Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
twa: The World Avatar Python package for dynamic knowledge graphs and its application in reticular chemistry†
Jiaru
Bai
 a,
Simon D.
Rihm
a,
Simon D.
Rihm
 a,
Aleksandar
Kondinski
ab,
Fabio
Saluz
a,
Xinhong
Deng
c,
George
Brownbridge
a,
Aleksandar
Kondinski
ab,
Fabio
Saluz
a,
Xinhong
Deng
c,
George
Brownbridge
 d,
Sebastian
Mosbach
d,
Sebastian
Mosbach
 ac,
Jethro
Akroyd
ac,
Jethro
Akroyd
 ac and
Markus
Kraft
ac and
Markus
Kraft
 *ace
*ace
aDepartment of Chemical Engineering and Biotechnology, University of Cambridge, Philippa Fawcett Drive, Cambridge, CB3 0AS, UK. E-mail: mk306@cam.ac.uk
bInstitute of Physical and Theoretical Chemistry, Graz University of Technology, Stremayrgasse 9, 8010 Graz, Austria
cCambridge Centre for Advanced Research and Education in Singapore, CARES Ltd, 1 Create Way, CREATE Tower #05-05, 138602, Singapore
dCMCL, No. 9 Journey Campus, Castle Park, Cambridge, CB30AX, UK
eDepartment of Chemical Engineering, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Room 66-350, Cambridge, Massachusetts 02139, USA
First published on 30th June 2025
Abstract
Data-driven discovery is crucial in scientific domains, yet the lack of standardised data management hinders reproducibility. In chemical science, this is exacerbated by fragmented data formats. The World Avatar (TWA) addresses these challenges via a dynamic knowledge graph historically provided in Java-based toolkits. We present twa, an open-source Python package that lowers the barrier to semantic data management. Its object-graph mapper (OGM) synchronises Python class hierarchies with RDF knowledge graphs, streamlining ontology-driven data integration and automated workflows. We demonstrate twa's capacity to unify fragmented chemical data and accelerate research through use cases in molecular design and AI-assisted synthesis protocol extraction for metal–organic polyhedra (MOPs). Our approach expands the existing OntoMOPs knowledge graph by adding 799 new MOPs derived from combinatorial assembly models. By abstracting complex SPARQL queries behind a user-friendly interface, twa fosters transparent, reproducible knowledge-driven discovery. The package is freely available via pip install twa or https://pypi.org/project/twa/.
1 Introduction
Modern scientific and industrial domains generate digital data at an unprecedented scale, yet interoperability remains hampered by fragmented formats and siloed architectures.1 These barriers limit data integration and reuse, undermining the potential of cross-domain collaboration. To address this, semantic web technologies systematise data into machine-readable formats that encode conceptual relationships, thus enabling cross-disciplinary interoperability.2 A core manifestation of the semantic web is the knowledge graph, which has become indispensable for scientific and industrial innovation, due to its ability to harmonise and automate data workflows.3,4A field that stands to benefit significantly from such innovation is chemistry, where researchers handle complex information on molecular structures, reaction pathways, and experimental protocols. Ontologies offer a powerful framework for encoding these data in a formal and computationally interpretable manner.5,6 Through adherence to semantic web standards, chemists can unify and automate disparate workflows, ultimately accelerating discovery.7,8 Despite the evident value of this approach, chemistry ontologies remain underutilised.5 Many researchers find them challenging to maintain and extend, given the delicate balance between stability and adaptability that is essential for effective knowledge representation.9,10
Managing the evolution of ontologies requires robust version control, comprehensive documentation, and consistent input from domain experts. While some software solutions and collaborative workflows support these needs,11,12 they are typically designed for expert programmers and require substantial knowledge of ontologies. This is particularly acute in AI-driven chemistry, where ontologies have immense potential to unify complex data but have seen limited adopted.13,14 As a result, user-friendly tools in widely adopted languages such as Python are sorely needed to lower entry barriers for non-experts, ensuring transparency, reproducibility, and broader adoption.
Over the past decade, object graph mappers (OGMs) have evolved from object-relational mappers (ORMs), which simplify database interactions by mapping object-oriented programming constructs to relational schemas.15 OGMs extend this approach to graph databases, allowing developers to work with structured knowledge while abstracting away query complexity. However, most existing Python-based OGMs are designed for property-graph databases, such as GQLAlchemy16 and neomodel,17 while resource description framework (RDF)-backed OGMs are predominantly developed in Java18–20 or TypeScript.21 In Python, RDF-focused OGMs remain scarce, with Owlready2 (ref. 22) being one of the few available options. Despite its utility, Owlready2, along with its extension for material science,23 is primarily designed for local ontology files rather than remote triple stores and lacks robust built-in type validation. Although Owlready2 offers an experimental quadstore approach that converts ontologies into SQL databases, this functionality does not fully support distributed or scalable SPARQL-based knowledge graphs. Consequently, its application remains limited in the context of scalable, distributed knowledge graph systems.8
The World Avatar (TWA) is a distributed, dynamic knowledge graph designed to create a digital replica of the physical world.24,25 It employs software agents to synchronise digital environments with geographically distributed physical systems.8 For seamless integration, modifying its states entirely through Python programmes is essential, necessitating an OGM solution tailored for distributed applications. To address this need, we introduce a Python-based OGM specifically designed for remote RDF-backed graph databases, featuring built-in type validation. This solution bridges modern scientific applications with the semantic web ecosystem and integrates directly with twa, the Python wrapper for The World Avatar project. By providing a vendor-neutral and standardised approach to managing RDF data structures, our OGM enhances accessibility and interoperability in chemical research.
Furthermore, as large language models (LLMs) gain traction in automating tasks within chemistry,26–29 integrating OGMs with AI-driven methods unlocks new opportunities for structured hypothesis generation and data analysis. This work aims to accelerate the adoption of graph-based data management in chemistry, fostering a globally connected research network through an accessible and open-source Python toolkit.
The remainder of this paper is structured as follows. Section 2 situates our work within the broader context of The World Avatar project. Section 3 details the technical underpinnings of the proposed OGM, while Section 4 demonstrates its utility through use cases in metal–organic polyhedra. Finally, Section 5 presents conclusions and perspectives for future development.
