Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
From in silico screening to in vivo validation in zebrafish – a framework for reeling in the right psychobiotics†
Benjamin
Valderrama‡
ab,
Isabelle
Daly‡
a,
Eoin
Gunnigle
a,
Kenneth J.
O'Riordan
a,
Maciej
Chichlowski
c,
Sagarika
Banerjee
d,
Alicja A.
Skowronski
e,
Neeraj
Pandey
 f,
John F.
Cryan
ab,
Gerard
Clarke
*ag and
Jatin
Nagpal
f,
John F.
Cryan
ab,
Gerard
Clarke
*ag and
Jatin
Nagpal
 *ah
*ah
aAPC Microbiome Ireland, University College Cork, T12 YT20, Cork, Ireland. E-mail: jatin.nagpal@ucc.ie; G.Clarke@ucc.ie
bDepartment of Anatomy and Neuroscience, University College Cork, T12 YT20, Cork, Ireland
cNutrition Science Platform, Reckitt|Mead Johnson Nutrition, Evansville, IN, USA
dMicrobiome Management Science Platform, Reckitt, Montvale, NJ, USA
eNutrition Science Platform, Reckitt|Mead Johnson Nutrition, Parsippany, NJ, USA
fNutrition Science Platform, Reckitt|Mead Johnson Nutrition, Slough, UK
gDepartment of Psychiatry & Neurobehavioural Sciences, University College Cork, T12 YT20, Cork, Ireland
hDepartment of Pharmacology & Therapeutics, School of Medicine, and School of Pharmacy, University College Cork, T12 YT20, Cork, Ireland
First published on 18th February 2025
Abstract
The potential of gut bacteria to interact with the nervous system is now well known. Therefore, the characterization of bacterial strains that can modulate signalling pathways of the nervous system is a topic of growing interest, as it represents a potential alternative therapeutic target to treat central nervous system disorders. However, a streamlined screening framework is required to guide the rational identification and selection of such bacteria, known as psychobiotics. In this work, we introduce a framework that integrates in silico, in vitro and in vivo approaches to identify psychobiotic candidates capable of both metabolising prebiotics of interest and producing neuroactive molecules. To prove the effectiveness of the approach, we characterized a bacterial strain, Lactiplantibacillus plantarum APC2688, for its capacity to modulate the GABAergic system and alter the stress-related behaviour of zebrafish larvae. In brief, in silico analyses of the genomic content of APC2688 identified it as capable of degrading different prebiotics and producing neuroactive compounds known to modulate the stress response in animal models. Then, in vitro results confirmed the ability of this strain to produce GABA, tryptophan and acetate, while growing with the candidate prebiotics of interest, fructooligosaccharides (FOS), galactooligosaccharides (GOS) and inositol. In vivo experiments demonstrated that the administration of bacterial supernatants induced changes in the expression of gad1 and gabra1 in zebrafish larvae, two essential genes in the GABAergic signalling pathway, and altered the anxiety-like behaviour of the larvae. These results highlight the efficiency of our framework in integrating orthogonal approaches to discover and characterise bacteria capable of modulating the microbiome–gut–brain axis.
Introduction
The bacteria in the gut microbiome have been identified as a key regulator of the communication between the gut and the brain.1 Many carbohydrate-degrading enzymes have been identified in the genomes of gut bacteria.2,3 This facilitates the degradation of complex prebiotic fibres such as fructooligosaccharides (FOS) and galactooligosaccharides (GOS), and simpler molecules, such as dextrose or myo-inositol, the most abundant isomer of inositol.4 Once in the gut, some bacteria metabolise the fibres undigested by the host, producing molecules that can modulate the gut–brain axis.5–7 These and other observations led to the coining of the term “psychobiotic”, including living beneficial bacteria (probiotics) and prebiotics that influence bacteria–brain relationships.8,9It has been proposed that the administration of psychobiotic bacteria represents an alternative avenue to support brain health and the management of disorders such as anxiety and depression.10–13 Indeed, bacterial strains have been reported to produce molecules involved in the stress response of animal models, such as GABA,14,15 acetate16,17 and tryptophan.18 Some bacterial strains may also modulate the expression of genes involved in the stress response, such as glutamic acid decarboxylase 1 (gad1) and γ-aminobutyric acid receptor subunit alpha-1 (gabra1),19,20 involved in the GABAergic signalling pathway; the aryl hydrocarbon receptor 2 (ahr2),21 involved in the sensing of metabolites derived from tryptophan; and brain-derived neurotrophic factor (bdnf),20 relevant in neuronal growth and survival. Moreover, administration of psychobiotics has been successfully used to reduce anxiety-like behaviour in animal models22 and to modulate stress,23,24 sleep25 and brain activity26 in healthy humans.
Although psychobiotics hold the potential to modulate gut–brain signalling, workflows for the rapid and rational screening and identification of promising candidates are still lacking although the potential benefits of their development and use have been highlighted in the literature.27,28 Current thinking advocates the earlier incorporation of in silico predictive tools29 and increased use of simpler genetic model organisms.12 This work presents a workflow that integrates in silico, in vitro and in vivo approaches, most often conducted in isolation, to unveil promising new psychobiotic candidates that fit a desired functional profile. This profile consists of defining prebiotics of interest that the bacterial strain can metabolise and neuroactive molecules of interest to be produced by the strain. To test its effectiveness, we defined a profile of interest, which combined the capability to degrade three selected prebiotics of different complexities (FOS, GOS and inositol) with the ability to produce three molecules known to modulate stress response in animals (GABA, acetate and tryptophan). Publicly available genomes were leveraged, and four Lactiplantibacillus plantarum strains were found to fit that profile. Therefore, we characterized a novel closely related strain, L. plantarum APC2668,30 isolated in previous studies conducted in our research centre (NCBI assembly accession code: GCA_040183155.1). In silico analysis of its genome demonstrated that the strain also fits the desired profile. In vitro experiments confirmed APC2668's potential to degrade the prebiotics of interest while producing the neuroactive molecules of interest. Additional in vivo experiments conducted in zebrafish larvae showed that the supernatant of APC2668 grown on GOS, FOS, or inositol can alter the expression of relevant genes (including gad1 and gabra1), and the stress-related behaviour of the model organism.
