Open Access Article
Open Access ArticlePolynuclear transition metal complexes: emerging agents for bacterial imaging and antimicrobial therapy
Bishnu
Das
 *,
Sooraj
Sathyanarayan
and
Parna
Gupta
*,
Sooraj
Sathyanarayan
and
Parna
Gupta
 *
*
Department of Chemical Sciences, Indian Institute of Science Education and Research, Kolkata, 741246, India. E-mail: parna@iiserkol.ac.in
First published on 13th May 2025
Abstract
Polynuclear transition metal complexes (PTMCs) represent a promising class of compounds with significant potential for advancing microbial diagnostics and treatment due to their multifunctional properties. This perspective highlights recent progress in the design of PTMCs for detecting and combating microbial infections. Complexes with multiple metal centers, such as silver(I), rhenium(I), iron(II), cobalt(II), nickel(II), copper(II), zinc(II), cadmium(II), ruthenium(II), iridium(III), gold(I), and gold(III), exhibit a wide range of structural motifs and are effective against a broad spectrum of multidrug-resistant bacterial infections. PTMCs show their antimicrobial effects through several mechanisms that include the generation of reactive oxygen species, which cause oxidative stress and damage bacterial cells, disrupt bacterial membranes, bind selectively to bacterial biomolecules, and interfere with critical cellular functions. Additionally, luminescent PTMCs are ideal for real-time imaging and tracking of bacterial cells during infection. In this perspective, we discuss their various applications, safety concerns, and emerging trends in the clinical use of PTMCs due to their enormous possibilities for future medical applications.
1. Introduction
The rise of multidrug-resistant bacterial diseases threatens global public health, compromising the efficacy of existing antibiotics and necessitating the development of new antimicrobial medicines.1 Antibiotics, due to their indiscriminate use, are gradually losing efficiency against resistant bacterial infections.2 This concerning trend emphasizes the need for novel therapeutic options to effectively combat these persistent diseases. Polynuclear transition metal complexes (PTMCs) are a class of molecules distinguished by the combination of several metal ions into a single molecule.3PTMCs have attracted a lot of attention due to their impressive structural variety and multifunctional characteristics. These complexes often contain metals such as iron, cobalt, nickel, copper, zinc, ruthenium, palladium, silver, cadmium, osmium, rhenium, iridium, platinum, gold, each with unique physicochemical properties that increase their potential for theranostic uses.4 The presence of at least one emissive metal centre in PTMCs enables them to be used as luminescent probes for bacterial imaging.5 The capacity for imaging, along with chemical or photoinduced antibacterial activity, is useful for tackling the difficulties faced by multidrug-resistant microorganisms.
Bacteria are typically categorized into two classes based on the cell wall composition: Gram-positive and Gram-negative.6 These structural differences have a substantial impact on bacterial sensitivity to antimicrobial drugs, with Gram-negative bacteria often being more resistant due to their complex cell wall design.7 The complicated structure of bacterial cell walls, the mechanisms underlying bacterial resistance, which are frequently linked to enzymatic systems involved in cell wall formation, present significant hurdles for effective treatment.8 Traditional bacterial detection technologies, such as culturing and the polymerase chain reaction (PCR), frequently fall short in terms of speed, sensitivity, and the ability to detect non-culturable bacteria, necessitating the development of alternative detection methodologies.9
Luminescent PTMCs enable rapid and selective bacterial detection through bacterial imaging, while their innate or photoinduced antibacterial action provides an excellent way to tackle resistant strains. The potential of PTMCs to function as theranostic agents makes them useful assets in the ongoing fight against infectious diseases. Their function as luminescent probes enables real-time bacterial imaging, providing vital insights into infection dynamics and treatment efficiency. Furthermore, the antimicrobial mechanisms used by PTMCs, such as microbial membrane rupture, the formation of reactive oxygen species (ROS), and specific binding to bacterial macromolecules, reveal their ability to overcome resistance mechanisms and improve clinical results.
This perspective highlights the potential of PTMCs as new agents in microbial imaging and antimicrobial therapy (Fig. 1) with a special emphasis on their structural variation and synthesis strategies. We discuss their structural and functional properties, intrinsic antimicrobial activities, bacterial imaging capabilities, strategies to overcome antimicrobial resistance, and the role of PTMCs in antimicrobial photodynamic therapy (aPDT).
 | ||
| Fig. 1 PTMCs enable bacterial imaging for diagnosis (left) and in the dark (middle), as well as killing bacteria through ROS production when exposed to light (right). Created with BioRender.com. | ||
2. Structural and functional properties
PTMCs are a remarkable class of compounds characterized by the insertion of multiple metal centers into a single molecular framework.3 This structural complexity gives PTMCs a distinct edge over typical mononuclear metal complexes, particularly in their structural flexibility and multifunctionality, which may be tuned for a variety of biological applications.2.1 Structural diversity
The structural diversity of PTMCs stems from the coordination of various metal ions within a single complex. These metal centres are strategically positioned to allow for a range of coordination geometries, ranging from octahedral to square planar, which significantly influence the physical and chemical properties of the complexes. The interactions between metals and ligands, the oxidation states of the metals, and the adaptable geometries achieved by modifying the attached ligands contribute to this variability. Such structural flexibility improves PTMCs’ biological activity by allowing specific interactions with bacterial components, thus boosting antimicrobial efficacy. For example, ruthenium(II) complexes often use octahedral coordination, which is suitable for interacting with bacterial membranes and DNA, making them effective at disrupting bacterial biological activities.10Polynuclear complexes containing two or more metal ions have higher antibacterial potency than mononuclear complexes as multiple metal centers can interact with different sites within bacterial cells at the same time, effectively targeting bacterial membranes, enzymes, and DNA, and can efficiently inhibit bacterial growth.11 Emissive PTMCs can be designed to produce intense luminescence in the visible range, allowing for real-time imaging of bacterial cells.12 Such customizable photophysical qualities are critical in diagnostic imaging, where high-contrast pictures are required to track bacterial behaviour and treatment effects efficiently.
2.2 Multifunctionality
One of the most notable characteristics of PTMCs is their capacity to execute many biological activities simultaneously.3 This multifunctionality is especially important in the context of bacterial infections, when prompt detection and tailored treatment are required. PTMCs, with their dual functions, provide significant advantages over traditional imaging probes and antibacterial drugs. Their capacity to function as both luminescent probes and antimicrobial agents makes them useful for creating integrated theranostic techniques.PTMCs have significant antibacterial activity against a wide range of bacterial infections, including multidrug resistant forms. Metals like gold, platinum, and ruthenium contribute to their antibacterial characteristics through a variety of mechanisms, including membrane rupture, the production of ROS, and DNA binding.13 For example, gold-based PTMCs have shown the ability to limit bacterial growth by disrupting bacterial cell membranes, resulting in cell lysis.14 Platinum(II)-based PTMCs can attach to bacterial DNA, causing irreparable damage and preventing bacterial multiplication.11 Zinc(II) complexes, particularly those coordinated with dipicolylamine, can selectively bind to bacterial membranes, allowing scientists to track bacterial cell division and infection progress in real time.15 Ruthenium(II)-based PTMCs, for example, have high fluorescence, allowing real-time imaging of bacterial cells.10 This characteristic is very useful in advanced microscopy techniques, such as super-resolution microscopy, where high-resolution imaging of bacterial structures is required.5c The capacity of PTMCs to selectively stain bacterial cells while avoiding mammalian cells increases their usefulness in diagnostic applications.
Furthermore, the multifunctionality of PTMCs extends to theragnostic applications, in which they can diagnose and treat bacterial infections simultaneously. This combined action streamlines the treatment process and enables real-time monitoring of therapeutic efficacy. By combining theranostic capabilities into a single compound, PTMCs streamline bacterial infection care, making them promising candidates for future clinical applications.
Biofilms, which are resistant to many conventional antibiotics, present a substantial difficulty in the treatment of persistent infections.16 PTMCs have shown potential for entering biofilms, altering their structure, and permitting simultaneous imaging. For example, copper(II)-based PTMCs have shown efficacy for biofilm penetration, allowing for high-resolution imaging of biofilm architecture and stressing its potential for treating biofilm-associated infections.17
Given these promising structural and functional features, it is crucial to understand the general antimicrobial mechanisms and strategic design principles that enhance the therapeutic performance of PTMCs.
3. General antimicrobial mechanisms and design strategies for PTMCs
Polynuclear transition metal complexes display antimicrobial activity through a variety of mechanisms, many of which differ from those employed by traditional antibiotics.10 A primary mode of action involves the generation of reactive oxygen species, either through redox cycling or upon photoactivation. These ROS, including singlet oxygen, superoxide, and hydroxyl radicals, induce oxidative stress that damages vital bacterial components such as membranes, proteins, and nucleic acids, ultimately leading to cell death. In addition to oxidative damage, many PTMCs directly disrupt bacterial membranes.18 Their structural features, such as hydrophobic or amphiphilic regions and cationic charges, promote strong electrostatic and lipophilic interactions with the negatively charged bacterial cell walls. This interaction results in membrane destabilization and leakage of intracellular contents. Another key mechanism involves interactions with bacterial DNA or enzymes.19 Certain metal centers and ligand frameworks enable selective binding to nucleic acids or inhibit the action of critical bacterial enzymes, thereby impairing replication and metabolism. Furthermore, PTMCs have demonstrated the ability to target and disrupt bacterial biofilms, which are protective communities that often make bacteria resistant to conventional therapies. Some PTMCs also modulate quorum sensing (QS) pathways, suppressing bacterial communication, virulence, and biofilm formation.The design of PTMCs for antimicrobial applications involves several strategic considerations. Firstly, the choice of metal center is fundamental to their biological activity.20 Transition metals such as copper, zinc, ruthenium, silver, iridium, and gold are commonly used because of their redox properties, coordination versatility, and inherent antimicrobial potential. Secondly, ligands are carefully selected to enhance solubility, cellular take up, target specificity, and photophysical properties. For example, Schiff base ligands, polypyridyl frameworks, and dipicolylamine derivatives can be used to tune lipophilicity and charge, which influences the complex's ability to interact with bacterial membranes. The polynuclear architecture itself contributes significantly to antimicrobial potency by enabling multivalent interactions and cooperative effects across multiple bacterial targets. Additionally, many PTMCs with luminescent or photoactive components serve as imaging probes along with antimicrobial activity. This theranostic approach supports simultaneous real-time visualization and infection treatment of bacterial cells. These combined design strategies enhance the ability of PTMCs to overcome conventional resistance mechanisms and establish them as promising candidates for next-generation antimicrobial theranostics.
While the design strategies and general antimicrobial mechanisms of PTMCs lay the groundwork for their effectiveness, it is their intrinsic antimicrobial activity that truly shows their potential as novel therapeutic agents. By leveraging unique metal–ligand interactions, PTMCs can disrupt bacterial membranes, generate reactive oxygen species, and bind to essential bacterial macromolecules, leading to potent antimicrobial effects. The following section delves deeper into the specific antimicrobial properties of various PTMCs, highlighting their capacity to combat multidrug-resistant bacteria and their role in addressing the growing threat of antimicrobial resistance.
4. Intrinsic antimicrobial activity
Traditional antibiotics are becoming increasingly ineffective against resistant bacteria, forcing experts to investigate other treatments.21 One interesting path is the development of PTMCs, which, because of their cooperative interactions, provide novel solutions to antimicrobial resistance (AMR). These complexes are distinguished by the presence of many metal centres that work together to enhance their potential to interact with and damage biological targets via novel mechanisms such as DNA binding and cellular membrane breakdown. In this section, we examine the potential of PTMCs as emerging agents in the fight against antimicrobial resistance, with a focus on several metal-based complexes that have demonstrated strong antibacterial properties. A summary of these metal-based complexes, namely, their metal centers, target bacteria, minimum inhibitory concentrations (MICs), and mechanisms of action, is provided in Table 1.| Complex | Metal center | Target bacteria | MIC (μM μg−1 mL−1) | Mechanism of action | Ref. |
|---|---|---|---|---|---|
| a Denotes calculated value (μM) converted from reported μg mL−1 using the molecular weight of the compound; not explicitly provided in the original reference. | |||||
| 1–6, 7–12 | Fe(II) | Mycobacterium tuberculosis (H37Rv) | 19.2 μM (4) | Penetration of biological membranes | 23 |
| 13–16 | Fe(II) | M. tuberculosis | 125 μM (13), 47.0 μM (14), 41.7 (16 μM) | Membrane interaction | 24 |
| 17 | Co(II) | Agrobacterium tumefaciens | Cell wall disruption | 26 | |
| 18, 19 | Ni(II) | Salmonella typhi | Lipophilic penetration | 28 | |
| 20 | Ni(II) | M. tuberculosis H37Rv | 8 μg mL−1 (10.34 μMa) | Fatty acid biosynthesis inhibition | 29 |
| 24–29 | Cu(II) | Gram-positive and Gram-negative | Protein synthesis inhibition | 32 | |
| 30–32 | Cu(II) | Pseudomonas aeruginosa, Bacillus proteus, Escherichia coli, Staphylococcus aureus | Pseudomonas aeruginosa (1 μg mL−1, 2 μg mL−1), Bacillus proteus (0.5 μg mL−1, 1 μg mL−1), Escherichia coli (64 μg mL−1), Staphylococcus aureus (2 μg mL−1) | 33 | |
| 33–37 | Cu(II) | P. aeruginosa, S. aureus | Biofilm inhibition | 34 | |
| 38, 39 | Cu(II) | Bacillus subtilis | Membrane disruption | 4f | |
| 40, 41 | Cu(II) | MRSA | ROS-mediated lipid peroxidation | 17 | |
| 42–46 | Zn(II) | S. aureus | 1 μg mL−1 (42) (1.18 μMa) | Disrupt bacterial cell membranes through depolarization | 15b |
| 47–51 | Zn(II) | S. aureus, B. subtilis, P. aeruginosa | DNA intercalation | 36 | |
| 52, 53 | Zn(II) | S. aureus | <0.5 mg mL−1 (<0.565 μMa for 52 and <0.47 μMa for 53) | Penetration into the lipid bilayer membrane | 37 |
| 54–57 | Ru(II) | S. aureus, MRSA, E. coli and P. aeruginosa | 2–4 μg mL−1 against S. aureus and MRSA; 8–16 μg mL−1 against E. coli and P. aeruginosa | 39 | |
| 58–60 | Ru(II) | S. aureus and E. coli. | For S. aureus, MICs were 8 μg mL−1 (58), 1 μg mL−1 (59), and 8 μg mL−1 (60); for E. coli, MICs were 8 μg mL−1 (58), 2 μg mL−1 (59), and 8 μg mL−1 (60) | Target intracellular proteins | 41 |