2 The World Avatar
The World Avatar (TWA) is an open, interoperable digital ecosystem that integrates dynamic knowledge graphs and autonomous software agents to create a scalable digital twin of the physical world. It bridges domain silos using ontologies and linked data principles to enable cross-disciplinary interoperability. Emphasising collaborative and agent-based intelligence,30 TWA supports applications from molecular and material discovery7 to urban resilience planning.25 Specifically, in the chemistry domain, TWA supports chemical species ontology,10 rational design of novel reticular and porous materials,31,32 natural language question-answering system,33 and distributed self-driving laboratories.8,34A key challenge across projects was the steep learning curve of Java and the inherent complexity of ontologies, making it difficult to onboard new team members. To improve accessibility, we developed a Python wrapper as a more accessible alternative. However, team members often had to write repetitive SPARQL boilerplate code to access graph data, particularly when working with the same ontology across different projects. This not only increased development time but also introduced inconsistencies when modifications to the same ontology were needed for cross-domain applications. Moreover, developers had to manually update their SPARQL scripts to ensure meaningful results as ontologies evolved. To address these issues, we set out to develop a reusable software package that simplifies access to knowledge graphs by replacing repetitive SPARQL queries and complex Java-based workflows with a more efficient, consistent, and Python-native approach. This integration also enhances ontology version control for better change tracking and streamlined updates.
3 Object graph mapper
Building on years of experience in knowledge graph development, we present the twa Python package as a comprehensive solution for more intuitive and efficient knowledge graph management. By abstracting interactions with the underlying graph database, twa simplifies ontology development and population. A central feature of this package is its object-graph mapper (OGM), which provides a unified and scalable way to interact with knowledge graphs using object-oriented principles. This section introduces the core component of twa, the OGM, highlighting its role in streamlining ontology management and enhancing interoperability in modern scientific applications.
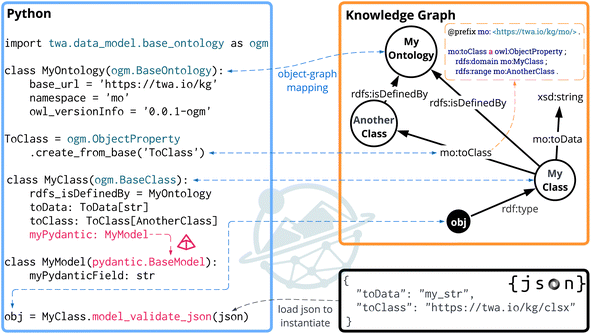
Fig. 1 presents an overview of the OGM, showcasing how it enables semantic translation between Python objects, RDF triples, and JSON data – both when utilising existing knowledge graphs and when constructing new ones. By leveraging Pydantic35 for structured data modelling and RDFLib36 for representing RDF triples, OGM bridges object-oriented programming with semantic web technologies. Python classes (left, blue box) explicitly map onto OWL classes and properties within the RDF graphs (top right, orange box). JSON objects (bottom right, black box) can be validated and directly instantiated into OGM objects through the Pydantic JSON validation method, which enforces data validation and ensures schema compliance. Notably, compared to the default instantiation behaviour of Pydantic when dealing with nested JSON, which creates a new model instance for every occurrence regardless of its contents, our OGM maintains an in-memory registry keyed by  . Repeated IRIs are resolved to the same Python object, preserving graph integrity and eliminating duplicate nodes without compromising standard validation semantics.
. Repeated IRIs are resolved to the same Python object, preserving graph integrity and eliminating duplicate nodes without compromising standard validation semantics.
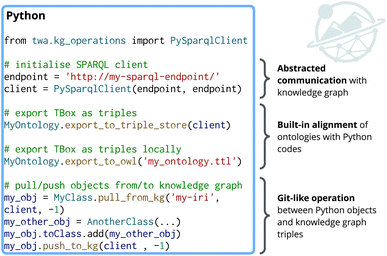
Fig. 2 exemplifies how OGM in the twa Python package simplifies both ontology management and data-level interactions by abstracting complexities of querying and updating knowledge graphs (boilerplate of SPARQL and RDF). Developers only need to specify the endpoint, without worrying about manually writing SPARQL queries. Ontologies emerge naturally as a byproduct of defining relationships in Python, with a single function called exporting them as structured graph data. Similarly, pulling and pushing objects to and from the knowledge graph is streamlined through intuitive Python functions. These abstractions shift the developers' focus from “how do I write SPARQL” to “how do I model my domain”, making semantic web technologies more accessible and efficient for rapid prototyping of complex and interlinked data workflows in chemistry (and beyond).
The core components of OGM include BaseOntology, BaseClass, ObjectProperty, and DatatypeProperty, which provide a direct mapping between Python classes and ontological concepts in the terminology component (TBox) of a knowledge graph, while instances correspond to assertion component (ABox). Designed to follow the standard subclassing mechanism in Python, these base classes can be extended by users to define domain-specific ontologies. Since they inherit from  , they seamlessly integrate semantic functions while remaining compatible with native Pydantic features, such as JSON parsing and validation. This enables structured JSON data, including outputs from large language models (LLMs), to be directly instantiated as Python objects while preserving alignment with formal ontologies. Additionally, OGM provides utility functions for exporting defined ontologies as description logic, ensuring interoperability with standard semantic reasoning tools.
, they seamlessly integrate semantic functions while remaining compatible with native Pydantic features, such as JSON parsing and validation. This enables structured JSON data, including outputs from large language models (LLMs), to be directly instantiated as Python objects while preserving alignment with formal ontologies. Additionally, OGM provides utility functions for exporting defined ontologies as description logic, ensuring interoperability with standard semantic reasoning tools.
