Open Access Article
Open Access ArticleVirulence gene profiling and cytotoxicity of Vibrio spp. isolated from treated wastewater effluent and receiving surface waters in Durban, South Africa†
Kerisha
Ramessar
and
Ademola O.
Olaniran
 *
*
Discipline of Microbiology, School of Life Sciences, University of KwaZulu-Natal (Westville Campus), Durban, 4000, South Africa. E-mail: olanirana@ukzn.ac.za; Tel: +27-31-260-7400
First published on 30th June 2025
Abstract
Untreated or partially treated wastewater often harbours virulent Vibrio species that threaten environmental and public health. This study aimed to characterize the virulence gene profiles and cytotoxic effects of Vibrio species isolated from treated effluents and downstream rivers at four wastewater facilities in Durban, KwaZulu-Natal province, South Africa. A total of 200 Vibrio spp., isolated from treated effluent and surface waters of four wastewater treatment facilities in Durban, KwaZulu-Natal, were screened, with Vibrio vulnificus isolates (n = 178) showing high prevalence of iron acquisition genes such as viuB (72.47%), feoB (56.74%) and fbpC (55.06%) while other virulence genes like ompU, apxIB, and hlyB were also detected. Vibrio alginolyticus isolates (n = 15) exhibited rtx (66.67%) and pvuA (46.67%), among others. Five representative isolates caused a progressive decline in cell viability in both HepG2 and HEK293 cells over 72 h, with final viability dropping below 3% in multiple instances. Morphological damage confirmed strong cytotoxic activity. Statistical analysis showed significant associations between specific genes detected among the isolates. These findings demonstrate that treated wastewater still contains highly virulent Vibrio strains capable of harming human cells, posing ongoing risks in regions with compromised water infrastructure.
Environmental significanceWastewater treatment plants are vital for reducing microbial contamination in aquatic environments. The persistence of pathogenic bacteria such as Vibrio spp. in treated wastewater effluents and environmental waters is a public health concern. Consequently, this study profiled virulence genes in 200 Vibrio spp. isolates obtained from treated effluent and surface waters at four wastewater treatment plants in Durban, KwaZulu-Natal. Additionally, the cytopathic effects of five selected isolates were assessed in HepG2 and HEK293 cell lines. The results reveal the presence of various virulence genes among Vibrio spp., their significant associations, and the isolates' strong cytotoxic effects on human cells. These findings emphasize the need for continuous monitoring and improvement of wastewater treatment processes to effectively eradicate pathogenic bacteria. |
1. Introduction
The global burden of waterborne diseases remains a pressing concern, driven largely by the discharge of inadequately treated wastewater into surface waters, which serves as a conduit for pathogenic microorganisms. This scenario poses severe public health threats, particularly in communities that depend on these contaminated water bodies for domestic purposes.1–3 The challenge is most acute in developing regions where wastewater infrastructure is often under-resourced or poorly maintained.4 South Africa exemplifies this crisis, with an estimated 40 to 50% of its 1400 wastewater treatment facilities failing to meet acceptable operational standards.3 Although existing studies have documented the occurrence of pathogenic bacteria in treated effluents,5,6 limited attention has been given to the virulence potential and cytotoxicity of Vibrio species within these systems. This gap is concerning, considering the growing recognition of Vibrio spp. as significant environmental and clinical pathogens.Vibrio species are Gram-negative, facultatively anaerobic bacteria belonging to the Vibrionaceae family and are increasingly identified as threats in wastewater-contaminated aquatic ecosystems.7 Pathogenic strains of Vibrio have been documented in treated effluents from wastewater treatment plants (WWTPs) in the Eastern Cape of South Africa and other areas.2 Among the 147 recognized species, twelve are known to cause disease in humans.8 Notable among these are Vibrio cholerae, which accounted for over 1.2 million cases, globally, in 2017,9V. vulnificus, associated with severe septicaemia and high mortality rates,10 and V. alginolyticus, implicated in wound infections and otitis.11,12 The pathogenicity of these organisms is attributed to a suite of virulence factors, including iron acquisition systems such as viuB, feoB, and fbpC, cytolytic hemolysins (hlyA–D), adhesion and colonization factors like ompU and tcp; and motility-related flagellin genes (flaA–C).13–21 These virulence determinants not only facilitate environmental persistence but also enhance host invasion and tissue damage, underscoring the need for targeted surveillance of Vibrio spp. in treated effluents.
While previous studies have documented the presence of pathogenic Vibrio spp. in wastewater effluents and receiving surface waters in South Africa,2,6 there remains a critical gap in understanding the virulence potential of these strains in treated waters and their implications for human health. This study uniquely addresses this gap by combining molecular profiling of virulence genes with statistical correlation analyses to elucidate the pathogenicity of Vibrio isolates from wastewater effluents. Additionally, we investigate the cytopathic effects of selected V. vulnificus strains on human hepatoblastoma (HepG2) and human embryonic kidney (HEK) cell lines, providing novel insights into their potential health risks upon exposure.
While most prior research has focused on marine or clinical isolates of Vibrio, our study addresses a critical gap by investigating strains that persist through municipal wastewater treatment processes and their receiving surface waters. We hypothesize that these environmental isolates harbour enriched virulence gene repertoires and exhibit heightened cytotoxicity toward human cells. Ultimately, this approach offers novel insights into the environmental persistence and health risk potential of Vibrio spp., with direct implications for water quality monitoring and public health policy in resource-limited settings.
2. Materials and methods
2.1 Isolation and molecular identification of the Vibrio isolates
Wastewater samples were obtained from four wastewater treatment facilities in Durban, KwaZulu-Natal, South Africa. The geographical coordinates of the four wastewater treatment plants are 29°47′43′′S 30°59′52′′E (WWTP1), 29°40′43′′S 31°02′01′′E (WWTP2), 29°59′25′′S 30°54′21′′E (WWTP3), and 29°50′42′′S 30°53′27′′E (WWTP4). Samples were collected seasonally from various locations at each wastewater treatment plant (WWTP), including (1) influent, (2) pre-chlorination, (3) activated sludge or biofilter, (4) post-chlorination, (5) upstream, and (6) downstream of the receiving rivers, during the sampling period from September 2020 to August 2021. Samples were collected in 5 L sterile bottles, subsequently carried on ice to the University of KwaZulu-Natal (Westville campus), where they were maintained at 4 °C and analysed within 48 h of collection. The materials underwent serial dilution with sterile distilled water and were subsequently filtered using a 0.45 μm cellulose nitrate filter. The filters were applied to thiosulfate-citrate-bile salts-sucrose agar (Sigma-Aldrich, St. Louis, MO, USA) and incubated at 37 °C for 24 h. Green and yellow colonies were recognized as probable Vibrio isolates. Two hundred putative Vibrio spp. were isolated from the treated effluents, as well as from upstream and downstream locations of the river. PCR amplification of the V16s housekeeping gene was conducted to verify the identity of the organisms as Vibrio spp. utilizing the primers V16s-700F: CGGTGAAATGCGTAGAGAT and V16s-1325R: TTACTAGCGATTCCGAGTTC.22 Species identification for V. vulnificus was conducted utilizing the primers Vv.hsp-326F: GTCTTAAAGCGGTTGCTGC and Vv.hsp-697R: CGCTTCAAGTGCTGGTAGAAG.22 A semi-nested PCR was conducted to identify V. alginolyticus using the primers: VA16F1: ATTGAAGAGTTTGATCATGGCTCAGA, VA16F2: CCTTCGGGTTGTAAAGCACT, and VA16R2: TCCTCCCGTAGTTGAAACTACCT.232.2 Virulence gene profiling of Vibrio spp.
