryptophan | 0.30 (0.02)b | 0.43 (0.03)a | 0.14 (0.03)c | 0.08 (0.02)d | 0.12 (0.02)cd | 0.12 (0.02)cd |
| Lysine | 3.0 (0.3)a | 1.1 (0.3)d | 2.80 (0.14)ab | 2.3 (0.2)bc | 1.4 (0.2)d | 2.15 (0.09)c |
| Isoleucine | 3.68 (0.13)a | 2.9 (0.2)b | 3.5 (0.3)a | 2.8 (0.3)b | 2.6 (0.2)b | 2.89 (0.04)b |
| Leucine | 7.32 (0.07)a | 7.13 (0.07)a | 7.16 (0.07)a | 5.6 (0.5)b | 5.53 (0.06)b | 5.8 (0.5)b |
| Phenylalanine | 4.3 (0.3)a | 4.21 (0.02)a | 4.36 (0.08)a | 3.60 (0.02)b | 3.22 (0.12)b | 3.6 (0.2)b |
| Total EAA | 36.5 (0.5)a | 34.9 (0.3)ab | 34.5 (0.4)b | 28.6 (0.2)c | 27.9 (1.4)c | 29.5 (0.9)c |
| Non-essential amino acids (NEAA) | ||||||
| Aspartic acid | 9 (2)ab | 5.8 (0.5)bc | 10 (2)a | 7 (2)abc | 4.31 (0.02)c | 6.55 (0.02)abc |
| Serine | 4.0 (0.3)a | 3.9 (0.2)a | 4.03 (0.12)a | 3.29 (0.13)b | 3.06 (0.05)b | 3.4 (0.3)b |
| Glutamic acid | 11 (3)a | 12 (2)a | 11 (4)a | 9 (3)a | 9 (2)a | 10 (3)a |
| Glycine | 2.4 (0.3)ab | 2.8 (0.3)a | 2.4 (0.2)ab | 2.0 (0.2)b | 1.9 (0.2)b | 2.4 (0.3)ab |
| Alanine | 3.28 (0.13)b | 4.28 (0.02)a | 4.24 (0.02)a | 2.68 (0.12)c | 3.25 (0.06)b | 3.1 (0.2)b |
| Proline | 0.35 (0.02)b | 0.56 (0.05)a | 0.35 (0.04)b | 0.25 (0.02)c | 0.21 (0.02)c | 0.19 (0.02)c |
| Cystine | 0.47 (0.07)d | 1.15 (0.07)a | 0.52 (0.03)d | 0.41 (0.02)d | 0.84 (0.06)b | 0.71 (0.04)c |
| Tyrosine | 3.85 (0.06)a | 4.2 (0.3)a | 3.91 (0.09)a | 2.41 (0.15)c | 2.96 (0.07)b | 2.87 (0.16)b |
| Asparagine | <0.05 (0.00) | <0.05 (0.00) | <0.05 (0.00) | <0.05 (0.00) | <0.05 (0.00) | <0.05 (0.00) |
| Glutamine | <0.05 (0.00) | <0.05 (0.00) | <0.05 (0.00) | <0.05 (0.00) | <0.05 (0.00) | <0.05 (0.00) |
| Total NEAA | 35 (2)a | 34.9 (1.5)a | 35.8 (1.9)a | 27.6 (1.3)b | 26 (3)b | 29.3 (1.3)b |
First, to highlight that the total protein content of the isolated samples ranged between 70 g per 100 g of product (IPP: 71.19 ± 0.14, IRP: 67.88 ± 0.86, IPRP: 72.89 ± 0.89), on the contrary, the texturized proteins had a lower protein content around 55 g per 100 g of texturized (TPP: 56.15 ± 1.81, TRP: 53.83 ± 0.01, TPRP: 55.59 ± 0.70). Extrusion conditions, such as high temperature, pressure, and shearing can reduce the protein content due to complete denaturation and protein–protein aggregation, as well as chemical reactions such as Maillard's reaction.35 Although the texturization process produced a decrease in protein content, these products still presented a high protein content, allowing to cover the daily recommended protein requirements of the population (40–60 g protein per day, 0.83 g protein per kg body weight per day).1
As for essential amino acids (EAA), the greatest discrepancies were observed between the different vegetable protein samples, especially between methionine and lysine, which may influence their nutritional quality and their ability to meet daily protein requirements. Rice protein isolate and texturized rice protein had the highest methionine content (p < 0.05), while pea protein isolate (IPP) had the lowest methionine content and the highest lysine content compared to the other proteins evaluated (p < 0.05). These results are in agreement with those obtained by other authors, since legumes, although rich in essential amino acids such as lysine, are limiting in methionine, while grasses such as rice have a complementary amino acid profile, richer in methionine.13,15,36 For the amino acids tryptophan and isoleucine, significant differences were also observed between rice and pea protein isolate (p < 0.05), with pea protein isolate having the highest isoleucine and the lowest tryptophan content. In the texturized proteins these differences were not so evident between TPP, TRP, and TPRP.