Fig. 3 illustrates the core functionalities of OGM for enabling object-level interaction between Python and the knowledge graph. These rely on two key algorithms that ensure consistent and efficient data synchronisation. The pull_from_kg function retrieves data from the graph and instantiates or updates corresponding Python objects. It dynamically resolves the appropriate Python class for a given node based on its rdf:type label and the class inheritance hierarchy in Python. The algorithm supports recursive loading of linked objects, with a recursion depth parameter that controls whether nested structures are fetched to a specified depth or infinitely. To optimise performance and prevent redundant operations, it maintains a cache of object states, mitigating race conditions during concurrent pulls. The push_to_kg function propagates local changes from in-memory objects back to the knowledge graph. It computes the differences between the cached graph state and the current Python values to determine which triples need to be added or removed. To prevent infinite loops when traversing cyclic structures, it tracks processed nodes during traversal. Both algorithms are detailed in ESI A.1.† The core features of the framework are detailed in the following subsections, and a comparative summary with other commonly used Python RDF libraries is provided in Table 1.
| Feature | RDFLib36 | SuRF37 | Owlready2 (ref. 22) | twa (this work) |
|---|---|---|---|---|
| Schema validation | No built-in schema validation or type safety; requires external tools | Dynamically generates Python proxies without built-in validation | Basic type alignment provided through OWL datatypes, but limited runtime validation | Uses Pydantic-based models for automatic schema validation and type safety |
| Remote SPARQL endpoint support | Full support for local and remote SPARQL querying via plugins | Supports remote SPARQL queries via RDFLib backend, without direct mapping of results to object | Local SPARQL queries through SQLite engine but no direct remote endpoint support | Provides querying abstraction for remote SPARQL endpoints integrated directly into Python objects |
| Reasoning capabilities | No native reasoning, but possible via additional tool, e.g. OWL-RL | No reasoning or inference support; solely provides RDF-to-object mapping | Native OWL reasoning capabilities via integrated reasoners (HermiT and Pellet) for consistency checks and inference | Currently lacks built-in reasoning capabilities; planned future integration |
| Object-oriented design | Triple-level RDF manipulation requiring manual class mappings for OOP designs | Provides ORM-like object-oriented proxy design for intuitive RDF-to-object interaction | Provides a highly Pythonic, ontology-driven programming interface for seamless OOP integration | Offers object-oriented Python classes closely aligned with Pydantic best practices |
| Performance and scalability | All triples are hosted in-memory, with external tools required to work with database backends | Performance reliant on chosen backend with minimal overhead but lacks bulk optimisation | Excellent local performance (up to billions of triples) through optimized SQLite storage, but single-node only | Possible to work with distributed graphs, further optimisation could be implemented to work with large graphs and deep ontologies |
| Ease of use | Straightforward for RDF-savvy Python users, but steep learning curve for RDF novices | Simple ORM-like interface, but outdated and less beginner-friendly due to inactivity | User-friendly OOP design, straightforward for Python developers, but requires OWL familiarity for advanced use | Python-friendly abstraction lowers semantic web entry barriers, improved from the initial complexity with Java setups |
| Integration with LLMs | No native AI or LLM integration | No integration as its design predates current AI workflows | No native AI or LLM integration; external development necessary for AI applications | Explicitly integrates LLM outputs into knowledge graphs via structured JSON conversion |
| Development status | Actively maintained with regular updates and strong community support | Inactive since 2016, minimal support and no recent updates | Actively maintained with frequent updates and robust community usage | Actively developed, with ongoing improvements as part of The World Avatar ecosystem |
3.1 Recursive synchronisation
Fig. 3(a) illustrates the Git-like recursive push and pull operations, where changes in the graph, such as SPARQL insertions of nodes i4 and i5, and modifications in Python objects, such as the removal of i2 or the addition of i6, are propagated in both directions. This mechanism ensures that Python objects remain aligned with the knowledge graph, even when updates are made externally.To achieve this, OGM maintains three sets of values for each node connection (i.e., object or datatype properties). The local state represents the current state of Python objects, the cached state stores the last known version retrieved from the knowledge graph, and the fetched state reflects the latest values obtained from the graph if the pull is flagged before the push. When synchronising, OGM first compares the fetched and cached states to identify external modifications that should be instantiated in Python. It then compares the cached and local states to detect changes made within Python that should be pushed to the graph. Users can provide a flag to enforce an overwrite of local modifications when pulling from the remote graph, ensuring that externally introduced changes take precedence in case of conflicts. This design enables multiple clients to operate concurrently on the same knowledge graph, similar to how developers collaborate on software projects using Git.
The recursive synchronisation mechanism operates at configurable depths, allowing users to control the extent of traversal during pull and push operations. A depth of 0 limits synchronisation to direct relationships, while a positive integer n restricts recursion to n levels. A depth of −1 enables full recursion, ensuring that updates propagate through all relationship links in the graph. This flexibility ensures that OGM can handle complex knowledge graphs efficiently while maintaining consistency between Python objects and the graph database.
Listing S1 in ESI† presents an example demonstrating how recursive synchronisation is performed as shown in Fig. 3(a). The process begins by pulling an instance and its connected objects from the knowledge graph into Python. After external modifications are made directly in the graph, a local deletion is performed in Python. The push operation then ensures that all changes, including those made externally and locally, are properly reconciled. Finally, a new instance is created and linked to the existing object, and another push is executed to propagate the update recursively.
3.2 Multi-inheritance resolution
In ontologies with subclass hierarchies, a single entity can often be viewed through multiple facets, leading to multiple class labels (i.e., rdf:type) for the same instance. This phenomenon, known as multiple inheritance,38 is common in knowledge representation systems. For example, the Climate Resilience Demonstrator (CReDo) project39 develops a digital twin of infrastructure networks to assess their resilience to climate events such as flooding and extreme heat. In their implementation, each node in the infrastructure network is instantiated both as an asset, which stores essential information, and as a site, which is used for visualisation. When performing analyses, it is crucial to retrieve only the relevant aspects of the data to maintain computational efficiency, especially when the digital twin is operated at the national level.Fig. 3(b) illustrates how OGM resolves such multiple-inheritance scenarios. When pulling an instance from the knowledge graph, OGM determines its Python instantiation based on the method resolution order (MRO) of the class hierarchy. The system identifies the deepest subclass at the intersection of the class used for pulling and the instance's assigned types in the knowledge graph. For example, as node i is labelled with multiple classes, it will always be instantiated as the most specific subclass, such as leaf class B when pulled using either A or B. If multiple parallel leaf classes exist, such as when pulling i using T and encountering both B and C, OGM raises an error to prevent ambiguity. The user must explicitly use either B or C to prevent the conflict. An error is also raised if an instance is pulled using a class that is not assigned as its type in the knowledge graph, even if it exists in the class hierarchies, such as attempting to pull i using D.