Detection of virulence genes in V. vulnificus, V. alginolyticus, and Vibrio spp. was conducted by monoplex PCR. Primers for the virulence genes were developed utilizing the DNA sequences from V. vulnificus (ATCC 27562) and V. alginolyticus (ATCC 17749) received from NCBI GenBank (https://www.ncbi.nlm.nih.gov/) and were subsequently extracted and aligned for the specific virulence genes. The primer-BLAST tool facilitated primer design, and the resulting primers (Table 1) were manufactured by Inqaba Biotech, Pretoria, South Africa. V. vulnificus was screened for apxIB, fbpC, feoB, ompU, sodB, tldD, viuB, and hlyB, whereas V. alginolyticus was checked for asp, pvuA, tlh, and rtx. All amplifications were conducted in 25 μL reactions, comprising 12.5 μL of 2× master mix (Thermo-Fisher, Waltham, MA, USA), 0.2 μM of each primer, 12 μL of nuclease-free water, and 2 μL of template DNA. The PCR cycling parameters for V. vulnificus genes consist of an initial denaturation at 96 °C for 5 min, succeeded by 35 cycles of 94 °C for 60 s, annealing (refer to Table 1), and 72 °C for 60 s, culminating in a final extension at 72 °C for 7 min. The PCR cycling parameters for V. alginolyticus genes consist of an initial denaturation at 93 °C for 15 min, succeeded by 35 cycles of denaturation at 92 °C for 40 s, annealing (Table 1), elongation at 72 °C for 1 min and 30 s, and a final elongation step at 72 °C for 7 min following the completion of 30 cycles. PCR products were resolved using gel electrophoresis at 100 V for 45 min on a 1.5% agarose gel, stained with 5 μg mL−1 ethidium bromide, and visualized under a UV transilluminator utilizing the Chemigenius Bioimaging System (Syngene, Cambridge, UK). The multi-virulence gene index (MVGI) was computed using the following equation MVGI = VGD/VGT24 where VGD denotes the virulence genes found, and VGT signifies the total number of virulence genes.| Target gene | Sequence (5′–3′) | Product length (bp) | Annealing temperature |
|---|---|---|---|
| apxIB | F: GCCAACTGCGTGATGTGTTT | 298 | 55.7 °C for 60 s |
| R: ATTTGGCCGGTTTGACGTTG | |||
| fbpC | F: GGAAGTGGAACAAGGGCAGA | 159 | 62.8 °C for 60 s |
| R: TCAGGTGGAACCCAATGCTC | |||
| feoB | F: AAGCGTTTCGTTTTGGGAGC | 237 | 64.3 °C for 40 s |
| R: ACCAACTACCGCCTCTTTGG | |||
| ompU | F: TGGCGTCTGTCATCGTTTCA | 254 | 55.7 °C for 60 s |
| R: ACCAGCACCGTTATCACCTG | |||
| sodB | F: TTGAGTTCCACCACGGCAAA | 370 | 58 °C for 40 s |
| R: GTGATTGGTGTTGCCGCATT | |||
| tldD | F: GGCAACAAACGCTGTAGCTC | 766 | 64.3 °C for 40 s |
| R: CATGTCTTTGCCCACCATGC | |||
| viuB | F: GCCTGAACACAAACCCGTTC | 449 | 62.8 °C for 60 s |
| R: CCGCGATAAAAGCAGACAGC | |||
| hlyB | F: GAACGCGAGTAAATCACGCC | 481 | 58 °C for 40 s |
| R: AAGCGCACGTTATCGAATGC | |||
| asp | F: AGACCGCATGTCTGGTTTGT | 475 | 64.3 °C for 30 s |
| R: GCTCAATACTCTCACGCCGA | |||
| pvuA | F: GATGCGCCGCTGAATGATTT | 343 | 64.3 °C for 30 s |
| R: CACTGTCGGCAAATGGCAAA | |||
| tlh | F: GTGGTTAGCGCGCAAGAAAA | 770 | 64.3 °C for 30 s |
| R: GGTGCTTTGGTTGCATCAGG | |||
| rtx | F: CCGACAGCTTGTTTGTTGGG | 473 | 64.3 °C for 30 s |
| R: CACGTCAACACCGTCTGGTA |
2.3 Cytotoxicity assays
Human embryonic kidney (HEK293) and human hepatoblastoma (HepG2) cells were cultured in tissue culture (T25) flasks using Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin (ThermoFisher, Waltham, MA, USA), and incubated at 37 °C in 5% carbon dioxide.The selected concentration balances several critical factors: (1) ecological relevance by reflecting potential environmental exposures, (2) reproducibility across experimental replicates, and (3) generation of measurable cytotoxic effects suitable for quantitative analysis. Furthermore, in vitro infection models commonly employ similar concentrations to assess bacterial cytotoxicity and virulence factor activity.30,31
To validate that the cytotoxic effects were attributable to virulence factors rather than the bacterial load alone, additional dose-response experiments were performed. Serial dilutions of V. vulnificus suspensions, ranging from 100 to 10−6, were prepared and used to treat HepG2 and HEK293 cells. Cell viability was then assessed following exposure to each dilution to observe changes in cytotoxicity.
Bacterial cells were cultured in nutritional broth to mid-log phase and centrifuged, the pellet was rinsed with phosphate-buffered saline (pH 7.4) and standardized to approximately 1.5 × 108 cells per mL using DMEM.
Twenty microliters of the standardized bacterial suspension were applied to each well of the 96-well microtiter plate and incubated for 24, 48, and 72 h. Uninfected cells were used as a negative control, whereas V. vulnificus ATCC 27562 served as a positive control. Each experimental condition was performed in triplicate (three technical replicates), and each experiment was repeated three times independently (three biological replicates) to ensure reproducibility and statistical reliability.
Following incubation of V. vulnificus with the cells, 20 μL of MTT (1 mg mL−1) was introduced to each well and incubated for 3 h at 37 °C in 5% carbon dioxide. Following incubation, the supernatant was carefully removed, and 100 μL of pure dimethyl sulfoxide (DMSO) was introduced to the wells to solubilize the purple formazan crystals that developed in the cell cytoplasm. Optical absorbance was measured with the plate reader at 570 nm.
It is hypothesized that differential cytotoxic responses between HepG2 and HEK-293 cells may result from their distinct cellular origins, with HepG2 cells (liver-derived) potentially exhibiting different detoxification mechanisms and cellular defence pathways compared to HEK-293 cells (kidney-derived), leading to varying susceptibilities to V. vulnificus virulence factors.
2.4 Evaluation of cell morphology
The cell morphology and overall health of HepG2 and HEK293 cells were assessed by capturing multiple field views for each experimental condition using the Invitrogen EVOS FLoid EN61326 high-resolution microscope, which is equipped with phase-contrast and fluorescence capabilities that ensured a representative and detailed analysis (ThermoFisher Scientific, Waltham, MA, USA).2.5 Statistics
The associations between various virulence genes among isolates were assessed by comparing Pearson chi-square values (χ2), determined using descriptive analysis of crosstabs for contingency tables, at a significance level of p < 0.05. Furthermore, the odds ratio (OR) for each pair of virulence genes was employed to measure the intensity of their correlation. An odds ratio (OR) greater than 1 indicates a positive correlation, implying that the presence of one gene enhances the likelihood of the other being present; an OR less than 1 indicates a negative association, while an OR equal to 1 shows no association.32 Analyses were conducted using IBM SPSS Statistics 28.0. The cytotoxicity results were analysed using a completely randomized design (CRD) with three replicates. The data underwent analysis of variance, and means were differentiated using the Tukey test at p ≤ 0.0001. Additionally, certain data were illustrated graphically as mean ± standard deviation.3. Results
3.1 Virulence gene signatures of the Vibrio isolates
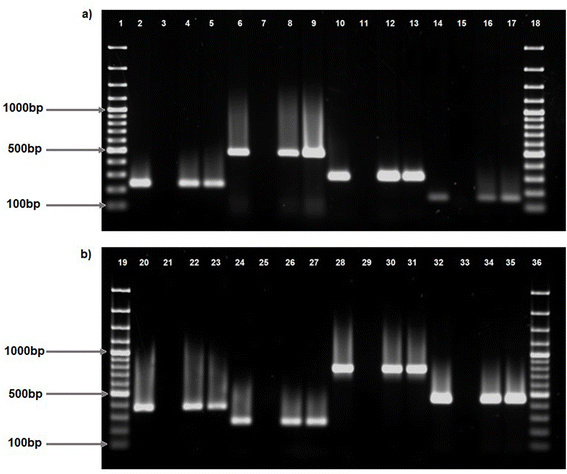
Two hundred presumptive Vibrio isolates were collected from the discharge point of treated effluent, as well as from upstream and downstream locations of receiving surface waters at four wastewater treatment plants in Durban, KwaZulu-Natal. The isolates were identified as Vibrio spp. by PCR amplification of the Vibrio 16s rRNA gene. Out of the two hundred isolates, 178 were identified as Vibrio vulnificus and 15 as Vibrio alginolyticus via PCR amplification of the Vv.hsp and VA16 genes, respectively, while 8 remained unidentifiable at the species level and were categorized as Vibrio spp.PCR amplification of virulence genes: apxIB, ompU, sodB, hlyB, fbpC, viuB, tldD and feoB demonstrated their presence in some V. vulnificus isolates (Fig. 1a, b and Table 2). Out of 178 V. vulnificus isolates, viuB was discovered in 72.47% (n = 129) of isolates, followed by feoB in 56.74% (n = 101) and fbpC in 55.06% (n = 98). Furthermore, ompU was identified in 35.39% (n = 63), apxIB in 24.72% (n = 44), and hlyB in 13.48% (n = 24) of the isolates. The tldD gene was identified in 6.74% (n = 12) of isolates, while sodB was identified in 1.69% (n = 3) of isolates. Additionally, asp, tlh, pvuA, and rtx were identified in certain V. alginolyticus isolates.rtx was predominantly identified in 66.67% (n = 10) of the isolates, while pvuA was recognized in 46.67% (n = 7) of the isolates. asp and tlh were identified in 13.33% (n = 2) and 6.67% (n = 1) of isolates, respectively. In Vibrio spp., viuB was the most common gene detected in 42.86% (3) of isolates followed by ompU and feoB, both, detected in 28.57% (n = 2) isolates. fbpC was only detected in 14.29% (n = 1) isolates.
