 Open Access Article
Open Access ArticleAcquisition of absorption and fluorescence spectral data using chatbots†
Masahiko
Taniguchi
 * and
Jonathan S.
Lindsey
* and
Jonathan S.
Lindsey

Department of Chemistry, North Carolina State University, Raleigh, NC 27695-8204, USA. E-mail: mtanigu@ncsu.edu
First published on 16th December 2024
Abstract
The field of photochemistry underpins broad scientific endeavors, encompasses diverse molecular substances, and incorporates descriptions of qualitative and quantitative properties, all of which together may be representative of many scientific disciplines. Yet finding absorption and fluorescence spectra along with companion values of the molar absorption coefficient (ε) and fluorescence quantum yield (Φf) for a given compound is an arduous task even with the most advanced search methods. To gauge whether chatbots could be used to reliably search the literature, the absorption and fluorescence spectra and quantitative parameters (ε and Φf) for 16 popular dyes and fluorophores were sought using ChatGPT 3.5, ChatGPT 4o, Microsoft Copilot, Google Gemini, Gemini advanced, and Meta AI. In most cases, the values of ε and Φf returned by the chatbots accurately cohered with known values from established resources, whereas the retrieval of spectra was only marginally successful. The chatbots were further challenged to find data for fictive compounds (e.g., rhodamine 7G). The results from each chatbot were categorized as follows: “fabricated” (provides numbers that do not exist in the context queried), “fooled” (mis-identifies the compound but does not return any data), “feigned” (acts as if the fictive compound is real but does not provide any data), or “faithful” (responds that the compound is not known or is not available). In summary, the present shortcomings should not cloud the view that chatbots – judiciously used – already provide a valuable resource for the challenging scientific task of finding granular data, and to lesser degree, spectral traces for known compounds.
1. Introduction
The first step in photochemistry is the absorption of light, and accordingly, knowledge of the wavelengths and intensity of absorbed light of a given compound is of utmost importance. Many compounds also emit light, which can be desired or undesired; regardless, knowledge of the wavelengths of the emitted light informs about the energy of the excited state, and the intensity of emitted light provides information about competitive excited-state processes. Accordingly, knowledge of the absorption/fluorescence spectra, the molar absorption coefficient (ε), and the fluorescence quantum yield (Φf) are of fundamental value across the photosciences. Knowledge of these parameters for a given compound impinges on the fields of medical imaging, fluorescence microscopy, photodynamic therapy, photocatalysis, natural and artificial photosynthesis, organic solar cells, and organic light emitting diodes. These photophysical parameters are also central for identification and quantification of diverse species in biochemistry and medicinal chemistry.Over the years, we have been working to assemble a curated database of absorption and fluorescence spectra along with computational modules for carrying out quantitative evaluations commonly encountered in the field.1–4 The term “curated” refers to the presence of considered spectral traces including values of ε and Φf (where available), solvent information, and references to the originating literature. Spectra databases have been prepared that include 339 common compounds,1,2,4 12 natural porphyrins,5 150 chlorophylls,6 14 tolyporphins,7 324 synthetic chlorins,8 73 phyllobilins,9 177 flavonoids10 and 220 bilins;11 altogether for the 1309 compounds there are >2000 absorption and fluorescence spectra in the databases.