In the current implementation, the OGM enforces a single perspective per query to simplify the object representation and maintain clarity, whereas attributes from other perspectives remain available but are not merged automatically. When a different view is required, the user can re-instantiate the same IRI as an instance of the alternative class, and all view-specific attributes will be accessible through that class. This design ensures that Python objects inherit only the relevant properties for each context while preserving the underlying graph structure. Additionally, when pushing local changes back to the graph, OGM updates only the pulled portions and leaves untouched any unpulled data, thereby preventing unintended data loss. Listing S2 in ESI† provides a minimal example demonstrating this behaviour. If simultaneous merging of multiple views becomes a common requirement, we will explore support for that feature in future iterations.
3.3 Transitive property
Transitive properties play a fundamental role in ontology modelling by enabling efficient reasoning over hierarchical and dependency-based relationships. They allow relationships to be inferred across multiple levels, ensuring a more comprehensive and structured representation of complex systems. Fig. 3(c) illustrates the retrieval of transitive properties using native SPARQL queries and OGM in Python, highlighting the difference in approach.In SPARQL, transitive properties are retrieved using property paths, which efficiently traverse hierarchical relationships directly within the query. OGM achieves this capability by implementing recursion, allowing Python objects to interact dynamically with connected entities. This recursive traversal is particularly useful in scientific domains where hierarchical relationships are prevalent, such as laboratory setups, reaction networks, or material dependency structures.
For instance (see Listing S3 in ESI†), in a lab setup, if Beaker A is part of Reaction Setup X, which itself is part of Experiment Y, transitive reasoning infers that Beaker A is part of Experiment Y. Similarly, if Clamp B is part of Stand C, and Stand C is part of Reaction Setup X, the entire component hierarchy can be traced. Such reasoning ensures precise representation and management of complex equipment arrangements, enhancing the efficiency and reliability of lab operations.40
By leveraging transitive reasoning, OGM enhances the usability of knowledge graphs by providing structured access to indirect relationships while preserving the interpretability of object-oriented representations in Python. This integration of SPARQL's declarative querying with OGM's recursive traversal ensures that complex hierarchical structures can be navigated and manipulated seamlessly in a programmatic environment.
4 Use cases for reticular chemistry
Metal–organic polyhedra (MOPs) can be described as structures formed by metal- and organic-based chemical building units (CBUs) that resemble regular polyhedra.31,41–43 The rational design of MOPs, as well as other cage-like assemblies, requires domain experts to carefully consider both the chemical compatibility and geometric complementarity of the constituent CBUs.31,44 Didactical research involving children's construction of polyhedral models from toys has shown that no formal geometric training is necessary to assemble such shapes,45,46 indicating that some form of cognitive visualisation is involved in the reasoning process. Motivated by these insights, the concepts of assembly models (AMs) and generic building units (GBUs) were introduced as mental “blueprints” for systematically designing MOPs from available CBUs.31 These ideas were first formalised in the OntoMOPs ontology,31 in which the CBUs were further classified as species using the OntoSpecies ontology.10The MOP discovery agent employs an algorithm based on set operations to identify which CBUs can be combined without generating undesirable strain.31 In an analysis of 151 experimentally reported MOPs constructed from 137 unique CBUs, the dataset was effectively organised into 18 AMs and 7 GBUs. According to the discovery agent, as many as 1418 new MOPs could be rationally designed,31 and several of these predicted structures have been confirmed by experimental synthesis.47 This targeted approach substantially narrows down the potential design space, originally estimated to be approximately 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 possibilities, thus allowing more focused and efficient computational and experimental investigations.31
000 possibilities, thus allowing more focused and efficient computational and experimental investigations.31
Fig. 4 depicts a MOP [(C6H3)((C6H4)2)3(CO2)3]4[V6O6(C5H4NPO3)(CH3O)9]48−, which follows the AM topology (3-planar)4(3-pyramidal)4_Td. The assembly model (AM) prescribes two types of generic building units (GBUs), “3-planar” and “3-pyramidal”, each appearing four times and identified by their spatial coordinates. Instances of the “3-planar” GBU connect to three “3-pyramidal” GBUs, forming the polyhedral framework. This hierarchical representation was implemented via the OGM method to encode both chemical structure and geometric relationship.31 Beyond rational design, the OGM method also includes automated assembly modelling of MOPs.48
 | ||
| Fig. 4 Representation of MOPs, CBUs, GBUs, AMs as part of OntoMOPs,31 key geometric concepts used in the assembly modelling of MOPs,48 and additional concepts added for semantic construction using the OGM. Class and relation names are abbreviated for clarity. | ||
4.1 Semantic geometry construction
In our previous work, we introduced an automated rational design framework for MOPs, leveraging assembly models (AMs) and generic building units (GBUs).31 Building on that foundation, subsequent developments extended the process of assembly modelling to generate structural information about MOPs, thereby enabling computational analyses of cavity and pore sizing.48 This work generalises these earlier methods and integrates them into the OGM infrastructure, providing a single, modular pipeline for constructing computation-ready 3D geometries of MOPs.Fig. 5 illustrates our semantic assembly workflow, which generalises and refines the vector-transformation approach from prior work.48 The process begins with identifying binding sites for the chemical building units (CBUs), as shown in Fig. 5(a). Each binding site is defined as the centroid of a user-labeled binding fragment, corresponding to the atomic group(s) involved in bonding, inspired by “connection points” as implemented in geometry-based assembly for metal–organic framework.50 A 2D circle is fitted through these binding sites, defining a plane whose normal vector serves as the “fingerprint vector” of the CBU. To ensure a unique orientational reference, we compute the cross-product of this fingerprint vector with a secondary vector extending from the circle's centre to the nearest binding site. For CBUs functioning as 4-planar GBUs with an ideal D2h symmetry, the secondary vector is instead defined from the centre to the shortest edges between two binding sites. The centroid of the CBU's atoms, projected onto the normal vector of this plane, is designated as the assembly centre.