| Isolates | Virulence genes | |||||||
|---|---|---|---|---|---|---|---|---|
| apxIB | ompU | sodB | hlyB | fbpC | viuB | tldD | feoB | |
| V. vulnificus (n = 178) | 24.72% (44) | 35.39% (63) | 1.69% (3) | 13.48% (24) | 55.06% (98) | 72.47% (129) | 6.74% (12) | 56.74% (101) |
| Vibrio spp. (n = 7) | 0% (0) | 28.57% (2) | 0% (0) | 0% (0) | 14.29% (1) | 42.86% (3) | 0% (0) | 28.57% (2) |
| asp | tlh | pvuA | rtx | |
|---|---|---|---|---|
| V. alginolyticus (n = 15) | 13.33% (2) | 6.67% (1) | 46.67% (7) | 66.67% (10) |
3.2 Virulence genes combinations detected in isolates
Multiple virulence genes were present in all three isolate types: V. vulnificus, V. alginolyticus, and Vibrio spp. (Tables 2 and S1†). Six virulence genes (MVGI = 0.75), namely apxIB, ompU, hlyB, fbpC, viuB and feoB, were simultaneously identified in V. vulnificus isolates. apxIB, hlyB, fbpC, viuB, tldD and feoB were the predominant virulence genes identified in two samples. Fifteen isolates were reported to contain the presence of five virulence genes (MVGI = 0.625) with the most prevalent pattern of apxIB, ompU, fbpC, viuB and feoB. Among the thirty-five isolates exhibiting four virulence genes (MVGI = 0.5), ten isolates possessed the combination of ompU, fbpC, viuB and feoB; seven isolates included the combination of apxIB, ompU, fbpC and viuB; and five isolates included the combination of hlyB, fbpC, viuB and feoB. The predominant virulence gene profile identified was fbpC, viuB, and feoB, observed in twenty-five isolates. Furthermore, eleven isolates exhibited the combination of fbpC and viuB, whereas 10 isolates demonstrated the combination of viuB and feoB. In V. alginolyticus, three virulence genes (MVGI = 0.75) were found in two isolates with combinations of asp, pvuA and rtx, and asp, tlh and rtx. The predominant combination of virulence genes identified was pvuA and rtx, observed in five isolates. In Vibrio spp., two isolates exhibited three virulence genes (MVGI = 0.375): ompU, viuB, and feoB, with fbpC, viuB, and feoB present in each sample. One isolate had a combination of ompU and viuB (MVGI = 0.25).3.3 The positive and negative associations of virulence genes detected in isolates
The Pearson χ2 values at P < 0.05 (unless specified otherwise) are derived from cross-tabulation data illustrating the association between the virulence genes in V. vulnificus, as shown in Table 3. The data reveal that viuB had a significant association with hlyB (χ2: 10.537, OR = 0.682), fbpC (χ2: 50.080, OR = 17.820), feoB (χ2: 32.393, OR = 7.972) and tldD (χ2: 4.888, OR = 1.103). tldD exhibited an association with apxIB (χ2: 4.419, OR = 3.368), ompU (χ2: 7.049, OR = 0.896), and hlyB (χ2: 8.762, OR = 5.526). feoB demonstrated a strong correlation with fbpC (χ2 16.592, OR = 3.569). fbpC had a significant association with apxIB (χ2: 5.601, OR = 2.385) and sodB (χ2: 6.517, OR = 3.608). The virulence gene hlyB revealed a significant relationship with sodB (χ2: 7.399, OR = 13.909). The cross-tabulation results demonstrated a significant correlation between the virulence genes tlh and asp (χ2: 6.964, OR = 0.071) in V. alginolyticus (Table 3). No positive relationships were discovered between virulence genes detected in Vibrio spp. (Table 3).| V. vulnificus | apxIB | ompU | sodB | hlyB | fbpC | feoB | tldD |
|---|---|---|---|---|---|---|---|
| a Positive association at p < 0.05, χ2-chi-square value, OR-odds ratio with 95% confidence interval, ND-not determined. | |||||||
| viuB | χ 2: 1.466; OR 1.655 (0.728–3.760) | χ 2: 0.676; OR 1.343 (0.664–2.718) | χ 2: 1.159; OR 0.720 (0.656–0.790) | χ 2: 10.537a; OR 0.682 (0.612–0.759) | χ 2: 50.080a; OR 17.820 (6.992–45.414) | χ 2: 32.393a; OR 7.972 (3.695–17.202) | χ 2: 4.888a; OR 1.103 (1.043–1.165) |
| tldD | χ 2: 4.419a; OR 3.368 (1.027–11.051) | χ 2: 7.049a; OR 0.896 (0.841–0.953) | χ 2: 3.432; OR 7.455 (0.626–88.738) | χ 2: 8.762a; OR 5.526 (1.594–19.157) | χ 2: 2.068; OR 2.596 (0.678–9.930) | χ 2: 3.707; OR 4.121 (0.876–19.389) | |
| feoB | χ 2: 0.000; OR 1.004 (0.505–1.997) | χ 2: 0.006; OR 1.026 (0.552–1.907) | χ 2: 0.681; OR 0.375 (0.033–4.213) | χ 2: 2.244; OR 2.024 (0.794–5.158) | χ 2: 16.592a; OR 3.569 (1.914–6.655) | ||
| fbpC | χ 2: 5.601a; OR 2.385 (1.148–4.952) | χ 2: 0.532; OR 1.260 (0.677–2.344) | χ 2: 6.517a; OR 3.608 (1.282–10.151) | ||||
| hlyB | χ 2: 1.106; OR 1.639 (0.649–4.142) | χ 2: 0.470; OR 0.721 (0.282–1.844) | χ 2: 7.399a; OR 13.909 (1.210–159.851) | ||||
| sodB | χ 2: 2.885; OR 6.333 (0.560–71.609) | χ 2: 1.305; OR 3.738 (0.332–42.055) | χ 2: 2.491; OR 0.543 (0.474–0.622) | ||||
| ompU | 14.354a; OR 3.787 (1.861–7.705) | ||||||
| V. alginolyticus | asp | pvuA | tlh |
|---|---|---|---|
| rtx | χ 2: 1.151; OR 1.250 (0.917–1.704) | χ 2: 2.143; OR 6.00 (0.478–75.344) | χ 2: 0.536, OR 1.111 (0.904–1.366) |
| tlh | χ 2: 6.964a; OR 0.071 (0.011–0.472) | χ 2: 0.938; OR 0.500 (0.296–0.844) | |
| pvuA | χ 2: 0.010; OR 1.167 (0.059–22.937) |
| Vibrio spp. | apxIB | ompU | sodB | hlyB | fbpC | feoB | tldD |
|---|---|---|---|---|---|---|---|
| viuB | ND | χ 2: 3.733; OR 0.2 (0.035–1.154) | ND | ND | χ 2: 1.556; OR 0.333 (0.10–1.034) | χ 2: 3.733; OR 0.053 (0.606–14.864) | ND |
| tldD | ND | ND | ND | ND | ND | ND | |
| feoB | ND | χ 2: 0.630; OR 4.000 (0.117–136.957) | ND | ND | χ 2: 2.917; OR 0.167 (0.28–0.997) | ||
| fbpC | ND | χ 2:0.467; OR 0.8 (0.516–1.240) | ND | ND | |||
| hlyB | ND | ND | ND | ||||
| sodB | ND | ND | |||||
| o mpU | ND |
In V. vulnificus, the odds ratio and confidence intervals (CI) indicated relationships (Table 3) involving viuB and fbpC (OR 17.820, CI: 6.992–45.414), viuB and feoB (OR 7.92, CI: 3.695–17.202), and hlyB and sodB (OR 13.909, CI: 1.210–159.851). The elevated OR values indicate a robust correlation between these gene pairs. However, a high confidence interval indicates variability in the data. Robust relationships were identified between fbpC and sodB (OR 3.608, CI: 1.282–10.151), ompU and apxIB (OR 3.787, CI: 1.861–7.705), and fbpC and apxIB (OR 2.385, CI: 1.148–4.952), as indicated by significant associations with confidence intervals that do not include one. Nonetheless, a negligible connection was noted between the gene pairs rtx and asp (OR 1.250, CI: 0.917–1.704) in V. alginolyticus, and feoB and apxIB (OR 1.004, CI: 0.505–1.997) in V. vulnificus, as the odds ratios were proximate to 1 and accompanied by extensive confidence intervals. In the Vibrio spp., the odds ratios were approximately one or lower, accompanied by wide confidence intervals, indicating no meaningful connections between the gene pairs.