The accumulation of curated databases has been a tedious task because the existing search methods are woefully inadequate for finding spectral traces and companion values of ε and Φf.12 A further challenge is assessing the appropriateness of values reported in the published literature. As one example, the reported values of ε and Φf for the benchmark compounds zinc(II)tetraphenylporphyrin and free base tetraphenylporphyrin are known to vary widely among hundreds of published papers.13 Another area of concern is whether light-scattering corrections have been applied upon acquisition of spectra.14 After appropriate spectra are identified in the existing literature, digitization is required to generate the requisite XY dataset of intensity versus wavelength (or wavenumber) that describes a spectrum.15 Collections of spectral traces are more valuable than tabulations of wavelength maxima12,15–17,19 in enabling important assessments such as molecular brightness18 and calculation of the spectral overlap term19 in Förster resonance energy transfer (FRET) processes.20
An alternative to seeking spectral traces is to calculate spectral properties. The in silico prediction of properties of organic molecules is not yet fully satisfactory. For example, density functional theory (DFT) with use of appropriate basis sets and parameters (typically divined by testing against a battery of known members in the target family) can now provide deep insight into electronic structure and the origin of molecular transitions as well as reasonably accurate excitation and emission energies, but not the bandwidths, vibronic progressions, and tails that are part and parcel of spectra of organic compounds in the condensed phase. Knowledge of the full spectra – not merely tabulated wavelengths – is essential for the creation and understanding of photoactive materials; the imaginative design of zero-overlap fluorophores by Flood and coworkers21 and the identification of fascinating pigments in plants by Bastos and coworkers22 may comprise ideal examples. The de novo prediction of a value for the fluorescence quantum yield, which is a consequence of competitive photophysical relaxation processes, is generally beyond the scope of present calculational methods. The question arises, however, concerning the extent to which prediction of spectra of organic molecules can be achieved by artificial intelligence (AI) technologies. For instance, natural language processing (NLP) text mining techniques can be utilized to “scrape” a massive amount of absorption and fluorescence spectral data from the literature.23,24 On the basis of acquired experimental data,25–31 in conjunction with experimental and DFT calculated data32–34 or solely DFT calculated data,35–41 machine learning (deep learning) has been used to predict spectra for organic molecules. The significant challenges to mining the extraordinary wealth of information in the chemistry literature, and possible resolutions to present limitations, have been articulated by Risko and coworkers, taking the venerable task of laboratory recrystallization as a case study.42
Chatbots, of which ChatGPT is perhaps the most popular and representative, are human-like conversational-styled AI software packages that rely on large language models and are integrated into web-based graphical user interfaces. Here, we report the capability of six chatbots for (i) data retrieval of absorption and fluorescence spectral parameters, and (ii) finding absorption and fluorescence spectral traces. The chatbots are ChatGPT (version 3.5 and 4o, OpenAI), Copilot (Microsoft), Gemini and Gemini advanced (Google), and Meta AI (Meta). The spectral traces and data are sought for compounds that are well-known in the fields of photochemistry and photobiology. The core issue is whether chatbots can ameliorate the tedious tasks of finding the spectral traces and critical companion granular information of ε and Φf for specific well-known compounds and thereby accelerate the assembly of curated spectral databases (Fig. 1). A surprising outcome is that regardless of the present shortcomings, we likely are standing at the dawn of chatbots, which already comprise innovative tools for appropriately chosen applications. The integration of AI technologies into photochemistry research should accelerate development of organic molecule-based dyes and fluorophores.
 | ||
| Fig. 1 The development of curated databases of spectra (e.g., PhotochemCAD) has required meticulous searching in the literature, which may be ameliorated through the use of chatbots. | ||
2. Materials and methods
2.1 General
The questions were addressed to four major chatbots and their variants (ChatGPT 3.5, and 4o; Copilot; Gemini and Gemini advanced; Meta AI) and the resulting responses were analyzed manually. ChatGPT 3.5, Copilot, Gemini, and Meta AI are freely accessible through web interfaces, while ChatGPT 4o and Gemini advanced are subscription-based paid platforms (∼$20 per month). Copilot provides control over conversation styles with three different levels depending on the nature of the answer (creative, balanced, precise); the precise style was chosen for this study. All the questions were made from a single user account in each platform in the period May 24–26, 2024. Each question was fed to chatbots only once, which is referred to as a zero-shot prompt.43,442.2 Retrieval of the molar absorption coefficient (ε) and the fluorescence quantum yield (Φf)