Methods
In silico search for candidate bacteria with neuroactive potential and the potential to degrade prebiotics
A manually curated list of enzymes that catalyse catabolic reactions of prebiotics of different complexities was generated by searching the KEGG47 and MetaCyc48 databases. The complete list of enzymes and prebiotics can be found in ESI Table 1.† Then, using the R package KEGGREST v.1.40.0, the 6963 bacterial genomes available in the KEGG database were screened to identify bacteria carrying any of those enzymes, as a reflection of their potential to degrade the prebiotics of interest, subsequently discarding genomes lacking them. Next, genomes were further screened to identify Gut Brain Modules (GBMs), a list of 56 metabolic pathways and their respective enzymes previously defined.7 These enzymes are involved in synthetic pathways of neuroactive molecules. To determine the presence of GBMs from the enzymes annotated in each genome, the OmixerRpm R package v. 0.3.3 was used.31 As there are several enzymes in each pathway, the genes for all the enzymes per pathway had to be identified in any given genome to consider the GBM as present. Finally, the list of bacteria with the potential to degrade prebiotics and to synthesise neuroactive molecules was further filtered to only include bacteria previously isolated from human gastrointestinal tracts using an in-house catalogue of bacteria isolated from different studies performed at APC Microbiome Ireland. This final list was then used to determine which species could be of interest to look for new strains with the three desired characteristics: (1) the ability to degrade all three named prebiotics of interest, as they covered a range of different molecular complexities, (2) the ability to produce the three neuroactive molecules of interest, and (3) the ability to live in the human gut. The script used to generate the list of bacteria with the desired characteristics can be accessed from https://github.com/Benjamin-Valderrama/kegg_probiotics, and a visual representation of the workflow along with the number of bacterial genomes identified as successful candidates in each step is provided (ESI Fig. 1†).Genomic analysis of a new strain from the best psychobiotic candidate species identified (L. plantarum APC 2688)
Whole genome sequencing was performed on L. plantarum APC2688 as described previously.30 First, DNA was extracted using the Qiagen PowerFecal Pro DNA kit (Qiagen) according to the manufacturer's guidelines and libraries were prepared using the Nextera XT DNA Library Preparation Kit. Library preparation steps included DNA fragmentation, adapter ligation, amplification, and size selection steps. Sequencing was performed using an Illumina NextSeq2000 (2 × 150 bp). The quality of the raw sequencing data was visualized using FastQC v0.11.9.32 Shotgun genome sequencing data were then processed through an analysis workflow that utilizes the KneadData v0.9.0 wrapper to obtain quality-controlled reads. Quality filtering was performed using Trimmomatic v0.3333 with the following parameters: ILLUMINACLIP:/NexteraPE.fa:2:30:10:8:True SLIDINGWINDOW:5:25 MINLEN:110 TRAILING:25 AVGQUAL:25. Genome assembly was performed using Unicycler v0.5.0.34 The completeness and the contamination percentage of the assembled genome sequence was determined using CheckM v1.1.3,35 and GTDB-Tk v1.3.036 was used for taxonomic classification of the genome. Finally, the annotated genome was queried for the presence of the enzymes involved in the catabolism of the prebiotics of interest and the enzymes in the GBMs pathways.7 The GBM abundance and coverage analysis was performed using OmixerRpm R package v. 0.3.3 with a coverage cutoff of 1, as the goal of the analysis was to identify complete biosynthetic pathways within the genomes of the bacteria.Bacterial growth curves
The growth media used in the experiment consisted of either the basal modified MRS medium with no dextrose (hereafter referred to as mMRS-C) (Condalab MRS Broth w/o Dextrose; catalogue number: 1295), or the same mMRS-C supplemented with 2% (w/v) FOS, GOS, inositol (provided by Reckitt Inc.) or dextrose. Basal mMRS-C used to prepare other media was autoclaved at 121 °C for 5 min prior to the addition of another carbon source (either a prebiotic or dextrose), which was dissolved in dH2O and sterilised with 0.2 μm filters before addition to mMRS-C. All media were allowed to deoxygenate and stored under anaerobic conditions for at least 12 h before use.For growth curves, Lactiplantibacillus plantarum APC2688 was grown from a glycerol stock on modified MRS (mMRS) agar for 24–48 h. Single colonies were inoculated into liquid mMRS broth and grown until OD600 0.5–0.7. Experimental media were inoculated to an OD600 of 0.01 in 3 biological replicates. A volume of 200 μL of each biological replicate was added in technical triplicates to a 96 well plate. The OD600 was monitored in a 96-well automated plate reader (Cerillo™ Stratus) for 36 hours under anaerobic conditions in the PLAS-LABS Simplicity 888 Automatic Atmosphere Chamber (N2: 80%, H2: 10%, CO2: 10%), at 37 °C with shaking at 400 rpm. OD measurements were taken every 10 min. The averages of the technical replicates were used for data analysis and representation.
Bacterial supernatant harvesting
L. plantarum APC2688 was grown from a glycerol stock on mMRS agar for 24–48 h. Single colonies were inoculated into mMRS broth and grown until OD600 0.5–0.7, from which experimental media were inoculated to an OD600 of 0.01 in 3 biological replicates. The OD600 value was monitored until the supernatant was collected at the late stationary phase of growth (30 h post-inoculation). Bacterial cultures were centrifuged at 5000 rpm for 10 minutes. Thereafter, bacterial cells were removed using 0.2 μm filters and the supernatants were then aliquoted and stored at −80 °C.Metabolomics analysis of semi-polar metabolites
Metabolomics analysis of harvested L. plantarum APC2688 supernatant samples was performed by MS-Omics (Clinical Microbiomics; Copenhagen, Denmark) as follows. The analysis was carried out using a Thermo Scientific Vanquish LC coupled to an Orbitrap Exploris 240 MS (Thermo Fisher Scientific). An electrospray ionization interface was used as the ionization source. Analysis was performed in positive and negative ionization modes under polarity switching. Ultra-performance liquid chromatography (UPLC) was performed.37 Peak areas were extracted using Compound Discoverer 3.3 (Thermo Scientific). Identification of compounds was performed at four levels; Level 1: identification by retention times (compared against in-house authentic standards), accurate mass (with an accepted deviation of 3 ppm), and MS/MS spectra, Level 2a: identification by retention times (compared against in-house authentic standards) and accurate mass (with an accepted deviation of 3 ppm), Level 2b: identification by accurate mass (with an accepted deviation of 3 ppm) and MS/MS spectra, and Level 3: identification by accurate mass alone (with an accepted deviation of 3 ppm). Metabolites annotated at the two highest levels of confidence (levels 1 and 2a) were used for further analysis. The output tables with the abundance of each metabolite per sample were CLR-transformed to account for compositionality38 using the clr_c function from the Tjazi v. 0.1.0.0 R package. Principal Component Analysis (PCA) was performed using the prcomp function from the stats v. 4.3.0 R package. The PCA plot was prepared using the CLR-transformed count table of the semi-polar metabolites identified.Metabolomics analysis of short chain fatty acids
Metabolomics analysis of the harvested L. plantarum APC2688 supernatant samples was also carried out by MS-Omics. In this approach, samples were acidified using hydrochloride acid, and deuterium labelled internal standards were added. All samples were analyzed in a randomized order. Analysis was performed using a high polarity column (Zebron™ ZB-FFAP, GC Cap. Column 30 m × 0.25 mm × 0.25 μm) installed in a GC (7890B, Agilent) coupled with a time-of-flight MS (Pegasus® BT, LECO). The system was controlled using a ChromaTOF® (LECO). Raw data were converted to netCDF format using Chemstation (Agilent), before the data were imported and processed in MATLAB R2021b (MathWorks, Inc.) using the PARADISe software. The statistical analysis of these metabolomics data was done in the same manner as described in the section on semi-polar metabolites.Zebrafish hatching and survival analysis
Wild-type AB zebrafish strain maintenance was performed under standard conditions with 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 light/dark cycles and fed with dry food and brine shrimp,39 in accordance with the European Directive 2010/63/EC and meeting the requirements of the S.I. No. 543 of 2012. This is in accordance with the Guidelines for Care and Use of Laboratory Animals at University College Cork and approved by the Health Products Regulatory Authority, Ireland. Aliquots of each bacterial supernatant were diluted with egg water (3 g sea salt per 10 L ultrapure water) in concentrations of 0.01 and 0.02% v/v. Supernatant pH was measured before and after dilution to find the highest administrable concentration of the supernatant to which zebrafish larvae can be exposed without any developmental malformations. Ten fertilized eggs at the same developmental stage were sorted and allocated to each well of a 6 well plate. Each well of the plate had 8 mL of either egg water, as a negative control, or the bacterial supernatant diluted with egg water. Embryos and larvae were incubated at 28 °C with 14
10 light/dark cycles and fed with dry food and brine shrimp,39 in accordance with the European Directive 2010/63/EC and meeting the requirements of the S.I. No. 543 of 2012. This is in accordance with the Guidelines for Care and Use of Laboratory Animals at University College Cork and approved by the Health Products Regulatory Authority, Ireland. Aliquots of each bacterial supernatant were diluted with egg water (3 g sea salt per 10 L ultrapure water) in concentrations of 0.01 and 0.02% v/v. Supernatant pH was measured before and after dilution to find the highest administrable concentration of the supernatant to which zebrafish larvae can be exposed without any developmental malformations. Ten fertilized eggs at the same developmental stage were sorted and allocated to each well of a 6 well plate. Each well of the plate had 8 mL of either egg water, as a negative control, or the bacterial supernatant diluted with egg water. Embryos and larvae were incubated at 28 °C with 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 light/dark cycles for 5 days. From day 1 to 5, fish larvae were visually inspected through a stereomicroscope to assess spinal malformations, the day of hatching and the proportion of surviving larvae. Dead embryos and larvae were removed under sterile conditions after checking each day.