3.4 Cell morphology of infected and uninfected HEK293 and HepG2 cells
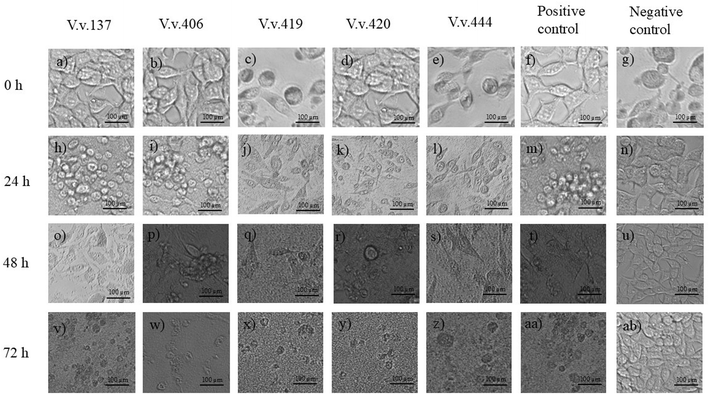
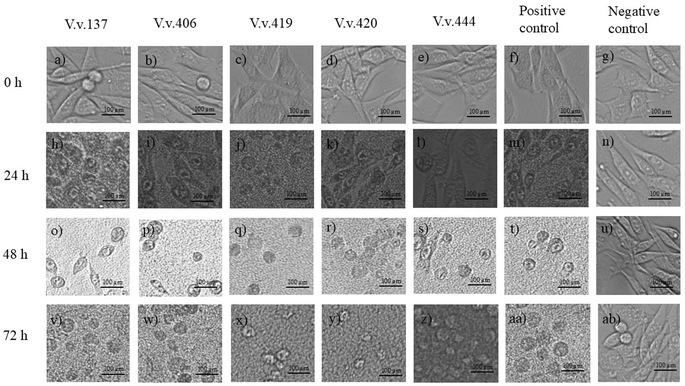
Cytopathic effects of V. vulnificus on HEK293 and HepG2 cells are depicted in Fig. 3 and 4, respectively, following infection and incubation with strains V.v.137, V.v.406, V.v.419, V.v.420, V.v.444, positive and negative controls over 24, 48, and 72 h. Prior infection with V. vulnificus the HEK293 cells exhibited a spindle morphology (Fig. 3a–g); however, after 24 h of incubation (Fig. 3h–n), the HEK293 cells began to deviate from their characteristic shape and lost their orderly structure. By 48 h (Fig. 3o–u), the cells started to appear rounder in shape and shrivelled, indicating symptoms of cytopathic stress with detachment of cells from the surface of the wells. Following 72 h incubation (Fig. 3v–z, aa, and ab), widespread breakdown of the monolayer was observed with significant distortion of the cells and cell loss being obvious. In HepG2 cells, prior to infection with V. vulnificus, cells exhibited an epithelial shape (Fig. 4a–g). At 24 h post-infection, the morphology began transitioning from epithelial shape to showing early symptoms of deformation (Fig. 4h–n). At 48 h (Fig. 4o–u), the cytopathic effects were more prominent as cells appeared rounder, shrivelled, and indications of cell-to-cell contact were observed. By 72 h (Fig. 4v–z, aa, and ab), cells were substantially distorted with substantial disintegration of the monolayer, and some cells were dislodged from the surface of the wells, exhibiting serious cellular damage and cell death. The progressive morphological alterations from spindle-shaped (HEK293) and epithelial-shaped (HepG2) to rounded, shrivelled, and deformed, alongside the gradual degradation of the cellular monolayer, underscore the cytopathic effects of V. vulnificus strains on both HEK293 and HepG2 cells over a 72-h period.Uninfected HEK293 and HepG2 cells are demonstrated throughout the 72-h period, exhibiting the development of cells. During the initial 24 h of incubation (Fig. 3g), the HEK293 cell culture demonstrated spheroid and spindle growth patterns. At 48 h (Fig. 3n), most cells adopted a spindle-shaped adherence, followed by the formation of a dense cell layer over the 72-h incubation period (Fig. 3u). Throughout the 72-h incubation period (Fig. 3u), the HEK293 cells remained adhere to the surface of the wells in the plate displaying no changes in morphology or any stress-related detachment. As demonstrated in Fig. 4g, post-24-h incubation, HepG2 cells are epithelial in nature and maintain their polygonal form throughout the 72-h incubation period. During the 48-h incubation period (Fig. 4n), HepG2 cells exhibited distinct boundaries and established cell-to-cell contact; by the 72-h incubation period (Fig. 4u), HepG2 cells formed compact clusters. Like the HEK293 cells, the uninfected HepG2 cells maintained their epithelial morphology, exhibiting no indications of rounding, detachment, or damage during the 72-h incubation period. Over the 72-h timeframe, both HEK293 and HepG2 cells retained their unmodified shape as well as established increasing cell concentrations. No sign of contamination was detected during the experiment, confirming that external variables did not affect the results and maintaining the baseline viability and integrity of the assay.
3.5 Cell viability assay
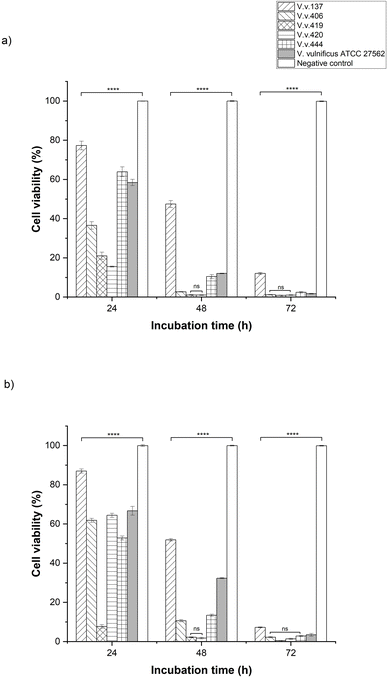
The cell viability of HEK293 and HepG2 cells (Fig. 5a and b) was assessed over 24-, 48-, and 72-h following exposure to five Vibrio vulnificus isolates (V.v.137, V.v.406, V.v.419, V.v.420 and V.v.444), with comparisons made to V. vulnificus ATCC 27562 (positive control) and uninfected cells (negative control). Following 24-h incubation, HEK293 cells infected with isolate V.v.420 exhibited the lowest cell viability (15.53% ± 0.26) while those infected with V.v.137 showed the highest (77.34% ± 2.2). By 48 h, cell viability had notably decreased for all isolates exposed to V.v. 420, reducing to 1.01% ± 0.16 and V.v. 137 to 47.44% ± 2.2. By 72 h, the cell viability continued to decline, with HEK293 cells infected with V.v.406, V.v.419, and V.v.420 showing cell viabilities close to or below 1% compared to those exposed to V.v.137, which exhibited the highest cell viability of 12.07% ± 0.5. HEK293 cells treated with V. vulnificus ATCC 27562 (positive control) also showed a marked decrease in cell viability over time, from 58.44% ± 1.63 at 24 h to 1.70% ± 0.15 at 72 h whereas the negative control maintained near-complete cell viability across all time points (from 99.99% to 99.88%). HepG2 infected with V.v.419 demonstrated the lowest cell viability at 24 h (7.7% ± 0.89) as compared to V.v.137 (87% ± 1.03). By 48 hours, cell viability decreases markedly with cells exposed to V.v.419 reducing to 2.18% ± 0.2 and V.v.137 to 51.89% ± 0.65. By 72 h, substantial reduction in cell viability was observed with V.v.137 infected HepG2 cells decreasing to 7.25% ± 0.22, while cells infected with V.v.406, V.v.419, and V.v.420 displayed minimal viability levels below 2%. HEPG2 cells treated with the positive control followed a similar trend, reducing from 66.73 ± 2.23% at 24 h to 3.43 ± 0.73% by 72 h. The negative control consistently showed no notable reduction in cell viability (99.99% to 99.88%) throughout the assay. Analysis of variance (ANOVA) revealed significant differences among all samples in comparison to the negative control (P ≤ 0.0001). However, no significant difference was detected between V.v.419 and V.v.420 at the 48-h incubation period for HEK293 and HepG2 cells, as well as among V.v.406, V.v.419, and V.v.420 at the 72-h incubation period for HEK293, and between V.v.406 and V.v.444 for HepG2. Furthermore, dose-response experiments, using serial dilutions of V. vulnificus suspensions demonstrated a proportional decrease in cell viability, supporting the role of virulence factors in cytotoxicity (Fig. S1–S6†).4. Discussion
This study investigates the virulence gene profiling and cytotoxicity of Vibrio spp. isolated from treated wastewater effluent and receiving surface waters in South Africa, as inadequate wastewater management results in the contamination of environmental water sources and the proliferation of waterborne diseases.33 The predominant pathogenic species, including Vibrio cholerae, Vibrio parahaemolyticus, Vibrio vulnificus, and Vibrio alginolyticus, are implicated in human infections associated with contaminated aquatic habitats and seafood.34The PCR amplification of virulence genes in V. vulnificus and V. alginolyticus isolates elucidated the presence and distribution of essential virulence components. Of the eight virulence genes examined (apxIB, ompU, sodB, hlyB, fbpC, viuB, tldD, and feoB) in V. vulnificus (Fig. 1a and b), viuB, fbpC, and feoB were identified as the most frequent, indicating a significant frequency of these genes within the V. vulnificus population. Table 2 indicates that the virulence gene, viuB, was found in 72.47% (n = 129) of cases. viuB encodes vulnibactin, a siderophore that facilitates iron uptake in V. vulnificus.10viuB was detected in all clinical and environmental V. vulnificus isolates documented in a prior investigation.35 Virulence genes fbpC and feoB were identified in 55.06% (n = 98) and 56.74% (n = 101) of isolates, respectively. These genes encode ferric (fbpC) and ferrous (feoB) iron transport systems, which are essential for Vibrio spp. survival as iron plays a crucial role in bacterial metabolism and gene regulation.36 The occurrence of ompU in 35.39% (n = 63), apxIB in 24.72% (n = 44), and hlyB in 13.48% (n = 24) of isolates demonstrates a heterogeneous distribution of these virulence genes. Outer membrane protein U (ompU) is a conserved main outer membrane protein, which is extensively prevalent in pathogenic Vibrio spp.37 This protein is identified as a significant adhesin in V. cholerae and V. vulnificus. Although the apxIB gene has been documented in Actinobacillus pleuropneumoniae, our results also identified the apxIB gene in V. vulnificus isolates.38 It is crucial to mention that the primer employed for this detection was created based on the genomic information collected from the Vibrio vulnificus ATCC 27562. The hemolysin region has been found to be highly conserved39 with studies reporting hlyB exclusively in V. cholera40–42 however in contrast with previous studies, this study reported hlyB in some V. vulnificus isolates. Conversely, tldD was identified in merely 6.74% (n = 12) of isolates, while sodB was detected in only 1.69% (n = 3) of isolates, indicating a diminished prevalence of these genes among the examined V. vulnificus isolates. A prior study, similarly, indicated the low incidence of tldD in another Vibrio spp.43 For example, V. anguillarum, the sodB gene, which encodes superoxide dismutase (SOD), is essential for oxidative stress tolerance in Vibrio vulnificus. The low incidence of sodB contrasted with the previous findings where sewage exposure elevated the transcription of sodB in Vibrio isolates.44
Virulence gene detection in V. alginolyticus isolates (n = 15) (Fig. 2) revealed rtxA, encoding for a holotoxin45 was the most prevalent, detected in 66.67% (n = 10) of isolates followed by pvuA which was detected in 46.67% (n = 7) of isolates. The prevalence of pvuA in this study was higher than V. alginolyticus isolated from seafood samples in Mexico city which reported 17.9% (n = 285).46 A low prevalence of asp (13.33%, n = 2) and tlh (6.67%, n = 1) was observed in the V. alginolyticus isolates. These genes, asp (encodes for alkaline serine protease) and tlh (encodes for thermolabile hemolysins), are typically identified in V. alginolyticus isolated from seafood products47 and were previously detected in silver sea bream (Sparus sarba) erythrocytes.48 Overall, this analysis underscores the diversity in the distribution of virulence genes among V. vulnificus and V. alginolyticus isolates, providing valuable information for understanding the pathogenicity and virulence potential of these bacteria. Furthermore, the co-existence of two or more virulence genes in an isolate suggests that a subset of strains can exhibit multiple virulence factors simultaneously (Table S1†). Moderate to high multi-virulence gene index (MVGI) displayed among some strains reflects distinct virulence gene compositions highlighting pathogenic risks of these species especially in immunocompromised individuals.24,49 Further investigation is warranted into how these virulence factors interact.
