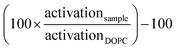Open Access Article
Open Access ArticleMelting point matters: designing lipid nanocarriers for improved T cell activation†
Carina S.
Fedosejevs
ab,
Lariana
Cline
 ab and
Neha P.
Kamat
*abc
ab and
Neha P.
Kamat
*abc
aDepartment of Biomedical Engineering, Northwestern University, Evanston, IL 60208, USA. E-mail: nkamat@northwestern.edu
bCenter for Synthetic Biology, Northwestern University, Evanston, IL 60208, USA
cChemistry of Life Processes Institute, Northwestern University, Evanston, IL 60208, USA
First published on 1st April 2025
Abstract
Surface-modified lipid nanocarriers are increasingly used as artificial antigen-presenting cells for therapeutic applications in immunotherapy. Within these nanocarriers, the role of the lipids is typically limited to providing structure/stability of the particle, to anchoring a targeting moiety, and/or to altering the biodistribution of the nanocarriers in vivo. However, lipid membranes also possess special thermodynamic properties that impact their function. Here, we investigate the effect of the melting transition temperature of lipid nanocarriers on the activation efficiency of an immortalized line of T lymphocytes. Using an established in vitro activation assay and αCD3-functionalized lipid nanocarriers, we screened a variety of lipid nanocarriers with respect to their capacity to activate T cells. We observed a correlation between T cell activation efficiency and proximity of the melting transition temperature of the lipid nanocarrier to the temperature at which the activation study was conducted (37 °C). This relationship held across a variety of lipid compositions and appeared to be more important than the lipid headgroup or chain length. This trend was preserved when the activation temperature was shifted to 30 °C, supporting the role of the nanocarrier membrane state for target cell activation and the potential impact of phase-transition-related effects on nanocarrier activity. We conclude that lipid composition is indeed an important parameter for lipid-based nanocarrier design, not only for the more explored biochemical roles of the lipids but also for the thermodynamic properties the lipid mixtures generate. Our results provide a new consideration in therapeutic nanocarrier design that could significantly improve the efficacy of targeted nanocarrier formulations.
Introduction
The design of artificial antigen-presenting cells (aAPCs) that target and activate T cells, either ex or in vivo, has become an important approach for T cell immunotherapy. Among the various nanoparticles used to design aAPCs, lipid-based nanoparticles are a popular choice due to their high biocompatibility and low systemic toxicity in vivo.1–5 While several design principles have been identified that enhance the biodistribution and compatibility of lipid-based particles in vivo,6–9 there remain a surprisingly low number of approved aAPC therapies for the clinic.10 This dearth of approved therapies for immune-cell activation suggests a greater need for improved lipid nanocarriers that robustly and efficiently activate their target cells.To better improve the design of aAPCs, it is useful to consider the sequence of events that T cells undergo during activation by an antigen-presenting cell. Specifically, for T lymphocytes, there have been a number of studies uncovering the complex processes that occur in the plasma membrane during activation.11–21 Upon binding APC antigens, an immunological synapse is formed between the T cell and antigen-presenting cell, triggering a downstream signaling cascade activated by the influx of Ca2+ ions.11,12 The formation of the immunological synapse is accompanied by a complex reorganization of the T cell membrane, referred to as “bullseye formation”, during which the T cell receptors (TCRs) cluster in the center of the bullseye.13,14 The asymmetry of the membrane in this region is reduced as phosphatidylserine (PS) lipids are relocated to the outer leaflet, creating a negative charge sink at the synapse and a local increase in lipid order.15–19 Simultaneously, local deformations in the T cell membrane are necessary to bring antigens on the aAPC and receptors on the T cell close enough to bind and initiate this process, requiring local areas with increased elasticity.20,21 Altogether, the activation of T cells requires rapid remodeling and reorganization of lipids and proteins on the T cell surface.
The dynamic rearrangements on the T cell membrane during synapse formation and activation suggest that the properties of the aAPCs facilitating this reorganization should enhance the efficiency of synapse formation and T cell activation. Along these lines, several properties of aAPCs have been shown to be important for T cell activation, including the biochemical identity and presentation of antigens and co-stimulatory molecules, as well as physical characteristics of the APCs and aAPCs themselves. Toward the former feature, the density of activating antibodies, such as the commonly used anti-CD3 (TCR stimulus) and anti-CD28 (costimulatory cue), has been shown to impact T cell activation.22,23 Toward the latter feature, it has been found that the size,24–26 shape,27–29 and membrane fluidity23,30 of the aAPC can impact aAPC efficacy. For example, the rigidity of the aAPC membrane is an important parameter for the activation process.14,20,31 Since TCRs cluster during activation, a corresponding mobility of the activating antigen on the antigen-presenting cell is advantageous. Indeed, spatial clustering of the targeting moiety has been shown to enhance its binding efficiency to cellular targets.32–34 Together, these studies suggest a critical role of the physical properties of functionalized aAPCs in effectively activating T cells.
While previous work has largely focused on the role of static physical properties of aAPCs in activating immune cells, perhaps a more important feature of aAPCs is their capacity to undergo dynamic changes in their properties. Lipid membranes and heterogenous cell membranes are able to undergo phase transitions, also known as melting transitions. Depending on their environment (temperature, pH, pressure, salt concentration, etc.), lipids adopt distinct orientations and phases within a bilayer, ranging from a less dense, disordered chain orientation (liquid disordered phase, LD) to a more crystalline, highly ordered one (gel phase, LO). Synthetic lipid membranes assembled from mixtures of lipids can undergo melting transitions between phase states. At these transition points, the membrane structure becomes highly dynamic, exhibiting large intrinsic fluctuations. The fluctuations can lead to unusual or unexpected events, as properties like compressibility, heat capacity, and area expansion coefficient diverge relative to points below or above the phase transition point.35–38 While most studies on lipid membrane transitions have focused on synthetic lipid mixtures, recent studies have shown these transitions exist in biological membranes as well.39–41 These phase transitions are expected to play an important role in cooperatively regulating various cellular functions such as transport processes, protein organization, and enzyme activities.42,43 For example, a membrane might get very soft at the phase transition point with an associated increased permeability relative to points below and above the transition point, leading to enhanced signal transduction.35–38,44 These prior results point to a potentially important but overlooked property in the design of nanocarrier membranes for activating signaling cascades in a target cell. Specifically, nanocarriers designed with a melting transition close to that of the target cell membrane's transition point might best support dynamic rearrangements of lipids and proteins on both membranes to efficiently drive cell synapse formation and activation.
Here, we set out to investigate the effect of the melting transition temperature of nanocarrier membranes on the activation of T cells. Using an in vitro platform to assemble aAPCs and assess T cell activation, we explored the effect of the nanocarrier melting transition temperature and its proximity to the cell growth temperature on T cell activation. Our results point to a new design consideration of surface functionalized, lipid-based aAPCs to effectively promote target cell activation.