 | ||
| Fig. 5 Automated semantic assembly of MOPs following the OGM implementation, inspired by previously developed geometry assembly protocol:48 (a) assignment of binding sites and calculation of the fingerprint vector for CBUs, (b) quaternion-based alignment of CBUs and GBUs via their fingerprint vectors, (c) calculation of translation vectors to shift CBUs such that their binding bonds are equidistant from the origin of MOPs, and (d) transformation of CBUs into the final 3D MOP geometry, re-centred at the origin. | ||
Next, Fig. 5(b) illustrates the quaternion-based alignment of the CBU fingerprint vector with that of its corresponding GBU in the AM. The fingerprint vectors of these GBUs are computed using the same procedure. Once rotational alignment is achieved, a translation vector is computed to place the rotated CBU at the appropriate distance for bonding (Fig. 5(c)). This step adjusts the distance between each CBU's centre and binding site by half the bond length for calculation, preserving structural fidelity.48 These adjustments prevent bonds from being too short, which would otherwise be difficult (though not impossible) to correct in future geometry optimisations.51 Scaling factors are determined per GBU type to maintain the proportional relationships dictated by the symmetric AM topology, ensuring consistency across all CBU transformations. A 3D MOP geometry derived by the OGM implementation is shown in Fig. 5(d), whole details on the construction steps are provided as part of ESI A.4.†
 | ||
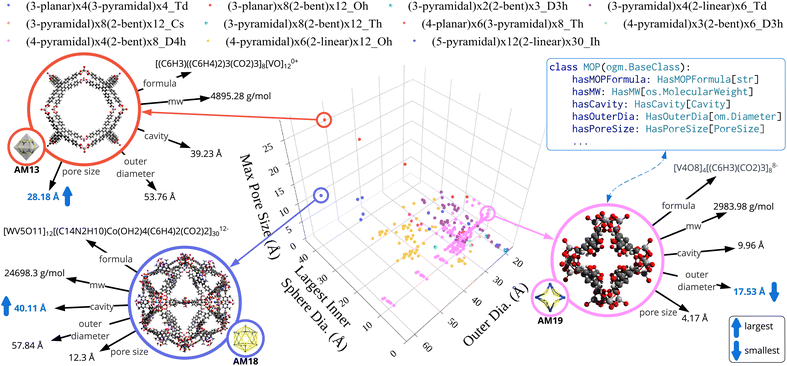
| Fig. 6 Examples of expanded chemical space resulting from the new AMs and MOPs introduced in this work. (a) A MOP [V5O9]4[(C6H4)(CO2)2]84−, added from the literature49 and assembled with existing CBUs under an unseen AM topology (4-pyramidal)4(2-bent)8_D4h (AM19). (b) Rational design of new MOPs, showing internal combinatorial CBU assembly for (4-planar)6(3-pyramidal)8_Th (AM20), and broader CBU swaps across compatible AMs (e.g., (3-planar)4(3-pyramidal)4_Td, AM2). The ontologised versions of both algorithms are provided in ESI A.3.† | ||
To systematically broaden the design space, we applied the algorithms in ESI A.3† to interchange compatible metal and organic CBUs across the newly added AMs. In contrast to Listing S4,† which strictly assembles CBUs already proven compatible with the same AM, Listing S5† allows for inter-AM CBU exchanges, provided both AMs share at least one common CBU. Fig. 6(b) illustrates how this generates novel combinations, such as leveraging AM20 ((4-planar)6(3-pyramidal)8_Th) and AM2 ((3-planar)4(3-pyramidal)4_Td). Overall, these expansions yielded 799 newly designed MOPs derived from a base set of 1584. A summary of the new structures is listed in Table S3†, with further details in Table S4 in ESI A.5.†
Fig. 7 offers an overview of these 799 new MOPs, based on structural properties computed from the 3D geometries. Each MOP is re-centred on its geometric midpoint to streamline calculations. The largest inner sphere diameter measures the distance from the centre to the closest atom (adjusted by covalent radii), while outer diameter captures the farthest atom. The maximum pore size is obtained by projecting atomic positions onto vectors connecting the MOP centre to the centroid of ring-forming GBUs. The mathematical details behind these estimations are given in ESI A.4.†
Among the newly generated structures, certain AM19-based MOPs stand out for their compactness, exhibiting relatively small cavities. This is primarily due to the “4-pyramidal” metallic CBUs and “2-bent” organic CBUs with narrow dihedral angles. Conversely, other AMs that incorporate bulkier organic linkers display significantly larger pore sizes, emphasising how AM geometry and CBU composition drive structural properties.
These findings validate our OGM workflow implementation. By leveraging semantic data representation and automated assembly, developers can focus on high-level design logic while leaving the recursive data retrieval and consistency checks to the OGM. This eliminates extensive scripting and facilitates the pre-screen of promising candidates before expensive density functional theory (DFT) optimisations. This continuing effort builds on the rational design concepts outlined in Kondinski et al.31,48 and highlights how a semantic-driven approach can streamline large-scale molecular design. Future work will explore the application of structural viability, based on computational chemistry protocols as proposed by Hoffmann et al.,52 and expand this approach to other reticular materials.