As observed in Table 3, descriptive analysis of crosstabs for virulence genes in V. vulnificus using Chi-squared analysis highlighted significant associations between these virulence factors and their contribution to the bacteria's pathogenicity. feoB and fbpC are both iron-transport related genes that showed significant association with each other indicating that iron acquisition is an important strategy contributing to the virulence of V. vulnificus.50 The association between the vulnibactin encoding gene, viuB, and hlyB, fbpC, feoB, and tldD suggests the relationship between iron acquisition systems, iron transport and hemolysins provide a source of iron for these bacterial strains through lysis of the red blood cells contributing to the survival and pathogenicity of V. vulnificus in iron-limited environments.51,52 The metalloprotease encoding gene, tldD, which is known to degrade host proteins and facilitate tissue invasion, was found to be associated with apxIB, ompU, and hlyB.53 The association between tldD and ompU implies these adhesion and invasion mechanisms work together to enhance bacterial pathogenicity as well as a suggested coordinated response that results in cell lysis, through hemolysin activity, and tissue invasion.54,55 The apxIB encodes for a transport protein that is involved with the transport of the RTX toxin leading to haemolytic and cytotoxic effects.56 Significant association between tldD and apxIB suggests that once the metalloproteases have degraded the host's proteins such as the extracellular matrix components, it will allow apxIB to transport the RTX toxins to access and cause greater damage in the host tissues.57,58 Additionally, the association between fbpC and sodB reflect a dual strategy in the bacterium's survival in hostile environments as not only does it scavenge iron to promote growth but also enhances its defence against oxidative stress.36,59 Despite a lower frequency of asp and tlh detected in V. alginolyticus, Table 3 showed a significant association was observed between these genes suggesting they play a coordinated role in the pathogenicity of the bacterium as the alkaline serine protease and thermolabile hemolysin assist with tissue degradation and breaking down of the host cell membranes, respectively.60 Despite their low prevalence, the co-occurrence of these virulence genes could enhance the bacterium's ability to establish infection by potentially facilitating acquisition and immune evasion. The significant association between these virulence genes highlight the multifaceted nature of bacterial pathogenicity which contribute to the bacterium's survival, proliferation and invasion within a host.61 Understanding the genetic associations between virulence factors in V. vulnificus, V. alginolyticus and another Vibrio spp. may help predict their pathogenic potential resulting in the development of targeted treatments or preventative measures, particularly for immunocompromised individuals who are vulnerable to severe infections.62
The odds ratio (OR) analysis presented in Table 3 elucidates potential correlations among several virulence genes within the examined bacterial strains that may enhance their pathogenicity.32 In V. vulnificus, the high odds ratio between ratios between viuB and fbpC (OR 17.820, CI: 6.992–45.414) and viuB and feoB (OR 7.972, CI: 3.695–17.202) suggests the presence of these gene pairs may synergistically enhance pathogenicity in the bacterial strains even though broad confidence intervals suggest variability in the data that may be influenced by sample size or other factors requiring further validation in larger studies.63 The odds ratio indicated that the associations between ompU and apxIB (OR 3.787, CI: 1.861–7.705), fbpC and apxIB (OR 2.385, CI: 1.148–4.952), and fbpC and sodB (OR 3.608, CI: 1.282–10.151) were more robust, as their confidence intervals do not encompass one, suggesting functional links between these genes that may enhance virulence.32,64 In contrast, minimal association was observed between rtx and asp (OR 1.250, CI: 0.917–1.704) in V. alginolyticus and feoB and apxIB (OR 1.004, CI: 0.505–1.997) in V. vulnificus as the odds ratios were close to one and confidence intervals overlapped one suggesting that these gene pairs do not contribute to increased virulence.63 In Vibrio spp., the odds ratios were approximately one or lower, accompanied by wide confidence intervals, indicating no significant associations between the gene pairs. However, due to the limited sample size of these bacterial strains, further research with a larger dataset is necessary to elucidate the relationship between the gene pairs in these isolates.65
V. vulnificus, a bacterium present in warm coastal waters, represents a considerable risk to human health, especially for those with liver illness or immunocompromised conditions.30,66 It may lead to severe sepsis and necrotizing infections with fatality rates of 50%.30 In this work, exposure to V. vulnificus resulted in severe cytotoxic effects on HEK293 and HepG2 cell lines, defined by morphological alterations and decreased cell viability, underlining the pathogen's potential clinical significance. V. vulnificus also elicits robust inflammatory responses, resulting in cellular damage and death.67,68 The cytotoxicity observed at a selected concentration of 108 CFU mL−1, while higher than average environmental levels, provides important insights into potential health risks during peak exposure scenarios. The dose-response experiments (Fig. S1–S6†) confirmed that cytotoxic effects occur proportionally even at lower concentrations, indicating that environmental Vibrio strains may pose health risks under real-world conditions, particularly for immunocompromised individuals.12,69–72 These findings suggest that Vibrio strains present in wastewater, even at concentrations lower than those used in this study, may pose significant health risks, especially through recreational water activities or consumption of seafood from contaminated waters. The rounding and detachment of HEK293 cells (Fig. 3) and the shrinkage and detachment of HepG2 cells (Fig. 4), transitioning from spindle- and epithelial-shaped forms, indicate that the cytotoxic effects are not confined to a single cell type but impact multiple cellular targets. This extensive cytotoxicity corresponds with prior research emphasizing the virulent characteristics of V. vulnificus in host tissues.73 The disparate reactions noted between HEK293 and HepG2 cells, with a more significant cytotoxic effect in HEK293 cells, may be ascribed to intrinsic variations in cellular pathways and vulnerabilities. HEK293 cells, generated from human embryonic kidney cells, and HepG2 cells, derived from human liver carcinoma cells, feature unique metabolic and signalling pathways, which may influence their response to bacterial toxins and virulence factors. Additional examination of these cellular pathways may yield profound insights into the precise connections between V. vulnificus and other cell types.
Renal impairment elevates the probability of V. vulnificus infection, highlighting the necessity of comprehending its cytotoxic consequence.74 This study revealed V. vulnificus's capacity to lethally affect HEK293 cells, signifying its potential to harm renal tissue. While V. vulnificus infections are mostly linked to gastrointestinal disorders, they may also result in systemic infections and consequences, such as renal failure.75 Chronic conditions such as liver disease and hemochromatosis elevate the likelihood of V. vulnificus infections.76 Iron is crucial for bacterial physiological functions, including transcription, DNA replication, metabolic activities, and energy production through respiration. Consequently, many pathogenic bacteria, such as V. vulnificus, have evolved siderophores and iron acquisition mechanisms to extract iron from their hosts.77 Kidney and liver cells necessitate iron for metabolic functions, with the liver significantly coordinating iron homeostasis by storing, using intracellularly, or mobilizing it for systemic distribution.78,79 HepG2 and HEK293 cells, being abundant in iron due to their origin from human liver and kidney tissue, respectively, serve as optimal targets for bacterial iron acquisition systems. As previously stated, viuB encodes the siderophore vulnibactin, which sequesters iron from the host, hence altering cellular processes and enhancing cytotoxicity through hepatocyte damage.80 The ferric and ferrous iron transport systems encoded by fbpC and feoB, respectively, augment the capacity of V. vulnificus to assimilate iron, leading to oxidative stress and ultimately host cell mortality.81 Reactive oxygen species (ROS) are generated as part of the host's immunological response to infections; however, bacteria employ enzymes like superoxide dismutase to mitigate this response.82 The generation of ROS may elicit an immunological response in HepG2 and HEK293 cells against bacterial infection; however, sodB produces superoxide dismutase (SOD), which may facilitate the survival of V. vulnificus, resulting in prolonged infection and harmful effects.83 The hemolysins encoded by hlyB may create pores in the membranes of HepG2 and HEK293 cells, compromising membrane integrity and leading to cell lysis, while the outer membrane protein encoded by ompU may facilitate the adhesion of V. vulnificus to HepG2 and HEK293 cells, thereby augmenting the bacteria's capacity to colonize host tissues.37,84 The metalloproteases produced by tldD may be responsible for the breakdown of essential cellular proteins, including structural components or immunological proteins, in HepG2 and HEK293 increasing cytotoxicity.43 Finally, the inclusion of apxIB enhances the cytotoxicity of the Vibrio isolates, as the RTX-I toxin it carries creates gaps in the host cell membranes, leading to cell lysis and apoptosis.85 This will cause cellular damage and reduce cell viability, as illustrated in Fig. 5, hence enhancing the virulence of these bacterial strains. Additionally, analysis of variance (ANOVA) revealed significant differences among all samples compared to the negative control (P ≤ 0.0001), indicating that infection with the Vibrio vulnificus strains impacted cell viability.