Experimental
Materials
The following lipids were purchased from Avanti Polar Lipids: 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), cholesterol, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[benzylguanine(polyethylene glycol)-2000] (18:1 PE PEG BG), 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (ammonium salt) (18:0 PE PEG 2000), 1,2-di-(9Z-octadecenoyl)-sn-glycero-3-phospho-L-serine (sodium salt) (18:1 PS; DOPS), 1,2-dilauroyl-sn-glycero-3-phospho-L-serine (sodium salt) (12:0 PS; DLPS), 1,2-dimyristoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt) (14:0 PG; DMPG), 1,2-dimyristoyl-sn-glycero-3-phosphocholine (14:0 PC; DMPC), 1-stearoyl-2-myristoyl-sn-glycero-3-phosphocholine (18:0-14:0 PC), 1-myristoyl-2-palmitoyl-sn-glycero-3-phosphocholine (14:0-16:0 PC), 1,2-dimyristoyl-sn-glycero-3-phospho-L-serine (sodium salt) (14:0 PS; DMPS), 1,2-dielaidoyl-sn-glycero-3-phosphoethanolamine (18:1 (Δ9-trans)PE), 1-myristoyl-2-stearoyl-sn-glycero-3-phosphocholine (14:0-18:0 PC), 1,2-dipalmitoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt) (14:0 PG; DPPG), 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (16:0 PC; DPPC), 1,2-diheptadecanoyl-sn-glycero-3-phosphocholine (17:0 PC), 1,2-dipalmitoyl-sn-glycero-3-phospho-L-serine (sodium salt) (16:0 PS; DPPS), 1,2-distearoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt) (18:0 PG, DSPG).The fluorescent dyes, 2-dimethylamino-6-lauroylnaphthalene (LAURDAN) and 3-((3-((9-(diethylamino)-5-oxo-5H-benzo[a]phenoxazin-2-yl)oxy)propyl)(dodecyl)(methyl)ammonio)propane-1-sulfonatem (NR12S), were purchased from Fisher Scientific.
DNA preparation and protein expression
The αCD3 scFv-SNAP-tag plasmid was purchased from Twist Bioscience. The plasmid is designed with a T7 promoter and terminator, a ribosomal binding site and an N-terminal FLAG-tag, and a SNAP-tag based on a pJL1 vector. The DNA sequence is provided in Table S1.† DNA was amplified and purified with a PureLink™ HiPure Plasmid Midiprep Kit (Invitrogen). The protein was expressed cell-free with a PURExpress In Vitro Protein Synthesis kit (New England Biolabs) and additional PURExpress Disulfide Bond Enhancers (New England Biolabs). The reaction was assembled on ice with 3.3 nM plasmid DNA and run for 16 h at 30 °C. Afterwards, the protein was stored at 4 °C.Liposome preparation
Liposomes were prepared via thin-film hydration: 70 mol% of a phospholipid of interest, 29 mol% cholesterol, 0.9 mol% 18:0 PE PEG 2000, and 0.1 mol% 18:1 PE PEG BG were mixed in a glass vial in chloroform and gently dried under a flow of nitrogen to create a thin film of the lipid mixture. The films were stored in a vacuum overnight to remove residual solvent. Films were rehydrated with phosphate-buffered saline (PBS) to a final lipid concentration of 4 mM at 60 °C for 30–60 minutes and vortexed in between. The vesicle suspensions were extruded through 100 nm filters (Whatman Nuclepore polycarbonate membranes) with a Mini-Extruder set (Avanti) with 11 passes through the membrane to form homogeneous large unilamellar vesicles. For functionalization, 4 μl of the αCD3 cell-free-expression crude mixture was incubated with 4 μl of liposomes for 2 h at room temperature to allow for conjugation of the αCD3-SNAP-tag to liposomes containing BG-lipids and was afterwards stored at 4 °C.GUV preparation
Giant unilamellar vesicles (GUVs) were prepared via spontaneous swelling. Briefly, 60 μl of a 2.5 mM lipid solution was diluted in 200 μl chloroform in a 20 ml glass vial. Under a stream of nitrogen, the chloroform was evaporated and a thin film formed under constant rotation of the glass by hand. The vial was placed in a vacuum for 2 h to remove residual solvent. 2 ml sucrose solution (295 mOsm) was gently pipetted into the vial, which was then placed in a 60 °C oven overnight. Without further vortexing of the lipid suspension, 1 ml of the volume was taken out the next morning. To concentrate the GUVs, 200 μl of the suspension was carefully pipetted into 1 ml glucose (295 mOsm). After 20–30 min, 20 μl of the bottom fraction was collected and transferred to a new tube to either add the GUVs directly to an imaging dish for quality control or to further conjugate the GUVs with 4 μl of the αCD3 antibody fragment for 2 h at room temperature.T cell activation
An in-house engineered Jurkat nuclear factor of activated T cells (NFAT) reporter cell line (described in detail in ref. 34) was used for this study. Cells were cultured in Roswell Park Memorial Institue (RPMI) 1640 medium (Gibco) with 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin–streptomycin (Gibco) at 37 °C with 5% CO2. For the activation study, 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Jurkat cells in 100 μl full RPMI were seeded into a well of a 96 well plate, per each condition and replicate. 8 μl of the functionalized liposome suspension was added, and the plate was incubated overnight at 37 °C with 5% CO2. As a positive control, phorbol 12-myristate 13-acetate (PMA) (0.1 ng ml−1 final) and ionomycin (2 ng ml−1 final) (both purchased from Sigma Aldrich) were added. For activation at 30 °C, the cells were incubated in an incubator at 30 °C and 5% CO2. In total, for each lipid type at least 3 biological replicates have been measured at 37 °C with at least 2 technical replicates for each biological replicate. For studies conducted at 30 °C, 3 measurements with 2 technical replicates each were conducted.
000 Jurkat cells in 100 μl full RPMI were seeded into a well of a 96 well plate, per each condition and replicate. 8 μl of the functionalized liposome suspension was added, and the plate was incubated overnight at 37 °C with 5% CO2. As a positive control, phorbol 12-myristate 13-acetate (PMA) (0.1 ng ml−1 final) and ionomycin (2 ng ml−1 final) (both purchased from Sigma Aldrich) were added. For activation at 30 °C, the cells were incubated in an incubator at 30 °C and 5% CO2. In total, for each lipid type at least 3 biological replicates have been measured at 37 °C with at least 2 technical replicates for each biological replicate. For studies conducted at 30 °C, 3 measurements with 2 technical replicates each were conducted.