10 light/dark cycles for 5 days. From day 1 to 5, fish larvae were visually inspected through a stereomicroscope to assess spinal malformations, the day of hatching and the proportion of surviving larvae. Dead embryos and larvae were removed under sterile conditions after checking each day.
Behavioural analysis of zebrafish larvae
At 08:00 AM of day 5 post fertilisation (5 dpf), a single larva was transferred from each well of the 6 well plates to 1 mL of egg water in a 24 well plate. The protocol used was modified from that previously described.40 Briefly, 4 × 24-well plates were filled, with a total of 4 zebrafish per treatment group in each plate (i.e., zebrafish larvae exposed to egg water, or to the supernatant of L. plantarum APC2688 grown in the mMRS-C, FOS, GOS or inositol media described above). Each 24-well plate was placed in a box to avoid light exposure and allowed to habituate in the dark at 28 °C for 2 hours. After that period, the 24-well plate was introduced to the Zantiks MWP system (also kept at 28 °C), and allowed to habituate for 20 min, followed by a 10 minute tracking calibration step, followed by initiation of the behavioural assays.The larvae spent 10 minutes in darkness, followed by 10 minutes of light exposure (92.07 ± 2.5 Lux), and another 10 minutes of darkness. During the assay, the position of the fish within the well and the time were recorded automatically. Two analyses were performed using these measurements: total distance travelled and thigmotaxis. The total distance travelled measured the distance within each phase of the experiment, or through the entire test. For thigmotaxis, two circular zones of the same total area were computed within each well of the 6-well plate, the inner and outer zone. The position of the fish and the time spent within each zone were determined throughout the experiment. Output data with the total distance travelled and the time spent within each zone were then aggregated into bins of 1 minute. The total distance travelled by the fish during each phase was then calculated, and the delta between the distance travelled during the light phase and the second dark phase was also calculated for each fish. Additionally, the ratio of the time each fish spent in the outer zone over the total length of the second dark phase was also calculated as a proxy of anxiety-like behaviour.
RT-qPCR with whole larval zebrafish homogenates
An independent cohort of zebrafish larvae not used in behavioural assays was culled on day 5 post fertilisation. Larvae of each well were pooled, so each well of the plate (i.e., 10 larvae) constitutes an n of 1. Experimental groups for RT-qPCR are the same as described for behavioural analysis. RNA was extracted from pools of roughly 10 larvae using the Qiagen RNeasy Plus Micro® kit following a modified protocol after disrupting the tissue with an electric homogenizer, in which the initial centrifugation and supernatant removal step was not carried out to increase the final RNA yield. cDNA was prepared using the MultiScribe™ Reverse Transcriptase Thermo Fisher Scientific® kit using 20 μL of RNA at a concentration of 25 ng μL−1. The following protocol was performed: 25 °C for 10 minutes, 37 °C for 120 minutes and 85 °C for 5 minutes using the MiniAmp thermal cycler. qPCR was performed using 2 μL of cDNA and 8 μL of PowerUp SYBR Green® Mastermix from Applied Biosystems in a Lightcycler 480 II (Roche) with the following protocol: 50 °C for 2 minutes (UDG activation), 95 °C for 2 minutes (polymerase activation), and 40 cycles of 95 °C for 15 seconds (denaturation) and 60 °C for 1 minute (annealing). Expression levels were calculated as the average of three technical replicates relative to a stably expressed housekeeper gene, β-actin. The relative mRNA expression was calculated using the 2−ΔΔCt method.41 Forward and reverse primers for the actb1,42gad1,43gabra1,43ahr244 and bdnf45 genes can be found in ESI Table 3.†Data analysis and statistics
All statistical analyses of the data were conducted using R version 4.1.1. For data manipulation and visualization, the Tidyverse ecosystem of packages was used.46 Analysis of the differences in the means between groups was performed using ANOVA for normally distributed data, and the Kruskal–Wallis test for non-normally distributed data. The Shapiro–Wilk test was used to assess the normality of the data. When differences between groups were deemed statistically significant by the corresponding tests, contrast analysis was performed using t-tests followed by False Discovery Rate (FDR) correction using the Benjamini–Hochberg method. For metabolomics analysis, the abundance of each metabolite was transformed using the Centred Log Ratios (CLR) transformation as it has been suggested previously for compositional data.38 The three metabolites of interest, acetate, tryptophan and GABA, were analysed in a targeted manner using ANOVA. When statistically significant differences between groups were identified, pairwise t-tests with FDR correction were applied to perform contrasts. For untargeted metabolomics analysis, Generalized Linear Models (GLMs) were implemented for each metabolite to determine differences between means of the groups using the formula “metabolite ∼ growth media”, as the growth media was the only relevant explanatory variable in the experimental design. The resulting p values obtained were corrected for multiple comparisons using the Benjamini–Hochberg method. Principal Component Analysis (PCA) for an untargeted metabolomics analysis was performed by calculating Aitchison distances between samples (defined as Euclidean distances on CLR-transformed data).Results
In silico: Screening of the prebiotic-degradation profile and potential to synthesise neuroactive molecules in bacterial genomes
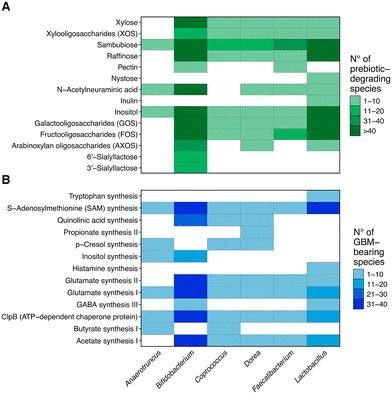
A list of prebiotic fibres with varying levels of complexity was compiled (ESI Table 1†), and a list of enzymes with experimental evidence of their potential to degrade them was curated from the KEGG47 and MetaCyc48 databases. Bacterial genomes in the KEGG database were screened to select only those carrying at least one enzyme from the list. Next, the list of genomes was further filtered to select those in which enzymes involved in synthetic pathways of neuroactive compounds were identified. Finally, the list was filtered again to identify bacterial strains previously isolated from the human intestine (see the Methods section for details on each filtering step). As a result of applying these three filters, a final list of bacteria with three key features was generated: (1) predicted potential to catabolise prebiotics of different complexities, (2) the presence of enzymes required for the synthesis and degradation of neuroactive molecules linked to the modulation of the stress response, and (3) isolation from the human gut. A summary of the in silico workflow employed and the number of genomes identified as successful candidates at each step is provided (ESI Fig. 1†).For further evaluation, bacterial candidates from the KEGG database were aggregated into six genera: Anaerotruncus, Bifidobacterium, Coprococcus, Dorea, Faecalibacterium and Lactobacillus. For each genus, the number of species with genes encoding enzymes required for the degradation of the prebiotics (Fig. 1A) and GBM-associated enzymes (Fig. 1B) was quantified. While prebiotics such as sambubiose and inositol could be degraded by species in all the six candidate genera, fibres such as arabinoxylan oligosaccharides and 3′- and 6′-sialyllactose could only be degraded by species within the genus Bifidobacterium. This suggests that species from different genera encode different prebiotic degrading enzymes, depicting genus specific prebiotic-degrading potential. On the other hand, while GBMs such as glutamate synthesis I and S-adenosylmethionine synthesis were detected across all the six genera, other GBMs were present only in specific genera, such as tryptophan synthesis that was identified only within the genus Lactobacillus. This also suggests genus-specific neuroactive potential.