The values of ε and Φf were sought for the 16 organic dyes and fluorophores shown in Chart 1 [naphthalene, anthracene, 8-anilinonaphthalene-1-sulfonic acid (ANS), 9,10-diphenylanthracene, quinine, acridine orange, coumarin 1, fluorescein, rhodamine 6G, tetraphenylporphyrin (TPP), chlorophyll a, chlorophyll b, chlorophyll d, chlorophyll f, indocyanine green (ICG), Alexa Fluor 488]. The questions to the chatbots were made as simple as possible without providing details concerning solvents, experimental conditions, and instrumental settings. The following questions were given to the chatbots: (i) What is the molar absorption coefficient of “compound name”? (ii) What is the fluorescence quantum yield of “compound name”? No limitation to the number of words was set so as to gain broader responses from the chatbots. All the chatbot responses are displayed in the ESI.† All values of ε are listed herein with implicit units of cm−1 M−1 unless noted otherwise; the units have been omitted for clarity.2.3 Questions pertaining to fictive dyes and fluorophores
ChatGPT has a somewhat notorious reputation for making up facts and data from relevant information on some occasions due to a lack of deep understanding of the subject.45 To challenge whether chatbots can distinguish fake compounds that at first glance have a veneer of correctness but are not real, the ε or Φf of each of the following six fictive compounds was queried: 10,10-diphenylanthracene (wrong chemical bond structure), coumarin 808, chlorophyll k, Lucifer Red, rhodamine 7G, and Alexa Fluor 850 (Table 1).| Q1 | What is the molar absorption coefficient of 10,10-diphenylanthracene? |
| Q2 | What is the molar absorption coefficient of coumarin 808? |
| Q3 | What is the molar absorption coefficient of chlorophyll k? |
| Q4 | What is the fluorescence quantum yield of Lucifer Red? |
| Q5 | What is the fluorescence quantum yield of rhodamine 7G? |
| Q6 | What is the fluorescence quantum yield of Alexa Fluor 850? |
2.4 Retrieval of absorption and fluorescence spectral traces
To gauge the current chatbot capability of handling graphical images, questions 7–10 were fed to chatbots (Table 2). Questions 7–9 are relatively simple tasks to identify the spectrum of a popular compound (i.e., published in many articles), while question 10 requires domain specific knowledge.| Q7 | Please display the absorption spectrum of beta-carotene |
| Q8 | Please display the absorption spectrum of tetraphenylporphyrin |
| Q9 | Please display the fluorescence spectrum of chlorophyll a |
| Q10 | Please display the spectral overlap integral of the absorption spectrum of Nile Blue green and the fluorescence spectrum of fluorescein |
Data from reliable sources are used for comparison with the responses from the chatbots. The data sources listed in the chatbots upon the question are categorized into four groups: (i) database freely accessible on the internet,46,47 (ii) data from chemical vendors,48–57 (iii) miscellaneous web site58–60 and (iv) published journal articles.61–88 Many chatbots refer to PhotochemCAD databases hosted in the Oregon Medical Laser Center (OMLC)46 as the sources. The program PhotochemCAD and accompanying spectral database of 125 compounds were conceived around 1980 at The Rockefeller University by one of us (J. S. L.),12,15 originated and developed by our group in the mid-late 1980s at Carnegie Mellon University, and first published in 19981 following a move to NC State University in the mid-1990s.3 The goal has always been that the spectral data can be freely downloaded for use by others. Sometime thereafter, the near-entirety of the PhotochemCAD spectral data and companion references were republished on a website at Oregon Medical Laser Center (OMLC). The chatbots may access the latter but not download the spectral data from the original PhotochemCAD site, hence the often-incomplete referencing by the chatbots concerning the true origin of the data (the PhotochemCAD spectral databases were expanded to 150 compounds and published in 2005 as PhotochemCAD 2.2 PhotochemCAD 3 database was expanded to 336 compounds in 2018 (ref. 4) and expanded further to >2000 absorption and fluorescence spectra in the databases as stated in the Introduction.)
3. Results and discussion
3.1 General responses from chatbots
The responses from ChatGPT (3.5 and 4o) are relatively short and are provided without any source or web link. The responses from Copilot provide the source and multiple web links, and the responses can be exported as word, pdf, or txt files. The web links of Copilot were not very well organized at the time of this research. The responses of Gemini (and Gemini advanced) are precise and somewhat wordy, which come with web links whenever sources are available. Gemini (and Gemini advanced) are also equipped with a “double-check response” button, which initiates a Google search to find relevant content from web resources; however, information germane to the response depends on the contents of the responses and is often not found. The responses from Meta AI are well organized with web links as sources of the information in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 relationship.
1 relationship.