Flow cytometry
Cells positive for green fluorescent protein (GFP) were detected via flow cytometry. Samples were diluted into 1 ml flow buffer (2 mM ethylenediamine tetraacetic acid (EDTA), 1% FBS) and washed (centrifugation at 500 g for 5 min, removal of supernatant, and resuspension in 300 μl flow buffer). Measurements were conducted on a BD64 LSRFortessa cell analyzer with sample excitation at 488 nm and GFP signal detection with a FITC filter. Events were gated for cells and single cells. The GFP-positive gate was based on the αCD3 only control (no liposomes) to account for background activation from unconjugated protein, although nearly no background activation has been observed with the soluble protein. Subsequent data analysis and creation of data plots was conducted in Python.Laurdan GP spectroscopy
Liposomes containing 0.5 mol% Laurdan were diluted to a final concentration of 400 μM into the medium of choice (PBS or RPMI) and transferred into a cuvette (UV-cuvette, Brandtech). Measurements were conducted on a Cary Eclipse fluorescence spectrophotometer (Agilent technologies) equipped with a multicell holder, allowing up to 4 samples to be measured at the same time. Temperature ramps were conducted with 0.5 °C min−1 steps and data points were acquired in 0.5 °C steps with an integration time of 1 s. The temperature was detected with a sensor placed in a cuvette filled with water allowing a feedback loop for the spectrometer to adjust its temperature during the ramp. The Laurdan signal was excited at 380 nm and emission detected in two channels (I1 = 450 nm and I2 = 500 nm) in order to calculate the generalized polarization (GP) value: GP = (I1 − I2)/(I1 + I2).Dynamic light scattering
Liposome size and polydispersity were measured with dynamic light scattering (DLS) in a 100 μM dilution using a Malvern Zetasizer Nano. The results display the mean values of at least two biological replicates and each replicate is the average of 3 runs with 3 reads each.Microscope activation study
Fluorescence images were recorded with confocal scanning microscopy on a Nikon Ti Eclipse microscope equipped with a confocal unit (Nikon C2 Si) and a 60× objective (Nikon Apo Lambda S 60× Oil). Activation was studied at 37 °C with a heated stage (PE100 Peltier system, Linkam). For imaging, cells were stained with 1 μM NR12S in RPMI and 1 × 105 cells in 130 μl full RPMI medium were transferred to a μ-Slide 18 Well imaging dish (#1.5, Ibidi). For activation, 20 μl of the GUV-αCD3 conjugation mixture was added to the cells. Imaging was started shortly after, as soon as the GUVs sank and started to attach to the Jurkat cells. GUVs were stained with free NR12S that was still in the cell solution. The background fluorescence of NR12S in the medium was not detectable. NR12S was excited at 488 nm and emission read in two channels (I1 = 565–595 nm and I2 = 600–630 nm). The emission spectrum is sensitive to changes in cholesterol and membrane state and accordingly spectral shifts were detected by measuring the ratio of two characteristic wavelengths (similarly to Laurdan). To quantify emission signals, the intensities were measured over regions of interest (ROIs) containing a GUV or a cell. Pixel-by-pixel analysis was not used for quantification as the slight movement of vesicles leads to small displacements and the edges of the membranes did not reflect correct ratio values. Ratio images and ROI analysis was conducted in the NIS-Elements software (Nikon).Results and discussion
Lipid nanocarrier Tm affects the activation of Jurkat cells more so than lipid identity
To investigate the effect of nanocarrier melting transition temperature on T cell activation, we first assembled aAPCs that bind and activate T cells. aAPCs were assembled from 100 nm large unilamellar vesicles (LUVs) containing 70 mol% of a phospholipid of interest (18:1 PC as a reference), 29 mol% cholesterol, 0.9 mol% 18:0 PE PEG 2000, and 0.1 mol% 18:1 PE PEG BG. The latter presents the covalent binding partner of the SNAP-tag coupled to a PEGylated lipid anchor. We next used a simplified one-pot workflow to assemble aAPCs by conjugating a cell-free-expressed αCD3–SNAP-tag single-chain variable fragment (scFv) to LUVs, as we have previously described.34 CD3 is an associated subunit of the TCR and presents a suitable target for this kind of study. An αCD3 scFv antibody fragment was designed as a fusion protein with a SNAP-tag and expressed in a bacterial cell-free expression system. Upon addition of the fusion protein to liposomes containing a benzylguanine (BG) lipid, the protein was covalently attached to liposomes and used for subsequent activation studies (Fig. 1A).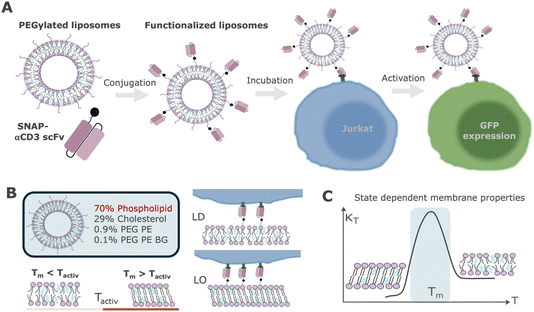 | ||
| Fig. 1 Schematic overview of the assembly of lipid-based artificial antigen-presenting cells (aAPCs) and activation of Jurkat cells. (A) aAPCs are assembled from large unilamellar vesicles (LUVs) containing 70% phospholipids, 29% cholesterol, 0.9% PEG-lipids, and 0.1% benzylguanine (BG)-modified lipids, in which % refers to mol%. BG is a substrate for the self-labeling protein SNAP-tag and allows a SNAP-tag–αCD3 scFv fusion protein to be conjugated to LUVs via attachment of the SNAP-tag to the BG groups on the LUV surface. These particles are incubated overnight at 37 °C with an engineered Jurkat cell line expressing GFP upon activation. Activation is confirmed via microscopy and quantified with flow cytometry. This assay is based on the one-pot approach of LUV assembly introduced previously.34 (B) The standard liposome formulation contains 18:1 PC (DOPC, Tm = −17 °C) keeping the liposome in the liquid disordered phase during interaction with the membrane of Jurkat cells. Phospholipids with Tm > Tactiv form more highly ordered membranes in the liquid ordered phase at Tactiv when mixed with cholesterol. (C) Phase states in lipid bilayers are associated with distinct membrane properties, such as compressibility, heat capacity or diffusivity, density and permeability. More highly ordered states, such as the gel phase or cholesterol-induced liquid ordered phase, are more rigid and display lower diffusivity rates than the liquid disordered state. During the transition between two phases, membrane fluctuations are maximized and diverging physical properties and nonlinear behavior emerge. We hypothesize that this nonlinear behavior should promote membrane rearrangement and corresponding activation in target Jurkat cells. | ||
For one biological replicate, we expressed one batch of protein and divided the protein between different lipid LUV samples to ensure a similar protein concentration on each LUV and so that differences in activation efficiencies relate to the lipid composition and not to protein amount. Due to varying protein yields between batches, the absolute % activation of the cells varied between biological replicates. The activation levels generated by the different lipid/LUV types are noted as relative values compared to the activation of the reference sample 18:1 PC within each biological replicate. In the following, if not stated otherwise, relative values in % refer to the difference in activation between the sample and the reference (DOPC liposomes):
The means of these relative activation values per lipid condition are presented in the following results. Data points represent the mean of at least 3 biological replicates.