In silico: Lactiplantibacillus plantarum APC2688 was identified as a promising probiotic candidate based on its prebiotic-degradation profile and its potential to synthesize neuroactive molecules
The specific goal of the in silico analysis was to identify a bacterial strain fitting a profile of interest: isolated from human stool samples, and degrading prebiotics of different complexities while also producing molecules known to modulate the stress-response in animals. Therefore, bacterial candidates capable of degrading FOS, GOS and inositol and synthesizing GABA, acetate and tryptophan were deemed most relevant for this study. From our results, only the genus Lactobacillus showed multiple candidates with the desired profile (Fig. 1A and B). Notably, within that genus, the species Lactiplantibacillus plantarum (formerly known as Lactobacillus plantarum) was found to have the highest number of strains with the desired profile, with a total of four genomes. Hence, the hypothesis was that a new strain from the same species will maximize the chances of finding a new psychobiotic candidate capable of degrading FOS, GOS and inositol and synthesizing GABA, acetate and tryptophan. Therefore, a previously uncharacterised strain of the species L. plantarum, the strain APC2688, was selected from the APC Microbiome Ireland culture collection for further analysis (NCBI assembly accession code: GCA_040183155.1). The genome of L. plantarum APC2688 was analysed, and it showed functional coherence with the profiles of the other four L. plantarum genomes available in the KEGG database, supporting our hypothesis (Fig. 2). Additionally, the average nucleotide identity (ANI) between the novel strain and the genomes available in KEGG was calculated (ESI Table 2†).In vitro: L. plantarum APC2688 was able to grow in media supplemented with the candidate prebiotics of interest (FOS, GOS, and inositol) and produce the neuroactive molecules of interest (GABA, tryptophan and acetate)
In vitro experiments were performed to confirm in silico results suggesting that L. plantarum APC2688 could grow in the presence of the prebiotics of interest. The growth of the strain was characterized using different media: mMRS with dextrose available at 2% w/v (mMRS group), mMRS without dextrose (MRS-C), and MRS-C supplemented with FOS, GOS, or inositol at 2% w/v referred to as FOS, GOS and inositol, respectively. Although bacterial growth was supported in every media, statistically significant differences were observed among groups (F(4,10) = 127.99; p < 1.5 × 10−8). In particular, while the FOS, GOS, and mMRS media showed a higher OD when compared to the mMRS-C group (T(4) = −5.04, p < 0.05; T(4) = −22.3, p < 1 × 10−4; and T(4) = −13.6, p < 1 × 10−4, respectively), inositol supplemented media showed no statistically significant differences when compared to the mMRS-C group (T(4) = 1.04; p < 0.4) (Fig. 3A).Next, metabolomics analysis confirmed that L. plantarum APC2688 could produce the neuroactive molecules GABA, acetate and tryptophan when grown in every media described above (Fig. 3B–D). In each experimental group, the CLR-transformed abundance of the metabolite was above 0, suggesting that the metabolites were enriched in the bacterial supernatant. Differences in the abundance of each metabolite were significant according to ANOVA: GABA (F(4,10) = 703.99; p < 1 × 10−4), acetate (F(4,10) = 3.54; p < 0.05), and tryptophan (F(4,10) = 139.55; p < 1 × 10−4). Contrasts were conducted to evaluate differences between the mMRS-C and mMRS groups when compared to the prebiotic-treatment groups (see the Methods section). Based on the assessment of the overall metabolic profile of the groups using PCA (see the Methods section), a unique metabolic profile was suggested for the mMRS, FOS and GOS groups, while the mMRS-C and inositol groups were grouped together (ESI Fig. 2†). Finally, untargeted metabolomics analysis was performed to analyse differences in the abundances of every metabolite detected across the different growth media (ESI Table 4†). For this final analysis, GABA, acetate and tryptophan were excluded as they were analysed previously in a targeted manner.
In vivo: The supernatant of L. plantarum APC2688 induced changes in the expression of genes related to the GABAergic signalling pathway and a reduction in anxiety-like behaviour in zebrafish larvae
After the ability of L. plantarum APC2688 to degrade FOS, GOS and inositol while producing GABA, acetate and tryptophan was assessed in silico and in vitro, the next objective was to test if the psychobiotic candidate could affect the stress-related behaviour in an animal model. We assessed whether the strain supernatant could induce changes in MGB axis signalling pathways or in the anxiety-like behaviour of zebrafish larvae. Therefore, zebrafish embryos were exposed to egg water, or to a cell-free supernatant of L. plantarum APC2688 grown in mMRS-C media or mMRS-C media supplemented with FOG, GOS or inositol. The effects of water supplementation with 0.01% and 0.02% v/v of bacterial supernatant were assessed on egg hatching (ESI Fig. 3A†) and larval survival (ESI Fig. 3B†). The results support the selection of the 0.01% v/v concentration, as at higher concentrations a tendency of reduced egg hatching and delayed larval survival was detected.Zebrafish larvae were exposed to 0.01% v/v of bacterial supernatant for five days and then divided into two experimental groups. One group underwent a molecular analysis and the other behavioural assessments. Expression of genes relevant for GABA signalling (gad1 and gabra1), tryptophan signalling (ahr2), and neuronal growth and survival (bdnf) was assessed for the first group. The hypothesis was that bacterial-derived metabolites present in the filtered supernatant will induce changes in the larvae's expression of these genes. The Kruskal–Wallis test shows statistically significant differences in the expression of gad1 (X(4)2 = 10.74, p < 0.05) and gabra1 (X(4)2 = 12.71; p < 0.05) but no differences in the expression of ahr2 (X(4)2 = 0.43; p = 0.98) or bdnf (X(4)2 = 4.22; p = 0.38). Contrasts were performed to compare the egg water treated group against every other group (see the Methods section). The analysis shows that while gad1 was upregulated in every group of zebrafish larvae treated with any supernatant, gabra1 was upregulated in each group except the group treated with the supernatant from APC2688 grown with inositol. Although not significant, our results show a trend of upregulated bdnf expression in the three bacterial supernatants supplemented with prebiotics (Fig. 4A).
In parallel, the second group of zebrafish larvae underwent a behavioural assessment to test the hypothesis that bacterial-derived metabolites present in the filtered supernatant will induce changes in anxiety-like behaviours (Fig. 4B). The Kruskal–Wallis tests showed no statistically significant differences in any of the three phases of the experiment (see the Methods section for details of the phases in behavioural assessments): habituation (X(5)2 = 2.88; p = 0.81), light (X(5)2 = 6.86; p = 0.792), and dark (X(5)2 = 2.26; p = 0.81) (Fig. 4C), or in the change in locomotion during the transition from the light to dark phases of the experiment (X(5)2 = 5.17; p = 0.79) (Fig. 4D). Interestingly, significant differences between groups were found for the thigmotactic behaviour (X(5)2 = 12.5; p < 0.05) (Fig. 4E). Contrasts were performed to compare the egg water-treated group with the groups treated with the bacterial supernatant. The only statistically significant difference was detected for the group of zebrafish larvae treated with the supernatant from APC2688 grown in mMRS-C (Fig. 4E).