4.2 Extraction of synthesis protocols using LLMs
Following the automated rational design and geometry construction, the chemical relevance of newly designed MOPs ultimately depends on their experimental synthesis. However, such validation often faces bottlenecks due to the inefficiency of traditional trial-and-error synthesis methods. The self-assembly of these structures offers the potential to reliably predict synthesis procedures based on building units and assembly models. Connecting computational design with experimental realisation requires the systematic inference of synthesis protocols from literature procedures. Unfortunately, such protocols are often documented inconsistently across scientific publications and in non-machine-readable formats. Large language models (LLMs) show promise in addressing these challenges by extracting detailed synthesis information from the scientific literature to infer new synthesis routes.53–55
Fig. 8 illustrates the integration of LLMs with OGM to bridge unstructured literature with computable chemical knowledge. The pipeline automates the extraction of synthesis protocols from scientific literature, such as ESI† in journal articles, using the OpenAI Python API56 with domain-specific prompts. The LLM-generated structured JSON outputs are then loaded into the knowledge graph through OGM. This integration is made possible by the OGM's foundation on Pydantic, which natively supports converting structured JSON into strongly-typed Python objects by validating both the structure and data types against predefined schemas. In the OGM framework, all user-defined classes inherit from a common BaseClass, which itself extends  . This design allows JSON outputs from LLMs—when conforming to the expected schema—to be directly parsed into in-memory Python objects. These objects can then be seamlessly converted into RDF triples, ensuring that only schema-compliant, semantically valid data enters the knowledge graph and reducing the likelihood of inconsistencies. This integration establishes a bidirectional connection between unstructured text and semantic triples, enabling the instantiation of new knowledge extracted from LLMs while leveraging existing structured knowledge to guide LLM prompts. By simplifying the transformation of data between JSON, Python objects, and graph nodes, this approach minimises coding overhead for researchers while ensuring compatibility with existing workflows. Full technical details of this use case are provided in Rihm et al.57
. This design allows JSON outputs from LLMs—when conforming to the expected schema—to be directly parsed into in-memory Python objects. These objects can then be seamlessly converted into RDF triples, ensuring that only schema-compliant, semantically valid data enters the knowledge graph and reducing the likelihood of inconsistencies. This integration establishes a bidirectional connection between unstructured text and semantic triples, enabling the instantiation of new knowledge extracted from LLMs while leveraging existing structured knowledge to guide LLM prompts. By simplifying the transformation of data between JSON, Python objects, and graph nodes, this approach minimises coding overhead for researchers while ensuring compatibility with existing workflows. Full technical details of this use case are provided in Rihm et al.57
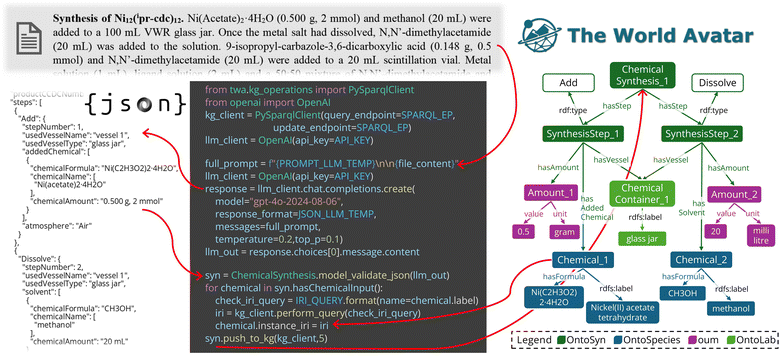 | ||
| Fig. 8 Automated extraction of MOPs synthesis protocols from literature using LLMs and integration into knowledge graphs using OGM. ESI† is processed through the OpenAI Python API to generate structured JSON outputs, which are then loaded by OGM to expand the knowledge graph. | ||
4.3 Natural language question answering
As OGM recursively pushes all generated triples to the knowledge graph, newly generated MOPs can be seamlessly queried by the chatbot Marie,32,58 enabling interactive exploration of calculated properties and their interrelationships. This functionality allows researchers to connect computational results to practical applications, such as screening MOPs with specific properties. Fig. 9 illustrates this through chained queries: the user first queries AMs with octahedral geometry and then identifies the largest pore size among MOPs associated with a selected AM. Next, observing the presence of a specific CBU within the identified MOP inspires a query for other MOPs formed from the same CBU. Marie retrieves detailed information, including structural visualisation and data from both literature and computational predictions. Future work includes integrating OGM into Marie's codebase to enable dynamic data retrieval and modification during interactions. | ||
| Fig. 9 Illustration of Marie's natural language interface for exploratory queries on MOPs data via chained questions. | ||
5 Conclusions
In this work, we introduced twa (The World Avatar) Python package, a comprehensive framework that democratises the creation and management of dynamic knowledge graphs. By abstracting SPARQL queries and ontology manipulation, twa enables developers to build and maintain RDF-based knowledge structures in a familiar Python environment. Its modular design facilitates seamless integration with existing Python ecosystems, while its core object graph mapper (OGM) provides a versatile means of synchronising semantic data between Python classes and RDF-backed graph databases. Through demonstrating how the OGM facilitates the automation of geometry assembly and synthesis protocol extraction for metal–organic polyhedra, we have shown that domain-specific abstractions can significantly streamline the handling of hierarchical and relational data, thus lowering barriers to the broader adoption of knowledge-graph technologies in scientific research.Embedding semantic definitions directly into Python code with twa unlocks powerful automation opportunities in chemical research. As users define classes for molecules, reactions, and protocols, twa automatically generates and maintains the underlying RDF schema. Combined with large language models (LLMs)-driven protocol extraction, which is validated against Pydantic-enforced schemas, this framework ensures high data quality with minimal manual curation and lays the groundwork for agentic experimentation, where AI agents plan, execute, and record syntheses in real time under semantic constraints. By lowering the barrier to semantic web integration, twa accelerates data-centric discovery, promotes interoperability across laboratories, and moves the field toward robust self-driving labs.
Looking ahead, future developments will focus on addressing current limitations while expanding the framework's capabilities. Planned improvements include the integration of continuous integration/deployment (CI/CD) pipelines to ensure consistency across multi-namespace ontologies, optimisations for query performance at scale (such as batched SPARQL queries and asynchronous fetches), support for list-based RDF container (rdf:Seq) and collection (rdf:List) handling in addition to the current set-based default, reasoning capabilities,22 and convenience inverse relationships. Enhancing interoperability with standards—such as SHACL,59 LinkML,60 and the Object-Oriented Linked Data Schema61—is also a key priority, alongside extending SPARQL support to include property-path queries for more expressive graph traversal. To improve OGM usability, we aim to support the automatic generation of Python class hierarchies from existing ontologies. A user interface component is also under consideration to assist non-expert users in defining and navigating semantic schemas. As part of this effort, we are also exploring mechanisms to simplify the manual annotation of relationships in Python classes, with the goal of making schema definition more intuitive for researchers unfamiliar with ontology modelling. Ultimately, these efforts contribute toward the broader vision of autonomous and AI-driven research ecosystems that support more efficient and transparent data-driven digital discovery.
Data availability
All data covered in this research can be automatically queried through TWA-Marie interface (https://theworldavatar.io/demos/marie). The twa Python package is publicly available at the following links:• PyPI: https://pypi.org/project/twa/.
• GitHub: https://github.com/TheWorldAvatar/baselib/tree/main/python_wrapper.
• Documentation: https://theworldavatar.github.io/baselib/.
The OntoMOPs use case using twa is publicly available at: https://github.com/TheWorldAvatar/MOPTools/tree/main/twa_mops.
The above codes presented in this paper (twa Python package and OntoMOPs use case using OGM) are also publicly available on Zenodo at https://zenodo.org/records/15731397 and can be accessed via the DOI: https://doi.org/10.5281/zenodo.15731397.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the National Research Foundation, Prime Minister's Office, Singapore under its Campus for Research Excellence and Technological Enterprise (CREATE) programme. Financial support from the UK Engineering and Physical Sciences Research Council (EPSRC) grant EP/Y016076/1 is also gratefully acknowledged. The authors thank Yiqun Bian for fruitful discussions on geometric vector operations. For the purpose of open access, the author has applied a Creative Commons Attribution (CC BY) licence to any Author Accepted Manuscript version arising. During the preparation of this work the authors used ChatGPT in order to enhance the readability and language of the manuscript. After using this tool, the authors reviewed and edited the content as needed and take full responsibility for the content of the publication.References
- V. Marx, Nature, 2013, 498, 255–260 CrossRef CAS PubMed.