All the lipid particles in this study were composed of the same amounts of cholesterol, PE PEG 2000, and PE PEG BG, with only the type of phospholipid changed, which represents 70 mol% of the total composition. Note that since the cholesterol, PEG and BG amounts are kept constant throughout the measurements, we will not explicitly refer to these components. Instead, we refer to the LUV type by the variable phospholipid type alone (e.g. PE LUV). An overview of the variable phospholipids tested in this study and their properties can be found in Table 1.
| Phospholipid (PL) chain length | Abbreviation | PL name | Net charge | T m (°C) of PLa |
|---|---|---|---|---|
| a Values taken from ref. 45. | ||||
| 18:1 PC | DOPC | 1,2-Dioleoyl-sn-glycero-3-phosphocholine | 0 | −17 |
| 14:0 PC | DMPC | 1,2-Dimyristoyl-sn-glycero-3-phosphocholine | 0 | 24 |
| 18:0-14:0 PC | 1-Stearoyl-2-myristoyl-sn-glycero-3-phosphocholine | 0 | 30 | |
| 14:0-16:0 PC | 1-Myristoyl-2-palmitoyl-sn-glycero-3-phosphocholine | 0 | 35 | |
| 14:0-18:0 PC | 1-Myristoyl-2-stearoyl-sn-glycero-3-phosphocholine | 0 | 39 | |
| 16:0 PC | DPPC | 1,2-Dipalmitoyl-sn-glycero-3-phosphocholine | 0 | 41 |
| 17:0 PC | 1,2-Diheptadecanoyl-sn-glycero-3-phosphocholine | 0 | 50 | |
| 18:1 PS | DOPS | 1,2-Di-(9Z-octadecenoyl)-sn-glycero-3-phospho-L-serine | −1 | −11 |
| 12:0 PS | DLPS | 1,2-Dilauroyl-sn-glycero-3-phospho-L-serine | −1 | 14 |
| 14:0 PS | DMPS | 1,2-Dimyristoyl-sn-glycero-3-phospho-L-serine | −1 | 35 |
| 16:0 PS | DPPS | 1,2-Dipalmitoyl-sn-glycero-3-phospho-L-serine | −1 | 51 |
| 14:0 PG | DMPG | 1,2-Dimyristoyl-sn-glycero-3-phospho-(1′-rac-glycerol) | −1 | 23 |
| 16:0 PG | DPPG | 1,2-Dipalmitoyl-sn-glycero-3-phospho-(1′-rac-glycerol) | −1 | 41 |
| 18:0 PG | DSPG | 1,2-Distearoyl-sn-glycero-3-phospho-(1′-rac-glycerol) | −1 | 54 |
| 18:1(Δ9-t) PE | 1,2-Dielaidoyl-sn-glycero-3-phosphoethanolamine | 0 | 38 | |
We first assessed the potential of different lipids to modify nanocarrier-mediated stimulation of an immortalized line of T lymphocytes. Lipids with different features according to headgroup, charge, chain length and saturation were tested. To measure lymphocyte activation, we used a Jurkat reporter cell line,46 in which Jurkat cell activation is reported via GFP expression. Specifically, downstream activation of the NFAT transcription factor after antigen-mediated stimulation of Jurkat cells initiates GFP expression. The percentage of Jurkat cells that were activated was quantified via flow cytometry. After overnight incubation of liposome-conjugated protein with cells, the GFP signal was read out and quantified via flow cytometry. We confirmed that free SNAP-tag–αCD3 or blank liposomes without protein did not activate Jurkat cells in the investigated concentration regime of crude cell-free product and lipid vesicles, respectively (Fig. 2A). Treatment with ionomycin and PMA activates the cells without TCR stimulation and served as a positive control, yielding activation levels around 99% (Fig. 2A). However, due to varying yields of the αCD3–SNAP-tag protein in the cell-free-expression reactions, absolute activation levels for LUV-induced activation were found to differ between LUV batches and the variation is reflected in the error bars shown for each experiment. We first confirmed that Jurkat cells could be activated with our cell-free modified aAPCs by using 18:1 PC (DOPC) LUVs that were subsequently included in each measurement. We measured the % of cells activated by DOPC LUVs to be between 20% and 50%.
We evaluated the impact of the lipid headgroup and chain length on T cell activation by assembling LUVs with a variable phospholipid containing one of three different headgroups (PC, PS, PG) and two different chain lengths (14:0 and 16:0). We did not observe a correlation between a given lipid headgroup or chain length and Jurkat cell activation (Fig. 2B). In contrast, for PS LUVs, the 14:0 lipid performed better than the 16:0 lipid, whereas the trend was reversed for PG LUVs. In comparison to the reference sample (DOPC LUVs), 14:0 PS and 16:0 PG LUVs increase the activation significantly by +70 to 85%. While 14:0 PS is expected to be in the LD phase during activation, 16:0 PG is expected to be in the LO phase. Interestingly, we noticed that both lipids that induced these enhanced activations have a melting transition temperature closer to the temperature at which activation studies were conducted (Tactiv = 37 °C) with Tm (14:0 PS) = 35 °C and Tm (16:0 PG) = 41 °C compared to other samples (Tm (16:0 PS) = 54 °C and Tm (14:0 PG) = 23 °C). The theoretical melting temperatures reported (Table 1) for the variable phospholipid refer to pure lipid systems and do not account for either the amount of cholesterol, PEG and BG in the liposome, or the interaction of the LUVs with cells during activation studies. While these factors could shift the actual membrane transition point by some °C, this observation suggested a potential relationship between the lipid transition temperature and Jurkat cell activation.