Discussion
The integrated framework of in silico, in vitro and in vivo approaches allowed us to identify the potential psychobiotic properties of Lactiplantibacillus plantarum APC2688. The strain was selected based on its potential to both degrade prebiotics and produce neuroactive molecules previously associated with stress-response modulation in animal models. Additionally, we showed that the APC2688 supernatant can modulate the MGB axis, inducing changes in the expression of gad1 and gabra1, crucial genes within the GABAergic signalling pathway and in the anxiety-like behaviour of the zebrafish larvae. Thus, this study underscores the utility of leveraging public data from databases such as KEGG47 and MetaCyc48 for the targeted identification of candidate bacterial strains, offering a more efficient alternative to resource-intensive untargeted screening of numerous candidates.An overarching strength of this framework lies in the integration of orthogonal approaches, such as in silico screening, in vitro experiments, metabolomics analysis, gene expression assessments and behavioural tests in model organisms. This allowed the full characterisation of a bacterial strain and its potential psychobiotic effects. The results reported here demonstrate that computational approaches can be used to efficiently identify bacterial strains with psychobiotic potential, materializing previous propositions made to advance the rate of probiotic discovery.27,28
Our in silico analysis identified L. plantarum as a bacterial species from which potentially interesting novel psychobiotic strains may be further characterised. Indeed, through the analysis of the L. plantarum APC2688 genome, we showed a functional overlap in the biochemical profiles of the newly described APC2688 strain and four other strains within the same species (Fig. 2A and B), each previously described as bacterial probiotics.49–53 Additionally, our in silico analysis identified a repertoire of genes across various L. plantarum strains that confer their ability to synthesise enzymes capable of degrading prebiotics of different complexities. Previous analyses performed on a wider set of strains of L. plantarum also showed a wide repertoire of carbon-utilizing enzymes in different strains,54 which is consistent with our results (Fig. 2). Furthermore, our results on the growth profile of APC2688 in media supplemented with FOS, GOS and dextrose (Fig. 3A) are consistent with previous data from the L. plantarum strain ATCC14917.55 The distinct overall metabolic profile of the ATCC14917 strain when grown in media supplemented with FOS, GOS and dextrose55 was similar to that of APC2688 (ESI Fig. 2†). The metabolites identified in the bacterial supernatant derived from APC2688 are also consistent with previous studies on L. plantarum using either in silico or in vitro approaches. Genes required for the synthesis of tryptophan were identified in Lactobacillus (now named Lactiplantibacillus) plantarum LRCC-5314.56 Additionally, several L. plantarum strains have been previously reported as capable of synthesising acetate57,58 and GABA.59–61 Together, this previously reported evidence is consistent with our results and further emphasizes the benefits of incorporating an in silico screening step prior to initiating in vitro experiments (Fig. 3B–D).
For the in vivo validation of the psychobiotic potential of APC2688, zebrafish played a pivotal role. Zebrafish are now a powerful preclinical model for human diseases with conserved physiology, pharmacology and metabolism, which approach (and occasionally surpass) those of rodents.62,63 Laboratories across academia and industry around the world are leveraging zebrafish as a more cost-effective and scalable alternative to rodents for in vivo drug discovery.64 Numerous drug treatments that have recently progressed to the clinic or are in clinical trials have their genesis in the zebrafish model providing many aquarium-to-bedside success stories.62,63,65 In fact, for some drugs, zebrafish recapitulate the effects observed in humans better than mouse models do (e.g. thalidomide).63 Zebrafish have emerged as a powerful model to study neurobehavioral disorders affecting stress-related and social behaviour due to evolutionary conservation of neuroanatomical and chemical substrates and development of robust behavioural paradigms.12,66–71 Zebrafish may vary in their exact microbiota composition from humans, but due to the conservation of these functional metabolites and host pathways they interact with, zebrafish can be used as a discovery platform for translational science. For a more detailed discussion on the use and translational relevance of simpler model organisms like zebrafish, we refer the reader to a recent review on this topic.12
The Lactiplantibacillus plantarum APC2688-derived supernatant showed potential to target the MGB axis of zebrafish larvae (Fig. 4A), consistent with results where another L. plantarum strain modulated the gene expression of adult zebrafish when administered alive.72 Interestingly, while the APC2688 supernatant was sufficient to induce an upregulation of gabra1 and gad1, the administration of a living L. plantarum strain was previously reported to upregulate gabra1 but not gad1, which remained unaltered,72 and conversely, other two Lactobacillus species, L. delbruecki and L. casei, also induced the upregulation of gad1 but not gabra1 in the zebrafish brain.19 Therefore, this result with our administration strategy implies that changes in the expression of these genes may be modulated by metabolites produced by the strains instead of structural molecules. Furthermore, we found larger effect sizes for the differences between the control group and any supernatant-treated group for the two genes involved in the GABAergic signalling pathway, gad1 and gabra1, compared to previous reports.72 We hypothesise two, potentially complementary, possible explanations for these differences. On the one hand, it could be that early developmental stages are more susceptible to the modulatory effects of L. plantarum strains, reflecting the effect of time-sensitive developmental windows. On the other hand, the potential of APC2688 to induce changes in both genes is higher. Further experiments are warranted to understand the basis underpinning these observations.
No changes were detected in the total locomotion of the larvae treated with L. plantarum (Fig. 4C), consistent with previous reports using strains from the same species,72 and from different species from the same genus.19,73 Additionally, we detected a significant reduction in the thigmotaxis index during the dark phase of the assay in the group of fish treated with the supernatant of APC2688 grown in the media that did not have an additional source of carbon (mMRS-C in figures) (Fig. 4E), suggesting a reduced anxiety-like behaviour in that group. Interestingly, that supernatant showed the highest abundance of tryptophan (Fig. 3D), which was previously linked to the reduction of the stress response in zebrafish adults.74 Supporting our results showing that APC2688 could reduce anxiety-like behaviour, other L. plantarum strains72 and other species from the same genus19,73,75 have shown potential to reduce anxiety-like behaviours in zebrafish in other experimental setups. Similarly, FOS and GOS administration in mice has been shown to elicit a stress-reduction response.76 Therefore, we anticipated that the supernatant of the APC2688 strain grown in their presence would reduce the anxiety-like behaviour in zebrafish (Fig. 4D and E). However, despite the high abundance of tryptophan in the supernatant from bacterial growth in FOS (Fig. 3D), no statistically significant differences were found between the prebiotic-treated and egg water groups.
Despite the novelty of the approach we have proposed and its potential to identify and validate new psychobiotic candidates, this work is not without its limitations. Our framework relies on the access to reference bacterial genomes to screen and identify genes of interest and allow predictions to be made about the functional properties of candidate bacteria. However, ever-growing compendiums of cultured and uncultured bacterial genomes are available,77 and many allow querying of genomes filtered according to the environment where they were recovered, which includes the human gut.78 This opens opportunities to identify new psychobiotic candidates from gut-associated bacterial strains, as well as bacteria from other environmental niches capable of producing neuroactive compounds of interest.7 Similarly, specialized databases like CAZy79 allow for thorough annotation of enzymes involved in the degradation of carbohydrates, which may be of interest to researchers interested in applying this framework using less characterised fibres.
In conclusion, this work introduces a framework for the identification and validation of bacteria with a biochemical profile of interest by integrating in silico, in vitro and in vivo approaches. Notably, the framework described guided the successful discovery and characterization of L. plantarum APC2688 as a bacterial strain capable of modulating the microbiome–gut–brain axis.