For the rest of essential amino acids, no significant differences were observed between rice protein, pea protein, and the combination of both (p ≥ 0.05), but there was a significant reduction (p < 0.05) in EAA due to extrusion cooking, except for histidine, threonine in all protein types evaluated, in valine, methionine, lysine, and isoleucine in rice protein (IRP and TRP), and in valine and tryptophan between IPRP and TPRP (p ≥ 0.05). Therefore, pea protein was more affected by the extrusion process. During the extrusion cooking process, several factors intervene that can affect the protein content, such as higher cooking temperature, residence time, mechanical forces, and lower protein moisture.37 In other words, the higher the intensity of the process, the greater the loss of amino acids due to complete denaturation of the protein and protein–protein aggregation. It should also be noted that the higher values of certain amino acids in isolated proteins could be attributable to purification processes that concentrate these amino acids.35 Pea protein is more susceptible to be affected by extrusion processing and transformed into disordered structures. As energy input increases, the β-sheet and α-helix structures decrease, because the intramolecular hydrogen bonds in pea protein are more vulnerable to proteolysis or aggregation of new disulfide (S–S) bonds compared to those in rice and soybean proteins.38 In addition, previous research indicates that non-covalent interactions are more significant than S–S bonds in maintaining the rigid pea structure.37,39
In the total EAA and non-essential amino acids (NEAA) content, no significant differences (p ≥ 0.05) were observed between the different types of protein (rice, pea, and combination of both), except between IPP and IPRP where the IPRP had lower total EAA content than IPP (p < 0.05). Therefore the combination of pea and rice protein did not improve the total free EAA content of the protein isolates. However, there were significant differences between the isolated proteins (IPP, IRP, IPRP) and the texturized proteins (TPP, TRP, TPRP). Texturized proteins shows the lowest total amino acid content (p < 0.05), both EAA and NEAA, with losses of around 20% for both pea and rice and 15% for the mixture of the two. Previous studies have reported that among all amino acids, lysine is the most easily affected during extrusion. Lysine is prone to participate in the Maillard reaction under high temperature conditions, due to the presence of a highly reactive free ε-amino group.37,40 The loss of lysine content usually ranges from 12 to 49%, depending on the extrusion conditions.40 In this work, the losses of lysine content were 23.4% in TPP and 23.2% in TPRP, with no difference for TRP as discussed above (p < 0.05), validating that optimal processing conditions could cause a less severe reduction of amino acid content.
As for the total NEAA, a higher proline and cysteine content was found in the rice protein isolate (IRP) and alanine content in the IRP and IPRP (p < 0.05), while the pea-textured samples showed the lowest proline and alanine content, and the TPP and IPP showed the lowest cysteine content. Differences due to the extrusion process were therefore shown for the amino acid serine, alanine, proline, and tyrosine (p < 0.05). This is in agreement with general knowledge, where it is stated that rice protein is richer in NEAA compared to pea.36 Furthermore, as previously commented, high feed moisture conditions and lower energy input (temperature and pressure) have a protective effect on amino acid denaturation during the texturization process, as especially moisture reduces shear stress and mechanical energy dissipation in the extruder.37,41 Beck et al.39 saw how the extrusion of pea protein with a moisture content of 35% caused proteolysis of the protein fraction. Although when working with higher humidities of around 60%, protein denaturation, Maillard reactions, and the formation of cross-links between proteins that can make some protein fractions insoluble or less accessible are minimal, and the formation of cross-links between proteins that may cause some protein fractions to become insoluble or less accessible are minimal. Pea protein is still more vulnerable to the extrusion process, regardless of whether it is high or low moisture, than rice and soy proteins.35,38
A Principal Component Analysis (PCA) was made to a better understand the behavior of amino acids (Fig. 1). The first main factor (F1) explained 46.98% of the variation among samples, while the second main factor (F2) explained 27.81% of the variance, with a cumulative variance contribution of F1 and F2 of 74.79%, which explains much of the variability of the amino acid content of the samples evaluated.