To assess this potential relationship, we displayed our data as a function of increasing melting temperature of the variable phospholipid in our LUVs (from left to right in Fig. 2C). We observed that Jurkat cell activation relative to activation with DOPC LUVs displays a parabolic curve. LUVs containing the variable phospholipid with a Tm closest to the temperature of the activation studies resulted in the highest % of activated cells. For LUVs with a variable phospholipid Tm much lower or much higher than Tactiv, the % activation decreased. This observation is independent of the headgroup type, chain length, saturation, or charge of the lipids we tested. In general, lipids with Tm < 37 °C should form disordered membranes during the activation studies at 37 °C, allowing increased mobility of the targeting moiety on the LUV surface, which could facilitate the clustering of the TCR receptor. Rigidity or stiffness of the substrate has been reported to enhance T cell activation before, but seems here to be limited to the LD phase.31 However, while many LD LUVs in our study seem to activate better than LO samples (Tm > 37 °C), membrane order alone does not appear to explain Jurkat cell activation. The LUV offering the highest mobility is the reference sample (DOPC or 18:1 PC) as it is composed of unsaturated lipids and has the furthest distance between Tm (−17 °C) and Tactiv (37 °C). Accordingly, it should remain in the fluid LD phase during activation. Since nearly all LUVs containing a majority LD phase showed enhanced activation compared to DOPC (except for negatively charged 18:1 PS and 12:0 PS), lipid fluidity alone does not dictate Jurkat cell activation. Instead, we observe that the LUV melting temperature appears to be a better predictor of Jurkat cell activation. When the data in Fig. 2C is plotted as a function of lipid Tm, we observe that LUVs with Tm values closest to the activation study temperature, Tactiv, induced the most Jurkat cell activation (Fig. 2D). This trend holds regardless of lipid head group and suggests that LUV proximity to a melting transition plays an important role in Jurkat cell activation.
Jurkat cell activation correlates with the measured melting transitions in LUVs
To better confirm the trend between LUV Tm and Jurkat cell activation, we experimentally measured the Tm of the LUVs used in our studies. The structure and transition behavior of lipid bilayers can be affected by ions47 as well as interactions with components in cell culture media. To evaluate the impact of these conditions on the LUV Tm, we determined LUV Tm values for LUVs expected to have a Tm close to 37 °C. To determine the Tm, we measured the Laurdan generalized polarization (GP) of LUVs as a function of temperature (Fig. 3). Laurdan is a fluorescent dye displaying a shift in emission spectrum between two phase states due to its solvatochromic character.48 The GP value is determined by measuring Laurdan emission intensity at two characteristic wavelengths (I1 = 450 nm and I2 = 500 nm with GP = (I1 − I2)/(I1 + I2)). The GP shifts nonlinearly when a membrane in which it is integrated undergoes a phase transition, providing an experimental readout of the melting temperature of LUVs under our experimental conditions.We measured the melting transition temperature of LUVs under three conditions: 100% phospholipid in PBS, 70% phospholipid/29% cholesterol/1% PEG in PBS and 70% phospholipid/29% cholesterol/1% PEG in full RPMI cell medium. Fig. 3 shows GP measurements as functions of temperature for 16:0 PG (A), 14:0 PS (B) and 16:0 PC (C) LUVs for these conditions. A nonlinearity in the GP signal as a function of temperature represents a transition between the ordered and fluid phase. This nonlinearity is visible as a maximum in the pointwise derivative of GP with respect to temperature and occurs at the temperature with the greatest GP shift. For pure phospholipid liposomes, the measured Tm is in good agreement with the literature values (16:0 PC: Tmeas = 41.8 °C and Ttheory = 41 °C; 14:0 PS: Tmeas = 34.8 °C and Ttheory = 35 °C; 16:0 PG: Tmeas = 40.8 °C and Ttheory = 41 °C). With the addition of cholesterol and PEGylated lipids to the LUVs, the transition gets less pronounced, which is typically observed with cholesterol addition; however, the melting temperature does not shift for all LUV types tested (Fig. 3 middle panel). In RPMI medium, the melting of 16:0 PC LUVs is not affected by the change in environment, whereas the 16:0 PG displays a strong increase in the transition temperature by ∼6 °C. PG is highly susceptible to divalent ions, which probably interferes with its transition behavior and membrane properties.49 For 14:0 PS LUVs, the transition is indicated as a point where the slope of the GP function changes significantly and a transition in the structural arrangement of the lipids happens. The inflection point of the derivative is at Tinflect = 37.7 °C and is increased compared to the transition temperature in PBS. Overall, while we observed some variations in the actual transition temperature of LUVs relative to the transition temperature of the major variable phospholipid, most transition temperatures were close to the values reported in Table 1.
In summary, PS and PG headgroups show modified melting temperatures and shapes of the GP curve when measured in cell media, as they are especially sensitive to ions. While incubation in media shifts the Tm of 14:0 PS LUVs closer to 37 °C, it shifts the 16:0 PG LUV Tm further away, presenting an anomaly from our other data showing a correlation between LUV Tm and cell activation. As expected, PC lipids are not affected by the RPMI conditions and similar trends hold for 18:1(Δ9-trans) PE, which has a Tm we measured at 35 °C in both PBS and RPMI (Fig. S1†). With these experimentally derived LUV Tm values, we could evaluate the activation data again and found that 14:0 PS LUVs have a higher Tm than 18:1 PE LUVs (reversed from when we presented Tm data according to the variable phospholipid alone in Fig. 2C). Hence, a reorganization of lipids when ordered according to their corrected transition points in the LUVs yields a cleaner correlation between the capacity to enhance activation and proximity of Tactiv to LUV Tm, with DPPG as a clear exception (Fig. 4A).
Activation efficiencies at a different Tactiv temperature
To test our hypothesis that the melting transition of LUVs is a driving factor for enhancing Jurkat cell activation, we repeated our study at an alternate temperature. Specifically, we reduced the incubation temperature to 30 °C during activation. Since PC headgroups do not show significant changes in the melting temperature, these lipids are the most promising candidates to investigate the direct influence of the incubation temperature on the activation. An exemplary, direct comparison of activation levels of cells treated at the same time with the same batch of PC LUVs at 30 °C and 37 °C is shown in Fig. 4B. Indeed, 18:0-14:0 PC LUVs (Tm = 30 °C) resulted in the highest Jurkat cell activation at 30 °C, while 14:0-16:0 PC LUVs (Tm = 35 °C) resulted in the highest activation at 37 °C. We applied statistical analysis to this data and activation levels of lipids relative to 14:0-18:0 PC LUVs (Tm = 39 °C) were plotted as a function of melting temperature at a given temperature. This ratio of both activation levels over n = 3 biological replicates underscores the trend observed in the previous analysis (Fig. 4B). The performance of different lipids at two different incubation temperatures shows a clear shift based on which lipid is closer to its transition at the given temperature (Fig. 4C). Adaption of the cells towards the 30 °C incubation can be excluded as they have been grown at 37 °C prior to the overnight activation at 30 °C and the time of incubation (16 h) is shorter than the doubling time of the Jurkat cells (20.7 ± 2.2 h).50 These results clearly corroborate that transition-related effects in the membrane of the liposomes promote the activation process.Due to the predicted anomalous behavior of the LUV in all properties during a melting transition, an exact mechanism responsible for the facilitated activation would be merely speculation.35,36 However, nonlinear effects, like an increased amplitude of area-density fluctuations, as well as membrane undulations, are likely to increase the likelihood of antigen-receptor binding, to facilitate clustering and enhanced presentation of the antibody to the Jurkat cell surface, and to induce a mechanical signal stimulation.14,20 During synapse formation, antigens and antibodies must overcome the distance of the immunological synapse (∼15 nm) in order to bind. Thermal membrane undulations of the T cell membrane have been indeed proposed to initiate this binding.21 Additional transition-induced undulations of the artificial antigen-presenting cell should further increase this interaction, especially for membranes in close proximity to their transition where they are expected to show maximal undulations.36
Besides PC-based liposomes, PG-based liposomes were also investigated at 30 °C. Surprisingly, in an overall comparison, 16:0 PG LUVs were still the best activating LUVs in absolute numbers compared to all tested lipids, even though the difference between Tm and Tactiv has further increased (Fig. S2†). The origin of the special activation potential of 16:0 PG is not known and we therefore set out to more closely examine this composition via microscopy.