| 62–67 | Ru(II) | S. aureus, MRSA and P. Aeruginosa | DNA interactions | 43 | |
| 68 | Ru(II) | E. coli | >256 μg mL−1 (>121.27 μMa) | Binds bacterial DNA | 44 |
| 74–76 | Ru(II) | E. coli and E. faecalis | Membrane disruption | 46 | |
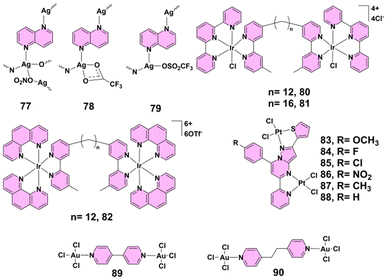
| 77–79 | Ag(I) | Candida spp. | 0.78–6.25 μg mL−1 (2.6–20.8 μM) | Biofilm inhibition | 4a |
| 80, 81 | Ir(III) | S. aureus, E. coli | For S. aureus, MICs were 16 μg mL−1 (80) and 2 μg mL−1 (81); for E. coli, 8 μg mL−1 (80) and 4 μg mL−1 (81). | Bacteriostatic action | 41 |
| 83–88 | Pt(II) | S. aureus, B. substilus and S. marcescens | Inhibit bacterial growth by disrupting essential cellular functions | 50 | |
| 89, 90 | Au(III) | Escherichia coli, Pseudomonas aeruginosa PAO1, Salmonella typhimurium, Staphylococcus aureus, Micrococcus luteus, Listeria monocytogenes | 3.9–62.5 μg mL−1 | 52 | |
| 91–98 | Au(I), Au(I)–Ag(I), Au(I)–Cu(II) | S. typhimurium, E. coli, B. cereus and S. aureus | 10–1 μg mL−1 | 54 | |
| 99 | Ru(II)–Pt(II) | E. coli | DNA crosslinking | 55 | |
| 100, 101 | Fe(II)–Mn(I)–Re(I), Ru(II)–Mn(I)–Re(I) | Methicillin-resistant Staphylococcus aureus | Disrupted essential cellular processes such as respiration and cell wall biosynthesis by altering membrane architecture | 56 | |
| 102–105 | Fe(II)–Mn(I)–Re(I), Fe(II)–Re(I), Mn(I)–Re(I), Re(I) | Methicillin-resistant Staphylococcus aureus | Membrane-targeting | 57 | |
4.1 Iron(II)-based complexes
Iron(II)-based complexes are particularly effective against Mycobacterium tuberculosis (Mtb).22 Ferrocenyl-based compounds, in particular, stand out for their capacity to be changed into a variety of forms, increasing bioactivity and opening up new avenues for therapeutic interventions. Smith's group made a significant contribution to this field in 2016 by synthesizing and evaluating a series of ferrocenyl mono- and polynuclear complexes against the Mycobacterium TB H37Rv strain. The synthesis employed Schiff base condensation and reductive amination to produce ferrocenylimine and ferrocenylamine molecules. The ferrocenylimine complexes 1–6 (Fig. 2) exhibited higher antimycobacterial activity than their ferrocenylamine counterparts 7–12 (Fig. 2). The trinuclear ferrocenylimine complex 4, produced from tris(2-amino)ethylamine, showed maximum efficacy, with a minimum inhibitory concentration (MIC90) of 19.2 μM. Mononuclear complexes 1–3 showed promising action, but were slightly less effective. This shows that the side chains of mononuclear complexes have no effect on their antibacterial activity. The increased activity of the ferrocenylimine series is most likely due to the Schiff base moiety, which allows for greater interaction with biological targets and hence increases the complexes’ antimicrobial potential. In contrast, the ferrocenylamine complexes had considerably lower activity, highlighting the importance of Schiff base structures in increasing the efficacy of these molecules.23 In parallel, Smith's group explored the role of sulfur-containing ligands by synthesizing ferrocenylthiosemicarbazone complexes 13–16 (Fig. 2). This series included complexes of varying nuclearity, from mononuclear complex 14 to octanuclear complex 16. Both complex 14 and complex 16 exhibited potent antimycobacterial activity, with MIC90 values of 47.0 μM and 41.7 μM, respectively. However, tetranuclear complex 15 and ferrocenyl dithiocarbamate complex 13 showed much lower activity, with MIC90 values exceeding 125 μM.24 These findings highlight the importance of both nuclearity and ligand composition in modulating the antimicrobial properties of iron(II)-based complexes.4.2 Cobalt(II)-based complexes
In addition to iron(II)-based PTMCs, cobalt(II) complexes have also been studied for their selective antibacterial capabilities.25 One notable example is homobimetallic cobalt(II) complex 17 (Fig. 2), which has demonstrated strong antibacterial activity. The cobalt(II) ions, coordinated by the bpt ligand, create a rigid and planar dinuclear structure, which may enhance the complex's ability to penetrate the lipid bilayer of microbial membranes due to its lipophilic character. Upon interaction with microbial cells, complex 17 could induce oxidative stress by promoting the generation of reactive oxygen species, a well-established mechanism for metal-based antimicrobial agents. Additionally, the metal centers may disrupt essential enzymatic processes or interfere with metal ion homeostasis within the microbes. This compound proves to be highly efficient against diverse strains of Agrobacterium tumefaciens, a phytopathogen that causes crown gall disease, which is a serious issue in crop production. The compound exhibited strong antibacterial activity against strains A281, C58, and Ach5, indicating its potential as a targeted antibacterial agent for crop protection. However, this cobalt(II) complex has minimal efficacy against Pseudomonas syringae pv. tabaci and Pseudomonas syringae pv. syringae, both of which cause diseases like wildfire in tobacco and other crops. The resistance of these bacteria is most likely due to their capacity to build biofilms by producing exopolysaccharides, which act as a permeability barrier and impede penetration of the cobalt(II) complex. This differential sensitivity demonstrates the selective character of cobalt(II) polynuclear complexes, making them interesting candidates for developing pathogen-specific therapies in agriculture. The selective action of complex 17, notably against A. tumefaciens, opens up new possibilities for antimicrobial research.26 By exploiting the unique features of polynuclear structures, it may be possible to design more efficient, tailored crop disease management strategies. Such focused therapies may reduce the demand for broad-spectrum antibiotics, lowering the likelihood of resistance development and encouraging sustainable agriculture practices.4.3 Nickel(II)-based complexes
Following cobalt(II)-based PTMCs, nickel(II)-based PTMCs have emerged as another promising class, demonstrating considerable promise as antibacterial agents through enhanced lipophilicity, membrane penetration, and disruption of vital cellular processes.27 The coordination of metal centers in these PTMCs facilitates penetration into bacterial membranes and effectively disrupts cellular functions. Trinuclear nickel(II) complex 18 (Fig. 2) and mononuclear nickel(II) complex 19 (Fig. 2), synthesized from the ligand 5-methyl-4-phenyl-1,2,4-triazole-3-thione (Hmptt) (Fig. 2), exhibit promising antibacterial activity against various human pathogens. The structures of complexes 18 and 19 were unambiguously confirmed by single crystal X-ray diffraction analysis. Among these, complex 18 demonstrated the highest antibacterial efficacy, showing a maximum zone of inhibition of 2.3 cm against Salmonella typhi at 50 μg per disc, outperforming both the free ligand and complex 19. The complexes showed greater inhibitory effects compared to the ligand at all tested concentrations, affirming that metal coordination significantly enhances antibacterial potency. The observed antibacterial activities can be rationalized by chelation theory, where the formation of metal–ligand complexes reduces metal ion polarity and increases lipophilicity, facilitating bacterial membrane penetration and disrupting normal cellular processes. The activity order is complex 18 > complex 19 > Hmptt, indicating the potential of complex 18 as a moderate antibacterial agent with therapeutic applications. So, the complexes exhibit significant biological activity, making them promising candidates for further development in antimicrobial therapy.28 Additionally, the tetradentate monoanionic N2O2 chelator HL (Fig. 2) and its dinuclear nickel(II) complex 20 (Fig. 2) also demonstrate noteworthy antibacterial properties, particularly against Mycobacterium tuberculosis strains. Both HL and complex 20 show significant activity against drug-resistant and drug-susceptible strains of M. tuberculosis H37Rv and H37Ra, with MICs ranging from 4 to 32 μg mL−1 and minimum bactericidal concentrations (MBCs) between 8 and 64 μg mL−1. Notably, ligand HL exhibits higher efficacy, achieving lower MIC and MBC values compared to complex 20, particularly against drug-resistant strains. The presence of complex lipids and mycolic acids in the bacterial cell wall poses a formidable hydrophobic barrier. However, both HL and complex 20 effectively penetrate this defense, exerting bactericidal effects. The compounds exhibit potent antibacterial action even against clinical strains, underscoring their potential as effective antimycobacterial agents. Furthermore, molecular docking studies reveal that HL forms stable hydrogen bonds with key residues in the enoyl acyl carrier protein reductase enzyme of M. tuberculosis H37Rv, supporting its role as a promising antimicrobial agent targeting fatty acid biosynthesis in the bacterial pathogen.29 These findings highlight the significant potential of nickel(II)-based complexes and their ligands as promising candidates for the development of new antimicrobial therapies, particularly against drug-resistant bacterial strains.4.4 Copper(II)-based complexes
Similar to nickel(II)-based PTMCs, copper(II)-based complexes have attracted attention for their potent antimicrobial and antibiofilm activities, particularly against multidrug-resistant pathogens.30 The presence of multiple copper centers enhances these effects by facilitating strong interactions with bacterial membranes and intracellular components. In 2013, Cunha-Silva's group reported the Schiff base methyl 2-pyridyl ketone semicarbazone, and its binuclear copper(II) complexes 21, 22, and 23 (Fig. 3) were evaluated for their antimicrobial activity against a range of bacterial and fungal pathogens, including Bacillus subtilis, Staphylococcus aureus, Escherichia coli, and Erwinia carotovora, as well as Candida kefyr, Candida krusei, and Aspergillus niger. The disc diffusion method revealed that all complexes exhibited superior antibacterial activity compared to the free ligand, with complex 23, containing a bridging chlorido co-ligand, demonstrating the highest efficacy. This increased antimicrobial activity can be attributed to the chelation effect, which enhances lipophilicity and reduces metal ion polarity, thus facilitating better interaction with microbial cell walls. The minimum inhibitory concentration (MIC) values indicated that complex 23 had significant activity against all tested organisms, making it a promising candidate for further antimicrobial development. So, the results highlight the importance of metal coordination for enhancing the biological properties of ligands, highlighting the potential of copper(II) complexes for combating microbial infections.31 In 2014, Rahiman's group reported a series of dinuclear nickel(II) and copper(II) complexes 24–29 (Fig. 3) of hexaaza macrocycles derived from 2,6-diformyl-4-methylphenol and varying benzoyl pendant-arms were synthesized and characterized for their antibacterial properties. The complexes exhibited notable activity against both Gram-positive and Gram-negative bacteria, with the order of efficacy being 29 > 28 > 27 > 26 > 25 > 24. Particularly, complex 29 demonstrated the highest antibacterial activity, attributed to the presence of dinitro substituents in its pendant-arm, which enhanced its lipophilicity and facilitated membrane permeation. This complexation effect reduces metal ion polarity, promoting interactions with bacterial cell membranes and disrupting normal cellular processes, thereby inhibiting protein synthesis and leading to microbial cell death. In vitro antibacterial screening at a fixed concentration of 150 μg mL−1 showed that the complexes significantly outperformed the standard antibiotic ciprofloxacin, emphasizing the potential of these copper(II) complexes as effective antimicrobial agents. The findings suggest that the incorporation of various functional groups into the pendant-arms can fine-tune the biological activity of macrocyclic complexes, leading the way for the design of new therapeutic agents.32 In the same year, Li and co-workers reported dinuclear copper(II) complexes 30–32 (Fig. 3), which exhibited significantly enhanced antibacterial efficacy compared to their mononuclear counterparts and standard drugs such as Chloromycin. Notably, complex 30 demonstrated potent activity against Pseudomonas aeruginosa and Bacillus proteus with MIC values of 1 μg mL−1 and 0.5 μg mL−1, respectively. Complex 31 showed comparable potency, with an MIC of 1 μg mL−1 for B. proteus and 64 μg mL−1 for Escherichia coli. Additionally, complex 32 exhibited broad-spectrum activity, inhibiting both Gram-positive and Gram-negative strains, including Staphylococcus aureus (MIC = 2 μg mL−1) and P. aeruginosa (MIC = 2 μg mL−1). These findings highlight the crucial role of the dinuclear architecture and the incorporation of aromatic linkers, particularly the m-xylyl moiety, for enhancing antimicrobial activity.33 In 2016, Glišić's group reported that copper(II) complexes 33–37 (Fig. 3) demonstrated notable activity in modulating QS and biofilm formation, though they showed limited direct antibacterial activity. These complexes were not potent antimicrobial agents, with MIC values generally in the millimolar range, and exhibited little effect on the growth of pathogens like Pseudomonas aeruginosa and Staphylococcus aureus. Despite their limited growth inhibition, complexes 33–37 significantly disrupted bacterial QS in Chromobacterium violaceum and P. aeruginosa, particularly inhibiting biofilm formation in P. aeruginosa PAO1, with quinazoline-containing complex 35 being the most effective. The disruption of QS was primarily through affecting the production of acyl-homoserine lactones and 2-alkyl-4-quinolones, which were key signaling molecules. Additionally, complexes 33 and 35 exhibited synergistic effects with antibiotics such as piperacillin and ceftazidime against a multidrug-resistant clinical isolate of P. aeruginosa, highlighting their potential as co-therapies for addressing bacterial resistance without directly promoting bacterial growth inhibition. These findings open new avenues for antivirulence therapies that could minimize the risk of resistance development.34Research on copper(II)-based complexes continued to advance, with a particular focus on their antibacterial potential against Gram-positive pathogens. In 2019, Powell's group reported tetranuclear chiral copper(II)–Schiff-base complexes 38 and 39 (Fig. 4), which were synthesized using enantiomerically pure (S)- and (R)-H2vanPheol ligands, exhibiting notable antibacterial activity, particularly against Gram-positive Bacillus subtilis. Both enantiomers show comparable growth inhibitory effects on B. subtilis, completely preventing bacterial growth at concentrations of 50 and 100 μM, while a lower concentration of 10 μM causes a delay in growth. However, these copper(II) complexes demonstrate no bactericidal activity against Gram-negative Escherichia coli. The antibacterial effects of complexes 38 and 39 are independent of chirality, as both enantiomers exhibit similar levels of efficacy against B. subtilis. These results suggest that the chiral Schiff-base copper(II) complexes have promising potential as antimicrobial agents against Gram-positive bacteria, although further investigation is necessary to enhance their activity against Gram-negative strains.4f In 2024, Mandal and Bera's group reported multicopper clusters 40 and 41 (Fig. 4), which were investigated for their antibacterial and antibiofilm properties. Both clusters demonstrated potent antibacterial and antibiofilm activities against methicillin-resistant Staphylococcus aureus (MRSA BAA1717) and a clinically isolated strain, MRSA CI1. Mechanistic investigations revealed that both clusters significantly enhanced the generation of ROS, leading to lipid peroxidation and disruption of bacterial cell membranes. Additionally, they exhibited synergistic effects with antibiotics such as vancomycin, further amplifying their antibacterial effectiveness. Notably, cluster 40 showed superior antibacterial and antivirulence actions compared to cluster 41.17 These studies highlight the significant potential of copper(II)-based complexes as versatile antimicrobial and antivirulence agents, offering promising strategies for combating both drug-resistant infections and biofilm-associated complications.