In the PCA factor loading plot (Fig. 1a), the amino acids methionine, tyrosine, serine, phenylalanine, glycine, histidine, alanine, tryptophan, leucine, threonine, and proline were positively correlated with F1, while lysine, isoleucine, aspartic, and glutamic acids, arginine, and valine were negatively correlated with F2. Therefore, the non-proportional distribution of factor loadings indicates that vectors F1 and F2 are capturing different directions of variability in the data. Suggesting that the amino acids in the lower right hand side show similarities in their chemical or structural characteristics, the same is true for those in the upper right hand side. Highlighting amino acids such as threonine and proline, and serine and phenylalanine which are closely related to each other, indicating that they tend to vary together in protein samples.
As for the amino acids cysteine and tyrosine, it is observed that they have a strong positive influence on F2 (r = 0.967 and r = 0.725, respectively). In general, the nonessential amino acids were the ones that exhibited the heaviest factor loadings, suggesting that they are important in distinguishing between the different types of proteins evaluated. In addition, it can be seen how cysteine and lysine are clearly separated from the other amino acids in terms of F2 and F1, respectively, and to a lesser extent methionine, suggesting that these amino acids have unique characteristics that are not shared with the others.
Lysine has a longer and more flexible side chain compared to other amino acids, composed of four carbons plus a terminal amino group (–NH2) which allows it to participate in ionic interactions and hydrogen bridges with other molecules or with parts of the same protein, providing greater stability and functionality of proteins.42 Both cysteine and methionine contain sulphur atoms in their structure, this is a distinctive feature that differentiates them from the rest of the amino acids. Cysteine has a thiol group (–SH) that can form disulphide bonds (–S–S–) with another cysteine residue, which is crucial for the stabilization of the three-dimensional structure of proteins and methionine has a thioether group (–S–), which also confers greater structural stability.43 Therefore, covalent disulphide bonds are essential to maintain the three-dimensional conformation of the protein by linking different parts of the polypeptide chain, providing rigidity and resistance to denaturation during heating,44 thus these amino acids are more resistant to the extrusion process. Although as discussed in pea protein it appears that non-covalent interactions play a more significant role in protein stability.39
On the other hand, Fig. 1b allowed grouping the samples according to their characteristics by projecting them on the principal component axes. Samples TPRP, TPP, and TRP are grouped in the upper right quadrant and are positively influenced by the F1 component where the highest variability of amino acids is explained, suggesting that they have relatively similar amino acid profiles due to the processing method, extrusion cooking, which affected their composition. Showing a greater distancing the TPRP sample especially with the TRP sample, possibly due to its lower percentage of amino acid loss during the extrusion process, although as shown in Table 2 no significant differences were observed between textured proteins. Therefore, the extrusion process is key in the final amino acid profile of plant proteins.35
In contrast, the distance of IPP, IRP, and IPRP samples in different quadrants suggests a greater variability in their amino acid content, indicative of their different biological nature.36 The location of IPP on the F1 axis suggests that certain specific amino acid characteristics are dominant in these samples, especially the amino acid lysine, since legumes such as pea are rich in this amino acid.13,36 The IRP sample is isolated in the lower left quadrant, suggesting that it has a unique amino acid profile, mainly negatively influenced by F2 where a higher content of cysteine stands out. While the greatest scatter is observed in the IPRP which could imply significant variability in its composition due to the mixture of its amino acid profiles from both rice and pea.13,15 Proteins from legumes/pulses, which typically have lower levels of sulphur-containing amino acids and higher levels of lysine, can be complementary to cereal proteins, which contain lower levels of lysine and higher levels of sulphur-containing amino acids.45 Thus, consuming a food containing a mixture of two protein sources can meet the indispensable amino acid requirements of humans.