Visualization of activation with state-sensitive dye NR12S
To investigate the potential differences/conspicuous features of DPPG lipids and other intermediately activating lipid types in the cell activation process, we used microscopy to evaluate lipid-membrane–Jurkat-cell interactions. Instead of LUVs (d = 100–200 nm), we prepared giant unilamellar vesicles (GUVs) (d = 5–30 μm) and attached the SNAP-tag–αCD3 protein similarly to the conjugation scheme used for LUVs (Fig. 5B). We confirmed that GUVs prepared in this way were able to activate Jurkat cells, as GFP fluorescence was observable after an overnight incubation of Jurkat cells with GUV-SNAP-tag–αCD3 conjugates (Fig. S3†). Cells and GUVs were stained with the state-sensitive membrane dye NR12S, and we measured the dye signal during the early activation period of the cells when the immunological synapse formed at 37 °C (30 to 60 minutes after addition of the GUVs to the cells). We measured the emission intensity ratio of NR12S at two wavelengths (I1 = 565–595 nm and I2 = 600–630 nm, with emission ratio parameter I1/I2). This ratio reflects the relative position of the emission spectrum, which is sensitive to cholesterol content and phase-state changes in the lipid and cell membranes.51Three different LUVs were compared during their antigen-mediated stimulation of the Jurkat cells: DOPC LUVs, which serve as the standard reference in our study, DMPS LUVs, which have a melting temperature close to 37 °C, and DPPG LUVs, which displayed the highest activation potential in our previous experiments. The GUVs have a similar composition to the LUVs we used for the activation studies (70 mol% PL, 29 mol% cholesterol, 0.9 mol% 18:0 PE PEG 2000, and 0.1 mol% 18:1 PE PEG BG). The NR12S signals for the different GUV types differ because of the different levels of order in these membranes at 37 °C (Fig. 5C). DOPC GUVs, far in the LD phase, display the lowest ratio, which can clearly be distinguished from the ratio of NR12S in the cell membrane at 37 °C (∼1.5). DMPS is close to the optical ratio generated in the cell membrane and DPPG GUVs appear strikingly dark with a ratio around 1.6. Interestingly, small bright spots can be observed close to the cellular contact site (Fig. 5D).
The NR12S dye is known to have a reduced quantum yield (QY) in the LO phase and an even more reduced QY in the gel phase. This feature helps explain the previous result, in which DPPG GUVs displayed a lower optical ratio, and indicates DPPG membranes are in an ordered state at 37 °C. The bright dots present in local areas of the membrane, on the other hand, indicate areas of less order and suggest these areas experience higher segregation of membrane components. Indeed, a low miscibility of cholesterol in PG membranes has been reported, which can lead to the appearance of a pure cholesterol phase.52 With the likely segregation of cholesterol, the remaining membrane could indeed be closer to the gel phase than the LO phase, leading to additional clustering of the unsaturated BG-conjugated PE anchored protein in the bright areas. Segregation of αCD3, which is attached to an unsaturated lipid, could better facilitate the Jurkat activation process. Interestingly, domains are already visible for the DPPG GUVs in 300 mOsm glucose before the dilution into RPMI medium (Fig. 5A), which was not observed for DMPS and DOPC GUVs. The presence of domains can also indicate coexistence of two phases in binary mixtures. NR12S segregation was observed at room temperature as well as 37 °C, and might provide an explanation for the observed enhanced activation induced by DPPG LUVs at both tested temperatures in our studies.
To further analyze differences in the interaction of cells and different GUVs, we investigated the optical signal of NR12S in the cellular contact sites, the potential immunological synapses (Fig. 5D). The ratiometric images present shifts of the color-coded NR12S emission ratio locally at the contact sites of the cells and DOPC and DMPS GUVs, which account for the local reorganization of the cell membrane in the synapse. For DOPC, the ratio of the interface between the GUV and cell is increased compared to the rest of the GUV and decreased compared to the cell. For DMPS, the immunological synapse has a decreased ratio compared to the cell and GUV, while DPPG shows no change compared to the cell. The interpretation of a decrease in the ratio of NR12S is not straight-forward, and either relates to a higher degree of disorder and a constant cholesterol amount or a decrease in the cholesterol amount itself.51 For membranes in the solid gel phase, additional cholesterol decreases the order, while for membranes in the LD phase, cholesterol increases order. For cell membranes, there are several examples showing that a depletion of cholesterol leads to an increase in membrane tension, which would lead to the observed local decrease in ratio,31,53,54 supporting reports that the immunological synapse is more ordered compared to the rest of the cell.15,17,18,55 A local accumulation of lipids with higher Tm and potentially local displacement of cholesterol in the Jurkat cell membrane at the site of the synapse could lead to the observed decrease in ratio and could explain the observed, local membrane state changes. At present, however, we do not have an explanation for the enhanced performance of DPPG-based vesicles.
Conclusion
Our results demonstrate that the interaction between T cells and substrates is not solely determined by the choice of targeting moiety, and neither is it determined by membrane fluidity alone. Rather, the choice of the phospholipid in nanocarrier design can either support the interaction efficiency of the targeting moiety with the target cell or even suppress it as a result of the modulation of properties of the functionalized membrane interface.LUVs residing close to their transition regime during incubation tended to activate best in this study, independent of their individual features, such as headgroup, chain length, saturation, or even charge. Laurdan GP measurements indicate that there is indeed still a measurable melting transition present in the tested LUV compositions, despite their relatively high cholesterol content of 29 mol%. Our results suggest that phase-transition-related effects and the associated anomalous behavior can enhance the capacity of designed nanocarrier substrates to activate target cells. Specifically, we hypothesized that as the Tm of nanocarrier membranes comes in close proximity to the temperature at which activation studies are conducted, density fluctuations in both membranes should promote local accumulations of the antibody and increased membrane undulations should facilitate antigen–receptor binding.