These findings have significant public health implications for regions where V. vulnificus is endemic in coastal and estuarine waters. Enhanced surveillance of water quality parameters and pathogen monitoring in high-risk areas is essential, particularly during warmer months when Vibrio populations typically increase. Public health authorities should prioritize targeted education campaigns for vulnerable populations, including immunocompromised individuals and those with chronic liver disease, regarding risks associated with exposure to potentially contaminated waters. The identification of key virulence determinants also provides molecular targets for developing novel therapeutic interventions, particularly protease and haemolysing inhibitors that could supplement current antibiotic treatments. This study provides important insights into the virulence gene profiles and cytotoxic potential of environmental Vibrio spp. isolates, though several limitations warrant consideration. Our analysis focused primarily on gene presence and in vitro cytotoxicity assessments, which establish a foundation for understanding pathogenic potential but do not fully elucidate the functional mechanisms of virulence factor expression or host–pathogen interactions. Future investigations incorporating functional assays for haemolytic activity, inflammatory cytokine induction, and iron acquisition mechanisms will be necessary to comprehensively characterize the pathogenic mechanisms of these environmental isolates.
5. Conclusion
Based on virulence gene profiling and cytotoxic data, this work examined the pathogenicity of 200 Vibrio isolates from treated wastewater effluent and receiving surface waters of four wastewater treatment plants in Durban, KwaZulu-Natal. In V. vulnificus isolates (n = 178), the most frequently detected virulence gene was viuB, followed by feoB, fbpC, ompU, apxIB, hlyB, tldD, and sodB whereas highest virulence gene detection rates were observed for rtx, followed pvuA, asp and tlh in V. alginolyticus (n = 15). In the Vibrio spp. (n = 7), the most common virulence gene detected was viuB, followed by ompU, feoB and fbpC. Significant associations between virulence genes were identified in V. vulnificus such as between viuB and fbpC, feoB and fbpC, and hlyB and sodB whereas in V. alginolyticus, significant association were observed between tlh and asp emphasizing the interactions and robust relationships among virulence genes. Infection of HEK293 and HepG2 cells with five selected V. vulnificus strains, V.v.137, V.v.406, V.v.419, V.v.420 and V.v.444, highlighted the pathogenic potential of these isolates. HEK293 and HepG2 exhibited morphological changes including, detachment, shrinkage and cell death following infection with all five V. vulnificus isolates. Additionally, exposure of these cells to V.v.419 and V.v.420 resulted in rapid declines in cell viability falling below 2% within 72 h, indicating severe cytotoxic effects. In contrast, less virulent strains like V.v.137 enabled HEK293 and HepG2 cells to retain higher cell viability despite demonstrating a progressive decline over time. These findings highlight the prevalence and diversity of virulence genes among Vibrio spp. isolated from wastewater and environmental waters, their significant associations, and their marked cytotoxic effects on human cell lines. The study underlines the importance of constant monitoring and enhancement of wastewater treatment processes to ensure successful elimination of pathogenic bacteria. Furthermore, public health programs should raise awareness about the dangers of exposure to contaminated water sources as well as promoting safe practices for recreational water use and seafood intake.Ethical approval
This research did not contain any studies involving animal or human participants, nor did it take place on any private or protected areas. No specific permissions were required for corresponding locations.Consent to participate
All the authors are aware and agree to participating in this manuscript.Consent to publish
All the authors agree to publish this manuscript.Data availability
Data are contained within the article or ESI Material.†Author contributions
Conceptualization, K. R. and A. O.·O.; methodology, K. R. and A. O.·O.; validation, K. R. and A. O.·O.; formal analysis, K. R.; investigation, K. R. and A. O.·O.; resources, A. O.·O.; data curation, K. R.; writing K. R. and A. O.·O.; visualization, K. R.; supervision, A. O.·O.; project administration, A. O.·O.; funding acquisition, K. R. A. O. O. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
National Research Foundation of South Africa (NRF grant no. UID 118324 and 139095). We appreciate the eThekwini Municipality for granting us permission to collect samples from the wastewater treatments plants in the Durban area, KwaZulu-Natal, South Africa.References
- A. Murei, I. Kamika, A. Samie and M. N. B. Momba, Assessment of the water sources for potential channels of faecal contamination within Vhembe District Municipality using sanitary inspections and hydrogen sulphide test, Sci. Rep., 2023, 13(1), 1–13, DOI:10.1038/s41598-023-33551-y.
- O. Osuolale and A. Okoh, Assessment of the physicochemical qualities and prevalence of Escherichia coli and Vibrios in the final effluents of two wastewater treatment plants in South Africa: Ecological and public health implications, Int. J. Environ. Res. Public Health, 2015, 12(10), 13399–13412, DOI:10.3390/ijerph121013399.
- P. Verlicchi and V. Grillini, Surface water and groundwater quality in South Africa and Mozambique—analysis of the most critical pollutants for drinking purposes and challenges in water treatment selection, Water, 2020, 12(1), 305, DOI:10.3390/W12010305.
- I. George, P. Crop and P. Servais, Fecal coliform removal in wastewater treatment plants studied by plate counts and enzymatic methods, Water Res., 2002, 36(10), 2607–2617, DOI:10.1016/S0043-1354(01)00475-4.
- C. O. Osunmakinde, R. Selvarajan, B. B. Mamba and T. A. M. Msagati, Profiling bacterial diversity and potential pathogens in wastewater treatment plants using high-throughput sequencing analysis, Microorganisms, 2019, 7(11), 506, DOI:10.3390/MICROORGANISMS7110506.
- O. Osuolale and A. Okoh, Isolation and antibiotic profile of Vibrio spp. in final effluents of two wastewater treatment plants in the Eastern Cape of South Africa, bioRxiv, 2018330456, DOI:10.1101/330456.
- K. Zhou, K. Y. Tian and X. Q. Liu, et al., Characteristic and otopathogenic analysis of a Vibrio alginolyticus strain responsible for chronic otitis externa in China, Front. Microbiol., 2021, 12, 750642, DOI:10.3389/FMICB.2021.750642/FULL.
- A. Sampaio, V. Silva, P. Poeta and F. Aonofriesei, Vibrio spp.: life strategies, ecology, and risks in a changing environment, Diversity, 2022, 14(2), 97, DOI:10.3390/D14020097.
- World Health Organization/Department of Control of Neglected Tropical Diseases, Weekly epidemiological record Relevé épidémiologique hebdomadaire. Weekly epidemiological record, 2018, http://www.who.int/wer2018,93,649-660No48%0Ahttp://www.who.int/wer Search PubMed.
- C. D'souza, K. S. Prithvisagar, V. K. Deekshit, I. Karunasagar, I. Karunasagar and B. K. Kumar, Exploring the pathogenic potential of Vibrio vulnificus isolated from seafood harvested along the Mangaluru Coast, India, Microorganisms, 2020, 8(7), 1–15, DOI:10.3390/MICROORGANISMS8070999.
- W. A. Norfolk, C. Shue, W. M. Henderson, D. A. Glinski and E. K. Lipp, Vibrio alginolyticus growth kinetics and the metabolic effects of iron, Microbiol. Spectrum, 2023, 11(6), e02680, DOI:10.1128/spectrum.02680-23.
- C. Baker-Austin, J. D. Oliver and M. Alam, et al., Vibrio spp. infections, Nat. Rev. Dis. Primers, 2018, 4(1), 1–19, DOI:10.1038/s41572-018-0005-8.
- H. Byun, I. J. Jung, J. Chen, J. L. Valencia and J. Zhu, Siderophore piracy enhances Vibrio cholerae environmental survival and pathogenesis, Microbiology, 2020, 166(11), 1038, DOI:10.1099/MIC.0.000975.
- M. A. Echazarreta and K. E. Klose, Vibrio flagellar synthesis, Front. Cell. Infect. Microbiol., 2019, 9, 131, DOI:10.3389/FCIMB.2019.00131.
- H. A. Ganie, A. Choudhary and S. Baranwal, Structure, regulation, and host interaction of outer membrane protein U (OmpU) of Vibrio species, Microb. Pathog., 2022, 162, 105267, DOI:10.1016/J.MICPATH.2021.105267.
- H. Gao, J. Xu and X. Lu, et al., Expression of hemolysin is regulated under the collective actions of HapR, Fur, and HlyU in Vibrio cholerae El Tor serogroup O1, Front. Microbiol., 2018, 9, 352724, DOI:10.3389/FMICB.2018.01310/BIBTEX.
- S. J. Krebs and R. K. Taylor, Protection and attachment of Vibrio cholerae mediated by the toxin-coregulated pilus in the infant mouse model, J. Bacteriol., 2011, 193(19), 5260, DOI:10.1128/JB.00378-11.
- M. Mavhungu, T. O. Digban and U. U. Nwodo, Incidence and virulence factor profiling of Vibrio species: a study on hospital and community wastewater effluents, Microorganisms, 2023, 11(10), 2449, DOI:10.3390/MICROORGANISMS11102449.
- H. Moeinizadeh and M. Shaheli, Frequency of hlyA, hlyB, hlyC and hlyD genes in uropathogenic Escherichia coli isolated from UTI patients in Shiraz, GMS Hyg. Infect. Control, 2021, 16, Doc25, DOI:10.3205/DGKH000396.
- N. Mohamad, M. N. A. Amal and M. Z. Saad, et al., Virulence-associated genes and antibiotic resistance patterns of Vibrio spp. isolated from cultured marine fishes in Malaysia, BMC Vet. Res., 2019, 15(1), 1–13, DOI:10.1186/S12917-019-1907-8/TABLES/4.