In vitro digestibility (IVD), and protein and AAs bioaccessibility
The percentage digestibility of pea protein was significantly (p < 0.05) higher than that of the other samples (Fig. 2). The texturization process favours the digestibility of pea protein, which showed the highest IVD% value among studied samples. However, this effect was not observed in rice protein.Several researchers have proposed that extrusion may be an effective way to improve the digestibility of plant protein.46Fig. 3 shows the protein and amino acid bioaccessibility of each of the samples studied before and after texturization. The significant increase in protein bioaccessibility after texturization of PP, RP, and PRP is evident (Fig. 3a). This confirms the trends shown by other authors. Wang et al.47 described the effect of extrusion with the transformation of plant protein from spherical to fibrous shapes, and the formation of larger aggregate particles. They also indicated that extrusion has the ability to modify the secondary structure of proteins by decreasing the a-helix/β-sheet ratio and impact on the tertiary structure by decreasing disulphide bonds and hydrophobic interactions. These alterations improve the digestibility and absorption of plant proteins. Zhang et al.48 also reported changes in protein digestibility in canola meal as a protein-rich food ingredient after extrusion. It has also been reported that extrusion resulted in a reduction of trypsin inhibitory activity and phytic acid content with an increase in protein digestibility.49,50Fig. 3b shows the same trend as above, but in this case the bioaccessibility quantified is for essential and non-essential amino acids and the sum of both. In addition, the figure shows that the samples with rice protein (RP and PRP) present this effect more markedly than PP. Extrusion is a process that has improved the bioaccessibility of amino acids, particularly those sulfur-containing amino acids,51 which are found to a greater extent in rice. In other matrices such as beans or lentils, also improvements in the bioaccessibility of amino acids by extrusion have been seen.51,52 The greatest differences by texturization in terms of amino acid bioaccessibility were found in NEAA. Isolated samples presented NEAA bioaccessibility values between 49 and 60% and texturized samples between 67 and 70%. However, the isolated samples presented bioaccessibility values of EAA between 42 and 59% and the texturized samples between 53 and 61%. PP showed no significant (p > 0.05) differences in the AA bioaccessibility between isolate and texturized samples.
Table 3 shows the effect of PP, RP, and PRP texturization on the bioaccessibility of individual amino acids. In general, an increase in the amino acids bioaccessibility by the extrusion process was observed, except for lysine where the opposite effect was detected. There were bioaccessibilities of amino acids that did not show a significant (p ≥ 0.05) effect by texturization such as histidine, arginine, isoleucine, glutamic acid, and cystine. However, tryptophan, leucine, serine, glycine, and tyrosine showed significantly (p < 0.05) higher bioaccessibilities after texturization for all samples studied. The bioaccessibilities of threonine, valine, and methionine were more affected by texturization in PP than in RP or RPR. However, the bioaccessibilities of phenylalanine, aspartic acid, alanine, and proline were more affected by texturization in samples containing RP.
| Amino acids | IPP | IRP | IPRP | TPP | TRP | TPRP |
|---|---|---|---|---|---|---|
| IPP: isolate pea protein; IRP: isolate rice protein; IPRP: isolate pea-rice protein; TPP: texturized pea protein; TRP: texturized rice protein; TPRP: texturized pea-rice protein. The same letter in superscript within row indicates homogeneous groups established by ANOVA (p ≤ 0.05). | ||||||
| Essential amino acids (EAA) | ||||||
| Histidine | 56 (10)a | 42 (6)a | 50 (9)a | 58 (12)a | 59.6 (0.3)a | 58.9 (0.9)a |
| Arginine | 63.8 (1.5)a | 60 (7)a | 49 (2)b | 67 (2)a | 64 (7)a | 64.3 (0.5)a |
| Threonine | 53 (3)bc | 50 (9)bc | 44 (2)c | 69.2 (1.2)a | 57 (2)b | 50 (2)bc |
| Valine | 52 (3)d | 57 (3)bc | 50.7 (0.6)d | 59.4 (0.2)b | 63.6 (0.5)a | 53 (2)cd |
| Methionine | 36.2 (1.2)c | 25.9 (1.2)d | 55 (2)ab | 60 (6)a | 46 (7)bc | 42 (3)c |
| Tryptophan | 52 (5)d | 75 (2)bc | 63 (5)cd | 87 (5)a | 85 (7)ab | 80 (2)ab |