 | ||
| Fig. 4 (a) Structures of complexes 38 and 39. (b) Molecular representations of complexes 38 and 39, with hydrogen atoms and nitrate counterions omitted for clarity. Hydrogen bonds are indicated by green dashed lines. Atom color scheme: Cu (light blue), N (blue), O (red), and C (black). Reproduced from ref. 4f with permission from the Royal Society of Chemistry, copyright 2019. (c) Structures of complexes 40 and 41. (d) ORTEP representations of the X-ray crystal structures of complexes 40 and 41, shown with 35% probability ellipsoids. For complex 40, all hydrogen atoms and crystallized water molecules are omitted for clarity. For complex 41, hydrogen atoms and counteranions are omitted. Color codes: Cu (brown), N (blue), O (red), S (yellow), Cl (light green), and C (black). Reproduced from ref. 17 with permission from the American Chemical Society, copyright 2024. | ||
4.5 Zinc(II)-based complexes
Beyond copper(II)-based PTMCs, zinc(II)-based complexes offer a unique, redox-inert alternative, demonstrating remarkable selectivity toward Gram-positive bacteria through membrane-targeted mechanisms.35 In 2008, Smith's group reported that zinc(II)-(2,2′-dipicolylamine) coordination complexes 42–46 (Fig. 5) exhibited notable antibacterial properties. Lipophilic analogues (e.g., complexes 44 and 45) show high membrane permeability, inducing phospholipid translocation and carboxyfluorescein leakage in vesicles. However, these complexes are moderately toxic to mammalian cells. In contrast, the hydrophilic analogue (complex 42) is highly selective against Gram-positive bacteria, such as Staphylococcus aureus, including resistant strains. It demonstrates a low minimum inhibitory concentration of 1 μg mL−1 and is notably less toxic to mammalian cells (LD50 > 50 μg mL−1). This selectivity is attributed to its ability to disrupt bacterial cell membranes through depolarization. The fluorescent conjugate of complex 42 enables effective bacterial imaging, indicating its potential as a bacterial probe.15b In 2018, Sedaghat's group reported that ternary complexes 47–51 (Fig. 5) exhibited significant antibacterial activity. It should be noted that the two molecules shown for the structure of complex 51 are crystallographically independent and occupy the asymmetric unit of a single crystal. Although chemically identical, they are not symmetry-related and display slight geometric differences. All complexes outperform the parent ligand for inhibiting Gram-positive (Staphylococcus aureus, Bacillus subtilis) and Gram-negative (Pseudomonas aeruginosa) bacteria. Complexes 49 and 50 show remarkable effectiveness against Pseudomonas aeruginosa, a notably resistant strain. The enhanced activity is attributed to increased lipophilicity and potential DNA intercalation by bipyridine ligands.36 Research conducted by Kargar and his group in 2022 highlighted the antibacterial activity of two binuclear zinc(II) Schiff base complexes, 52 and 53 (Fig. 5), synthesized from Schiff base ligands derived from 2,2-dimethyl-1,3-propanediamine and either 5-chlorosalicylaldehyde or 5-bromosalicylaldehyde. Antibacterial investigations revealed that these zinc(II) complexes, particularly complexes 52 and 53, exhibited significant activity against Gram-positive bacteria, namely, Staphylococcus aureus and Bacillus cereus. The zinc(II) complexes showed greater inhibitory effects than their corresponding free ligands, which displayed no significant antibacterial activity. This enhanced efficacy is attributed to the chelation effect, where coordination with the metal ion enhances lipophilicity, facilitating better penetration into bacterial cell membranes. Specifically, complex 52 showed higher zones of inhibition compared to complex 53, with complex 52 displaying up to 16 mm of inhibition against S. aureus and 15 mm against B. cereus at a concentration of 3 mg mL−1. Both complexes were less effective against Gram-negative bacteria such as Escherichia coli and Pseudomonas aeruginosa, with no significant activity observed. The minimum inhibitory concentration tests further supported these findings, where complexes 52 and 53 demonstrated potent activity against Gram-positive strains, with MIC values below 0.5 mg mL−1. The study concluded that the antibacterial activity of these complexes depended largely on the metal ion, with the zinc(II) complexes showing a synergistic effect in comparison with the free ligands.374.6 Ruthenium(II)-based complexes
Zinc(II)-based PTMCs excel at membrane targeting, whereas ruthenium(II)-based complexes offer a distinct advantage by combining DNA interactions, luminescent imaging properties, and potent antibacterial activity.38 These complexes have demonstrated significant efficacy against a broad range of pathogens, including both Gram-positive and Gram-negative bacteria. In 2011, Keene, Collins, and their team explored the antimicrobial properties of dinuclear polypyridylruthenium(II) complexes, highlighting their potential against Gram-positive and Gram-negative bacteria, including the drug-resistant strain MRSA. The dinuclear ruthenium(II) complexes ΔΔ/ΛΛ-54, 55, and 56 (Fig. 6), featuring flexible alkane-linking chains, demonstrated exceptional antibacterial activity with MICs as low as 1 μg mL−1 against both S. aureus and MRSA. Their efficacy was lower against Gram-negative bacteria such as E. coli and P. aeruginosa, showing MIC values between 2 and 16 μg mL−1. Notably, the mononuclear [Ru(Me4phen)3]2+ complex exhibited lower activity against MRSA and Gram-negative strains, emphasizing the importance of the dinuclear structure for better penetration or overcoming bacterial resistance. The study also correlated antibacterial activity with lipophilicity, measured through the octanol–water partition coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P), but found that overall charge distribution played a critical role. For instance, highly lipophilic mononuclear complex 57 (Fig. 6) was less effective than its dinuclear counterpart. In terms of cytotoxicity, complex ΔΔ-54 showed the best profile, being highly selective towards bacterial cells with minimal toxicity to human red blood cells and THP-1 cells. These results suggest that dinuclear ruthenium(II) complexes, especially complex 54, hold promise as potent antibacterial agents with selective toxicity, potentially addressing clinical challenges posed by antibiotic-resistant bacteria.39 In 2012, they further investigated the antimicrobial properties of dinuclear ruthenium(II) complexes, highlighting their significant activity against various bacterial strains. In particular, compounds 54 and 56 (Fig. 6) demonstrated potent bactericidal effects with low MIC and MBC values, especially against Gram-positive bacteria such as Staphylococcus aureus and methicillin-resistant Staphylococcus aureus, showing values as low as 1–2 mg L−1. Both complexes also displayed moderate activity against Gram-negative bacteria like Escherichia coli and Pseudomonas aeruginosa, although higher concentrations were required for P. aeruginosa. The study confirmed that these ruthenium(II) complexes killed bacteria within 2–6 h, and their activity was strongly correlated with cellular take up, with greater take up observed in Gram-positive bacteria compared to Gram-negative strains. Moreover, confocal microscopy indicated that the complexes, especially complex 56, were internalized within bacterial cells. The study also noted that cellular take up was not time-dependent, and dead cells exhibited a higher take up than live cells, suggesting passive diffusion as the mechanism of entry. So, complexes 54 and 56 show promise as novel antimicrobial agents, offering a potential alternative to conventional antibiotics due to their robust bactericidal action and ability to target bacterial cells over human cells. These findings provide a solid foundation for further optimization of these ruthenium(II)-based compounds for enhanced selectivity and efficacy against bacterial pathogens.40 In 2013, they highlighted the potential of chlorido-containing ruthenium(II) complexes 58–60 (Fig. 6) as effective antimicrobial agents. A key finding was the enhanced activity of the dinuclear ruthenium(II) complexes, particularly complex 59, which exhibited strong bactericidal activity against both Gram-positive (S. aureus and MRSA) and Gram-negative bacteria (P. aeruginosa and E. coli). The incorporation of chlorido ligands into the complexes, such as in [{Ru(tpy)Cl}2{μ-bbn}]2+ (Cl-Rubbn), appears to facilitate bacterial cell membrane penetration. This is achieved by lowering the complex's initial cationic charge, which can increase to a 4+ charge upon aquation inside the cell, thus regaining the ability to interact with biological targets like DNA or proteins. Notably, complex 59 demonstrated better antimicrobial activity than its inert counterpart 54, and other analogues such as complexes 58 and 60. The activity of these ruthenium(II) complexes varies with chain length, with complex 59 standing out as the most potent. The research emphasizes the importance of charge, charge separation, and lipophilicity in the antimicrobial efficacy of these metal complexes, suggesting that the inclusion of labile ligands like chlorido can enhance both cellular take up and bactericidal potency.41 In the same year, they investigated ruthenium(II)-based complexes, particularly dinuclear polypyridylruthenium(II) complexes (Rubbn), which showed significant promise as new antimicrobial agents against bacterial strains like Staphylococcus aureus and Escherichia coli. These complexes, including [{Ru(phen)2}2{μ-bbn}]4+ (complexes 54, 56, and 61) (Fig. 6), have been observed to accumulate in bacteria in a temperature-dependent manner, with increased take up at higher temperatures. Notably, accumulation is more pronounced in E. coli than in S. aureus, likely due to differences in membrane fluidity and composition. Despite their cationic nature, the take up of Rubbn complexes, particularly complex 54, appears to be energy-independent and is not driven by ATP production or the bacterial membrane potential, as demonstrated by experiments with metabolic inhibitors and membrane potential disruptors such as CCCP. In contrast, the mononuclear ruthenium(II) complex [Ru(Me4phen)3]2+ (Mono-Me4) behaves differently from the dinuclear complexes, showing significant membrane depolarization but without increasing membrane permeability. This suggests distinct modes of action, with Mono-Me4 possibly targeting intracellular proteins, while the Rubbn complexes act by depolarizing and permeabilizing bacterial membranes rapidly, particularly complex 56, which acts quickly, within 15–30 min. The study highlights the potential of these ruthenium(II) complexes to overcome antimicrobial resistance, with their ability to interfere with bacterial membranes providing a unique mechanism that differs from conventional antibiotics.42 In 2014, they reported that ruthenium(II) complexes, particularly multinuclear polypyridyl species, showed promising antibacterial activity against both Gram-positive and Gram-negative bacteria. They focused on synthesizing tri- and tetra-nuclear ruthenium(II) complexes linked by bis[4(4′-methyl-2,2′-bipyridyl)]-1,n-alkane ligands (Rubbn), evaluating their antimicrobial properties. Compounds 62–65 (Fig. 6) were identified as the most active, particularly against Gram-positive strains like S. aureus and MRSA. The linear tetranuclear complexes generally outperformed their non-linear counterparts, with MIC values below 1 μM for Gram-positive bacteria, suggesting that structural linearity enhanced antibacterial efficacy. Despite the higher lipophilicity and cellular accumulation of non-linear complexes 66 and 67 (Fig. 6), the linear forms exhibited greater activity, potentially due to their ability to closely associate with DNA. Against Gram-negative bacteria, particularly Pseudomonas aeruginosa (P. aeruginosa), the antibacterial activity was less pronounced, despite significant cellular take up, implying an intrinsic resistance mechanism. Dinuclear complexes like 54 and 56 also demonstrated good antimicrobial
properties, though higher nuclearity complexes, especially 64 and 65, were consistently more potent. The study highlights the potential of higher nuclearity ruthenium(II) complexes as bactericidal agents, though their efficacy appears to be influenced by both lipophilicity and molecular structure.43 In 2015, Crowley's group reported the synthesis of ruthenium(II) helicates 68–71 (Fig. 6), derived from bis-bidentate “click” pyridyl-1,2,3-triazole ligands and RuCl3. These helicates were thoroughly characterized using X-ray crystallography and IR, UV-visible, and NMR spectroscopy. Among them, the antibacterial properties of the racemic ruthenium(II) helicate 68 and Hannon's racemic iron(II) helicate 71 were investigated. Unlike iron(II) analogue 71, complex 68 exhibited excellent kinetic inertness and high stability under biologically relevant conditions, including in DMSO and in the presence of histidine. Antibacterial screening against Staphylococcus aureus and Escherichia coli revealed that complex 68 had modest activity, with MICs exceeding 256 μg mL−1, although small zones of inhibition were observed in agar-based disk diffusion assays. In contrast, the free ligand and helicate 71 showed no detectable activity under the same conditions. These findings suggest that while ruthenium(II) helicate 68 has limited antibacterial efficacy in its current form, enhancing its lipophilicity, an approach shown to improve activity in related dinuclear ruthenium(II) complexes, may significantly boost its potential as a promising antimicrobial agent.44 In 2018, Terbouche's research highlighted the synthesis and evaluation of a novel bis-[1-({2-[(2-hydroxynaphthalen-1-yl)methylidene]amino}ethyl)-1-ethyl-3-phenylthiourea] Schiff base ligand and its binuclear ruthenium(II) and palladium(II) complexes 72 and 73 (Fig. 6). The study particularly emphasizes the antibacterial efficacy of these complexes, focusing on their enhanced activity against various bacterial strains. Ruthenium(II) complex 72, in particular, demonstrated remarkable antibacterial activity, outperforming both the ligand and palladium complex 73. Specifically, ruthenium(II) complex 72 showed significant inhibition against methicillin-resistant Staphylococcus aureus (22 mm), E. coli (20 mm), and methicillin-sensitive Staphylococcus aureus (18 mm). In contrast, palladium(II) complex 73 exhibited slightly lower activity against these strains, with inhibition zones of 19 mm for MRSA and 16 mm for MSSA. The research suggests that the coordination of the Schiff base ligand with ruthenium(II) ions significantly enhances its antibacterial properties, making it a potent candidate for further development in antibacterial applications.45 In 2019, Thomas and his team evaluated the antibacterial activity of ruthenium(II) complexes 74, 75, and 76 (Fig. 6), with a focus on their efficacy against clinically critical bacteria. Compound 74 displayed poor solubility and minimal activity, while 75 and 76 exhibited significant antibacterial effects. Notably, 76 demonstrated superior activity in both glucose-defined minimal media and Mueller–Hinton II (MH-II), surpassing ampicillin against some strains. It was effective against multidrug-resistant E. coli EC958 and E. faecalis V583, with a notably high activity–toxicity ratio in human cells. Time–kill and MBC assays confirmed the potent bactericidal action of complex 76, which was attributed to membrane disruption, as supported by ICP-AES, stimulated emission depletion (STED) microscopy, and ATP leakage assays. The compound's selective targeting of bacterial membranes, coupled with low toxicity to human cells, emphasizes its potential as a novel antimicrobial agent. Further studies will refine our understanding of its mechanisms and optimize its therapeutic use.46 Together, these findings highlight the remarkable structural versatility and therapeutic potential of ruthenium(II)-based complexes, particularly dinuclear and multinuclear architectures, as promising alternatives to conventional antibiotics for combating multidrug-resistant bacterial infections.
P), but found that overall charge distribution played a critical role. For instance, highly lipophilic mononuclear complex 57 (Fig. 6) was less effective than its dinuclear counterpart. In terms of cytotoxicity, complex ΔΔ-54 showed the best profile, being highly selective towards bacterial cells with minimal toxicity to human red blood cells and THP-1 cells. These results suggest that dinuclear ruthenium(II) complexes, especially complex 54, hold promise as potent antibacterial agents with selective toxicity, potentially addressing clinical challenges posed by antibiotic-resistant bacteria.39 In 2012, they further investigated the antimicrobial properties of dinuclear ruthenium(II) complexes, highlighting their significant activity against various bacterial strains. In particular, compounds 54 and 56 (Fig. 6) demonstrated potent bactericidal effects with low MIC and MBC values, especially against Gram-positive bacteria such as Staphylococcus aureus and methicillin-resistant Staphylococcus aureus, showing values as low as 1–2 mg L−1. Both complexes also displayed moderate activity against Gram-negative bacteria like Escherichia coli and Pseudomonas aeruginosa, although higher concentrations were required for P. aeruginosa. The study confirmed that these ruthenium(II) complexes killed bacteria within 2–6 h, and their activity was strongly correlated with cellular take up, with greater take up observed in Gram-positive bacteria compared to Gram-negative strains. Moreover, confocal microscopy indicated that the complexes, especially complex 56, were internalized within bacterial cells. The study also noted that cellular take up was not time-dependent, and dead cells exhibited a higher take up than live cells, suggesting passive diffusion as the mechanism of entry. So, complexes 54 and 56 show promise as novel antimicrobial agents, offering a potential alternative to conventional antibiotics due to their robust bactericidal action and ability to target bacterial cells over human cells. These findings provide a solid foundation for further optimization of these ruthenium(II)-based compounds for enhanced selectivity and efficacy against bacterial pathogens.40 In 2013, they highlighted the potential of chlorido-containing ruthenium(II) complexes 58–60 (Fig. 6) as effective antimicrobial agents. A key finding was the enhanced activity of the dinuclear ruthenium(II) complexes, particularly complex 59, which exhibited strong bactericidal activity against both Gram-positive (S. aureus and MRSA) and Gram-negative bacteria (P. aeruginosa and E. coli). The incorporation of chlorido ligands into the complexes, such as in [{Ru(tpy)Cl}2{μ-bbn}]2+ (Cl-Rubbn), appears to facilitate bacterial cell membrane penetration. This is achieved by lowering the complex's initial cationic charge, which can increase to a 4+ charge upon aquation inside the cell, thus regaining the ability to interact with biological targets like DNA or proteins. Notably, complex 59 demonstrated better antimicrobial activity than its inert counterpart 54, and other analogues such as complexes 58 and 60. The activity of these ruthenium(II) complexes varies with chain length, with complex 59 standing out as the most potent. The research emphasizes the importance of charge, charge separation, and lipophilicity in the antimicrobial efficacy of these metal complexes, suggesting that the inclusion of labile ligands like chlorido can enhance both cellular take up and bactericidal potency.41 In the same year, they investigated ruthenium(II)-based complexes, particularly dinuclear polypyridylruthenium(II) complexes (Rubbn), which showed significant promise as new antimicrobial agents against bacterial strains like Staphylococcus aureus and Escherichia coli. These complexes, including [{Ru(phen)2}2{μ-bbn}]4+ (complexes 54, 56, and 61) (Fig. 6), have been observed to accumulate in bacteria in a temperature-dependent manner, with increased take up at higher temperatures. Notably, accumulation is more pronounced in E. coli than in S. aureus, likely due to differences in membrane fluidity and composition. Despite their cationic nature, the take up of Rubbn complexes, particularly complex 54, appears to be energy-independent and is not driven by ATP production or the bacterial membrane potential, as demonstrated by experiments with metabolic inhibitors and membrane potential disruptors such as CCCP. In contrast, the mononuclear ruthenium(II) complex [Ru(Me4phen)3]2+ (Mono-Me4) behaves differently from the dinuclear complexes, showing significant membrane depolarization but without increasing membrane permeability. This suggests distinct modes of action, with Mono-Me4 possibly targeting intracellular proteins, while the Rubbn complexes act by depolarizing and permeabilizing bacterial membranes rapidly, particularly complex 56, which acts quickly, within 15–30 min. The study highlights the potential of these ruthenium(II) complexes to overcome antimicrobial resistance, with their ability to interfere with bacterial membranes providing a unique mechanism that differs from conventional antibiotics.42 In 2014, they reported that ruthenium(II) complexes, particularly multinuclear polypyridyl species, showed promising antibacterial activity against both Gram-positive and Gram-negative bacteria. They focused on synthesizing tri- and tetra-nuclear ruthenium(II) complexes linked by bis[4(4′-methyl-2,2′-bipyridyl)]-1,n-alkane ligands (Rubbn), evaluating their antimicrobial properties. Compounds 62–65 (Fig. 6) were identified as the most active, particularly against Gram-positive strains like S. aureus and MRSA. The linear tetranuclear complexes generally outperformed their non-linear counterparts, with MIC values below 1 μM for Gram-positive bacteria, suggesting that structural linearity enhanced antibacterial efficacy. Despite the higher lipophilicity and cellular accumulation of non-linear complexes 66 and 67 (Fig. 6), the linear forms exhibited greater activity, potentially due to their ability to closely associate with DNA. Against Gram-negative bacteria, particularly Pseudomonas aeruginosa (P. aeruginosa), the antibacterial activity was less pronounced, despite significant cellular take up, implying an intrinsic resistance mechanism. Dinuclear complexes like 54 and 56 also demonstrated good antimicrobial
properties, though higher nuclearity complexes, especially 64 and 65, were consistently more potent. The study highlights the potential of higher nuclearity ruthenium(II) complexes as bactericidal agents, though their efficacy appears to be influenced by both lipophilicity and molecular structure.43 In 2015, Crowley's group reported the synthesis of ruthenium(II) helicates 68–71 (Fig. 6), derived from bis-bidentate “click” pyridyl-1,2,3-triazole ligands and RuCl3. These helicates were thoroughly characterized using X-ray crystallography and IR, UV-visible, and NMR spectroscopy. Among them, the antibacterial properties of the racemic ruthenium(II) helicate 68 and Hannon's racemic iron(II) helicate 71 were investigated. Unlike iron(II) analogue 71, complex 68 exhibited excellent kinetic inertness and high stability under biologically relevant conditions, including in DMSO and in the presence of histidine. Antibacterial screening against Staphylococcus aureus and Escherichia coli revealed that complex 68 had modest activity, with MICs exceeding 256 μg mL−1, although small zones of inhibition were observed in agar-based disk diffusion assays. In contrast, the free ligand and helicate 71 showed no detectable activity under the same conditions. These findings suggest that while ruthenium(II) helicate 68 has limited antibacterial efficacy in its current form, enhancing its lipophilicity, an approach shown to improve activity in related dinuclear ruthenium(II) complexes, may significantly boost its potential as a promising antimicrobial agent.44 In 2018, Terbouche's research highlighted the synthesis and evaluation of a novel bis-[1-({2-[(2-hydroxynaphthalen-1-yl)methylidene]amino}ethyl)-1-ethyl-3-phenylthiourea] Schiff base ligand and its binuclear ruthenium(II) and palladium(II) complexes 72 and 73 (Fig. 6). The study particularly emphasizes the antibacterial efficacy of these complexes, focusing on their enhanced activity against various bacterial strains. Ruthenium(II) complex 72, in particular, demonstrated remarkable antibacterial activity, outperforming both the ligand and palladium complex 73. Specifically, ruthenium(II) complex 72 showed significant inhibition against methicillin-resistant Staphylococcus aureus (22 mm), E. coli (20 mm), and methicillin-sensitive Staphylococcus aureus (18 mm). In contrast, palladium(II) complex 73 exhibited slightly lower activity against these strains, with inhibition zones of 19 mm for MRSA and 16 mm for MSSA. The research suggests that the coordination of the Schiff base ligand with ruthenium(II) ions significantly enhances its antibacterial properties, making it a potent candidate for further development in antibacterial applications.45 In 2019, Thomas and his team evaluated the antibacterial activity of ruthenium(II) complexes 74, 75, and 76 (Fig. 6), with a focus on their efficacy against clinically critical bacteria. Compound 74 displayed poor solubility and minimal activity, while 75 and 76 exhibited significant antibacterial effects. Notably, 76 demonstrated superior activity in both glucose-defined minimal media and Mueller–Hinton II (MH-II), surpassing ampicillin against some strains. It was effective against multidrug-resistant E. coli EC958 and E. faecalis V583, with a notably high activity–toxicity ratio in human cells. Time–kill and MBC assays confirmed the potent bactericidal action of complex 76, which was attributed to membrane disruption, as supported by ICP-AES, stimulated emission depletion (STED) microscopy, and ATP leakage assays. The compound's selective targeting of bacterial membranes, coupled with low toxicity to human cells, emphasizes its potential as a novel antimicrobial agent. Further studies will refine our understanding of its mechanisms and optimize its therapeutic use.46 Together, these findings highlight the remarkable structural versatility and therapeutic potential of ruthenium(II)-based complexes, particularly dinuclear and multinuclear architectures, as promising alternatives to conventional antibiotics for combating multidrug-resistant bacterial infections.