- K. Schwartz, J. A. Hammerl, C. Göllner and E. Strauch, Environmental and clinical strains of Vibrio cholerae non-O1, non-O139 from Germany possess similar virulence gene profiles, Front. Microbiol., 2019, 10, 438958, DOI:10.3389/FMICB.2019.00733/BIBTEX.
- C. L. Tarr, J. S. Patel, N. D. Puhr, E. G. Sowers, C. A. Bopp and N. A. Strockbine, Identification of Vibrio isolates by a multiplex PCR assay and rpoB sequence determination, J. Clin. Microbiol., 2007, 45(1), 134–140, DOI:10.1128/JCM.01544-06.
- C. H. Liu, W. Cheng, J. P. Hsu and J. C. Chen, Vibrio alginolyticus infection in the white shrimp Litopenaeus vannamei confirmed by polymerase chain reaction and 16S rDNA sequencing, Dis. Aquat. Org., 2004, 61(1–2), 169–174, DOI:10.3354/dao061169.
- O. E. Abioye, C. A. Osunla, N. Nontongana and A. I. Okoh, Occurrence of virulence determinants in Vibrio cholerae, Vibrio mimicus, Vibrio alginolyticus, and Vibrio parahaemolyticus isolates from important water resources of Eastern Cape, South Africa, BMC Microbiol., 2023, 23(1), 316, DOI:10.1186/S12866-023-03060-Z.
- J. Sahandi, P. Sorgeloos and L. Xiao, et al., The use of selected bacteria and yeasts to control Vibrio spp. in live food, Antibiotics, 2019, 8(3), 95, DOI:10.3390/antibiotics8030095.
- J. Sun, J. Zheng, G. Wang, Y. Li and H. Shen, Apoptotic effect of Vibrio vulnificus cytolysin on A549 human lung adenocarcinoma cells, Mol. Med. Rep., 2012, 5(3), 668–674, DOI:10.3892/mmr.2011.690.
- C. K. Constante, J. Rodríguez, S. Sonnenholzner and C. Domínguez-Borbor, Adaptation of the methyl thiazole tetrazolium (MTT) reduction assay to measure cell viability in Vibrio spp, Aquaculture, 2022, 560, 738568, DOI:10.1016/j.aquaculture.2022.738568.
- W. Chao, Z. Zhe and L. Yupeng, et al., Type III secretion 1 effector gene diversity among Vibrio isolates from coastal areas in China, Front. Cell. Infect. Microbiol., 2020, 10, 301, DOI:10.3389/fcimb.2020.00301.
- K. D. Brumfield, A. J. Chen and M. Gangwar, et al., Environmental Factors Influencing Occurrence of Vibrio parahaemolyticus and Vibrio vulnificus, Appl. Environ. Microbiol., 2023, 89(6), e0030723, DOI:10.1128/AEM.00307-23.
- M. A. Horseman and S. Surani, A comprehensive review of Vibrio vulnificus: An important cause of severe sepsis and skin and soft-tissue infection, Int. J. Infect. Dis., 2011, 15(3), e157–e166, DOI:10.1016/J.IJID.2010.11.003.
- M. K. Jones and J. D. Oliver, Vibrio vulnificus: Disease and pathogenesis, Infect. Immun., 2009, 77(5), 1723–1733, DOI:10.1128/IAI.01046-08.
- M. Szumilas, Explaining Odds Ratios, J. Can. Acad. Child Adolesc. Psychiatry, 2010, 19(3), 227 CrossRef PubMed , Accessed November 6, 2024, https://pmc.ncbi.nlm.nih.gov/articles/PMC2938757/.
- B. J. Singh, A. Chakraborty and R. Sehgal, A systematic review of industrial wastewater management: Evaluating challenges and enablers, J. Environ. Manage., 2023, 348, 119230, DOI:10.1016/J.JENVMAN.2023.119230.
- A. Castello, V. Alio and S. Sciortino, et al., Occurrence and molecular characterization of potentially pathogenic Vibrio spp. in seafood collected in Sicily, Microorganisms, 2022, 11(1), 53, DOI:10.3390/MICROORGANISMS11010053.
- N. Yokochi, S. Tanaka and K. Matsumoto, et al., Distribution of virulence markers among Vibrio vulnificus isolates of clinical and environmental origin and regional characteristics in Japan, PLoS One, 2013, 8(1), e55219, DOI:10.1371/JOURNAL.PONE.0055219.
- A. Pramanik and R. K. Vibhuti, Molecular mechanism of iron transport systems in Vibrio, J. Pure Appl. Microbiol., 2022, 16(1), 116–129, DOI:10.22207/JPAM.16.1.77.
- X. Liu, H. Gao, N. Xiao, Y. Liu, J. Li and L. Li, Outer membrane protein U (OmpU) mediates adhesion of Vibrio mimicus to host cells via two novel n-terminal motifs, PLoS One, 2015, 10(3), e0119026, DOI:10.1371/journal.pone.0119026.
- S. J. Foote, J. T. Bossé, A. B. Bouevitch, P. R. Langford, N. M. Young and J. H. E. Nash, The complete genome sequence of Actinobacillus pleuropneumoniae L20, J. Bacteriol., 2008, 190(4), 1495–1496, DOI:10.1128/JB.01845-07.
- J. L. Rock and D. R. Nelson, Identification and characterization of a hemolysin gene cluster in Vibrio anguillarum, Infect. Immun., 2006, 74(5), 2777–2786, DOI:10.1128/IAI.74.5.2777-2786.2006/ASSET/79A6BEA6-02D4-4BF6-AFB6-93E6ACA67758/ASSETS/GRAPHIC/ZII0050658670006.JPEG.
- R. A. Alm and P. A. Manning, Characterization of the hlyB gene and its role in the production of the El Tor haemolysin of Vibrio cholerae O1, Mol. Microbiol., 1990, 4(3), 413–425, DOI:10.1111/J.1365-2958.1990.TB00608.X.
- I. Altinoglu, G. Abriat and A. Carreaux, et al., Analysis of HubP-dependent cell pole protein targeting in Vibrio cholerae uncovers novel motility regulators, PLOS Genet., 2022, 18(1), e1009991, DOI:10.1371/journal.pgen.1009991.
- M. A. Matilla and T. Krell, The effect of bacterial chemotaxis on host infection and pathogenicity, FEMS Microbiol. Rev., 2018, 42(1), 40–67, DOI:10.1093/femsre/fux052.
- I. Vasquez, T. Cao and S. Chakraborty, et al., Comparative genomics analysis of Vibrio anguillarum isolated from lumpfish (Cyclopterus lumpus) in Newfoundland reveal novel chromosomal organizations, Microorganisms, 2020, 8(11), 1666, DOI:10.3390/microorganisms8111666.
- J. W. Conrad and V. J. Harwood, Sewage Promotes Vibrio vulnificus growth and alters gene transcription in Vibrio vulnificus CMCP6, Microbiol. Spectrum, 2022, 10(1), e0191321, DOI:10.1128/spectrum.01913-21.
- F. Håkonsholm, B. T. Lunestad, J. R. Aguirre Sánchez, J. Martinez-Urtaza, N. P. Marathe and C. S. Svanevik, Vibrios from the Norwegian marine environment: Characterization of associated antibiotic resistance and virulence genes, Microbiologyopen, 2020, 9(9), 1–19, DOI:10.1002/mbo3.1093.
- A. K. Álvarez-Contreras, E. I. Quiñones-Ramírez and C. Vázquez-Salinas, Prevalence, detection of virulence genes and antimicrobial susceptibility of pathogen Vibrio species isolated from different types of seafood samples at “La Nueva Viga” market in Mexico City, Antonie van Leeuwenhoek, 2021, 114(9), 1417–1429, DOI:10.1007/S10482-021-01591-X.
- H. Rui, Q. Liu, Y. Ma, Q. Wang and Y. Zhang, Roles of LuxR in regulating extracellular alkaline serine protease A, extracellular polysaccharide and mobility of Vibrio alginolyticus, FEMS Microbiol. Lett., 2008, 285(2), 155–162, DOI:10.1111/J.1574-6968.2008.01185.X.
- S. K. Wong, X. H. Zhang and N. Y. S. Woo, Vibrio alginolyticus thermolabile hemolysin (TLH) induces apoptosis, membrane vesiculation and necrosis in sea bream erythrocytes, Aquaculture, 2012, 330–333, 29–36, DOI:10.1016/j.aquaculture.2011.12.012.
- M. Restrepo-Benavides, D. Lozano-Arce and L. N. Gonzalez-Garcia, et al., Unveiling potential virulence determinants in Vibrio isolates from Anadara tuberculosa through whole genome analyses, Microbiol. Spectrum, 2024, 12(2), e0292823, DOI:10.1128/SPECTRUM.02928-23/SUPPL_FILE/SPECTRUM.02928-23-S0006.XLSX.
- K. Miyamoto, H. Kawano and N. Okai, et al., Iron-utilization system in Vibrio vulnificus M2799, Mar. Drugs, 2021, 19(12), 710, DOI:10.3390/MD19120710.
- C. Gómez-Garzón and S. M. Payne, Divide and conquer: genetics, mechanism, and evolution of the ferrous iron transporter Feo in Helicobacter pylori, Front. Microbiol., 2023, 14, 1219359, DOI:10.3389/FMICB.2023.1219359/BIBTEX.
- J. R. Sheldon, H. A. Laakso and D. E. Heinrichs, Iron acquisition strategies of bacterial pathogens, Microbiol. Spectrum, 2016, 4(2) DOI:10.1128/microbiolspec.VMBF-0010-2015.