An exception of the observed correlation with Tm was found for 16:0 PG LUVs, as this nanocarrier led to an increased Jurkat activation level compared to that induced by 14:0 PS LUVs, even though its Tm was measured to be ∼10 °C above Tactiv. Interestingly, 16:0 PG still performs best at 30 °C incubation temperature (Fig. S2†) and the activation capacity of this composition of LUV appears to be independent of the proximity of its Tm to Tactiv. Domains observed in the GUVs made from the 16:0 PG lipids suggest cholesterol accumulations in which the PE anchor of the BG group would cluster too. Hence, segregation-related clustering could explain the improved activation capabilities for the temperatures of 30 °C and 37 °C, opening up another potential interesting feature for the design of nanocarriers. Our findings set the stage for future investigations exploring the design of LUVs and the role of both LUV Tm and domain-segregation of targeting molecules.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its ESI.† Flow cytometry data can be requested from the authors.Author contributions
The study was designed by C. S. F. and N. P. K. C. S. F and L. C. performed measurements. C. S. F. conducted data analysis and prepared the figures. C. S. F. and N. P. K. wrote the manuscript.Conflicts of interest
C. S. F. and N. P. K. are inventors on a U.S. provisional patent (provisional US 63/746,157) submitted by Northwestern University that covers technology in this manuscript.Acknowledgements
This work was supported by the National Science Foundation's MRSEC program (DMR-2308691) and made use of its Shared Facilities at the Materials Research Center of Northwestern University. This work was funded in part by the Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust for C. S. F. and the International Institute for Nanotechnology (IIN) at Northwestern University (L. C.).References
- M. V. Maus, A. K. Thomas, D. G. B. Leonard, D. Allman, K. Addya, K. Schlienger, J. L. Riley and C. H. June, Nat. Biotechnol., 2002, 20, 143–148 CrossRef CAS PubMed.
- R. Zappasodi, M. Di Nicola, C. Carlo-Stella, R. Mortarini, A. Molla, C. Vegetti, S. Albani, A. Anichini and A. M. Gianni, Haematologica, 2008, 93, 1523–1534 CrossRef CAS PubMed.
- K. Perica, J. G. Bieler, C. Schütz, J. C. Varela, J. Douglass, A. Skora, Y. L. Chiu, M. Oelke, K. Kinzler, S. Zhou, B. Vogelstein and J. P. Schneck, ACS Nano, 2015, 9, 6861–6871 CrossRef CAS PubMed.
- S. Mandal, R. Hammink, J. Tel, Z. H. Eksteen-Akeroyd, A. E. Rowan, K. Blank and C. G. Figdor, ACS Chem. Biol., 2015, 10, 485–492 CrossRef CAS PubMed.
- T. R. Fadel, F. A. Sharp, N. Vudattu, R. Ragheb, J. Garyu, D. Kim, E. Hong, N. Li, G. L. Haller, L. D. Pfefferle, S. Justesen, K. C. Harold and T. M. Fahmy, Nat. Nanotechnol., 2014, 9, 639–647 CrossRef CAS PubMed.
- R. Nisini, N. Poerio, S. Mariotti, F. De Santis and M. Fraziano, Front. Immunol., 9, 155 CrossRef PubMed.
- W. Zhang, C. E. Callmann and C. A. Mirkin, ACS Appl. Mater. Interfaces, 2021, 13, 46325–46333 CrossRef CAS PubMed.
- D. Papahadjopoulos, S. Nir and S. Ohki, Biochim. Biophys. Acta, Biomembr., 1972, 266, 561–583 CrossRef CAS PubMed.
- J. Damen, J. Regts and G. Scherphof, Biochim. Biophys. Acta, Lipids Lipid Metab., 1981, 665, 538–545 CrossRef CAS PubMed.
- L. Sercombe, T. Veerati, F. Moheimani, S. Y. Wu, A. K. Sood and S. Hua, Front. Pharmacol, 2015, 6, 286 Search PubMed.
- J. B. Huppa and M. M. Davis, Nat. Rev. Immunol., 2003, 3, 973–983 CrossRef CAS PubMed.
- M. L. Dustin, Cancer Immunol. Res., 2014, 2, 1023–1033 CrossRef CAS PubMed.
- M. J. P. Biggs, M. C. Milone, L. C. Santos, A. Gondarenko and S. J. Wind, J. R. Soc. Interface, 2011, 8, 1462–1471 CrossRef CAS PubMed.
- M. Huse, Nat. Rev. Immunol., 2017, 17, 679–690 CrossRef CAS PubMed.
- W. Wu, X. Shi and C. Xu, Nat. Rev. Immunol., 2016, 16, 690–701 CrossRef CAS PubMed.
- J. A. Rudd-Schmidt, A. W. Hodel, T. Noori, J. A. Lopez, H. J. Cho, S. Verschoor, A. Ciccone, J. A. Trapani, B. W. Hoogenboom and I. Voskoboinik, Nat. Commun., 2019, 10, 5396 CrossRef PubMed.
- K. Gaus, E. Chklovskaia, B. Fazekas De St. Groth, W. Jessup and T. Harder, J. Cell Biol., 2005, 171, 121–131 CrossRef CAS PubMed.
- T. Zech, C. S. Ejsing, K. Gaus, B. De Wet, A. Shevchenko, K. Simons and T. Harder, EMBO J., 2009, 28, 466–476 CrossRef CAS PubMed.
- A. MacDonald, B. Lam, J. Lin, L. Ferrall, Y. J. Kung, Y. C. Tsai, T. C. Wu and C. F. Hung, Front. Immunol., 2021, 12, 755995 CrossRef CAS PubMed.
- C. Hivroz and M. Saitakis, Front. Immunol., 2016, 7, 46 Search PubMed.
- M. A. Al-Aghbar, A. K. Jainarayanan, M. L. Dustin and S. R. Roffler, Commun. Biol., 2022, 5, 40 CrossRef CAS PubMed.
- D. Hollyman, J. Stefanski, M. Przybylowski, S. Bartido, O. Borquez-Ojeda, C. Taylor, R. Yeh, V. Capacio, M. Olszewska, J. Hosey, M. Sadelain, R. J. Brentjens and I. Rivière, J. Immunother., 2009, 32, 169–180 CrossRef CAS PubMed.
- A. S. Cheung, D. K. Y. Zhang, S. T. Koshy and D. J. Mooney, Nat. Biotechnol., 2018, 36, 160–169 CrossRef CAS PubMed.