- T. Berners-Lee, J. Hendler and O. Lassila, Sci. Am., 2001, 284, 34–43 CrossRef.
- P. Hitzler, Commun. ACM, 2021, 64, 76–83 Search PubMed.
- N. Noy, Y. Gao, A. Jain, A. Narayanan, A. Patterson and J. Taylor, Commun. ACM, 2019, 62, 36–43 CrossRef.
- A. Kondinski, J. Bai, S. Mosbach, J. Akroyd and M. Kraft, Acc. Chem. Res., 2023, 56, 128–139 CrossRef CAS PubMed.
- P. Strömert, J. Hunold, A. Castro, S. Neumann and O. Koepler, Pure Appl. Chem., 2022, 94, 605–622 CrossRef.
- A. Kondinski, S. Mosbach, J. Akroyd, A. Breeson, Y. R. Tan, S. Rihm, J. Bai and M. Kraft, Chem, 2024, 10, 1071–1083 CAS.
- J. Bai, S. Mosbach, C. J. Taylor, D. Karan, K. F. Lee, S. D. Rihm, J. Akroyd, A. A. Lapkin and M. Kraft, Nat. Commun., 2024, 15, 462 Search PubMed.
- Directorate General for Research and Innovation (European Commission), Turning FAIR into Reality: Final Report and Action Plan from the European Commission Expert Group on FAIR Data, Publications Office, LU, 2018 Search PubMed.
- L. Pascazio, S. Rihm, A. Naseri, S. Mosbach, J. Akroyd and M. Kraft, J. Chem. Inf. Model., 2023, 63, 6569–6586 CrossRef CAS PubMed.
- A. Maedche, B. Motik and L. Stojanovic, VLDB J. Int. J. Very Large Data Bases, 2003, 12, 286–302 Search PubMed.
- A. Paschke and R. Schäfermeier, in OntoMaven – Maven-Based Ontology Development and Management of Distributed Ontology Repositories, Springer International Publishing, 2017, pp. 251–273 Search PubMed.
- K. M. Jablonka, L. Patiny and B. Smit, Nat. Chem., 2022, 14, 365–376 Search PubMed.
- J. Bai, L. Cao, S. Mosbach, J. Akroyd, A. A. Lapkin and M. Kraft, JACS Au, 2022, 2, 292–309 CrossRef CAS PubMed.
- M. Ledvinka and P. Křemen, Semant. Web, 2020, 11, 483–524 Search PubMed.
- Memgraph, GQLAlchemy, 2024, https://github.com/memgraph/gqlalchemy, accessed 29 January 2025 Search PubMed.
- Neo4j, neomodel, 2025, https://github.com/neo4j-contrib/neomodel, accessed 29 January 2025 Search PubMed.
- M. Grove, Empire: RDF for JPA, 2019, https://github.com/mhgrove/Empire, accessed 01 February 2025 Search PubMed.
- The Apache Software Foundation, Apache Jena, 2025, https://jena.apache.org/, accessed 01 February 2025 Search PubMed.
- A. Chadzynski, S. Li, A. Grišišiūtė, J. Chua, M. Hofmeister, J. Yan, H. Y. Tai, E. Lloyd, Y. K. Tsai, M. Agarwal, J. Akroyd, P. Herthogs and M. Kraft, Data-Centric Engineering, 2023, 4, e20 CrossRef.
- K. Klíma, R. Taelman and M. Nečaský, in LDkit: Linked Data Object Graph Mapping Toolkit for Web Applications, Springer Nature Switzerland, 2023, pp. 194–210 Search PubMed.
- J.-B. Lamy, Artif. Intell. Med., 2017, 80, 11–28 CrossRef PubMed.
- S. Clark, F. L. Bleken, S. Stier, E. Flores, C. W. Andersen, M. Marcinek, A. Szczesna-Chrzan, M. Gaberscek, M. R. Palacin, M. Uhrin and J. Friis, Adv. Energy Mater., 2021, 12, 2102702 CrossRef.
- J. Akroyd, S. Mosbach, A. Bhave and M. Kraft, Data-Centric Engineering, 2021, 2, e14 CrossRef.
- Y. R. Tan, M. Hofmeister, S. Z. Phua, G. Brownbridge, K. Rustagi, J. Akroyd, S. Mosbach, A. Bhave and M. Kraft, Beyond Connected Digital Twins – from GIS to The World Avatar, 2024, Preprint at https://como.ceb.cam.ac.uk/preprints/332/ Search PubMed.
- A. M. Bran, S. Cox, O. Schilter, C. Baldassari, A. D. White and P. Schwaller, Nat. Mach. Intell., 2024, 6, 525–535 CrossRef PubMed.
- Z. Ren, Z. Zhang, Y. Tian and J. Li, ChemRxiv, 2023, preprint, DOI:10.26434/chemrxiv-2023-tnz1x-v4.
- D. A. Boiko, R. MacKnight, B. Kline and G. Gomes, Nature, 2023, 624, 570–578 CrossRef CAS PubMed.
- K. Darvish, M. Skreta, Y. Zhao, N. Yoshikawa, S. Som, M. Bogdanovic, Y. Cao, H. Hao, H. Xu, A. Aspuru-Guzik, A. Garg and F. Shkurti, Matter, 2024, 8, 101897 CrossRef.
- J. Bai, K. F. Lee, M. Hofmeister, S. Mosbach, J. Akroyd and M. Kraft, Future Gener. Comput. Syst., 2024, 152, 112–126 CrossRef.
- A. Kondinski, A. Menon, D. Nurkowski, F. Farazi, S. Mosbach, J. Akroyd and M. Kraft, J. Am. Chem. Soc., 2022, 144, 11713–11728 CrossRef CAS PubMed.
- A. Kondinski, P. Rutkevych, L. Pascazio, D. N. Tran, F. Farazi, S. Gangulya and M. Kraft, Digital Discovery, 2024, 3, 2070–2084 RSC.