- X. Tang, Q. Yang, Z. Nan, T. Wang, Y. Li and Z. Wu, The potential compensatory mechanism between pmbA and tldD governing the virulence of Aeromonas veronii and its implications on the immune response in freshwater bivalve (Hyriopsis cumingii), Aquaculture, 2025, 595, 741524, DOI:10.1016/J.AQUACULTURE.2024.741524.
- Z. Li, Y. Fan and Z. Li, et al., Nonhemolysis of epidemic El Tor biotype strains of Vibrio cholerae is related to multiple functional deficiencies of hemolysin A, Gut Pathog., 2019, 11(1), 1–10, DOI:10.1186/S13099-019-0316-7/TABLES/3.
- D. Meparambu Prabhakaran, T. Ramamurthy and S. Thomas, Genetic and virulence characterisation of Vibrio parahaemolyticus isolated from Indian coast, BMC Microbiol., 2020, 20(1), 62, DOI:10.1186/S12866-020-01746-2.
- J. Liu, X. Chen and C. Tan, et al., In vivo induced RTX toxin ApxIVA is essential for the full virulence of Actinobacillus pleuropneumoniae, Vet. Microbiol., 2009, 137(3–4), 282–289, DOI:10.1016/J.VETMIC.2009.01.011.
- K. Dzobo and C. Dandara, The extracellular matrix: Its composition, function, remodeling, and role in tumorigenesis, Biomimetics, 2023, 8(2), 146, DOI:10.3390/BIOMIMETICS8020146.
- M. Kamali, M. Carossino and F. Del Piero, et al., Pathological features and genomic characterization of an Actinobacillus equuli subsp. equuli bearing unique virulence-associated genes from an adult horse with pleuropneumonia, Pathogens, 2023, 12(2), 224, DOI:10.3390/PATHOGENS12020224/S1.
- H. Najmuldeen, R. Alghamdi, F. Alghofaili and H. Yesilkaya, Functional assessment of microbial superoxide dismutase isozymes suggests a differential role for each isozyme, Free Radical Biol. Med., 2019, 134, 215–228, DOI:10.1016/J.FREERADBIOMED.2019.01.018.
- B. Yang, S. Zhai and X. Li, et al., Identification of Vibrio alginolyticus as a causative pathogen associated with mass summer mortality of the Pacific Oyster (Crassostrea gigas) in China, Aquaculture, 2021, 535, 736363, DOI:10.1016/J.AQUACULTURE.2021.736363.
- J. Soni, S. Sinha and R. Pandey, Understanding bacterial pathogenicity: a closer look at the journey of harmful microbes, Front. Microbiol., 2024, 15, 1370818, DOI:10.3389/FMICB.2024.1370818.
- G. Muteeb, M. T. Rehman, M. Shahwan and M. Aatif, Origin of antibiotics and antibiotic resistance, and their impacts on drug development: a narrative review, Pharmaceuticals, 2023, 16(11), 1615, DOI:10.3390/PH16111615.
- S. Tenny and M. R. Hoffman, Odds Ratio, Encycl. Genet. Genomics Proteomics Inform., 2023, 1388, DOI:10.1007/978-1-4020-6754-9_11771.
- N. G. Djomgoue, L. J. Fonbah, A. I. Mbulli, K. Ousenu and T. C. Bonglavnyuy, Risk factors and associated outcomes of virulence genes eae, entB, and pipD Carriage in Escherichia coli, Klebsiella pneumoniae, and Salmonella spp. from HIV-1 and HIV-negative gastroenteritis patients in the Dschang Regional Hospital Annex, Cureus, 2023, 15(7), e42329, DOI:10.7759/CUREUS.42329.
- S. Nemes, J. M. Jonasson, A. Genell and G. Steineck, Bias in odds ratios by logistic regression modelling and sample size, BMC Med. Res. Methodol., 2009, 9(1), 1–5, DOI:10.1186/1471-2288-9-56/FIGURES/3.
- K. E. Phillips and K. J. F. Satchell, Vibrio vulnificus: From oyster colonist to human pathogen, PLOS Pathog., 2017, 13(1), e1006053, DOI:10.1371/JOURNAL.PPAT.1006053.
- K. B. Kwon, J. Y. Yang and D. G. Ryu, et al., Vibrio vulnificus cytolysin induces superoxide anion-initiated apoptotic signaling pathway in human ECV304 cells, J. Biol. Chem., 2001, 276(50), 47518–47523, DOI:10.1074/JBC.M108645200.
- C. Murciano, L. I. Hor and C. Amaro, Host-pathogen interactions in Vibrio vulnificus: responses of monocytes and vascular endothelial cells to live bacteria, Future Microbiol., 2015, 10(4), 471–487, DOI:10.2217/FMB.14.136.
- J. M. Pérez Martín, P. Fernández Freire, A. Peropadre and M. J. Hazen, Cytotoxic evaluation of a mixture of eight pollutants at environmental relevant concentrations, Rev. Toxicol., 2014, 31(2), 172–175 Search PubMed.
- C. Kataoka and S. Kashiwada, Ecological risks due to immunotoxicological effects on aquatic organisms, Int. J. Mol. Sci., 2021, 22(15), 8305, DOI:10.3390/ijms22158305.
- C. Viegas, P. Pena, B. Gomes, M. Dias, L. A. Caetano and S. Viegas, Are in vitro cytotoxicity assessments of environmental samples useful for characterizing the risk of exposure to multiple contaminants at the workplace? A systematic review, Toxics, 2022, 10(2), 72, DOI:10.3390/toxics10020072.
- J. D. Oliver, The biology of Vibrio vulnificus, Microbiol. Spectrum, 2015, 3(3), 0001–2014, DOI:10.1128/microbiolspec.VE-0001-2014.
- Y. R. Kim, S. E. Lee and H. Kook, et al., Vibrio vulnificus RTX toxin kills host cells only after contact of the bacteria with host cells, Cell. Microbiol., 2008, 10(4), 848–862, DOI:10.1111/J.1462-5822.2007.01088.X.
- C. S. Kim, E. H. Bae, S. K. Ma and S. W. Kim, Severe septicemia, necrotizing fasciitis, and peritonitis due to Vibrio vulnificus in a patient undergoing continuous ambulatory peritoneal dialysis: A case report, BMC Infect. Dis., 2015, 15(1), 1–4, DOI:10.1186/S12879-015-1163-X/PEER-REVIEW.
- N. Lerstloompleephunt, T. Tantawichien and V. Sitprija, Renal failure in Vibrio vulnificus infection, Renal Failure, 2000, 22(3), 337–343, DOI:10.1081/JDI-100100877.
- J. Wang, X. Weng, Y. Weng, Q. Xu, Y. Lu and Y. Mo, Clinical features and treatment outcomes of Vibrio vulnificus infection in the coastal city of Ningbo, China, Front. Microbiol., 2023, 14, 1220526, DOI:10.3389/FMICB.2023.1220526/BIBTEX.
- J. W. Kronstad and M. Caza, Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans, Front. Cell. Infect. Microbiol., 2013, 3, 80, DOI:10.3389/FCIMB.2013.00080.
- U. Abbasi, S. Abbina, A. Gill and J. N. Kizhakkedathu, Development of an iron overload HepG2 cell model using ferrous ammonium citrate, Sci. Rep., 2023, 13(1), 21915, DOI:10.1038/S41598-023-49072-7.
- T. Matsuoka, M. Abe and H. Kobayashi, Iron metabolism and inflammatory mediators in patients with renal dysfunction, Int. J. Mol. Sci., 2024, 25, 3745, DOI:10.3390/IJMS25073745.
- W. Tan, V. Verma and K. Jeong, et al., Molecular characterization of vulnibactin biosynthesis in Vibrio vulnificus indicates the existence of an alternative siderophore, Front. Microbiol., 2014, 5, 1, DOI:10.3389/FMICB.2014.00001.
- R. L. Gonciarz, E. A. Collisson and A. R. Renslo, Ferrous iron–dependent pharmacology, Trends Pharmacol. Sci., 2020, 42(1), 7, DOI:10.1016/J.TIPS.2020.11.003.
- Y. Chen, F. Wu, H. Pang, J. Tang, S. Cai and J. Jian, Superoxide dismutase B (sodB), an important virulence factor of Vibrio alginolyticus, contributes to antioxidative stress and its potential application for live attenuated vaccine, Fish Shellfish Immunol., 2019, 89, 354–360, DOI:10.1016/J.FSI.2019.03.061.
- L. Cavinato, E. Genise, F. R. Luly, E. G. D. Domenico, P. Del Porto and F. Ascenzioni, Escaping the phagocytic oxidative burst: The role of SodB in the survival of Pseudomonas aeruginosa within macrophages, Front. Microbiol., 2020, 11, 326, DOI:10.3389/FMICB.2020.00326/FULL.
- L. C. Ristow and R. A. Welch, Hemolysin of uropathogenic Escherichia coli: A cloak or a dagger?, Biochim. Biophys. Acta, Biomembr., 2016, 1858(3), 538–545, DOI:10.1016/J.BBAMEM.2015.08.015.
- H. J. Yoo, S. Lee and D. Y. Ryu, Role of the ApxIB/ApxID exporter in secretion of the ApxII and ApxIII toxins in Actinobacillus pleuropneumoniae, Korean J. Vet. Res., 2020, 60(4), 225–228, DOI:10.14405/KJVR.2020.60.4.225.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5em00083a |
| This journal is © The Royal Society of Chemistry 2025 |