- S. T. Reddy, A. Rehor, H. G. Schmoekel, J. A. Hubbell and M. A. Swartz, J. Controlled Release, 2006, 112, 26–34 CrossRef CAS PubMed.
- K. Perica, A. De León Medero, M. Durai, Y. L. Chiu, J. G. Bieler, L. Sibener, M. Niemöller, M. Assenmacher, A. Richter, M. Edidin, M. Oelke and J. Schneck, Nanomedicine, 2014, 10, 119–129 CrossRef CAS PubMed.
- L. E. Paulis, T. Geelen, M. T. Kuhlmann, B. F. Coolen, M. Schäfers, K. Nicolay and G. J. Strijkers, J. Controlled Release, 2012, 162, 276–285 CrossRef CAS PubMed.
- R. A. Meyer, J. C. Sunshine, K. Perica, A. K. Kosmides, K. Aje, J. P. Schneck and J. J. Green, Small, 2015, 11, 1519–1525 CrossRef CAS PubMed.
- J. C. Sunshine, K. Perica, J. P. Schneck and J. J. Green, Biomaterials, 2014, 35, 269–277 CrossRef CAS PubMed.
- A. C. Wauters, J. F. Scheerstra, I. G. Vermeijlen, R. Hammink, M. Schluck, L. Woythe, H. Wu, L. Albertazzi, C. G. Figdor, J. Tel, L. K. E. A. Abdelmohsen and J. C. M. Van Hest, ACS Nano, 2022, 16, 15072–15085 CrossRef CAS PubMed.
- Z. Liu, Y. R. Li, Y. Yang, Y. Zhu, W. Yuan, T. Hoffman, Y. Wu, E. Zhu, J. Zarubova, J. Shen, H. Nan, K. W. Yeh, M. M. Hasani-Sadrabadi, Y. Zhu, Y. Fang, X. Ge, Z. Li, J. Soto, T. Hsiai, L. Yang and S. Li, Nat. Biomed. Eng., 2024, 8, 1615–1633 CrossRef CAS PubMed.
- K. Lei, A. Kurum, M. Kaynak, L. Bonati, Y. Han, V. Cencen, M. Gao, Y. Q. Xie, Y. Guo, M. T. M. Hannebelle, Y. Wu, G. Zhou, M. Guo, G. E. Fantner, M. S. Sakar and L. Tang, Nat. Biomed. Eng., 2021, 5, 1411–1425 CrossRef CAS PubMed.
- T. Q. Vu, L. E. Sant'Anna and N. P. Kamat, bioRxiv, 2022, preprint, DOI:10.1101/2022.10.18.512758.
- T. Q. Vu, J. A. Peruzzi, L. E. Sant'Anna, E. W. Roth and N. P. Kamat, Nano Lett., 2022, 22, 2627–2634 CrossRef CAS PubMed.
- J. A. Peruzzi, T. Q. Vu, T. F. Gunnels and N. P. Kamat, Small Methods, 2023, 7, 2201718 CrossRef CAS PubMed.
- E. Evans and R. Kwok, Biochemistry, 1982, 21, 4874–4879 CrossRef CAS PubMed.
- A. Kuklin, D. Zabelskii, I. Gordeliy, J. Teixeira, A. Brûlet, V. Chupin, V. Cherezov and V. Gordeliy, Sci. Rep., 2020, 10, 5749 CrossRef CAS PubMed.
- B. Wunderlich, C. Leirer, A. Idzko, U. F. Keyser, A. Wixforth, V. M. Myles and T. Heimburg, Biophys. J., 2009, 96, 4592–4597 CrossRef CAS PubMed.
- T. Heimburg and A. D. Jackson, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 9790–9795 CrossRef CAS PubMed.
- T. Mužić, F. Tounsi, S. B. Madsen, D. Pollakowski, M. Konrad and T. Heimburg, Biochim. Biophys. Acta, Biomembr., 2019, 1861, 183026 CrossRef PubMed.
- J. H. Crowe, F. A. Hoekstra, L. M. Crowe, T. J. Anchordoguy and E. Drobnis, Cryobiology, 1989, 26, 76–84 CrossRef CAS PubMed.
- C. S. Fedosejevs and M. F. Schneider, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2117521119 CrossRef CAS PubMed.
- L. Thilo, P. Overath and H. Träuble, Biochemistry, 1977, 16, 1283–1290 CrossRef CAS PubMed.
- D. T. Hanisch and M. F. Schneider, Langmuir, 2024, 40, 3369–3375 CAS.
- W. V. Kraske and D. B. Mountcastle, Biochim. Biophys. Acta, Biomembr., 2001, 1514, 159–164 CrossRef CAS PubMed.
- D. Marsh, Handbook of Lipid Bilayers, CRC press, 2nd edn, 2013 Search PubMed.
- R. T. Abraham and A. Weiss, Nat. Rev. Immunol., 2004, 4, 301–308 CrossRef CAS PubMed.
- H. Träuble and H. Eibl, Proc. Natl. Acad. Sci. U. S. A., 1974, 71, 214–219 CrossRef PubMed.
- T. Parasassi, E. K. Krasnowska, L. Bagatolli and E. Gratton, J. Fluoresc., 1998, 8, 365–373 CrossRef CAS.
- P. W. M. Van Dijck, P. H. J. T. Ververgaert, A. J. Verkleij, L. L. M. Van Deenen and J. De Gier, Biochim. Biophys. Acta, Biomembr., 1975, 406, 465–478 CrossRef CAS PubMed.
- N. W. Schoene and K. S. Kamara, Free Radical Biol. Med., 1999, 27, 364–369 CrossRef CAS PubMed.
- O. A. Kucherak, S. Oncul, Z. Darwich, D. A. Yushchenko, Y. Arntz, P. Didier, Y. Mély and A. S. Klymchenko, J. Am. Chem. Soc., 2010, 132, 4907–4916 CrossRef CAS PubMed.
- N. Borochov, E. J. Wachtel and D. Bach, Chem. Phys. Lipids, 1995, 76, 85–92 CrossRef CAS PubMed.
- A. Biswas, P. Kashyap, S. Datta, T. Sengupta and B. Sinha, Biophys. J., 2019, 116, 1456–1468 CrossRef CAS PubMed.
- M. A. Ayee and I. Levitan, Front. Biosci., 2016, 1245–1259 CAS.
- D. M. Owen, S. Oddos, S. Kumar, D. M. Davis, M. A. A. Neil, P. M. W. French, M. L. Dustin, A. I. Magee and M. Cebecauer, Mol. Membr. Biol., 2010, 27, 178–189 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5fd00002e |
| This journal is © The Royal Society of Chemistry 2025 |