Author contributions
BV and ID drafted the manuscript. BV performed the bioinformatic analyses. BV and ID conducted the in vitro and in vivo experiments. BV analysed the data and performed statistical analyses. BV and KOR prepared the figures. GC, JN, JFC, EG, MC, SB, and NP co-designed the study. All authors revised and edited the manuscript. The final version of the manuscript was approved by all authors.Data availability
The 6963 genomes used for the in silico screening (Fig. 1 and 2) were obtained from the KEGG database, and analysed using the script available in https://github.com/Benjamin-Valderrama/kegg_probiotics. The genome of Lactiplantibacillus plantarum APC268830 is available in NCBI (assembly accession code: GCA_040183155.1). Other data from the in vivo and in vitro sections of this work can be found in the ESI,† available on Zenodo – open data repository (https://zenodo.org/records/13325351).Conflicts of interest
NP and AS are employed at Reckitt Benckiser Health Limited while MC and SB were previously employed at Reckitt Benckiser Health Limited. JFC has spoken at conferences organized by Mead Johnson, Ordesa, and Yakult and has received research funding from Reckitt, Nutricia, Dupont/IFF, Kerry Group and Nestle. GC has received honoraria from Boehringer Ingelheim, Janssen, Probi, and Apsen, research funding from Pharmavite, Fonterra, Kerry Group, Nestle, Tate and Lyle and Reckitt, and has provided consultancy for Bayer Healthcare, Yakult, Zentiva and Pharmavite. JN has received research funding from Reckitt. APC Microbiome Ireland has received research support from Mead Johnson, Cremo, 4D Pharma, Suntory Wellness and Nutricia. This support neither influenced nor constrained the content of this article.Acknowledgements
This study was funded by Reckitt Benckiser Health Limited. APC Microbiome Ireland is a research centre funded by Science Foundation Ireland (SFI), through the Irish Governments’ national development plan (Grant No. 12/RC/2273_P2). J. N. acknowledges the support of SFI-IRC Pathway Grant (22/PATH-S/10876). Research Ireland is the new Irish national funding agency, established through the amalgamation of the activities and functions of Science Foundation Ireland (SFI) and the Irish Research Council (IRC). The authors thank Caoimhe M. K. Lynch, Rie Matsuzaki and Carlos Pomares Diaz for their help in setting up the behavioural protocol and for helping with the maintenance of the zebrafish facility.References
- J. F. Cryan, K. J. O'Riordan, C. S. M. Cowan, K. V. Sandhu, T. F. S. Bastiaanssen and M. Boehme, et al., The Microbiota-Gut-Brain Axis, Physiol. Rev., 2019, 99(4), 1877–2013 Search PubMed.
- B. L. Cantarel, P. M. Coutinho, C. Rancurel, T. Bernard, V. Lombard and B. Henrissat, The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics, Nucleic Acids Res., 2009, 37(Database issue), D233–D238 CrossRef CAS PubMed.
- B. L. Cantarel, V. Lombard and B. Henrissat, Complex Carbohydrate Utilization by the Healthy Human Microbiome, PLoS One, 2012, 7(6), e28742 CrossRef CAS PubMed.
- K. Yoshida, M. Yamaguchi, T. Morinaga, M. Kinehara, M. Ikeuchi and H. Ashida, et al., myo-Inositol Catabolism in Bacillus subtilis*, J. Biol. Chem., 2008, 283(16), 10415–10424 CrossRef CAS PubMed.
- J. F. Cryan and T. G. Dinan, Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour, Nat. Rev. Neurosci., 2012, 13(10), 701–712 CrossRef CAS PubMed.
- L. M. T. Dicks, Gut Bacteria and Neurotransmitters, Microorganisms, 2022, 10(9), 1838 CrossRef CAS PubMed.
- M. Valles-Colomer, G. Falony, Y. Darzi, E. F. Tigchelaar, J. Wang and R. Y. Tito, et al., The neuroactive potential of the human gut microbiota in quality of life and depression, Nat. Microbiol., 2019, 4(4), 623–632 CrossRef CAS PubMed.
- T. G. Dinan, C. Stanton and J. F. Cryan, Psychobiotics: A Novel Class of Psychotropic, Biol. Psychiatry, 2013, 74(10), 720–726 CrossRef CAS PubMed.
- C. Long-Smith, K. J. O'Riordan, G. Clarke, C. Stanton, T. G. Dinan and J. F. Cryan, Microbiota-Gut-Brain Axis: New Therapeutic Opportunities, Annu. Rev. Pharmacol. Toxicol., 2020, 60(1), 477–502 CrossRef CAS PubMed.
- T. G. Dinan and J. F. Cryan, Regulation of the stress response by the gut microbiota: Implications for psychoneuroendocrinology, Psychoneuroendocrinology, 2012, 37(9), 1369–1378 CrossRef CAS PubMed.
- J. A. Foster, L. Rinaman and J. F. Cryan, Stress & the gut-brain axis: Regulation by the microbiome, Neurobiol. Stress, 2017, 7, 124–136 CrossRef PubMed.
- J. Nagpal and J. F. Cryan, Microbiota-brain interactions: Moving toward mechanisms in model organisms, Neuron, 2021, 109(24), 3930–3953 CrossRef CAS PubMed.
- A. Sarkar, S. M. Lehto, S. Harty, T. G. Dinan, J. F. Cryan and P. W. J. Burnet, Psychobiotics and the Manipulation of Bacteria-Gut-Brain Signals, Trends Neurosci., 2016, 39(11), 763–781 CrossRef CAS PubMed.
- H. Luo, Z. Liu, F. Xie, M. Bilal, L. Liu and R. Yang, et al., Microbial production of gamma-aminobutyric acid: applications, state-of-the-art achievements, and future perspectives, Crit. Rev. Biotechnol., 2021, 41(4), 491–512 CrossRef PubMed.
- A. Monteagudo-Mera, V. Fanti, C. Rodriguez-Sobstel, G. Gibson, A. Wijeyesekera and K. A. Karatzas, et al., Gamma aminobutyric acid production by commercially available probiotic strains, J. Appl. Microbiol., 2023, 134(2), lxac066 CrossRef PubMed.
- B. Dalile, L. Van Oudenhove, B. Vervliet and K. Verbeke, The role of short-chain fatty acids in microbiota–gut–brain communication, Nat. Rev. Gastroenterol. Hepatol., 2019, 16(8), 461–478 CrossRef PubMed.
- K. J. O'Riordan, M. K. Collins, G. M. Moloney, E. G. Knox, M. R. Aburto and C. Fülling, et al., Short chain fatty acids: Microbial metabolites for gut-brain axis signalling, Mol. Cell. Endocrinol., 2022, 546, 111572 CrossRef PubMed.
- A. Agus, J. Planchais and H. Sokol, Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease, Cell Host Microbe, 2018, 23(6), 716–724 CrossRef CAS PubMed.
- J. P. Olorocisimo, L. A. Diaz, D. E. Co, H. M. Carag, J. A. Ibana and M. C. Velarde, Lactobacillus delbrueckii reduces anxiety-like behavior in zebrafish through a gut microbiome – brain crosstalk, Neuropharmacology, 2023, 225, 109401 CrossRef CAS PubMed.
- V. Philip, D. F. Newton, H. Oh, S. M. Collins, P. Bercik and E. Sibille, Transcriptional markers of excitation-inhibition balance in germ-free mice show region-specific dysregulation and rescue after bacterial colonization, J. Psychiatr. Res., 2021, 135, 248–255 CrossRef PubMed.
- A. Korecka, A. Dona, S. Lahiri, A. J. Tett, M. Al-Asmakh and V. Braniste, et al., Bidirectional communication between the Aryl hydrocarbon Receptor (AhR) and the microbiome tunes host metabolism, npj Biofilms Microbiomes, 2016, 2(1), 1–10 CrossRef PubMed.
- J. A. Bravo, P. Forsythe, M. V. Chew, E. Escaravage, H. M. Savignac and T. G. Dinan, et al., Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(38), 16050–16055 CrossRef CAS PubMed.
- A. P. Allen, W. Hutch, Y. E. Borre, P. J. Kennedy, A. Temko and G. Boylan, et al., Bifidobacterium longum 1714 as a translational psychobiotic: modulation of stress, electrophysiology and neurocognition in healthy volunteers, Transl. Psychiatry, 2016, 6(11), e939 CrossRef CAS PubMed.