4.7 Silver(I)-based complexes
Alongside ruthenium(II)-based PTMCs, silver(I)-based complexes have proved particularly effective against fungal pathogens, especially in disrupting biofilm formation in Candida species.47 Silver(I) complexes 77–79 (Fig. 7) have demonstrated significant antimicrobial activity, particularly against various Candida species. These complexes were synthesized via reactions with 1,5-naphthyridine in ethanol and characterized using NMR, IR, and UV-visible spectroscopy and single-crystal X-ray diffraction. The complexes exhibited minimal inhibitory concentration (MIC) values ranging from 0.78 to 6.25 μg mL−1 (2.6–20.8 μM) against Candida spp., markedly outperforming their antibacterial activity against Gram-positive and Gram-negative bacteria, where MICs were generally ≥12.5 μg mL−1. Notably, complexes 77 and 78 effectively inhibited biofilm formation by C. albicans, with complex 77 also reducing mixed biofilm formation with Pseudomonas aeruginosa. Toxicity studies in zebrafish embryos revealed that complex 79 had the most favorable therapeutic index and safety profile, showing no myelosuppressive or inflammatory responses.4a These findings highlight silver(I) complexes as potent antifungal agents with promising therapeutic potential, especially in targeting biofilm-associated Candida infections with minimal toxicity.4.8 Iridium(III)-based complexes
Expanding the scope further, iridium(III)-based complexes, although primarily bacteriostatic, represent an intriguing direction for future antimicrobial development.48 In a 2013 study by Keene and Collins, the antimicrobial activity of iridium(III) complexes was investigated, highlighting some interesting findings. Dinuclear iridium(III) complexes 80 and 81 (Fig. 7) demonstrated antimicrobial activity but were primarily bacteriostatic rather than bactericidal. Complex 80 exhibited significant activity with MIC values of 16 μg mL−1 against S. aureus and 8 μg mL−1 against E. coli; however, it required higher MBC values, indicating a lack of bactericidal action. Inert dinuclear iridium(III) complex 82 (Fig. 7) showed no antimicrobial activity, likely due to its inability to penetrate bacterial membranes as a result of its high 6+ charge. The study also noted that complexes 80 and 81 aquated rapidly, with a pKa of 6.0 for iridium(III)-bound water, suggesting that iridium(III) complexes might enter bacterial cells as hydroxo species.41 These results suggest that while iridium(III) complexes currently exhibit primarily bacteriostatic activity, further structural modifications to enhance membrane permeability and optimize reactivity could open the way for the development of more potent bactericidal iridium(III)-based antimicrobial agents.4.9 Platinum(II)-based complexes
Similarly, platinum(II)-based PTMCs, traditionally known for anticancer applications, have shown impressive antibacterial activity by disrupting essential bacterial functions.49 A series of pyrazolo[1,5-a]pyrimidine-based binuclear platinum(II) complexes 83–88 (Fig. 7) demonstrated significant antibacterial activity. Among them, complexes 84, 85, and 86, which contain the electron-withdrawing fluorine, chlorine, and nitro groups, respectively, showed enhanced efficacy against both Gram-positive and Gram-negative bacteria. Particularly complex 86, with a nitro group, exhibited the strongest antibacterial and antituberculosis activity; this was attributed to its high lipophilicity and ability to penetrate bacterial cell membranes. These complexes inhibit bacterial growth by disrupting essential cellular functions, such as cell wall synthesis, nucleic acid synthesis, and protein production, showcasing their potential as effective antimicrobial agents.504.10 Gold(III)-based complexes
Gold(III)-based PTMCs further enrich this landscape, offering broad-spectrum antimicrobial effects and high selectivity, particularly against Gram-positive strains.51 The incorporation of gold(III) centers within PTMCs enhances their ability to interact with biological targets, presenting a novel approach to antimicrobial therapy. In 2016, Glišić's group reported that dinuclear gold(III) complexes 89 and 90 (Fig. 7) demonstrated significant antibacterial potential. Both complexes were tested against a panel of Gram-positive and Gram-negative bacteria, where complex 89 showed particularly strong activity with MIC values ranging from 3.9 to 31.2 μg mL−1, outperforming the standard gold(III) complex K[AuCl4] and even showing comparable results to the antibiotic kanamycin. Complex 89 was especially effective against Gram-positive strains like Micrococcus luteus and Listeria monocytogenes, where its activity was 8- and 6.4-fold higher than that of K[AuCl4], respectively. Complex 90, while slightly less active than complex 89, still exhibited noteworthy antibacterial efficacy. Importantly, selectivity index values highlighted a clear distinction between the two: complex 89 had high SI values (up to 19.2), indicating strong selectivity for bacterial cells over human fibroblasts, while complex 90 had SI values close to or below 1, suggesting lower therapeutic selectivity and potential off-target cytotoxicity. Both complexes were largely inactive against fungal strains such as Candida albicans and exhibited moderate cytotoxicity on MRC5 human fibroblasts.52 These findings emphasize the potential of gold(III) complexes as promising antibacterial agents with notable selectivity, although further optimization is needed to minimize cytotoxicity and broaden their therapeutic applications.4.11 Mixed-metal-based complexes
Mixed-metal complexes, where various metal ions are present within a single structure, offer unique advantages by combining the distinct properties of each metal center.53 This synergy can enhance antimicrobial efficacy, making them promising candidates for the development of new therapeutic agents. In 2012, Contel's group reported the dinuclear gold(I) organometallic complex 91 (Fig. 8), water-soluble complex 92 (Fig. 8) and their heterometallic derivatives 93–98 (Fig. 8), which exhibited significant antibacterial activity. These complexes, particularly those with dppe(1,2-bis(diphenylphosphano)ethane) ligands (complexes 93, 95 and 97), showed strong efficacy against both Gram-positive and Gram-negative bacteria, with MICs ranging from 10 to 1 μg mL−1. The gold(I)–silver(I) compounds (e.g., complex 93) displayed potent activity, with complex 93 demonstrating remarkable effectiveness against Gram-positive Bacillus cereus at nanomolar concentrations.54 Dinuclear complex 99 (Fig. 8), incorporating ruthenium(II) and cisplatin units, demonstrates notable antibacterial activity. Although the complex binds DNA more effectively than cisplatin, it requires higher concentrations for bacterial growth inhibition. Agarose gel electrophoresis confirms significant DNA binding, with a marked retardation of DNA migration compared to cisplatin. Despite its lower in vivo efficacy, the complex's selective DNA targeting and enhanced binding profile suggest potential as a novel antibacterial agent. Additional research is needed to elucidate its full mechanism of action and optimize its therapeutic application.55 In 2013, Metzler-Nolte, Bandow and their group reported that hetero-tri-organometallic compounds ferrocene peptide nucleic acid (complex 100) (Fig. 8) and ruthenocene peptide nucleic acid (complex 101) (Fig. 8) exhibited potent antibacterial activity against Gram-positive bacteria, including multi-resistant Staphylococcus aureus. These organometallics consist of a peptide nucleic acid backbone with an alkyne side chain substituted with cymantrene, a (dipicolyl)Re(CO)3 moiety, and either ferrocene or ruthenocene. Comparative analysis revealed that the bacterial membrane was the primary target, with complex 100 accumulating in the membrane, as confirmed by manganese tracing. Both compounds disrupted essential cellular processes such as respiration and cell wall biosynthesis by altering the membrane architecture. Complex 100, in particular, induced oxidative stress due to the redox-active ferrocene moiety, enhancing its antibacterial potency. While both complexes 100 and 101 depolarized bacterial membranes, only complex 100 triggered significant oxidative stress, explaining its superior antibacterial activity. These compounds demonstrated bactericidal activity without causing cell lysis and exhibited limited cytotoxicity within their solubility range. Given the strong antibacterial efficacy and membrane-targeting mechanism, complex 100 represents a promising lead for the development of novel antibiotics targeting resistant Gram-positive pathogens, though further studies are needed to improve solubility for clinical application.56 In 2015, they reported trimetallic compound 102 (Fig. 8), containing a ferrocenyl (Fc), a CpMn(CO)3 (cymantrene), and a [(dpa)Re(CO)3] residue, which demonstrated potent antibacterial activity, particularly against Gram-positive bacteria, including methicillin-resistant Staphylococcus aureus. Subsequent structure–activity relationship studies revealed that the [(dpa)Re(CO)3] moiety was essential for antibacterial efficacy, while the ferrocenyl and CpMn(CO)3 units could be replaced by organic moieties such as phenyl groups without loss of activity. Compounds 103 and 104 (Fig. 8), which contained the [(dpa)Re(CO)3] moiety alongside either a ferrocenyl or CpMn(CO)3 unit, were also highly effective, suggesting that the {Re(CO)3} core was crucial for activity. The mono-metallic derivative 105 (Fig. 8), containing only the [(dpa)Re(CO)3] fragment, showed antibacterial activity comparable to that of conventional drugs like amoxicillin and norfloxacin, and crucially, exhibited moderate cytotoxicity against mammalian cells. These active derivatives, including complex 105, were shown to disrupt bacterial membranes and compromise cell wall integrity, similarly to complex 102, but with enhanced membrane permeabilization. Additionally, complex 105 displayed significantly higher solubility and selective toxicity, being 15–30 times more toxic to bacteria than mammalian cells. Although inactive against Gram-negative bacteria, the promising selectivity and membrane-targeting mechanism of complex 105, combined with its favourable solubility and low mammalian toxicity, make it a strong candidate for further development in the fight against resistant Gram-positive pathogens.58 These findings highlight the immense potential of mixed-metal-based complexes as versatile and highly effective antibacterial agents, particularly against resistant Gram-positive pathogens, by leveraging unique metal synergies and membrane-targeting mechanisms.4.12 Biological functions of metal centers and mechanistic insights
The antimicrobial activity of PTMCs is closely related to the intrinsic biological properties of their constituent metal centers. Each metal contributes a distinct mode of action, such as redox reactivity, membrane interaction, or enzymatic inhibition, which often surpasses the mechanisms of traditional antibiotics. Understanding these individual roles is key to elucidating the mechanistic diversity of PTMCs and guiding the rational design of next-generation antimicrobial agents.Among these, iron(II), particularly in ferrocenyl-based architectures, exhibits redox activity that facilitates the generation of reactive oxygen species. This leads to oxidative damage in bacterial DNA, proteins, and membranes. For example, complex 4 (Fig. 2) demonstrated strong antimycobacterial activity (MIC90 = 19.2 μM), which was attributed to enhanced redox properties and membrane penetration facilitated by Schiff base coordination.23 Building on redox-active systems, cobalt(II) complexes, such as complex 17 (Fig. 2), exhibit selective efficacy against Agrobacterium tumefaciens.26 Their biological action is believed to stem from ROS generation and direct membrane interaction, aided by their planar geometry and lipophilic nature. These features make them especially suitable for agricultural pathogen control. In a similar fashion, nickel(II) complexes enhance antimicrobial potency by increasing lipophilicity and promoting chelation with biomolecular targets. Complex 18 (Fig. 2) was particularly effective against Salmonella typhi, while complex 20 (Fig. 2) showed activity against Mycobacterium tuberculosis, possibly by interfering with fatty acid biosynthesis pathways essential for bacterial viability.28,29 Copper(II) complexes, widely recognized for their broad-spectrum antimicrobial properties, exert their effects through multiple pathways. For example, complex 29 (Fig. 3) acts via membrane permeation and inhibition of protein synthesis.32 Complex 35 (Fig. 3) targets bacterial quorum sensing, while multicopper clusters 40 and 41 (Fig. 4) exhibit strong activity against MRSA by inducing lipid peroxidation through ROS generation.17,34 Unlike redox-active metals, zinc(II) is redox-inert but still exerts strong antimicrobial effects through other mechanisms. It readily interacts with anionic bacterial membranes, disrupting the membrane potential and integrity. Complex 42 (Fig. 5), for instance, selectively depolarizes Gram-positive bacterial membranes while sparing mammalian cells, lending itself to dual imaging and therapeutic applications.15b Other zinc(II) complexes, such as 49 and 52 (Fig. 5), demonstrate antibacterial effects through DNA intercalation and enhanced lipophilicity.36,37 Ruthenium(II)-based PTMCs provide another layer of multifunctionality. Complexes like 54–56 (Fig. 6) simultaneously bind DNA and ribosomes, disrupt membranes, and offer luminescence properties for imaging.39 Higher-nuclearity complexes, such as 64 and 65 (Fig. 6), further enhance DNA affinity and cellular take up, improving their efficacy against resistant strains.43 Silver(I) complexes (e.g., complexes 77–79) (Fig. 7) are particularly effective against fungal biofilms, notably those formed by Candida species.4a Their mechanism involves the disruption of thiol-containing enzymes, ROS production, and membrane destabilization, resulting in potent antifungal and antibiofilm activity. Iridium(III) complexes such as 80 and 81 (Fig. 7), although less explored, demonstrate bacteriostatic properties through aquation and membrane disruption.41 Future developments may further improve their efficacy by increasing nuclearity or lipophilicity. Platinum(II) complexes, often inspired by anticancer agents, work primarily by crosslinking bacterial DNA and disrupting essential biosynthetic pathways. Complexes 84–86 (Fig. 7) exemplify this dual interaction with nucleic acids and proteins, leading to cell death.50 Gold(III) complexes such as 89 and 90 (Fig. 7) leverage enzymatic inhibition and redox imbalance to exert antimicrobial effects across Gram-positive and Gram-negative bacteria.52 Their performance frequently exceeds that of gold salts and, in some cases, matches or surpasses traditional antibiotics. Finally, mixed-metal PTMCs integrate the benefits of multiple metal ions to achieve synergistic effects. Complexes such as 93 (Au–Ag) and 100–102 (Fe–Mn–Re or Fe–Ru–Re) (Fig. 8) combine redox activity, membrane targeting, and oxidative stress induction to combat multidrug-resistant pathogens effectively.56,57
These diverse metal-centered mechanisms emphasize the versatility of PTMCs in targeting bacterial membranes, DNA, enzymes, and signaling pathways. The integration of multiple antibacterial modalities within a single molecular framework offers a promising platform to overcome current resistance mechanisms and develop effective theranostic agents.