| Lysine | 77 (3)ab | 82 (3)a | 14.0 (0.8)d | 51 (6)c | 69 (6)b | 0 (0)e |
| Isoleucine | 66 (3)a | 52 (7)bc | 31 (5)d | 61 (2)ab | 59.4 (1.5)ab | 49.2 (0.9)c |
| Leucine | 54.9 (0.5)c | 42.0 (0.4)d | 35 (5)e | 62.3 (0.3)a | 60.4 (1.2)ab | 55 (3)c |
| Phenylalanine | 58 (4)b | 47 (5)c | 59 (2)b | 57.8 (0.9)b | 70 (6)a | 71 (5)a |
| Non-essential amino acids (NEAA) | ||||||
| Aspartic acid | 54 (3)b | 61 (7)b | 33 (7)c | 60 (6)b | 82 (9)a | 54 (2)b |
| Serine | 56 (9)bc | 50 (2)c | 66 (6)ab | 78 (8)a | 76 (9)a | 80.1 (0.2)a |
| Glutamic acid | 67 (4)ab | 63 (11)ab | 52 (4)b | 74.6 (1.4)a | 69 (11)a | 68 (4)ab |
| Glycine | 79.5 (1.2)b | 51 (3)c | 75 (3)b | 91 (4)a | 96.7 (1.4)a | 88 (6)a |
| Alanine | 57 (2)b | 42 (5)d | 48 (5)cd | 62 (2)ab | 68.7 (0.04)a | 54.5 (0.8)bc |
| Proline | 43 (2)c | 26 (2)d | 7.7 (0.2)e | 37 (6)c | 77 (2)a | 50.2 (0.5)b |
| Cystine | 38.7 (1.6)cd | 17 (5)e | 61.3 (1.2)a | 44 (2)c | 33.2 (0.6)d | 51.6 (0.2)b |
| Tyrosine | 49.1 (0.9)c | 34 (4)d | 49.8 (0.5)c | 61.4 (0.5)b | 47.33(0.05)c | 73.1 (1.3)a |
Conclusions
The extrusion process increased the in vitro digestibility of the proteins, with pea protein reaching the highest values compared to rice and the mixture of both. In terms of bioaccessibility, texturization improved the accessibility of EAA and NEAA compared to the isolated proteins, except for lysine, which showed a significant decrease due to its susceptibility to high temperatures during extrusion.Regarding the differences between proteins, rice protein showed greater stability to extrusion compared to pea protein, which was more susceptible to protein denaturation and aggregation. This is reflected in a lower loss of essential amino acids in rice, while pea experienced greater reductions in the amino acid methionine and lysine. The combination of pea and rice proteins had an intermediate performance, showing an improvement in tyrosine bioaccessibility. However, the combination of the two did not result in a significant improvement in the total bioaccessibility of essential amino acids.
These findings suggest that texturization can be a useful tool to improve digestibility and AA bioaccessibility, but negatively affects lysine, highlighting the importance of optimizing extrusion parameters in order to minimize losses of essential amino acids.
Author contributions
Conceptualization, P. G., M. I., I. P., M. D. G. and J. M.; conducted the study, M. I.; data curation, M. I., I. P.; formal analysis: I. P., M. I.; methodology, M. I. and I. P.; software M. I. and J. M.; validation, M. I., I. P., M. D. G., P. G. and J. M.; formal analysis, M. I., and I. P.; investigation, I. P., M. I. and J. M.; resources, M. D. G., J. M. and P. G.; writing—original draft preparation, M. I. and I. P.; writing—review and editing, I. P., M. I., M. D. G. and J. M. and P. G.; visualisation, M. I., I. P., M. D. G. and P. G.; supervision, M. I., I. P. and J. M.; project administration, M. D. G., P. G. and J. M.; funding acquisition, M. D. G. and P. G. All authors have read and approved the final manuscript.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors want to thank Juan Vicente Baldo from Food Technology Department, Universitat Politècnica de València, for his technical support and Dr Antonio Silva of the research support services of the Facility of Innovation and Analysis in Animal Source Foodstuffs (SiPA) of SAIUEx of the University of Extremadura for the amino acids analysis.References
- FAO, Assessment of dietary protein quality in human nutrition: report of an FAO expert consultation. 2013 in FAO Food and Nutrition Paper 92. https://www.fao.org.
- A. Hassoun, F. Boukid, A. Pasqualone, C. J. Bryant, G. G. García, C. Parra-López, S. Jagtap, H. Trollman, J. Cropotova and F. J. Barba, Emerging trends in the agri-food sector: Digitalisation and shift to plant-based diets, Curr. Res. Food Sci., 2022, 5, 2261–2269 CrossRef PubMed.
- S. M. Morán and C. M. D. Barrado, El objetivo de desarrollo sostenible 11 de la Agenda 2030: ciudades y comunidades sostenibles. Metas, desafíos, políticas y logros, Cuadernos de Estrategia, 2020, vol. 206, pp. 21–68 Search PubMed.
- FAO, Dried pulses, total: crops and livestock products. 2023, FAOSTAT. https://www.fao.org/faostat/es/#search/legumbres.
- K. Kołodziejczak, A. Onopiuk, A. Szpicer and A. Poltorak, Meat analogues in the perspective of recent scientific research: A review, Foods, 2021, 11, 105 CrossRef PubMed.