- P. Tian, K. J. O'Riordan, Y.-k. Lee, G. Wang, J. Zhao and H. Zhang, et al., Towards a psychobiotic therapy for depression: Bifidobacterium breve CCFM1025 reverses chronic stress-induced depressive symptoms and gut microbial abnormalities in mice, Neurobiol. Stress, 2020, 12, 100216 CrossRef PubMed.
- G. M. Moloney, C. M. Long-Smith, A. Murphy, D. Dorland, S. F. Hojabri and L. O. Ramirez, et al., Improvements in sleep indices during exam stress due to consumption of a Bifidobacterium longum, Brain, Behav., Immun.: Health, 2021, 10, 100174 Search PubMed.
- H. Wang, C. Braun, E. F. Murphy and P. Enck, Bifidobacterium longum 1714™ Strain Modulates Brain Activity of Healthy Volunteers During Social Stress, Am. J. Gastroenterol., 2019, 114(7), 1152 CrossRef PubMed.
- S. T. Karlsen, M. H. Rau, B. J. Sánchez, K. Jensen and A. A. Zeidan, From genotype to phenotype: computational approaches for inferring microbial traits relevant to the food industry, FEMS Microbiol. Rev., 2023, fuad030 CrossRef CAS PubMed.
- I. D. Kwoji, O. A. Aiyegoro, M. Okpeku and M. A. Adeleke, ‘Multi-omics’ data integration: applications in probiotics studies, npj Sci. Food, 2023, 7(1), 25 CrossRef PubMed.
- S. Spichak, T. F. S. Bastiaanssen, K. Berding, K. Vlckova, G. Clarke and T. G. Dinan, et al., Mining microbes for mental health: Determining the role of microbial metabolic pathways in human brain health and disease, Neurosci. Biobehav. Rev., 2021, 125, 698–761 CrossRef CAS PubMed.
- B. Valderrama, I. Daly, E. Gunnigle, M. C. Rea, J. Kenny and M. Coakley, et al., High-quality draft genome of Lactiplantibacillus plantarum strain APC2688 isolated from human feces, Microbiol. Resour. Announce., 2024, 13(11), e00641-24 CrossRef PubMed.
- Y. Darzi, G. Falony, S. Vieira-Silva and J. Raes, Towards biome-specific analysis of meta-omics data, ISME J., 2016, 10(5), 1025–1028 CrossRef CAS PubMed.
- A. S. Fastqc, A quality control tool for high throughput sequence data, 2010 Search PubMed.
- A. M. Bolger, M. Lohse and B. Usadel, Trimmomatic: a flexible trimmer for Illumina sequence data, Bioinformatics, 2014, 30(15), 2114–2120 CrossRef CAS PubMed.
- R. R. Wick, L. M. Judd, C. L. Gorrie and K. E. Holt, Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads, ed. A. M. Phillippy, PLoS Comput. Biol., 2017, 13(6), e1005595 CrossRef PubMed.
- D. H. Parks, M. Imelfort, C. T. Skennerton, P. Hugenholtz and G. W. Tyson, CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes, Genome Res., 2015, 25(7), 1043–1055 CrossRef CAS PubMed.
- D. H. Parks, M. Chuvochina, C. Rinke, A. J. Mussig, P. A. Chaumeil and P. Hugenholtz, GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy, Nucleic Acids Res., 2022, 50(D1), D785–D794 CrossRef CAS PubMed.
- Waters , UPLC/MS Monitoring of Water-Soluble Vitamin Bs in Cell Culture Media in Minutes. Available from: https://www.waters.com/waters/library.htm?lid=134636355 [cited 2024 Jul 11].
- G. B. Gloor, J. M. Macklaim, V. Pawlowsky-Glahn and J. J. Egozcue, Microbiome Datasets Are Compositional: And This Is Not Optional, Front. Microbiol., 2017, 8, 2224 CrossRef PubMed.
- M. Westerfield, The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio Rerio) [Internet], University of Oregon Press, 2000. Available from: https://books.google.ie/books?id=Iy8PngEACAAJ Search PubMed.
- S. J. Schnörr, P. J. Steenbergen, M. K. Richardson and D. L. Champagne, Measuring thigmotaxis in larval zebrafish, Behav. Brain Res., 2012, 228(2), 367–374 CrossRef PubMed.
- K. J. Livak and T. D. Schmittgen, Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method, Methods, 2001, 25(4), 402–408 CrossRef CAS PubMed.
- B. R. Keegan, J. L. Feldman, D. H. Lee, D. S. Koos, R. K. Ho and D. Y. R. Stainier, et al., The elongation factors Pandora/Spt6 and Foggy/Spt5 promote transcription in the zebrafish embryo, Development, 2002, 129(7), 1623–1632 CrossRef CAS PubMed.
- G. A. Hortopan, M. T. Dinday and S. C. Baraban, Spontaneous Seizures and Altered Gene Expression in GABA Signaling Pathways in a mind bomb Mutant Zebrafish, J. Neurosci., 2010, 30(41), 13718–13728 CrossRef CAS PubMed.
- H. Meyer-Alert, K. Ladermann, M. Larsson, S. Schiwy, H. Hollert and S. H. Keiter, A temporal high-resolution investigation of the Ah-receptor pathway during early development of zebrafish (Danio rerio), Aquat. Toxicol., 2018, 204, 117–129 CrossRef CAS PubMed.
- M. Pavlidis, A. Theodoridi and A. Tsalafouta, Neuroendocrine regulation of the stress response in adult zebrafish, Danio rerio, Prog. Neuropsychopharmacol. Biol. Psychiatry, 2015, 60, 121–131 CrossRef CAS PubMed.
- H. Wickham, M. Averick, J. Bryan, W. Chang, L. D. McGowan and R. François, et al., Welcome to the Tidyverse, J Open Source Software, 2019, 4(43), 1686 CrossRef.
- M. Kanehisa and S. Goto, KEGG: Kyoto Encyclopedia of Genes and Genomes, Nucleic Acids Res., 2000, 28(1), 27–30 CrossRef CAS PubMed.
- P. D. Karp, M. Riley, S. M. Paley and A. Pellegrini-Toole, The MetaCyc Database, Nucleic Acids Res., 2002, 30(1), 59–61 CrossRef CAS PubMed.
- M. C. de Vries, E. E. Vaughan, M. Kleerebezem and W. M. de Vos, Lactobacillus plantarum—survival, functional and potential probiotic properties in the human intestinal tract, Int. Dairy J., 2006, 16(9), 1018–1028 CrossRef CAS.
- N. Ivanovic, R. Minic, L. Dimitrijevic, S. Radojevic Skodric, I. Zivkovic and B. Djordjevic, Lactobacillus rhamnosus LA68 and Lactobacillus plantarum WCFS1 differently influence metabolic and immunological parameters in high fat diet-induced hypercholesterolemia and hepatic steatosis, Food Funct., 2015, 6(2), 558–565 RSC.
- C. Suo, Y. Yin, X. Wang, X. Lou, D. Song and X. Wang, et al., Effects of lactobacillus plantarumZJ316 on pig growth and pork quality, BMC Vet. Res., 2012, 8(1), 89 CrossRef PubMed.
- L. Zang, Y. Ma, W. Huang, Y. Ling, L. Sun and X. Wang, et al., Dietary Lactobacillus plantarum ST-III alleviates the toxic effects of triclosan on zebrafish (Danio rerio) via gut microbiota modulation, Fish Shellfish Immunol., 2019, 84, 1157–1169 CrossRef CAS PubMed.
- Z. Y. Zhang, C. Liu, Y. Z. Zhu, Y. Zhong, Y. Q. Zhu and H. J. Zheng, et al., Complete Genome Sequence of Lactobacillus plantarum JDM1, J. Bacteriol., 2009, 191(15), 5020–5021 CrossRef CAS PubMed.