5. Overcoming antimicrobial resistance
The diverse intrinsic antimicrobial activities of PTMCs highlight their immense potential as alternative agents against resistant pathogens. However, to fully realize their clinical promise, it is essential to understand how PTMCs can overcome the growing challenge of antimicrobial resistance, which remains a major hurdle in modern medicine.1c This increasing problem has prompted the hunt for new therapeutic techniques, with PTMCs appearing as a viable option. Because of their unique capacity to target bacteria via various, often unusual routes, PTMCs hold promise for fighting multidrug-resistant infection by potentially bypassing recognized defensive tactics of bacteria.5.1. Hydrophobicity and membrane penetration
The capacity of PTMCs to penetrate bacterial membranes is critical to their antibacterial action.11 This is especially essential in the case of Gram-negative bacteria, the outer membrane of which presents a severe barrier to many therapeutic medicines.58 Hydrophobicity is key in this process. Hydrophobic molecules prefer to connect with nonpolar substances rather than water, allowing them to integrate into and cross the hydrophobic core of bacterial lipid membranes. This feature enables PTMCs to permeate bacterial membranes, alter membrane integrity, and exert antibacterial actions.Hydrophobicity has a prominent impact on investigations involving lipophilic zinc(II) coordination complexes 42–45 (Fig. 5). Complexes 44 and 45, for example, have high hydrophobicity, allowing them to readily integrate into both zwitterionic and anionic vesicles (such as POPC and POPG/POPC vesicles). This integration is required for these complexes to interact with lipid membranes, resulting in phospholipid translocation and the leakage of intracellular components such as carboxyfluorescein, which indicates membrane rupture. In contrast, more hydrophilic complexes, such as 42 and 43, have limited ability to partition into zwitterionic membranes but selectively bind with anionic bacterial membranes. Despite its hydrophilic nature, compound 42 successfully targets bacterial membranes while causing minimal harm to mammalian cells. This selective targeting highlights the zinc(II) coordination complexes’ potential as both luminescent probes and antibacterial medicines for bacterial imaging. These findings demonstrate the importance of hydrophobicity in mediating PTMC–bacterial membrane interactions and regulating antibiotic effectiveness.15b
Hydrophobicity also proves essential in the action of ruthenium(II)-based PTMCs, such as compound 76 (Fig. 6), which demonstrates remarkable antimicrobial efficacy against multidrug-resistant Gram-negative bacteria like Escherichia coli strain EC958 ST131. The hydrophobic nature of compound 76 enhances its ability to penetrate bacterial membranes, as observed through super-resolution STED nanoscopy, which shows the complex initially localizing at bacterial membranes before migrating to cell poles, causing significant membrane damage. This membrane disruption is concentration-dependent, as demonstrated in time–kill assays and ATP leakage experiments, further confirming the role of hydrophobicity in the mechanism of action. Importantly, compound 76 exhibits no toxicity to human cells or Galleria mellonella larvae, highlighting its selectivity for bacterial cells and its potential as a therapeutic agent against resistant pathogens. This example shows the pivotal role of hydrophobicity in facilitating membrane penetration and enhancing the overall antimicrobial efficacy of PTMCs.46
5.2. Structural design for resistance evasion
While hydrophobicity plays a crucial role in enabling membrane penetration, the structural design of PTMCs is equally important in overcoming bacterial resistance mechanisms.58 Strategic design features such as nuclearity and charge distribution can have a substantial impact on PTMCs’ capacity to bypass bacterial defenses and exert strong antibacterial action.For instance, dinuclear ruthenium(II) complexes, such as ΔΔ/ΛΛ-54, 55, and 56 (Fig. 6), have high antibacterial action, notably against Gram-positive bacteria like Staphylococcus aureus and methicillin-resistant Staphylococcus aureus. These complexes have MICs as low as 1 μg mL−1. A critical factor in their efficacy is their lipophilicity, as measured by the octanol–water partition coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P). Higher lipophilicity correlates with enhanced membrane penetration, as seen in compound 56, which is notably potent compared to its less lipophilic counterparts. However, the activity of these dinuclear complexes is not solely dependent on hydrophobicity. Their dinuclear structure plays a crucial role, as evidenced by the lower efficacy of mononuclear analogs like complex 57 (Fig. 6). This demonstrates the importance of charge distribution and the specific architecture of the complexes in achieving effective membrane penetration and subsequent antimicrobial activity. Structural design, in this case, provides a way to bypass bacterial resistance mechanisms by combining lipophilicity with a dinuclear framework that facilitates interaction with bacterial membranes.39
P). Higher lipophilicity correlates with enhanced membrane penetration, as seen in compound 56, which is notably potent compared to its less lipophilic counterparts. However, the activity of these dinuclear complexes is not solely dependent on hydrophobicity. Their dinuclear structure plays a crucial role, as evidenced by the lower efficacy of mononuclear analogs like complex 57 (Fig. 6). This demonstrates the importance of charge distribution and the specific architecture of the complexes in achieving effective membrane penetration and subsequent antimicrobial activity. Structural design, in this case, provides a way to bypass bacterial resistance mechanisms by combining lipophilicity with a dinuclear framework that facilitates interaction with bacterial membranes.39
In addition to dinuclear structures, ruthenium(II) polypyridyl complexes 62–66 (Fig. 6) have shown promise in overcoming bacterial resistance. For example, trinuclear and tetranuclear ruthenium(II) complexes (such as complexes 62, 63, 64, and 65) display significant antimicrobial activity. These complexes exhibit varying degrees of hydrophobicity, again measured by their log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values, with higher nuclearity complexes generally being more lipophilic. Interestingly, tetranuclear linear complexes (e.g., complex 64) exhibit superior antimicrobial activity despite being less lipophilic than their trinuclear counterparts. This superior activity is likely due to their enhanced ability to penetrate bacterial membranes and align with cellular targets, such as DNA, contributing to their potent antimicrobial effects. In contrast, non-linear tetranuclear complexes (e.g., complex 66) have lower activity despite accumulating at higher levels within bacterial cells. This discovery emphasizes the important interaction between structural conformation and membrane penetration, as well as the need for aligning the structure of the complex with its cellular targets in order to maximize antimicrobial effectiveness. The findings highlight the importance of hydrophobicity and structural design in increasing the therapeutic potential of ruthenium(II)-based antibacterial medicines.43
P values, with higher nuclearity complexes generally being more lipophilic. Interestingly, tetranuclear linear complexes (e.g., complex 64) exhibit superior antimicrobial activity despite being less lipophilic than their trinuclear counterparts. This superior activity is likely due to their enhanced ability to penetrate bacterial membranes and align with cellular targets, such as DNA, contributing to their potent antimicrobial effects. In contrast, non-linear tetranuclear complexes (e.g., complex 66) have lower activity despite accumulating at higher levels within bacterial cells. This discovery emphasizes the important interaction between structural conformation and membrane penetration, as well as the need for aligning the structure of the complex with its cellular targets in order to maximize antimicrobial effectiveness. The findings highlight the importance of hydrophobicity and structural design in increasing the therapeutic potential of ruthenium(II)-based antibacterial medicines.43
PTMCs offer a novel and promising approach for reducing antimicrobial resistance, especially in light of the mounting challenges posed by multidrug-resistant bacteria. PTMCs have tremendous potential as antibacterial agents and diagnostic tools because they use hydrophobicity to improve membrane penetration and careful structural design to avoid bacterial resistance mechanisms. The combination of these factors, efficient membrane contact, selective targeting of bacterial cells, and avoidance of conventional resistance pathways, makes PTMCs a promising class of therapeutic medicines in the ongoing fight against antibiotic resistance. Additional research on enhancing these qualities could lead to new, more effective treatments for resistant infections.
Beyond combating antimicrobial resistance, real-time detection and monitoring of bacteria are essential for effective infection management. Polynuclear transition metal complexes, with their unique photophysical properties, offer promising tools for rapid bacterial imaging. The next section highlights recent advances in this area.
6. Bacterial imaging
Accurate bacterial imaging is critical for diagnosing infections and investigating bacterial activity.59 Traditional approaches, such as culturing and PCR, are time-consuming and impracticable for real-time analysis.60 PTMCs have recently emerged as a promising alternative, with unique luminescence characteristics and the ability to selectively target bacterial cells. These compounds improve luminescence, photostability, and structural flexibility, thereby overcoming the limits of traditional emissive dyes that stain both bacterial and mammalian cells. PTMCs provide for fast and precise imaging, making them excellent for detecting bacterial cells in complex situations such as mixed microbial populations or host tissues.PTMCs offer a wide range of applications, including medical diagnostics, environmental microbiology, and the study of bacterial processes like biofilm development and pathogen invasion. In this perspective, we look at significant breakthroughs in PTMC-based bacterial imaging, with an emphasis on different types of metal complexes and the particular features that make them useful tools in microbiology. A summary of these PTMCs, namely, their metal centers, target bacteria, imaging techniques, and key features is provided in Table 2.
| Complex | Metal center | Target bacteria | Imaging technique | Key features | Ref. |
|---|---|---|---|---|---|
| 106, 107 | Zn(II) | E. coli, P. aeruginosa, S. aureus | Epifluorescence microscopy | Selective bacterial membrane targeting, no DNA staining | 15c |
| 46 | Zn(II) | S. aureus, E. coli | Fluorescence imaging | Highlights binary fission in bacteria | 15b |
| 54, 56 | Ru(II) | E. coli, S. aureus, MRSA | Confocal microscopy | High bacterial take up, membrane and intracellular binding | 40 |
| 56 | Ru(II) | E. coli | Wide-field fluorescence microscopy | Binds ribosomes, inhibits protein synthesis | 61 |
| 108 | Zn(II) | B. subtilis, E. coli | CLSM (confocal laser scanning microscopy) | Wash-free imaging, highly luminescent in bacterial membranes | 62 |
| 103–105 | Re(I) | B. subtilis | Light microscopy | Targets bacterial membrane, induces depolarization | 57 |
| 109 | Zn(II) | B. subtilis, E. coli | Confocal microscopy | Aggregation-induced emission, selective for bacteria | 63 |
| 76 | Ru(II) | E. coli | SIM (structured illumination microscopy), STED | Accumulates in bacterial membranes, causes disruption | 46 |
| 110–112 | Ir(III)–Pt(II) | E. coli | Confocal microscopy | pH-sensitive imaging in acidic environments (e.g., the gut) | 64 |
Smith's group made one of the field's first breakthroughs in 2006, when they presented zinc(II) complexes coordinated to dipicolylamine. These complexes target bacterial cells specifically by attaching to anionic phospholipids on bacterial membranes, which distinguishes bacteria from mammalian cells. Smith's team developed two fluorophore–dipicolylamine–zinc(II) conjugates, compounds 106 and 107 (Fig. 9a), with exceptional selectivity for bacterial membranes. Compound 106, based on anthracene, showed a considerable increase in fluorescence when bound to bacterial membranes. This enabled efficient imaging of bacterial cells without the need for further washing processes. Compound 107, which included a dansyl fluorophore, stained bacterial membranes but produced more background fluorescence. Both compounds were tested on Gram-negative Escherichia coli and Pseudomonas aeruginosa, as well as Gram-positive Staphylococcus aureus (Fig. 9b). The results showed selective staining of bacterial membranes without targeting intracellular DNA, indicating their potential for distinguishing bacterial cells from mammalian cells in complex environments like saliva. This discovery laid the groundwork for selective bacterial imaging in theranostic applications.15c In 2008, the same research group reported the development of fluorescent complex 46, (Figs. 5 and 9c) which effectively stained both Gram-positive Staphylococcus aureus and Gram-negative E. coli (Fig. 9d). This complex localized to bacterial cell walls and was able to reveal dynamic cellular processes such as binary fission. The study highlighted the advantages of zinc(II) coordination complexes for bacterial detection and imaging, providing insights into bacterial structure and function.15b In 2012, Keene, Collins, and their team turned their attention to dinuclear ruthenium(II) complexes, which demonstrated both antibacterial activity and promising imaging capabilities. The researchers focused on two key complexes, 54 and 56, which were shown to be taken up by bacterial cells, with compound 56 demonstrating particularly high cellular take up. Using confocal microscopy, they observed that E. coli cells incubated with compound 56 (Fig. 6 and 9e) showed a strong presence of the complex on the cell surface or inside the cytoplasm (Fig. 9f). This take up correlated with the complex's strong antimicrobial activity, especially against Gram-positive strains like Staphylococcus aureus and methicillin-resistant S. aureus. The antimicrobial activity of these complexes was concentration-dependent, with 99.9% of bacterial cells being killed within 2–6 h. Interestingly, while the bacteria remained redox-active after exposure to compound 56, they lost the ability to divide, suggesting that the complex disrupted vital cellular processes. These results indicate that dinuclear ruthenium(II) complexes could represent a new class of antimicrobial agents, driven by their ability to penetrate bacterial cells and interfere with key metabolic activities.40 In 2014, Keene and Collins continued their investigation into ruthenium(II) complexes, revealing that compound 56 selectively bound to bacterial ribosomes, particularly when they were aggregated into polysomes (Fig. 9g). This selective binding halts protein synthesis, effectively inhibiting bacterial growth. Localization studies using RNA-binding dyes and ribosome-disrupting antibiotics confirmed that the complex bound preferentially to RNA in ribosomes rather than chromosomal DNA. This selective toxicity toward bacterial cells, along with reduced cytotoxicity in mammalian liver and kidney cells, suggests the potential for therapeutic applications targeting bacterial infections.61 In 2014, Liu's group introduced novel zinc(II) complex 108 (Fig. 9h), which exploited aggregation-induced emission and excited-state intramolecular proton transfer properties for selective bacterial imaging. This complex was shown to selectively stain both Gram-positive Bacillus subtilis and Gram-negative E. coli with strong fluorescence and low background noise (Fig. 9i). One of the key advantages of compound 108 is its ability to accumulate on bacterial membranes, activating its fluorescence without the need for additional washing steps. This feature allows for real-time imaging of bacterial cells directly in culture media, with mammalian cells remaining unstained. This selectivity makes compound 108 a powerful tool for distinguishing bacteria from healthy mammalian cells in mixed cultures, enhancing its potential for medical diagnostics and microbiological studies.62
 | ||
| Fig. 9 (a) Structures of complexes 106 and 107. (b) E. coli cells were stained with either complex 106 or complex 107 along with 7AAD. Panels A and C display the blue and green fluorescence from complex 106 and complex 107, respectively, while panels B and D show the overlay of 7AAD co-staining, indicating the membrane and DNA localization (1500× magnification). S. aureus cells were stained with complex 106 (panel E) and complex 107 (panel F). Reproduced from ref. 15c with permission from the Royal Society of Chemistry, copyright 2006. (c) Structure of complex 46. (d) Fluorescence images of E. coli and S. aureus (bottom) after treatment with zinc(II) complex 46, with the right panels zooming in on single cells in binary fission. Each image has a 2 μm scale bar. Reproduced from ref. 15b with permission from Wiley-VCH, copyright 2008. (e) Structure of complex 56. (f) Confocal microscopy images of E. coli incubated with 16 mg L−1 of complex 56 for 2 h and stained with BacLight RedoxSensor Green dye. Images show staining by complex 56 (A) and the BacLight RedoxSensor Green dye (B). Reproduced from ref. 40 with permission from Oxford Academic, copyright 2012. (g) Localization of complex 56 in E. coli MG1665 cells at MIC of 4 μg mL−1, with images showing (A) phase-contrast, (B) phosphorescence, and (C) a merged view. The scale bar is 5 μm. Reproduced from ref. 61 with permission from the Royal Society of Chemistry, copyright 2014. (h) Structure of complex 108. (i) CLSM images show bright field and fluorescent views of Gram-positive B. subtilis (A and B) and Gram-negative E. coli (C and D) treated with complex 108 (20 μM), with the fluorescence of complex 108 shown in green. Panel D presents I/I0 for B. subtilis and E. coli after treatment with complex 108 (20 × 10−6 M) under irradiation, where I0 and I refer to fluorescence intensities before and after light exposure (λex = 405 nm, λem = 515–560 nm). Reproduced from ref. 62 with permission from Wiley-VCH, copyright 2015. | ||
In 2015, Bandow and Metzler-Nolte's team explored the antimicrobial potential of organometallic complexes, particularly those based on the rhenium(I) tricarbonyl moiety {Re(CO)3} (103–105) (Figs. 8 and 10a). They focused on compound 105, which demonstrated significant antibacterial activity against Gram-positive pathogens, including MRSA. The mechanism of action involved targeting bacterial membranes and disrupting cell wall integrity. The team used advanced imaging techniques to observe that compounds 103, 104, and 105 caused depolarization of bacterial membranes and induced permeabilization, leading to bacterial cell death (Fig. 10b). Microscopy revealed membrane extrusions in treated cells, confirming the disruption of cell wall biosynthesis. Notably, compound 105 stood out for its enhanced solubility and lower cytotoxicity against mammalian cells, making it a promising candidate for further antibacterial development.57 In 2017, Liu's group introduced compound 109 (Fig. 10c), a zinc(II)-tetradentate probe that utilized aggregation-induced emission (AIE) for selective bacterial imaging. This probe binds to bacterial membranes, enhancing fluorescence without the need for post-staining washes (Fig. 11). Its selective interaction with negatively charged bacterial membranes allows for the clear differentiation of bacteria from mammalian cells, making it an effective tool for visualizing Gram-positive Bacillus subtilis and Gram-negative E. coli in real-time (Fig. 10d).63
 | ||
| Fig. 10 (a) Structures of complexes 103–105. (b) Effects of complex 103, complex 104, and complex 105 on B. subtilis membrane depolarization, membrane permeabilization, and cell wall integrity: (A) GFP-MinD localization post-antibiotic treatment, showing membrane depolarization; (B) light microscopy images of the corresponding cells; (C) antibiotic-treated cells stained with BacLight, where red/orange cells indicate pore formation; and (D) antibiotic-stressed cells fixed with acetic acid and methanol, revealing membrane excrescences from cell wall biosynthesis inhibition. Reproduced from ref. 57 with permission from the Royal Society of Chemistry, copyright 2015. (c) Structure of complex 109. (d) Confocal (A–C) and bright-field (D–F) images of E. coli (A and D), B. subtilis (B and E), and HeLa cells (C and F) incubated with probe 109 (20 μM) for 30 min without washing. Red fluorescence, excited at 458 nm and collected above 590 nm, is shown in all images with a 10 μm scale bar. Reproduced from ref. 63 with permission from the American Chemical Society, copyright 2018. | ||
 | ||
| Fig. 11 (a) Structure of complex 76. (b) SIM images of E. coli EC958 cells treated with complex 76 at 5, 20, and 60 min, showing emission from complex 76 (A), phase contrast (B), and a combined view (C). After treatment with 0.8 μM complex 76, cells were washed with nitric acid and fixed with paraformaldehyde (16% v/v). Reproduced from ref. 46 with permission from the American Chemical Society, copyright 2019. (c) Structures of complexes 110–112. (d) pH-dependent staining of E. coli (DH5α) with complexes 110–112 (scale bar: ∼5 μm; λ = 630 nm). Reproduced from ref. 64 with permission from the Royal Society of Chemistry, copyright 2023. | ||
In 2019, Thomas and his team focused on ruthenium(II) complex 76 (Figs. 6 and 12a), which exhibited potent antimicrobial activity against both Gram-negative and Gram-positive bacteria, particularly multidrug-resistant E. coli strains. Advanced imaging techniques, such as structured illumination microscopy (Fig. 12b) and STED microscopy, revealed that the complex accumulated rapidly in bacterial cells and disrupted membrane integrity. Transmission electron microscopy (TEM) confirmed the extent of membrane damage, which led to cell lysis. The complex also demonstrated low toxicity in human cells and proved to be effective in a Galleria mellonella model, showing the potential of ruthenium(II) complexes as antibacterial agents with exceptional imaging capabilities.46 In 2023, our research group developed heterobimetallic iridium(III)–platinum(II) complexes (110, 111, and 112) (Fig. 12c) that exhibited promising aggregation-induced photoluminescence emission for bacterial imaging. These complexes are water-soluble and feature a bridging Schiff base ligand, which results in enhanced fluorescence under acidic conditions (pH 1.2 and 3). These properties enable the complexes to selectively stain E. coli cells in low-pH environments, such as simulated gut fluids (Fig. 12d). While these complexes did not exhibit notable bactericidal activity, their ability to visualize bacteria under acidic conditions highlights their potential for non-invasive, pH-sensitive bacterial imaging in biological systems. Complex 112 maintained its staining capability even at neutral pH, further broadening its potential applications in bacterial imaging.64
 | ||
| Fig. 12 (a) Structure of complex 108. (b) Schematic illustration of selective targeting, imaging, and bactericidal activity, preferentially affecting bacteria over mammalian cells, of complex 108. Note: cell and bacterial sizes are not to scale. (c) Morphological changes in (A–C) B. subtilis and (D–F) E. coli following incubation with complex 108: (A and D) without complex 108 in the dark; (B and E) with complex 108 (20 × 10−6 M) in the dark; (C and F) with complex 108 (20 × 10−6 M) and white light irradiation (100 mW cm−2, 6 min). Reproduced from ref. 62 with permission from Wiley-VCH, copyright 2015. (d) Structure of complex 109. (e) SEM images of E. coli and B. subtilis after treatment with complex 109, with or without light irradiation. The scale bar for all images is 1 μm. Reproduced from ref. 63 with permission from the American Chemical Society, copyright 2017. | ||
Polynuclear transition metal complexes have revolutionized bacterial imaging by offering selective targeting, enhanced fluorescence, and photostability. From zinc(II) and ruthenium(II) complexes that target bacterial membranes to rhenium(I) complexes with antimicrobial properties, these complexes provide powerful tools for real-time, precise bacterial visualization. Their ability to distinguish bacterial cells from mammalian cells, coupled with their unique luminescence properties, positions PTMCs as indispensable assets in medical diagnostics, environmental microbiology, and bacterial process studies. As research continues, the potential for PTMCs in theranostic applications is vast, promising exciting advances in bacterial detection and treatment.