- S. Samard, T. T. Maung, B. Y. Gu, M. H. Kim and G. H. Ryu, Influences of extrusion parameters on physicochemical properties of textured vegetable proteins and its meatless burger patty, Food Sci. Biotechnol., 2021, 30, 395–403 CrossRef CAS PubMed.
- B. Mao, J. Singh, S. Hodgkinson, M. Farouk and L. Kaur, Conformational changes and product quality of high-moisture extrudates produced from soy, rice, and pea proteins, Food Hydrocolloids, 2024, 147, 109341 CrossRef CAS.
- S. R. Hertzler, J. C. Lieblein-Boff, M. Weiler and C. Allgeier, Plant proteins: assessing their nutritional quality and effects on health and physical function, Nutrients, 2020, 12, 3704 CrossRef CAS PubMed.
- I. Peñaranda, M. D. Garrido, P. García-Segovia, J. Martínez-Monzó and M. Igual, Enriched pea protein texturing: Physicochemical characteristics and application as a substitute for meat in hamburgers, Foods, 2023, 12, 1303 CrossRef PubMed.
- T. Zhang, W. Dou, X. Zhang, Y. Zhao, Y. Zhang, L. Jiang and X. Sui, The development history and recent updates on soy protein-based meat alternatives, Trends Food Sci. Technol., 2021, 109, 702–710 CrossRef CAS.
- L. Angiolillo, A. Conte and M. A. Del Nobile, Technological strategies to produce functional meat burgers, LWT, 2015, 62, 697–703 CrossRef CAS.
- L. Sha and Y. L. Xiong, Plant protein-based alternatives of reconstructed meat: Science, technology, and challenges, Trends Food Sci. Technol., 2020, 102, 51–61 CrossRef CAS.
- L. Day, J. A. Cakebread and S. M. Loveday, Food proteins from animals and plants: Differences in the nutritional and functional properties, Trends Food Sci. Technol., 2020, 119, 428–442 CrossRef.
- M. Di Cairano, N. Condelli, F. Galgano and M. Caruso, Experimental gluten–free biscuits with underexploited flours versus commercial products: Preference pattern and sensory characterisation by Check All That Apply Questionnaire. International, J. Food Sci. Technol., 2022, 57, 1936–1944 CrossRef CAS.
- J. S. Lee, H. Oh, I. Choi, C. S. Yoon and J. Han, Physico-chemical characteristics of rice protein-based novel textured vegetable proteins as meat analogues produced by low-moisture extrusion cooking technology, LWT, 2022, 157, 113056 CrossRef CAS.
- H. J. Lee, H. I. Yong, M. Kim, Y. S. Choi and C. Jo, Status of meat alternatives and their potential role in the future meat market—A review. Asian-Australas, J. Anim. Sci., 2020, 33, 1533 CAS.
- P. Kataria, M. Nautiyal, I. Tuteja, V. Sharma, F. Ahmad, S. Haque, M. Shahwan, E. Capanoglu, R. Vashishth and A. Gupta, Comprehensive review on the role of plant protein as a possible meat analogue: Framing the future of meat, ACS Omega, 2023, 8, 23305–23319 CrossRef PubMed.
- M. Immonen, A. Chandrakusuma, J. Sibakov, M. Poikelispää and T. Sontag-Strohm, Texturization of a Blend of Pea and Destarched Oat Protein Using High-Moisture Extrusion, Foods, 2021, 10, 1517 CrossRef CAS PubMed.
- H. Zheng, G. Yan, Y. Lee, C. Alcaraz, S. Marquez and E. G. de Mejia, Effect of the extrusion process on allergen reduction and the texture change of soybean protein isolate-corn and soybean flour-corn mixtures, Innovative Food Sci. Emerging Technol., 2020, 64, 102421 CrossRef CAS.
- M. G. Nosworthy, G. Medina, A. J. Franczyk, J. Neufeld, P. Appah, A. Utioh, P. Frohlich and J. House, Effect of processing on the in vitro and in vivo protein quality of beans (Phaseolus vulgaris and Vicia Faba), Nutrients, 2018, 10, 671 CrossRef PubMed.
- M. Igual, A. Păucean, D. C. Vodnar, P. García-Segovia, J. Martínez-Monzó and M. S. Chiş, In Vitro Bioaccessibility of Bioactive Compounds from Rosehip-Enriched Corn Extrudates, Molecules, 2022, 27, 1972 CrossRef CAS PubMed.