3.2 Retrieval of the molar absorption coefficient (ε)
The results for chatbot retrieval of ε values are summarized in Table 3. How to evaluate the accuracy of an ε value found by chatbots? Here, as long as the information provided by chatbots can be matched with a value from the sources, the data are judged as accurate regardless of any disparity from the true values (i.e., authentic, generally accepted values) found in reliable sources. The rationale for this approach is that chatbots are not culpable for errors (real or typographical) that contaminate the sources used for training. If discrepancies are found from the value provided by chatbots and that found in the sources provide by chatbots, the data are judged as inaccurate. For ChatGPT 3.5 and 4o, which do not provide data sources, the accuracy allowance of the data was set at 0.5–2.0 times the values from the reliable sources, a variance chosen given the typical variations of measurement conditions. The ε and absorption maxima (λmax) may vary depending on the solvents, but the choice of solvent was not included in the question. Chatbots may provide more accurate responses if the solvent is specified in the questions; however, doing so crimps the availability of the data. Hence, the questions here were intentionally made as simple as possible.| ChatGPT 3.5 | ChatGPT 4o | Copilot | Gemini | Gemini advanced | Meta AI | Data from reliable sourcesb | |
|---|---|---|---|---|---|---|---|
| a Reported values are significantly different from the authentic, generally accepted data. b Data from PhotochemCAD are curated, and original literature sources are provided therein. | |||||||
| Naphthalene | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400a (216 nm) 400a (216 nm) |
23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700a (220 nm) 700a (220 nm) |
600046 | 600046 | 6000 (275 nm)46 | 600046 | 133![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (220 nm)64 000 (220 nm)64 |
| 6000 (275 nm)47 | |||||||
| See ref. 61–63 and 65 | |||||||
| 1,8-ANS | 5000 (375 nm) | 4950 (350 nm) | 4.9a![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 48 48 |
NA | 4950 (350 nm)48 | NA | 4950 (350 nm)66 |
| 8000 (375 nm)49 | 4000 (375 nm)47 | ||||||
| Anthracene | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 (374 nm) 300 (374 nm) |
8600a (252 nm) | 970046 | 970046 | 9700 (356 nm)46 | 9700 (356.2 nm)46 | 180![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (256 nm)64 000 (256 nm)64 |
| 400a (350 nm) | 9700 (356 nm)47 | ||||||
| 9,10-DPA | 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000a (333 nm)a 000a (333 nm)a |
22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000a (354 nm) 000a (354 nm) |
14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (372.5 nm)46 000 (372.5 nm)46 |
NA | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (372.5 nm)46 000 (372.5 nm)46 |
14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (372.5 nm)46 000 (372.5 nm)46 |
4000 (338 nm) |
| 8740 (354 nm) | |||||||
14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (373 nm) 000 (373 nm) |
|||||||
| 9200 (392 nm)47 | |||||||
| Quinine | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (350 nm) 000 (350 nm) |
5810 (347 nm) | 5700 (349 nm)46 | 5700 (347.5 nm)46 | 5700 (347.5 nm)46 | 5700 (349 nm)46 | 5700 (349 nm)47 |
| Acridine orange | 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (495 nm) 000 (495 nm) |
70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (493 nm) 000 (493 nm) |
27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (430.8 nm)46 000 (430.8 nm)46 |
27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 00050 00050 |
27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000a (492 nm)a 000a (492 nm)a![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 50 |
27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (430.8 nm)46 000 (430.8 nm)46 |
27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (433 nm)47 000 (433 nm)47 |
| See ref. 67–76 | |||||||
| Coumarin 1 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (350 nm)a 000 (350 nm)a |
29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (350 nm)a 000 (350 nm)a |
23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 (373.2 nm)46 500 (373.2 nm)46 |
23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 (373.25 nm)46 500 (373.25 nm)46 |
23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 (373 nm)46 500 (373 nm)46 |
23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 (373.2 nm)46 500 (373.2 nm)46 |
23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 (373 nm)47 500 (373 nm)47 |
| Fluorescein | 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (494 nm) 000 (494 nm) |
83![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (494 nm) 000 (494 nm) |
70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (485 nm)51 000 (485 nm)51 |
70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (485 nm)51 000 (485 nm)51 |
92![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 (490 nm)46 300 (490 nm)46 |
92![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 (500.2 nm)46 300 (500.2 nm)46 |
92![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 (500 nm)47 300 (500 nm)47 |
| Rhodamine 6G | 108![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (525 nm) 000 (525 nm) |
116![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (530 nm) 000 (530 nm) |
116![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (529.8 nm)47 000 (529.8 nm)47 |
116![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (529.75 nm)47 000 (529.75 nm)47 |
116![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (530 nm)46 000 (530 nm)46 |
116![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (529.8 nm)46 000 (529.8 nm)46 |
116![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (530 nm)47 000 (530 nm)47 |
| Chlorophyll a | 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (430 nm) 000 (430 nm) |
117![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (430 nm) 000 (430 nm) |
71![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 (665.5 nm)58,77 400 (665.5 nm)58,77 |
117![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (427.8 nm)46 000 (427.8 nm)46 |
120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (430 nm) 000 (430 nm) |
117![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (427.8 nm)46 000 (427.8 nm)46 |
117![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (429 nm) 000 (429 nm) |
23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000a (660 nm) 000a (660 nm) |
86![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 (662 nm) 300 (662 nm) |
86![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (661 nm)47 000 (661 nm)47 |
|||||