- X. Zhou, D. Nurkowski, S. Mosbach, J. Akroyd and M. Kraft, J. Chem. Inf. Model., 2021, 61, 3868–3880 CrossRef CAS PubMed.
- S. D. Rihm, J. Bai, A. Kondinski, S. Mosbach, J. Akroyd and M. Kraft, Nexus, 2024, 1, 100004 CrossRef.
- Pydantic, pydantic, 2025, https://github.com/pydantic/pydantic, accessed 31 January 2025 Search PubMed.
- RDFLib, rdflib, 2025, https://github.com/RDFLib/rdflib, accessed 31 January 2025 Search PubMed.
- C. Basca, SuRF: A Python Object RDF Mapper (ORM), 2016, https://github.com/cosminbasca/surfrdf, accessed 13 May 2025 Search PubMed.
- N. F. Noy and D. L. McGuinness, Ontology Development 101: A Guide to Creating Your First Ontology, 2001, https://protegewiki.stanford.edu/images/6/61/First_ontology_guide.pdf, accessed 29 January 2025 Search PubMed.
- J. Akroyd, A. Bhave, G. Brownbridge, E. Christou, M. Hillman, M. Hofmeister, M. Kraft, J. Lai, K. F. Lee, S. Mosbach, D. Nurkowski and O. Parry, CReDo Technical Paper 1: Building a Cross-Sector Digital Twin, 2022, DOI:10.17863/CAM.81779, accessed 26 Jan 2025.
- S. D. Rihm, Y. R. Tan, W. Ang, H. Y. Quek, X. Deng, M. T. Laksana, J. Bai, S. Mosbach, J. Akroyd and M. Kraft, Nexus, 2024, 1, 100031 CrossRef.
- D. J. Tranchemontagne, Z. Ni, M. O'Keeffe and O. M. Yaghi, Angew. Chem., Int. Ed., 2008, 47, 5136–5147 CrossRef CAS PubMed.
- R. Chakrabarty, P. S. Mukherjee and P. J. Stang, Chem. Rev., 2011, 111, 6810–6918 CrossRef CAS PubMed.
- A. J. Gosselin, C. A. Rowland and E. D. Bloch, Chem. Rev., 2020, 120, 8987–9014 CrossRef CAS PubMed.
- A. Kondinski, ChemistryEurope, 2025, 3(3), e202400118 CrossRef CAS.
- A. Kondinski and T. N. Parac-Vogt, J. Chem. Educ., 2019, 96, 601–605 CrossRef CAS.
- A. Kondinski, J. Moons, Y. Zhang, J. Bussé, W. De Borggraeve, E. Nies and T. N. Parac-Vogt, J. Chem. Educ., 2020, 97, 289–294 CrossRef CAS.
- Y.-H. Wang, K.-W. Tong, C.-Q. Chen, J. Du and P. Yang, Chin. Chem. Lett., 2024, 35, 109066 CrossRef CAS.
- A. Kondinski, A. M. Oyarzún, S. D. Rihm, J. Bai, S. Mosbach, J. Akroyd and M. Kraft, Automated Assembly Modelling of Metal-Organic Polyhedra, 2024, Preprint at https://como.ceb.cam.ac.uk/preprints/332/ Search PubMed.
- Y. Zhang, X. Wang, S. Li, B. Song, K. Shao and Z. Su, Inorg. Chem., 2016, 55, 8770–8775 CrossRef CAS PubMed.
- D. A. Gómez-Gualdrón, Y. J. Colón, X. Zhang, T. C. Wang, Y.-S. Chen, J. T. Hupp, T. Yildirim, O. K. Farha, J. Zhang and R. Q. Snurr, Energy Environ. Sci., 2016, 9, 3279–3289 RSC.
- M. A. Addicoat, D. E. Coupry and T. Heine, J. Phys. Chem. A, 2014, 118, 9607–9614 CrossRef CAS PubMed.
- R. Hoffmann, P. v. R. Schleyer and H. F. Schaefer, Angew. Chem., Int. Ed., 2008, 47, 7164–7167 CrossRef PubMed.
- M. Suvarna, A. C. Vaucher, S. Mitchell, T. Laino and J. Pérez-Ramírez, Nat. Commun., 2023, 14, 7964 CrossRef CAS PubMed.
- Q. Zhu, F. Zhang, Y. Huang, H. Xiao, L. Zhao, X. Zhang, T. Song, X. Tang, X. Li, G. He, B. Chong, J. Zhou, Y. Zhang, B. Zhang, J. Cao, M. Luo, S. Wang, G. Ye, W. Zhang, X. Chen, S. Cong, D. Zhou, H. Li, J. Li, G. Zou, W. Shang, J. Jiang and Y. Luo, Natl. Sci. Rev., 2022, 9, nwac190 CrossRef CAS PubMed.
- S. X. Leong, S. Pablo-García, Z. Zhang and A. Aspuru-Guzik, Chem. Sci., 2024, 15, 17881–17891 RSC.
- OpenAI, OpenAI Python API Library, 2025, https://github.com/openai/openai-python, accessed 30 January 2025 Search PubMed.
- S. D. Rihm, F. Saluz, A. Kondinski, J. Bai, S. Mosbach, J. Akroyd and M. Kraft, Extraction of chemical synthesis information using The World Avatar, 2025, Preprint at https://como.ceb.cam.ac.uk/preprints/336/ Search PubMed.
- D. N. Tran, S. D. Rihm, A. Kondinski, L. Pascazio, F. Saluz, S. Mosbach, J. Akroyd and M. Kraft, Natural Language Access Point to Digital Metal-Organic Polyhedra Chemistry in The World Avatar, 2024, Preprint at https://como.ceb.cam.ac.uk/preprints/327/ Search PubMed.
- W3C, Shapes Constraint Language (SHACL), 2025, https://www.w3.org/TR/2017/REC-shacl-20170720/, accessed 28 January 2025 Search PubMed.
- LinkML, LinkML – Linked Open Data Modeling Language, 2025, https://github.com/linkml/linkml, accessed 13 May 2025 Search PubMed.
- S. Stier, OO-LD/schema: v0.1.0, 2024, DOI:10.5281/zenodo.11401727.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5dd00069f |
| This journal is © The Royal Society of Chemistry 2025 |