- S. Shaghaghian, D. J. McClements, M. Khalesi, M. Garcia-Vaquero and A. Mirzapour-Kouhdasht, Digestibility and bioavailability of plant-based proteins intended for use in meat analogues: A review, Trends Food Sci. Technol., 2022, 129, 646–656 CrossRef CAS.
- J. Auer, M. Alminger, M. Marinea, M. Johansson, G. Zamaratskaia, A. Högberg and M. Langton, Assessing the digestibility and estimated bioavailability/bioaccessibility of plant-based proteins and minerals from soy, pea, and faba bean ingredients, LWT, 2024, 115893 CrossRef CAS.
- M. Minekus, M. Alminger, P. Alvito, S. Balance, T. Bohn, C. Bourlieu, F. Carríere, R. Boutrou, M. Corredig, D. Dupont, D. Dufour, L. Egger, M. Golging, S. Karakaya, B. Kirkhus, S. L. Feunteun, U. Lesmes, A. Macierzanka, A. Mackie, S. Marze, D. J. McClements, O. Ḿenard, I. Recio, C. N. Santos, R. P. Singh, G. E. Vegarud, M. S. J. Wickham, W. Weitschies and A. Brodkorb, A standardized static in vitro digestion method suitable for food- an international consensus, Food Funct., 2014, 5, 1113–1124 RSC.
- S. Baugreet, C. Gomez, M. A. Auty, J. P. Kerry, R. M. Hamill and A. Brodkorb, In vitro digestion of protein-enriched restructured beef steaks with pea protein isolate, rice protein and lentil flour following sous vide processing, Innovative Food Sci. Emerging Technol., 2019, 54, 152–161 CrossRef CAS.
- Z. N. Uribe-Wandurraga, M. Igual, P. García-Segovia and J. Martínez-Monzó, Effect of microalgae addition on mineral content, colour and mechanical properties of breadsticks, Food Funct., 2019, 10, 4685–4692 RSC.
- A. P. Batista, A. Niccolai, P. Fradinho, S. Fragoso, I. Bursic, L. Rodolfi, N. Biondi, M. R. Tredici, I. Sousa and A. Raymundo, Microalgae biomass as an alternative ingredient in cookies: Sensory, physical and chemical properties, antioxidant activity and in vitro digestibility, Algal Res., 2017, 26, 161–171 CrossRef.
- AOAC International, Official methods 955.04 of analysis. Washington, Association of Official Analytical Chemists, Gaithersburg, USA, 2023 Search PubMed.
- F. Mariotti, D. Tomé and P. P. Mirand, Converting nitrogen into protein—beyond 6.25 and Jones’ factors, Crit. Rev. Food Sci. Nutr., 2008, 48, 177–184 CrossRef CAS PubMed.
- R. B. Khouzam, P. Pohl and R. Lobinski, Bioaccessibility of essential elements from white cheese, bread, fruit and vegetables, Talanta, 2011, 86, 425–428 CrossRef CAS PubMed.
- A. Sahuquillo, R. Barbera and R. Farre, Bioaccessibility of calcium, iron and zinc from three legume samples, Nahrung, 2003, 47, 438–441 CrossRef CAS PubMed.
- M. Utrera, D. Morcuende, J. G. Rodríguez-Carpena and M. Estévez, Fluorescent HPLC for the detection of specific protein oxidation carbonyls–α-aminoadipic and γ-glutamic semialdehydes–in meat systems, Meat Sci., 2011, 89, 500–506 CrossRef CAS PubMed.
- A. Dubey, A. Mateen and N. Singh, Exploring textural, solubility and rheological characteristics of high–moisture extruded meat analogues: effects of wheat gluten and rice protein incorporation in pea protein isolate and feed moisture levels, Int. J. Food Sci. Technol., 2024, 59, 5560–5575 CrossRef CAS.
- J. S. Lee, I. Choi and J. Han, Construction of rice protein based meat analogues by extruding process: effect of substitution of soy protein with rice protein on dynamic energy, appearance, physicochemical, and textural properties of meat analogues, Food Res. Int., 2022, 161, 111840 CrossRef CAS PubMed.
- T. Zhang, S. Yu, Y. Pan, H. Li, X. Liu and J. Cao, Properties of texturized protein and performance of different protein sources in the extrusion process: A review, Food Res. Int., 2023, 113588 CrossRef CAS PubMed.
- H. M. Bailey, N. S. Fanelli and H. H. Stein, Effect of heat treatment on protein quality of rapeseed protein isolate compared with non–heated rapeseed isolate, soy and whey protein isolates, and rice and pea protein concentrates, J. Sci. Food Agric., 2023, 103, 7251–7259 CrossRef CAS PubMed.