Building on their imaging capabilities, polynuclear transition metal complexes are increasingly being explored for integrated theranostic applications, where diagnosis and therapy are combined in a single platform. The following section discusses the evolving role of PTMCs in theranostics, offering innovative strategies for simultaneous bacterial detection and treatment.
7. Antimicrobial photodynamic therapy (aPDT)
Antimicrobial photodynamic treatment is a potential method that uses a photosensitizer (PS), light, and molecular oxygen to produce ROS, which can efficiently kill pathogens.65 Unlike typical antibiotics, aPDT targets various bacterial structures, both internal and external, lowering the risk of resistance development.66Recently, researchers have turned to PTMCs, which are known for their excellent photophysical characteristics and high ROS production capacity.3 In this section, we describe recent advances in PTMCs, with a particular emphasis on complexes comprising zinc(II), ruthenium(II), iridium(III), palladium(II), and platinum(II) and their applications in aPDT. A summary of these PTMCs, namely their metal centers, target bacteria, and key features, is provided in Table 3.
| Complex | Metal center | Target bacteria | Key features | Ref. |
|---|---|---|---|---|
| 108 | Zn(II) | B. subtilis, E. coli | Binds bacterial membranes, triggers ROS upon light exposure | 62 |
| 109 | Zn(II) | B. subtilis, E. coli | Aggregation-induced emission, membrane targeting, high ROS generation | 63 |
| 113 | Ru(II) | S. mutans, S. aureus | High phototherapeutic index, excellent bacterial take up | 67 |
| 114, 115 | Ru(II), Zn(II) | M. abscessus, M. fortuitum | Zinc(II) enhances singlet oxygen generation | 68 |
| 116, 117 | Pd(II) | M. abscessus, M. fortuitum, M. smegmatis, M. massiliense | Strong bactericidal activity, mycobacterial targeting | 69 |
| 118 | Ru(II)–Pt(II) | Multidrug-resistant bacteria | Dual action: ROS production and DNA damage | 70 |
| 119 | Ir(III) | MRSA | Effective against resistant bacteria, membrane oxidative damage | 71 |
| 120–122 | Pt(II) | E. coli | Self-assembly with TAT-peptide, enhances bacterial adhesion and killing | 72 |
One important problem in aPDT is to selectively target bacterial cells while causing minimal damage to mammalian cells. To address this issue, Liu's team developed compound 108 (Figs. 9h and 12a), a highly effective photosensitizer for aPDT. The positively charged zinc(II)–dipicolylamine unit was deliberately chosen to selectively target negatively charged bacterial membranes via electrostatic interactions, ensuring selectivity over mammalian cells. This compound exhibits minimal fluorescence in aqueous environments but becomes highly emissive when it aggregates on bacterial membranes, facilitating selective bacterial imaging. Its light-triggered generation of reactive oxygen species plays a key role in its antimicrobial efficacy. ROS production by compound 108 was confirmed through reduced absorbance of 2′,7′-dichlorofluorescin upon light irradiation, indicating its robust ROS generation capacity. It boasts a high singlet oxygen quantum yield, which significantly contributes to its potent bactericidal effects. In studies, compound 108 effectively killed both Gram-positive Bacillus subtilis and Gram-negative Escherichia coli, demonstrating strong phototoxicity against a broad range of bacteria (Fig. 12b and c). Notably, it remains non-toxic to mammalian cells, making it a highly selective antimicrobial agent.62 Building on the aggregation-induced emission principle, zinc(II)-tetradentate-coordinated probe 109 (Figs. 10c and 12d) was also developed. The design integrates a red-emissive AIE-active fluorogen with a high-affinity zinc(II)-binding cyclen-based ligand via CuAAC click chemistry. This structure ensures tight and stable coordination with zinc(II), overcoming the limitations of previously reported DPA-based systems with lower binding affinities. The strong electrostatic interactions between dicationic complex 109 and negatively charged bacterial membranes promote selective binding and aggregation on bacterial surfaces, leading to fluorescence activation and targeted imaging. Moreover, probe 109 exhibits exceptional ROS generation, achieving a singlet oxygen quantum yield of 77.6%. In bacterial inactivation studies, the probe showed potent effects under light irradiation, particularly against Gram-positive Bacillus subtilis and Gram-negative Escherichia coli, the latter of which was more resistant due to its protective outer membrane (Fig. 12e). Importantly, probe 109 demonstrated minimal toxicity to mammalian cells like HeLa cells, both under dark and light conditions. This combination of selective binding, strong ROS generation, and minimal toxicity highlights its potential in theragnostic applications, where bacterial imaging and treatment can be integrated.63
Building on the promising results from these AIE-based probes, polynuclear ruthenium(II) complexes have also emerged as highly efficient photosensitizers in aPDT due to their strong visible light absorption and high ROS production. For instance, studies on dinuclear and trinuclear ruthenium(II) complexes using the bis[pyrrolyl ruthenium(II)] triad scaffold have demonstrated potent photodynamic efficacy. One standout example is compound 113 (Fig. 13), which features a pyrenyl linker and has shown excellent antibacterial activity against strains such as Streptococcus mutans and Staphylococcus aureus. The complex exhibited a high phototherapeutic index (PI), indicating its strong selectivity and effectiveness.67 Additionally, the incorporation of ruthenium(II) into porphyrin structures has shown potential for further enhancing aPDT. Complex 114 and its zinc(II) derivative 115 (Fig. 13) were evaluated for their antimicrobial properties, with complex 115 outperforming its counterpart for both singlet oxygen generation and fluorescence. This difference highlights the importance of optimizing the photophysical properties of ruthenium(II)-based photosensitizers to achieve maximum efficacy in antimicrobial applications. While PTMCs are highly effective against a range of bacterial species, their application in the treatment of mycobacterial infections is particularly promising. Mycobacteria, especially Mycobacteroides abscessus and Mycolicibacterium fortuitum, are notoriously difficult to treat and are often misdiagnosed as tuberculosis.68 In this context, tetra-cationic porphyrins, such as meta-substituted isomer 116 (Fig. 13), have shown significant bactericidal effects under white LED irradiation. The effectiveness of compound 116 was evident in its substantial reduction of the minimum inhibitory concentration compared to para isomer 117 (Fig. 13). Singlet oxygen produced during photoinactivation caused severe damage to the bacterial cell walls, as confirmed by atomic force microscopy (AFM). In addition to altering the bacterial morphology, complex 116 affected the nanomechanical and electrostatic properties of the bacteria, enhancing adhesion and facilitating greater interaction with the AFM tip. These results suggest that incorporating aPDT into conventional antimycobacterial therapies could shorten treatment times and improve outcomes for patients.69 To further enhance the antibacterial effects of aPDT, researchers have explored the combination of different metal centers. Ruthenium(II)–platinum(II) bimetallic complex 118 (Fig. 13) represents a novel approach for improving aPDT efficacy. Compound 118 combines a ruthenium(II)-based chromophore with a cis-PtCl2 bioactive site, which allows it to target bacterial DNA, causing covalent modifications that lead to increased bacterial cell death. Upon light activation, the ruthenium(II)–platinum(II) complex generates ROS while also binding to bacterial DNA, making it highly effective against multidrug-resistant bacteria. This dual mechanism positions ruthenium(II)–platinum(II) complexes as powerful alternatives to traditional antibiotics.70
In addition to bimetallic complexes, trinuclear cyclometalated iridium(III) complex 119 (Fig. 14a) has also shown exceptional antibacterial properties. Compound 119 has demonstrated potent activity against MRSA (methicillin-resistant Staphylococcus aureus). The complex generates high levels of ROS upon light activation, which cause significant oxidative damage and bacterial cell death (Fig. 14b). Notably, complex 119 shows a high safety profile in mammalian cells, making it a strong candidate for future clinical applications in antimicrobial PDT.71
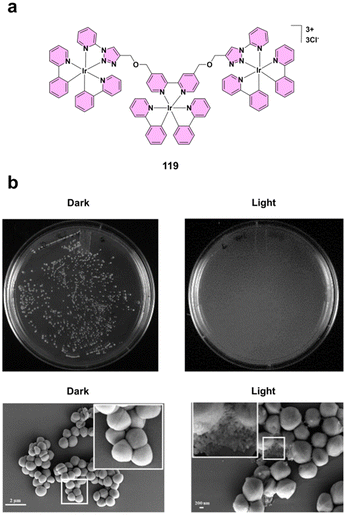 | ||
| Fig. 14 (a) Structure of complex 119. (b) Upper panel: impact on the viability of MRSA (as measured by colony-forming units, CFU) following treatment with complex 119, with and without photoirradiation. Without photoirradiation, complex 119 (25 μg mL−1) showed no significant effect on MRSA viability, confirming its “bacteria-safe” property. However, under photoirradiation, complex 119 (25 μg mL−1) caused a marked reduction in MRSA, demonstrating a positive photodynamic therapy (PDT) effect. Representative images of MRSA colonies post-treatment with complex 119 (25 μg mL−1) with or without photoirradiation are shown. All error bars represent the standard error of the mean (mean ± SEM) (n = 6) from two biological replicates. Lower panel: MRSA cells exhibited no significant morphological changes when treated with complex 119 (25 μg mL−1) without photoirradiation, but clear cellular damage was observed when photoirradiation was applied, indicating a successful PDT effect. Reproduced from ref. 71 with permission from Wiley-VCH, copyright 2024. | ||
Another exciting development in aPDT is the use of tetraphenylethylene-based metallacycles to enhance bacterial membrane targeting and ROS generation. Metallacycle 120 (Fig. 15a), a tetraphenylethylene-based organoplatinum(II) complex, self-assembles with TAT-decorated tobacco mosaic virus coat proteins 121 (Fig. 15b) to form assembly 122 (Fig. 15b). This system targets bacterial membranes through electrostatic interactions, and upon light irradiation, the complex generates ROS that lead to bacterial cell death (Fig. 15c). The combination of metallacycle 120 and the TAT peptide dramatically improves the photodynamic inactivation efficiency, especially against Gram-negative Escherichia coli, which is typically more resistant to aPDT due to its outer membrane (Fig. 15c). This innovative system also shows efficacy against Gram-positive bacteria, though to a lesser extent. The versatility of this assembly in targeting a broad spectrum of bacteria makes it a promising tool for both superficial infections and potential adaptation for deeper tissue infections using near-infrared light.72
 | ||
| Fig. 15 (a) Structure of complex 120. (b) (A) A schematic illustrating the self-assembly of structure 122, formed from the combination of metallacycle 120 and protein 121. (B) The antibacterial mechanism of assembly 122. (c) (A–H) SEM images of E. coli either untreated (A and E) or incubated with protein 121 (B and F), metallacycle 120 (C and G), or assembly 122 (D and H), in the dark (A–D) or following 420 nm light irradiation (25 mW cm−2) for 15 min (E–H). Scale bars for A–H are 1 μm. (I and J) TEM section images of E. coli untreated in the dark (I) or incubated with assembly 122 and irradiated with 420 nm light (25 mW cm−2) for 15 min (J). Panels K and L provide close-ups of the red boxes in panels I and J, respectively. Reproduced from ref. 72 with permission from the National Academy of Sciences, copyright 2019. | ||
PTMCs have demonstrated immense potential in advancing aPDT as a highly selective and effective approach for combating bacterial infections, particularly antibiotic-resistant strains. The ability of PTMCs, such as zinc(II), ruthenium(II), iridium(III), palladium(II), and platinum(II)-based complexes, to generate high levels of ROS, combined with their selective action against bacterial cells while sparing mammalian cells, highlights their promise in clinical applications.
Despite these advancements, further research is necessary to optimize their design, fully understand their mechanisms of action, and conduct clinical trials to ensure their safety and efficacy in real-world settings. Future directions should focus on developing new complex architectures, improving targeting mechanisms, and integrating aPDT with conventional antimicrobial therapies for enhanced treatment outcomes. These developments highlight the potential of PTMCs as versatile agents for simultaneous bacterial imaging and therapy.
While the therapeutic and diagnostic potential of PTMCs is promising, their successful clinical application also depends heavily on their safety and biocompatibility. The next section examines critical considerations regarding the biological interactions, toxicity profiles, and overall biocompatibility of PTMCs.