- Y. Cui, M. Wang, Y. Zheng, K. Miao and X. Qu, The Carbohydrate Metabolism of Lactiplantibacillus plantarum, Int. J. Mol. Sci., 2021, 22(24), 13452 CrossRef CAS PubMed.
- P. Cao, L. Wu, Z. Wu, D. Pan, X. Zeng and Y. Guo, et al., Effects of oligosaccharides on the fermentation properties of Lactobacillus plantarum, J. Dairy Sci., 2019, 102(4), 2863–2872 CrossRef CAS PubMed.
- J. Jeong, Y. Lee, S. Yoon, J. H. Kim and W. Kim, Lactiplantibacillus plantarum LRCC5314 includes a gene for serotonin biosynthesis via the tryptophan metabolic pathway, J. Microbiol., 2021, 59(12), 1092–1103 CrossRef CAS PubMed.
- P. Goffin, L. Muscariello, F. Lorquet, A. Stukkens, D. Prozzi and M. Sacco, et al., Involvement of Pyruvate Oxidase Activity and Acetate Production in the Survival of Lactobacillus plantarum during the Stationary Phase of Aerobic Growth, Appl. Environ. Microbiol., 2006, 72(12), 7933–7940 CrossRef CAS PubMed.
- F. Lorquet, P. Goffin, L. Muscariello, J. B. Baudry, V. Ladero and M. Sacco, et al., Characterization and Functional Analysis of the poxB Gene, Which Encodes Pyruvate Oxidase in Lactobacillus plantarum, J. Bacteriol., 2004, 186(12), 3749–3759 CrossRef CAS PubMed.
- R. Di Cagno, F. Mazzacane, C. G. Rizzello, M. De Angelis, G. Giuliani and M. Meloni, et al., Synthesis of γ-aminobutyric acid (GABA) by Lactobacillus plantarum DSM19463: functional grape must beverage and dermatological applications, Appl. Microbiol. Biotechnol., 2010, 86(2), 731–741 CrossRef CAS PubMed.
- L. Diez-Gutiérrez, L. S. Vicente, J. Sáenz, A. Esquivel, L. J. R. Barron and M. Chávarri, Biosynthesis of gamma-aminobutyric acid by Lactiplantibacillus plantarum K16 as an alternative to revalue agri-food by-products, Sci. Rep., 2022, 12(1), 18904 CrossRef PubMed.
- M. Rezaei, Y. Ghasemi, A. Sharifan and H. Bakhoda, Gamma-Aminobutyric Acid (GABA) Biosynthesis from Lactobacillus plantarum subsp. plantarum IBRC10817 Optimized and Modeled in Response to Heat and Ultrasonic Shock, Probiotics & Antimicro. Prot., 2024, 16, 1304–1312 Search PubMed.
- C. A. MacRae and R. T. Peterson, Zebrafish as tools for drug discovery, Nat. Rev. Drug Discovery, 2015, 14(10), 721–731 CrossRef CAS PubMed.
- E. E. Patton, L. I. Zon and D. M. Langenau, Zebrafish disease models in drug discovery: from preclinical modelling to clinical trials, Nat. Rev. Drug Discovery, 2021, 20(8), 611–628 CrossRef CAS PubMed.
- M. Cully, Zebrafish earn their drug discovery stripes, Nat. Rev. Drug Discovery, 2019, 18(11), 811–813 CrossRef CAS PubMed.
- J. B. Phillips and M. Westerfield, Zebrafish models in translational research: tipping the scales toward advancements in human health, Dis. Models Mech., 2014, 7(7), 739–743 CrossRef PubMed.
- A. V. Kalueff, A. M. Stewart and R. Gerlai, Zebrafish as an emerging model for studying complex brain disorders, Trends Pharmacol. Sci., 2014, 35(2), 63–75 CrossRef CAS PubMed.
- A. M. Stewart, J. F. P. Ullmann, W. H. J. Norton, M. O. Parker, C. H. Brennan and R. Gerlai, et al., Molecular psychiatry of zebrafish, Mol. Psychiatry, 2015, 20(1), 2–17 CrossRef CAS PubMed.
- Y. Geng and R. T. Peterson, The zebrafish subcortical social brain as a model for studying social behavior disorders, Dis. Models Mech., 2019, 12(8), dmm039446 CrossRef PubMed.
- M. S. de Abreu, K. A. Demin, A. C. V. V. Giacomini, T. G. Amstislavskaya, T. Strekalova and G. O. Maslov, et al., Understanding how stress responses and stress-related behaviors have evolved in zebrafish and mammals, Neurobiol. Stress, 2021, 15, 100405 CrossRef CAS PubMed.
- B. D. Fontana, T. E. Müller, M. Cleal, M. S. de Abreu, W. H. J. Norton and K. A. Demin, et al., Using zebrafish (Danio rerio) models to understand the critical role of social interactions in mental health and wellbeing, Prog. Neurobiol., 2022, 208, 101993 CrossRef PubMed.
- J. Nagpal, U. Herget, M. K. Choi and S. Ryu, Anatomy, development, and plasticity of the neurosecretory hypothalamus in zebrafish, Cell Tissue Res., 2019, 375(1), 5–22 CrossRef PubMed.
- D. J. Davis, H. M. Doerr, A. K. Grzelak, S. B. Busi, E. Jasarevic and A. C. Ericsson, et al., Lactobacillus plantarum attenuates anxiety-related behavior and protects against stress-induced dysbiosis in adult zebrafish, Sci. Rep., 2016, 6(1), 33726 CrossRef CAS PubMed.
- D. G. Valcarce, J. M. Martínez-Vázquez, M. F. Riesco and V. Robles, Probiotics reduce anxiety-related behavior in zebrafish, Heliyon, 2020, 6(5), e03973 CrossRef PubMed.
- A. C. V. V. Giacomini, A. S. Piassetta, R. Genario, C. D. Bonan, A. Piato and L. J. G. Barcellos, et al., Tryptophan alleviates neuroendocrine and behavioral responses to stress in zebrafish, Behav. Brain Res., 2020, 378, 112264 CrossRef PubMed.
- L. Borrelli, S. Aceto, C. Agnisola, S. De Paolo, L. Dipineto and R. M. Stilling, et al., Probiotic modulation of the microbiota-gut-brain axis and behaviour in zebrafish, Sci. Rep., 2016, 6(1), 30046 CrossRef CAS PubMed.
- A. Burokas, S. Arboleya, R. D. Moloney, V. L. Peterson, K. Murphy and G. Clarke, et al., Targeting the Microbiota-Gut-Brain Axis: Prebiotics Have Anxiolytic and Antidepressant-like Effects and Reverse the Impact of Chronic Stress in Mice, Biol. Psychiatry, 2017, 82(7), 472–487 CrossRef CAS PubMed.
- M. Hunt, L. Lima, D. Anderson, J. Hawkey, W. Shen, J. Lees, et al., AllTheBacteria – all bacterial genomes assembled, available and searchable [Internet], bioRxiv, 2024, pp. 2024.03.08.584059. Available from: https://www.biorxiv.org/content/10.1101/2024.03.08.584059v3 [cited 2024 Nov 19].
- L. Richardson, B. Allen, G. Baldi, M. Beracochea, M. L. Bileschi and T. Burdett, et al., MGnify: the microbiome sequence data analysis resource in 2023, Nucleic Acids Res., 2023, 51(D1), D753–D759 CrossRef CAS PubMed.
- E. Drula, M. L. Garron, S. Dogan, V. Lombard, B. Henrissat and N. Terrapon, The carbohydrate-active enzyme database: functions and literature, Nucleic Acids Res., 2022, 50(D1), D571–D577 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4fo03932g |
| ‡ These authors contributed equally to this project. |
| This journal is © The Royal Society of Chemistry 2025 |