| Chlorophyll b | 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000a (453 nm) 000a (453 nm) |
54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000a (453 nm) 000a (453 nm) |
159![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 (453 nm)46 100 (453 nm)46 |
62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (643.3 nm)78 000 (643.3 nm)78 |
56![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200a (453 nm) 200a (453 nm) |
159![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 (453 nm)56 100 (453 nm)56 |
159![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (453 nm) 000 (453 nm) |
22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000a (642 nm) 000a (642 nm) |
40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (642 nm) 000 (642 nm) |
46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 (642 nm) 900 (642 nm) |
57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 (643 nm)47 600 (643 nm)47 |
||||
| Chlorophyll d | NA | 63![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (402 nm) 000 (402 nm) |
63![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 (697 nm)58,79 700 (697 nm)58,79 |
63![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 680 (697 nm)79 680 (697 nm)79 |
63![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 680 (697 nm)79 680 (697 nm)79 |
NA | 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 740 (400 nm) 740 (400 nm) |
21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000a (662 nm)a 000a (662 nm)a |
44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 410 (455.5 nm) 410 (455.5 nm) |
||||||
63![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 680 (697 nm)79 680 (697 nm)79 |
|||||||
| Chlorophyll f | NA | 71![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (706 nm) 000 (706 nm) |
71![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 (707 nm)58,79 100 (707 nm)58,79 |
57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500a (705 nm)79 500a (705 nm)79 |
71![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000a (437 nm)a 000a (437 nm)a |
NA | 66![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 920 (406.5 nm) 920 (406.5 nm) |
48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000a (740 nm)a 000a (740 nm)a |
56![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800a (706 nm)79 800a (706 nm)79 |
71![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 110 (707 nm)79 110 (707 nm)79 |
|||||
| TPP | 250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (420 nm) 000 (420 nm) |
530![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (419 nm) 000 (419 nm) |
4450a (532 nm)a![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 59 59 |
18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 (515 nm)46 900 (515 nm)46 |
480![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (415 nm) 000 (415 nm) |
18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 (515 nm)46 900 (515 nm)46 |
443![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (419 nm) 000 (419 nm) |
4450a (532 nm)a![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 60 |
18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 (515 nm)47 900 (515 nm)47 |
||||||
| ICG | 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 25 000 to 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (780 to 805 nm) 000 (780 to 805 nm) |
136![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (780 nm) 000 (780 nm) |
NA | 230![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 00052 00052 |
78![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000a (780 nm) 000a (780 nm) |
230![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 00052 00052 |
194![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (789 nm)47 000 (789 nm)47 |
| Alexa 488 | 76![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (495 nm) 000 (495 nm) |
71![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (495 nm) 000 (495 nm) |
73![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 00053 00053 |
73![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 00055 00055 |
91![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 (495 nm) 400 (495 nm) |
73![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (495 nm)53 000 (495 nm)53 |
73![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (495 nm)55 000 (495 nm)55 |
Each chatbot generally retrieved a value for ε, but with accuracy dependent on the given chatbot and particular compound. The values in question have been flagged in Table 3. Chatbots recognize two chief elements of absorption spectra: (i) the ε value depends on the wavelength, and (ii) the absorption spectrum may consist of multiple peaks. Most of the responses from chatbots include information on the wavelength (at λmax) as well as values at other wavelengths (e.g., for multi-banded spectra), whenever applicable, without a specific request in the questions. In general, the data accuracies of ChatGPT 3.5 and 4o (9 out of 16 for both) were inconsistent and hence the two chatbots were unreliable as a sole source of a value of ε. On the other hand, Copilot, Gemini, Gemini advanced and Meta AI were surprisingly reliable. In particular, Copilot and Meta AI did not afford apparently fabricated data; indeed, those cases with wildly disparate data were reported faithfully from the cited source, but the data in the source itself were incorrect. The retrieval results are reported in detail along with our analyses as described below:
(i) The absorption spectra of (polycyclic) aromatic hydrocarbons typically exhibit strong ethylenic bands (E1 and E2 bands) and weak benzenoid bands (B bands).61,63 For example, the absorption spectrum of benzene consists of an E1 band (∼180 nm, ε = 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), E2 band (∼200 nm, ε = 8000), and tiny B band (255 nm, ε = 215).61 Although the E bands are documented in classical articles,61,63–65 such absorption features are often omitted in modern articles and authoritative treatises.62 The apparent rationale for the omission is perhaps not because the absorption is <250 nm, but because the E bands arise from an S0 → S2 transition. Immediate relaxation occurs therefrom to the S1 excited state; hence, E bands do not contribute directly to fluorescence. Some chatbots chose the E1 band for the wavelength for the ε value. Therefore, spectral traces of polycyclic aromatic hydrocarbons (naphthalene, anthracene, 1,8-ANS, and 9,10-DPA) were freshly measured here and are displayed from 200 nm to capture the E bands (Fig. 2). For the spectra in Fig. 2, the ε values of naphthalene and anthracene were applied from representative literature values,64 while those of 1,8-ANS and 9,10-DPA were redetermined herein.