- R. Osen, S. Toelstede, P. Eisner and U. Schweiggert-Weisz, Effect of high moisture extrusion cooking on protein–protein interactions of pea (Pisum sativum L.) protein isolates, Int. J. Food Sci. Technol., 2015, 50, 1390–1396 CrossRef CAS.
- B. Mao, J. Singh, S. Hodgkinson, M. Farouk and L. Kaur, Conformational changes and product quality of high-moisture extrudates produced from soy, rice, and pea proteins, Food Hydrocolloids, 2024, 147, 109341 CrossRef CAS.
- S. M. Beck, K. Knoerzer and J. Arcot, Effect of low moisture extrusion on a pea protein isolate's expansion, solubility, molecular weight distribution and secondary structure as determined by Fourier Transform Infrared Spectroscopy (FTIR), J. Food Eng., 2017, 214, 166–174 CrossRef CAS.
- F. H. Brishti, S. Y. Chay, K. Muhammad, M. R. Ismail-Fitry, M. Zarei and N. Saari, Texturized mung bean protein as a sustainable food source: Effects of extrusion on its physical, textural and protein quality, Innovative Food Sci. Emerging Technol., 2021, 67, 102591 CrossRef CAS.
- O. O. Adeleye, S. T. Awodiran, A. O. Ajayi and T. F. Ogunmoyela, Effect of high-temperature, short-time cooking conditions on in vitro protein digestibility, enzyme inhibitor activity and amino acid profile of selected legume grains, Heliyon, 2020, 6, e05419 CrossRef CAS PubMed.
- R. Nagai and N. Taniguchi, 2 Amino Acids and Proteins, in Medical Biochemistry: Medical Biochemistry E-Book, 2018, vol. 6 Search PubMed.
- R. Cecil, Intramolecular bonds in proteins. I. The role of sulfur in proteins, Proteins, 2012, 1, 379–476 Search PubMed.
- N. Darby and T. E. Creighton, Disulfide bonds in protein folding and stability, in Protein Stability and Folding: Theory and Practice, 1995, pp. 219–252 Search PubMed.
- C. P. Marinangeli, W. D. Mansilla and A. K. Shoveller, Navigating protein claim regulations in North America for foods containing plant-based proteins, Cereal Foods World, 2018, 63, 207–216 CAS.
- M. O. Omosebi, O. F. Osundahunsi and T. N. Fagbemi, Effect of extrusion on protein quality, antinutritional factors, and digestibility of complementary diet from quality protein maize and soybean protein concentrate, J. Food Biochem., 2018, 42, e12508 CrossRef.
- Y. Wang, Z. Zheng, C. Zhang, C. Wu, C. P. Tan and Y. Liu, Comparative structural, digestion and absorption characterization of three common extruded plant proteins, Food Res. Int., 2024, 177, 113852 CrossRef CAS PubMed.
- B. Zhang, G. Liu, D. Ying, L. Sanguansri and M. A. Augustin, Effect of extrusion conditions on the physico-chemical properties and in vitro protein digestibility of canola meal, Food Res. Int., 2017, 100, 658–664 CrossRef CAS PubMed.
- A. Rivera del Rio, R. M. Boom and A. E. M. Janssen, Effect of fractionation and processing conditions on the digestibility of plant proteins as food ingredients, Foods, 2022, 11, 870 CrossRef CAS PubMed.
- P. Duque-Estrad, K. Hardiman, A. Bøgebjerg Dam, N. Dodge, M. D. Aaslyng and I. L. Petersen, Protein blends and extrusion processing to improve the nutritional quality of plant proteins, Food Funct., 2023, 14, 7361–7374 RSC.
- M. Cotacallapa-Sucapuca, E. N. Vega, H. A. Maieves, J. D. J. Berrios, P. Morales, V. Fernández-Ruiz and M. Cámara, Extrusion process as an alternative to improve pulses products consumption. A review, Foods, 2021, 10, 1096 CrossRef CAS PubMed.
- M. G. Nosworthy, G. Medina, A. J. Franczyk, J. Neufeld, P. Appah, A. Utioh, P. Frohlich and J. D. House, Effect of processing on the in vitro and in vivo protein quality of beans (Phaseolus vulgaris and Vicia Faba), Nutrients, 2018, 10, 671 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4fo04829f |
| This journal is © The Royal Society of Chemistry 2025 |