8. Cytotoxicity and safety profiles
PTMCs are emerging as a promising frontier of antimicrobial therapies and medical imaging. However, their potential is dependent not only on therapeutic efficacy but also on safety profiles, specifically cytotoxicity. When designing these metal-based medicines, it is critical to achieve a precise balance between effectively targeting bacterial cells and being biocompatible with mammalian tissues. This balance is critical for the practical feasibility of PTMCs, particularly in medical applications that need sustained exposure.The ability of PTMCs to selectively target infections while preserving human cells is critical for therapeutic success. If these complexes injure healthy tissues, their clinical use may be challenged. As a result, comprehensive in vitro and in vivo studies are required to assess their cytotoxicity. These studies often evaluate their effects on various mammalian cell types, including fibroblast, liver, kidney, and immune cells. Researchers can fine-tune the therapeutic application of PTMCs by defining safe concentration limits and knowing how they interact with cell membranes.
One of the primary issues of metal-based complexes is their ability to generate reactive oxygen species.73 ROS, while effective at killing bacteria, can also cause oxidative stress in mammalian cells, resulting in damage, apoptosis (cell death), or necrosis.74 If left untreated, oxidative stress can impair tissue integrity and function. As a result, knowledge of the cytotoxic mechanisms of PTMCs, particularly how they produce ROS and interact with mammalian cell membranes, is critical for ensuring safety.
Another important element impacting the safety of PTMCs is their pharmacokinetics and biodistribution throughout the body. Certain metals, such as platinum, ruthenium, and gold, can accumulate in tissues, resulting in chronic exposure and toxicity.75 As a result, it is crucial to produce PTMCs that exit the body efficiently or accumulate minimally in non-target tissues. Structural changes, particularly in the ligands that bind to these metals, can greatly increase biocompatibility and minimize toxicity.
Zinc(II)-based complexes outperform other PTMCs in terms of cytotoxicity. Zinc(II)–(22′-dipicolylamine) complexes, such as compounds 42–45 (Fig. 5), have shown remarkable selectivity for bacterial cells while sparing mammalian organs. These complexes showed considerable promise in antimicrobial applications, with low toxicity in mammalian cells and suitability for bacterial imaging and therapy. One reason zinc(II)-based complexes are less hazardous is that zinc is an important metal in the human body. Because the body naturally processes and uses zinc, zinc(II)-based PTMCs are more biocompatible than those based on other metals. This natural compatibility minimizes the risk of adverse reactions, making zinc(II)-based complexes safer options for clinical use. Zinc(II)-based PTMCs show promise for next-generation antimicrobial therapeutics due to their selective targeting of bacterial cells and low toxicity in mammalian cells.15b
Ruthenium(II)-based PTMCs have received a lot of attention due to their high antibacterial activity and low toxicity in mammalian cells. One notable example is dinuclear ruthenium(II) complex 56 (Fig. 6), which preferentially targets bacterial cells by binding to ribosomal RNA within bacterial ribosomes, inhibiting protein synthesis and limiting bacterial growth. This specific action against bacterial cells, combined with minimal effects on human tissues, demonstrates the therapeutic promise of ruthenium(II)-based complexes. In addition to their antibacterial characteristics, ruthenium(II) compounds have structural flexibility, which allows for specific targeting of bacterial membranes. This flexibility increases their efficacy while retaining biocompatibility. Furthermore, ruthenium(II) complexes can be functionalized for targeted distribution, which reduces non-specific interactions with mammalian cells and thus total cytotoxicity. These characteristics make ruthenium(II)-based PTMCs ideal for clinical applications requiring both safety and therapeutic efficacy.61
Gold(I)-based PTMCs have also demonstrated significant potential, particularly when combined with silver(I). Gold(I) organometallic complexes, notably complexes 93–96 (Fig. 8), have high antibacterial action and low toxicity in mammalian cells. The combination of gold(I) and silver(I) boosts their antibacterial properties while reducing the likelihood of bacterial resistance. These gold complexes showed low cytotoxicity across a variety of mammalian cell lines, making them promising candidates for clinical usage. The biocompatibility of gold(I) complexes can be enhanced further through careful ligand design. Researchers can optimize the efficiency and safety of these complexes by reducing interactions with human cells and enhancing selectivity for bacterial targets. Furthermore, the photophysical properties of gold(I) complexes make them appropriate for imaging applications, allowing for the detection of bacterial infections while causing minimal injury to host tissues. Their dual role as theranostic agents increases their clinical appeal.54
The cytotoxicity and safety characteristics of PTMCs are crucial for their development as therapeutic agents. Researchers can improve the therapeutic efficiency of PTMCs by developing complexes that preferentially target bacterial cells while protecting mammalian organs. Ruthenium(II), gold(I), silver(I), and zinc(II)-based compounds showed great promise in establishing this balance. Their low toxicity in mammalian cells, combined with strong antibacterial action, make them promising candidates for future theranostic uses. With further investigation and refining, PTMCs could make a significant contribution to the development of next-generation antimicrobial medicines, providing hope in the fight against drug-resistant infections. Understanding the safety and biological interactions of PTMCs is vital for their translation into clinical practice.
Despite significant progress, several challenges remain in the development and application of PTMCs. The next section outlines the key obstacles and future directions needed to fully harness the potential of these complexes in antimicrobial therapy and diagnostics.
9. Future directions for safety optimization
While these examples show that PTMCs have the ability to achieve low cytotoxicity while also being highly selective for bacterial cells, more research is needed to refine their safety profiles. Continued efforts should be directed toward improving the design of PTMCs to increase their biocompatibility, notably by increasing selectivity for bacterial targets and lowering off-target effects. Investigating the pharmacokinetics, biodistribution, and clearance in vivo of these complexes will also be critical for determining their long-term safety and feasibility as therapeutics.Advances in nanotechnology and drug delivery methods may improve the safety of PTMCs by allowing for tailored distribution to infection locations while minimizing exposure to healthy tissue. Furthermore, the use of biocompatible ligands and coating materials can enhance the safety and efficacy of PTMC-based therapies. Ultimately, the goal is to create PTMCs that are not only effective at treating bacterial infections but also safe for long-term usage in patients, thus presenting a realistic answer to the rising problem of antibiotic resistance.
10. Conclusion and future directions
Polynuclear transition metal complexes are altering the landscape of antimicrobial tactics, providing novel solutions that combine theranostic functions. Their unique capacity to tackle multidrug-resistant bacterial strains while also allowing for precise imaging distinguishes them from traditional techniques. The strategic employment of metal such as silver(I), rhenium(I), iron(II), cobalt(II), nickel(II), copper(II), zinc(II), cadmium(II), ruthenium(II), iridium(III), gold(I) and gold(III) inside these complexes improves their antibacterial effectiveness via mechanisms such as reactive oxygen species production, membrane disruption, and specific interactions with bacterial cells. This multimodal action highlights their ability to handle the developing issue of antibiotic resistance more effectively than typical medicines. In diagnostics, the luminescent features of PTMCs enable real-time viewing of bacterial infections, giving them an advantage in both early diagnosis and monitoring of treatment efficacy. Their use in theragnostic applications, such as biofilm targeting, increases their utility, making them essential for developing integrated diagnostic and treatment strategies. These features make PTMCs viable tools for developing clinical microbiology and environmental applications.Nonetheless, in order to fully exploit the therapeutic promise of PTMCs, ongoing research must focus on addressing hurdles associated with their safety profile, biological interactions, and pharmacokinetics. Understanding their molecular mechanisms of action and reducing cytotoxicity to host cells are crucial steps toward transferring laboratory results into therapeutic practice. Addressing these problems will be critical to realizing their potential as next-generation antimicrobial agents. As we proceed in the fight against antimicrobial resistance, PTMCs serve as a source of inspiration for new ideas. They provide not only a revolutionary way to combat persistent infections, but also a transformative vision for future medicine, combining tailored treatment with precise diagnostic capabilities. The emergence of these complexes represents a huge step toward more tailored, effective, and long-lasting treatment options in infection management.
Data availability
This perspective does not present original research, include software or code, nor involve the generation or analysis of new data.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Parna Gupta (PG) acknowledges IISER Kolkata and CSIR grant no. 01/3028/21/EMR-II for financial support.References
- (a) Nat. Rev. Microbiol., 2024, 22, 589–590. ; (b) K. Ralhan, K. A. Iyer, L. L. Diaz, R. Bird, A. Maind and Q. A. Zhou, ACS Infect. Dis., 2024, 10, 1483–1519 CrossRef CAS PubMed; (c) C. S. Ho, C. T. H. Wong, T. T. Aung, R. Lakshminarayanan, J. S. Mehta, S. Rauz, A. McNally, B. Kintses, S. J. Peacock, C. de la Fuente-Nunez, R. E. W. Hancock and D. S. J. Ting, Lancet Microbe, 2024, 100947 Search PubMed.
- (a) M. S. Butler, W. Vollmer, E. C. A. Goodall, R. J. Capon, I. R. Henderson and M. A. T. Blaskovich, ACS Infect. Dis., 2024, 10, 3440–3474 CrossRef CAS PubMed; (b) J. D. Prajapati, U. Kleinekathöfer and M. Winterhalter, Chem. Rev., 2021, 121, 5158–5192 CrossRef CAS PubMed.
- (a) V. Balzani, A. Juris, M. Venturi, S. Campagna and S. Serroni, Chem. Rev., 1996, 96, 759–834 CrossRef CAS PubMed; (b) B. Das and P. Gupta, Coord. Chem. Rev., 2024, 504, 215656 CrossRef CAS; (c) B. Das, A. Chatterjee, P. Purkayastha and P. Gupta, J. Phys. Chem. C, 2023, 127, 12096–12106 CrossRef CAS; (d) B. Das, Small, 2025, 21, 2410338 CrossRef CAS PubMed; (e) B. Das, Chem. Commun., 2025, 61, 6391–6416 RSC.
- (a) S. Đurić, S. Vojnovic, A. Pavic, M. Mojicevic, H. Wadepohl, N. D. Savić, M. Popsavin, J. Nikodinovic-Runic, M. I. Djuran and B. Đ. Glišić, J. Inorg. Biochem., 2020, 203, 110872 CrossRef PubMed; (b) S. Ajay Sharma, N. Vaibhavi, B. Kar, U. Das and P. Paira, RSC Adv., 2022, 12, 20264–20295 RSC; (c) K. Burger, I. Zay and G. T. Nagy, Inorg. Chim. Acta, 1983, 80, 231–235 CrossRef CAS; (d) J.-L. Li, L. Jiang, B.-W. Wang, J.-L. Tian, W. Gu, X. Liu and S.-P. Yan, J. Inorg. Biochem., 2015, 145, 19–29 CrossRef CAS PubMed; (e) B. Das and P. Gupta, Chem. – Asian J., 2021, 16, 2495–2503 CrossRef CAS PubMed; (f) K. Peewasan, M. P. Merkel, K. Zarschler, H. Stephan, C. E. Anson and A. K. Powell, RSC Adv., 2019, 9, 24087–24091 RSC; (g) B. Das, S. T. Borah, S. Ganguli and P. Gupta, Chem. – Eur. J., 2020, 26, 14987–14995 CrossRef CAS PubMed; (h) Saswati, M. Mohanty, A. Banerjee, S. Biswal, A. H. Jr, G. Schenk, K. Brzezinski, E. Sinn, H. Reuter and R. Dinda, J. Inorg. Biochem., 2020, 203, 110908 CrossRef CAS PubMed; (i) J. González-García, C. Galiana, M. A. Dea-Ayuela, M. R. Stojković, S. López-Molina, C. Galiana-Roselló, S. Blasco, I. Piantanida and E. García-España, Dalton Trans., 2023, 52, 5478–5485 RSC; (j) A. A. Hassoon, S. J. Smith and R. G. Harrison, RSC Adv., 2024, 14, 31850–31860 RSC; (k) M. Wenzel, A. de Almeida, E. Bigaeva, P. Kavanagh, M. Picquet, P. L. Gendre, E. Bodio and A. Casini, Inorg. Chem., 2016, 55, 2544–2557 CrossRef CAS PubMed; (l) A. K. Gorle, A. J. Ammit, L. Wallace, F. R. Keene and J. G. Collins, New J. Chem., 2014, 38, 4049–4059 RSC; (m) S. Gupta, P. Biswas, B. Das, S. Mondal, P. Gupta, D. Das and A. I. Mallick, Gut Pathog., 2024, 16, 38 CrossRef CAS PubMed; (n) A. Sarkar, R. Kumar, B. Das, P. S. Ray and P. Gupta, Dalton Trans., 2020, 49, 1864–1872 RSC; (o) B. Das, S. Gupta, A. Mondal, K. J. Kalita, A. I. Mallick and P. Gupta, J. Med. Chem., 2023, 66, 15550–15563 CrossRef CAS PubMed; (p) J. H. Kim, E. Reeder, S. Parkin and S. G. Awuah, Sci. Rep., 2019, 9, 12335 CrossRef PubMed; (q) B. Das and P. Gupta, Dalton Trans., 2021, 50, 10225–10236 RSC; (r) C. G. Hartinger, A. D. Phillips and A. A. Nazarov, Curr. Top. Med. Chem., 2011, 11, 2688–2702 CrossRef CAS PubMed; (s) T. M. Silva, S. Andersson, S. K. Sukumaran, M. P. Marques, L. Persson and S. Oredsson, PLoS One, 2013, 8, e55651 CrossRef CAS PubMed.
- (a) A. Frei, A. D. Verderosa, A. G. Elliott, J. Zuegg and M. A. T. Blaskovich, Nat. Rev. Chem., 2023, 7, 202–224 CrossRef CAS PubMed; (b) T. W. Rees, P.-Y. Ho and J. Hess, ChemBioChem, 2023, 24, e202200796 CrossRef CAS PubMed; (c) L. C.-C. Lee and K. K.-W. Lo, Chem. Rev., 2024, 124, 8825–9014 CrossRef CAS PubMed.
- D. Megrian, N. Taib, J. Witwinowski, C. Beloin and S. Gribaldo, Mol. Microbiol., 2020, 113, 659–671 CrossRef CAS PubMed.
- (a) D. Saxena, R. Maitra, R. Bormon, M. Czekanska, J. Meiers, A. Titz, S. Verma and S. Chopra, npj Antimicrob. Resist., 2023, 1, 17 CrossRef PubMed; (b) D. Gurvic and U. Zachariae, npj Antimicrob. Resist., 2024, 2, 6 CrossRef PubMed.
- G. Kapoor, S. Saigal and A. Elongavan, J. Anaesthesiol., Clin. Pharmacol., 2017, 33, 300–305 CrossRef CAS PubMed.
- (a) P. Rajapaksha, A. Elbourne, S. Gangadoo, R. Brown, D. Cozzolino and J. Chapman, Analyst, 2019, 144, 396–411 RSC; (b) P.-E. Fournier, G. Dubourg and D. Raoult, Genome Med., 2014, 6, 114 CrossRef PubMed.
- F. Li, J. G. Collins and F. R. Keene, Chem. Soc. Rev., 2015, 44, 2529–2542 RSC.
- O. V. Salishcheva and A. Y. Prosekov, Foods Raw Mater., 2020, 8, 298–311 CAS.
- L. C.-C. Lee and K. K.-W. Lo, Small Methods, 2024, 2400563 CrossRef CAS PubMed.
- (a) Y. Wang, J. Wan, R. J. Miron, Y. Zhao and Y. Zhang, Nanoscale, 2016, 8, 11143–11152 RSC; (b) A. Jain, N. T. Garrett and Z. P. Malone, Photochem. Photobiol., 2022, 98, 6–16 CrossRef CAS PubMed; (c) A. Mondal, B. Das, S. Karmakar, S. Pani, S. Khan, P. Gupta and J. D. Sarma, J. Med. Chem., 2024, 67, 20559–20570 CrossRef CAS PubMed; (d) K. B. A. Ahmed, T. Raman and V. Anbazhagan, RSC Adv., 2016, 6, 44415–44424 RSC; (e) S. T. Borah, A. Mondal, B. Das, S. Saha, J. D. Sarma and P. Gupta, ACS Appl. Bio Mater., 2025, 8, 3331–3342 CrossRef CAS PubMed.
- C. Ratia, R. G. Soengas and S. M. Soto, Front. Microbiol., 2022, 13, 846959 CrossRef PubMed.
- (a) D. R. Rice, K. J. Clear and B. D. Smith, Chem. Commun., 2016, 52, 8787–8801 RSC; (b) K. M. DiVittorio, W. M. Leevy, E. J. O'Neil, J. R. Johnson, S. Vakulenko, J. D. Morris, K. D. Rosek, N. Serazin, S. Hilkert, S. Hurley, M. Marquez and B. D. Smith, ChemBioChem, 2008, 9, 286–293 CrossRef CAS PubMed; (c) W. M. Leevy, J. R. Johnson, C. Lakshmi, J. Morris, M. Marquez and B. D. Smith, Chem. Commun., 2006, 1595–1597 RSC; (d) B. Das and P. Gupta, Coord. Chem. Rev., 2025, 522, 216209 CrossRef CAS.
- A. Zhao, J. Sun and Y. Liu, Front. Cell. Infect. Microbiol., 2023, 13, 1137947 CrossRef CAS PubMed.
- S. Sk, S. Bandyopadhyay, C. Sarkar, I. Das, A. Gupta, M. Sadangi, S. Mondal, M. Banerjee, G. Vijaykumar, J. N. Behera, S. Konar, S. Mandal and M. Bera, ACS Appl. Bio Mater., 2024, 7, 2423–2449 CrossRef CAS PubMed.
- W. K. Kim, J. M. An, Y. J. Lim, K. Kim, Y. H. Kim and D. Kim, Mater Today Adv., 2025, 25, 100569 CrossRef CAS.
- (a) D. Desbouis, I. P. Troitsky, M. J. Belousoff, L. Spiccia and B. Graham, Coord. Chem. Rev., 2012, 256, 897–937 CrossRef CAS; (b) F. R. G. Bergamini, J. H. B. Nunes, M. A. de Carvalho, M. A. Ribeiro, P. P. de Paiva, T. P. Banzato, A. L. T. G. Ruiz, J. E. de Carvalho, W. R. Lustri, D. O. T. A. Martins, A. M. da C. Ferreira and P. P. Corbi, Inorg. Chim. Acta, 2019, 484, 491–502 CrossRef CAS.
- Q. Bugnon, C. Melendez, O. Desiatkina, L. F. de Chaptes, I. Holzer, E. Păunescu, M. Hilty and J. Furrer, Microbiol. Spectrum, 2023, 11, e00954–e00923 Search PubMed.
- G. Muteeb, M. T. Rehman, M. Shahwan and M. Aatif, Pharmaceuticals, 2023, 16, 1615 CrossRef CAS PubMed.
- C. B. Scarim, R. L. de Farias, A. V. de G. Netto, C. M. Chin, J. L. dos Santos and F. R. Pavan, Eur. J. Med. Chem., 2021, 214, 113166 CrossRef CAS PubMed.
- N. Baartzes, T. Stringer, R. Seldon, D. F. Warner, C. de Kock, P. J. Smith and G. S. Smith, J. Organomet. Chem., 2016, 809, 79–85 CrossRef CAS.
- N. Baartzes, T. Stringer, J. Okombo, R. Seldon, D. F. Warner, C. de Kock, P. J. Smith and G. S. Smith, J. Organomet. Chem., 2016, 819, 166–172 CrossRef CAS.
- (a) M. M. Risana, S. Balasubramaniyan, R. Govindharaju, A. M. Azharudeen, B. M. Juliet, V. MukilMeenakshi, A. A. Alsaiari, A. Roy, R. Verma and K. Sharma, Inorg. Chim. Acta, 2024, 569, 122125 CrossRef; (b) A. A. Nejo, G. A. Kolawole and A. O. Nejo, J. Coord. Chem., 2010, 63, 4398–4410 CrossRef CAS; (c) C. Villa-Pérez, J. F. Cadavid-Vargas, G. E. Camí, F. Giannini, M. E. C. Villalba, G. Echeverria, I. C. Ortega, G. C. Valencia-Uribe, S. B. Etcheverry and D. B. Soria, Inorg. Chim. Acta, 2016, 447, 127–133 CrossRef.
- A. Laachir, S. Guesmi, E. M. Ketatni, M. Saadi, L. E. Ammari, O. Mentré, S. Esserti, M. Faize and F. Bentiss, Polyhedron, 2020, 189, 114722 CrossRef CAS.
- (a) D. O. Maia, V. F. Santos, C. R. S. Barbosa, Y. N. Fróes, D. F. Muniz, A. L. E. Santos, M. H. C. Santos, R. R. S. Silva, C. G. L. Silva, R. O. S. Souza, J. C. S. Sousa, H. D. M. Coutinho and C. S. Teixeira, Chem.-Biol. Interact., 2022, 351, 109714 CrossRef CAS PubMed; (b) S. Bhatnagar, A. Jain, N. Sharma, K. K. Jhankal and N. Fahmi, New J. Chem., 2025, 49, 4132–4147 RSC.
- M. K. Bharty, S. Paswan, R. K. Dani, N. K. Singh, V. K. Sharma, R. N. Kharwar and R. J. Butcher, J. Mol. Struct., 2017, 1130, 181–193 CrossRef CAS.
- K. Das, A. Datta, S. Nandi, S. B. Mane, S. Mondal, C. Massera, C. Sinha, C.-H. Hung, T. Askun, P. Celikboyun, Z. Cantürk, E. Garribbaf and T. Akitsu, Inorg. Chem. Front., 2015, 2, 749–762 RSC.
- (a) P. Y. Chung, R. E. Y. Khoo, H. S. Liew and M. L. Low, Ann. Clin. Microbiol. Antimicrob., 2021, 20, 67 CrossRef CAS PubMed; (b) M. L. Beeton, J. R. Aldrich-Wright and A. Bolhuis, J. Inorg. Biochem., 2014, 140, 167–172 CrossRef CAS PubMed; (c) H. Bilal, C.-X. Zhang, M. I. Choudhary, S. Dej-adisai, Y. Liu and Z.-F. Chen, J. Inorg. Biochem., 2025, 266, 112835 CrossRef CAS PubMed; (d) U. Brahma, R. Kothari, P. Sharma and V. Bhandari, Sci. Rep., 2018, 8, 8050 CrossRef PubMed.
- B. Shaabani, A. A. Khandar, F. Mahmoudi, M. A. Maestro, S. S. Balula and L. Cunha-Silva, Polyhedron, 2013, 57, 118–126 CrossRef CAS.
- P. Arthi, A. Haleel, P. Srinivasan, D. Prabhu, C. Arulvasu and A. K. Rahiman, Spectrochim. Acta, Part A, 2014, 129, 400–414 CrossRef CAS PubMed.
- S. Li, J.-X. Chen, Q.-X. Xiang, L.-Q. Zhang, C.-H. Zhou, J.-Q. Xie, L. Yu and F.-Z. Li, Eur. J. Med. Chem., 2014, 84, 677–686 CrossRef CAS PubMed.
- B. Đ. Glišić, I. Aleksic, P. Comba, H. Wadepohl, T. Ilic-Tomic, J. Nikodinovic-Runic and M. I. Djuran, RSC Adv., 2016, 6, 86695–86709 RSC.
- (a) J. G. de O. Neto, J. A. O. Rodrigues, J. R. Viana, J. D. S. Barros, M. R. Lage, F. F. de Sousa, R. P. Dutra, E. B. Souto and A. O. dos Santos, J. Mol. Liq., 2024, 403, 124846 CrossRef; (b) E. Szunyogová, D. Mudroňová, K. Györyová, R. Nemcová, J. Kovářová and L. Piknová-Findoráková, J. Therm. Anal. Calorim., 2007, 88, 355–361 CrossRef.
- M. Azarkish, A. Akbari, T. Sedaghat and J. Simpson, J. Mol. Struct., 2018, 1156, 34–42 CrossRef CAS.
- H. Kargar, M. Fallah-Mehrjardi, R. Behjatmanesh-Ardakani, H. A. Rudbari, A. A. Ardakani, S. Sedighi-Khavidak, K. S. Munawar, M. Ashfaq and M. N. Tahir, Inorg. Chim. Acta, 2022, 530, 120677 CrossRef CAS.
- (a) K. L. Smitten, E. J. Thick, H. M. Southam, J. B. de la Serna, S. J. Foster and J. A. Thomas, Chem. Sci., 2020, 11, 8828–8838 RSC; (b) J.-L. Fillaut, Coord. Chem. Rev., 2024, 518, 216050 CrossRef CAS; (c) G. E. Giacomazzo, L. Conti, C. Fagorzi, M. Pagliai, C. Andreini, A. Guerri, B. Perito, A. Mengoni, B. Valtancoli and C. Giorgi, Inorg. Chem., 2023, 62, 7716–7727 CrossRef CAS PubMed.
- F. Li, Y. Mulyana, M. Feterl, J. M. Warner, J. G. Collins and F. R. Keene, Dalton Trans., 2011, 40, 5032–5038 RSC.
- F. Li, M. Feterl, Y. Mulyana, J. M. Warner, J. G. Collins and F. R. Keene, J. Antimicrob. Chemother., 2012, 67, 2686–2695 CrossRef CAS PubMed.
- M. Pandrala, F. Li, M. Feterl, Y. Mulyana, J. M. Warner, L. Wallace, F. R. Keene and J. G. Collins, Dalton Trans., 2013, 42, 4686–4694 RSC.
- F. Li, M. Feterl, J. M. Warner, F. R. Keene and J. G. Collins, J. Antimicrob. Chemother., 2013, 68, 2825–2833 CrossRef CAS PubMed.
- A. K. Gorle, M. Feterl, J. M. Warner, L. Wallace, F. R. Keene and J. G. Collins, Dalton Trans., 2014, 43, 16713–16725 RSC.
- (a) S. V. Kumar, W. K. C. Lo, H. J. L. Brooks and J. D. Crowley, Inorg. Chim. Acta, 2015, 425, 1–6 CrossRef CAS; (b) A. D. Richards, A. Rodger, M. J. Hannon and A. Bolhuis, Int. J. Antimicrob. Agents, 2009, 33, 469–472 CrossRef CAS PubMed.
- A. Terbouche, C. Ait-Ramdane-Terbouche, Z. Bendjilali, H. Berriah, H. Lakhdari, D. Lerari, K. Bachari, D. Mezaoui, N. E. H. Bensiradj, J.-P. Guegan and D. Hauchard, Spectrochim. Acta, Part A, 2018, 205, 146–159 CrossRef CAS PubMed.
- K. L. Smitten, H. M. Southam, J. B. de la Serna, M. R. Gill, P. J. Jarman, C. G. W. Smythe, R. K. Poole and J. A. Thomas, ACS Nano, 2019, 13, 5133–5146 CrossRef CAS PubMed.
- (a) M. Stanković, J. Kljun, N. Lj. Stevanović, J. Lazic, S. S. Bogojevic, S. Vojnovic, M. Zlatar, J. Nikodinovic-Runic, I. Turel, M. I. Djuran and B. Đ. Glišić, Dalton Trans., 2024, 53, 2218–2230 RSC; (b) M. Stanković, S. S. Bogojevic, J. Kljun, Ž. Milanović, N. Lj. Stevanović, J. Lazic, S. Vojnovic, I. Turel, M. I. Djuran and B. Đ. Glišić, J. Inorg. Biochem., 2024, 256, 112572 CrossRef PubMed.
- (a) M. Fallon, R. Lalrempuia, L. Tabrizi, M. P. Brandon, R. McGarry, A. Cullen, F. J. Fernández-Alvarez, M. T. Pryce and D. Fitzgerald-Hughes, J. Photochem. Photobiol., A, 2025, 462, 116218 CrossRef CAS; (b) A. Gautam, A. Gupta, P. Prasad and P. K. Sasmal, Chem. – Asian J., 2025, 20, e202401060 CrossRef CAS PubMed; (c) F. Chen, J. Moat, D. McFeely, G. Clarkson, I. J. Hands-Portman, J. P. Furner-Pardoe, F. Harrison, C. G. Dowson and P. J. Sadler, J. Med. Chem., 2018, 61, 7330–7344 CrossRef CAS PubMed.
- (a) S. Adhikari, P. Nath, A. Das, A. Datta, N. Baildya, A. K. Duttaroy and S. Pathak, Biomed. Pharmacother., 2024, 171, 116211 CrossRef CAS PubMed; (b) T. R. Panda and M. Patra, ChemMedChem, 2024, 19, e202400196 CrossRef CAS PubMed.
- M. V. Lunagariya, K. P. Thakor, B. N. Waghela, C. Pathak and M. N. Patel, Appl. Organomet. Chem., 2018, 32, e4222 CrossRef.
- (a) R. Büssing, B. Karge, P. Lippmann, P. G. Jones, M. Brönstrup and I. Ott, ChemMedChem, 2021, 16, 3402–3409 CrossRef PubMed; (b) M. Balsera-Manzanero, R. G. Soengas, M. Carretero-Ledesma, C. Ratia, M. J. Iglesias, J. Pachón, F. López-Ortiz, E. Cordero, S. M. Soto and J. Sánchez-Céspedes, Heliyon, 2024, 10, e27601 CrossRef CAS PubMed.
- B. Đ. Glišić, N. D. Savić, B. Warżajtis, L. Djokic, T. Ilic-Tomic, M. Antić, S. Radenković, J. Nikodinovic-Runic, U. Rychlewska and M. I. Djuran, MedChemComm, 2016, 7, 1356–1366 RSC.
- R. Bhowmik, A. Panwar, G. Bag and M. Roy, Coord. Chem. Rev., 2025, 537, 216689 CrossRef CAS.
- M. Frik, J. Jiménez, I. Gracia, L. R. Falvello, S. Abi-Habib, K. Suriel, T. R. Muth and M. Contel, Chem. – Eur. J., 2012, 18, 3659–3674 CrossRef CAS PubMed.
- A. Jain, B. S. J. Winkel and K. J. Brewer, J. Inorg. Biochem., 2007, 101, 1525–1528 CrossRef CAS PubMed.
- M. Wenzel, M. Patra, C. H. R. Senges, I. Ott, J. J. Stepanek, A. Pinto, P. Prochnow, C. Vuong, S. Langklotz, N. Metzler-Nolte and J. E. Bandow, ACS Chem. Biol., 2013, 8, 1442–1450 CrossRef CAS PubMed.
- M. Patra, M. Wenzel, P. Prochnow, V. Pierroz, G. Gasser, J. E. Bandow and N. Metzler-Nolte, Chem. Sci., 2015, 6, 214–224 RSC.
- A. H. Delcour, Biochim. Biophys. Acta, Proteins Proteomics, 2009, 1794, 808–816 CrossRef CAS PubMed.
- S. K. Jain, Semin. Nucl. Med., 2023, 53, 138–141 CrossRef PubMed.
- M. F. Anjum, E. Zankari and H. Hasman, Microbiol. Spectrum, 2017, 5, 17 Search PubMed.
- F. Li, E. J. Harry, A. L. Bottomley, M. D. Edstein, G. W. Birrell, C. E. Woodward, F. Richard Keene and J. G. Collins, Chem. Sci., 2014, 5, 685–693 RSC.
- M. Gao, Q. Hu, G. Feng, N. Tomczak, R. Liu, B. Xing, B. Z. Tang and B. Liu, Adv. Healthcare Mater., 2015, 4, 659–663 CrossRef CAS PubMed.
- G. Feng, C.-J. Zhang, X. Lu and B. Liu, ACS Omega, 2017, 2, 546–553 CrossRef CAS PubMed.
- S. T. Borah, B. Das, P. Biswas, A. I. Mallick and P. Gupta, Dalton Trans., 2023, 52, 2282–2292 RSC.
- W. C. M. A. de Melo, R. Celiešiūtė-Germanienė, P. Šimonis and A. Stirkė, Virulence, 2021, 12, 2247–2272 CrossRef PubMed.
- Y. Liu, R. Qin, S. A. J. Zaat, E. Breukink and M. Heger, J. Clin. Transl. Res., 2015, 1, 140–167 Search PubMed.
- D. A. Smithen, S. Monro, M. Pinto, J. Roque, R. M. Diaz-Rodriguez, H. Yin, C. G. Cameron, A. Thompson and S. A. McFarland, Chem. Sci., 2020, 11, 12047–12069 RSC.
- P. J. Gonçalves, F. C. Bezzerra, A. V. Teles, L. B. Menezes, K. M. Alves, L. Alonso, A. Alonso, M. A. Andrade, I. E. Borissevitch, G. R. L. Souza and B. A. Iglesias, J. Photochem. Photobiol., A, 2020, 391, 112375 CrossRef.
- G. G. Rossi, K. B. Guterres, K. S. Moreira, T. A. L. Burgo, M. M. A. de Campos and B. A. Iglesias, Photodiagn. Photodyn. Ther., 2021, 36, 102514 CrossRef CAS PubMed.
- S. L. Hopkins, L. Stepanyan, N. Vahidi, A. Jain, B. S. J. Winkel and K. J. Brewer, Inorg. Chim. Acta, 2017, 454, 229–233 CrossRef CAS.
- B. Das, P. Biswas, A. I. Mallick and P. Gupta, Chem. – Eur. J., 2024, 30, e202400646 CrossRef CAS PubMed.
- S. Gaoa, X. Yanc, G. Xie, M. Zhu, X. Jua, P. J. Stangd, Y. Tian and Z. Niu, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 23437–23443 CrossRef PubMed.
- (a) M. Khalid, S. Hassani and M. Abdollahi, Curr. Opin. Toxicol., 2020, 20–21, 55–68 CrossRef; (b) A. A. Dayem, M. K. Hossain, S. B. Lee, K. Kim, S. K. Saha, G.-M. Yang, H. Y. Choi and S.-G. Cho, Int. J. Mol. Sci., 2017, 18, 120 CrossRef PubMed.
- M. Sharifi-Rad, N. V. A. Kumar, P. Zucca, E. M. Varoni, L. Dini, E. Panzarini, J. Rajkovic, P. V. T. Fokou, E. Azzini, I. Peluso, A. P. Mishra, M. Nigam, Y. E. Rayess, M. E. Beyrouthy, L. Polito, M. Iriti, N. Martins, M. Martore, A. O. Docea, W. N. Setzer, D. Calina, W. C. Cho and J. Sharifi-Rad, Front. Physiol., 2020, 11, 694 CrossRef PubMed.
- A. Singh and I. Kostova, Inorg. Chim. Acta, 2024, 568, 122068 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2025 |