000), E2 band (∼200 nm, ε = 8000), and tiny B band (255 nm, ε = 215).61 Although the E bands are documented in classical articles,61,63–65 such absorption features are often omitted in modern articles and authoritative treatises.62 The apparent rationale for the omission is perhaps not because the absorption is <250 nm, but because the E bands arise from an S0 → S2 transition. Immediate relaxation occurs therefrom to the S1 excited state; hence, E bands do not contribute directly to fluorescence. Some chatbots chose the E1 band for the wavelength for the ε value. Therefore, spectral traces of polycyclic aromatic hydrocarbons (naphthalene, anthracene, 1,8-ANS, and 9,10-DPA) were freshly measured here and are displayed from 200 nm to capture the E bands (Fig. 2). For the spectra in Fig. 2, the ε values of naphthalene and anthracene were applied from representative literature values,64 while those of 1,8-ANS and 9,10-DPA were redetermined herein.
The absorption spectrum of naphthalene comprises a strong E1 band (221 nm, ε = 133![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), multiple E2 bands (∼275 nm, ε = ∼6000), and a weak B band (311 nm, ε = ∼300) (Fig. 2, panel a). ChatGPT 3.5 and 4o chose the E1 band (216 and 220 nm, respectively) for the wavelength; however, the corresponding ε values were ∼5 to ∼9 times less than the actual values. All other chatbots culled the E2 band maxima and quote 6000 as the ε value of naphthalene.
000), multiple E2 bands (∼275 nm, ε = ∼6000), and a weak B band (311 nm, ε = ∼300) (Fig. 2, panel a). ChatGPT 3.5 and 4o chose the E1 band (216 and 220 nm, respectively) for the wavelength; however, the corresponding ε values were ∼5 to ∼9 times less than the actual values. All other chatbots culled the E2 band maxima and quote 6000 as the ε value of naphthalene.
The absorption spectrum of 1,8-ANS exhibits solvent effects; for example, the ε value in water and ethanol vary depending on the sources from 4000 to 8000.47–49,66 The new measurement of the ε value here is 3780 at 354 nm in water and 5810 at 375 nm in ethanol (Fig. 2, panel b). ChatGPT 3.5 applied the data in ethanol (375 nm),66 while ChatGPT 4o adopted the data in water 4950 at 350 nm.48,64 The molar absorption coefficient is listed as EmM unit (equal to cm−1 mM−1) in a catalogue from the dye vendor,48,49 which requires conversion to cm−1 M−1. Gemini advanced successfully converted and responded in proper units, while Copilot was incapable of the unit conversion, in which case the value is listed as is.
The absorption spectrum of anthracene also comprises a strong E1 band (252 nm, ε = 180![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), multiple E2 bands (maxima at 356 nm, ε = 7400 measured here; 9700 in the literature47), and the B band is submerged into the region of the E2 bands (Fig. 2, panel c). ChatGPT 3.5 picked the longest wavelength E2 band (374 nm, ε = 10
000), multiple E2 bands (maxima at 356 nm, ε = 7400 measured here; 9700 in the literature47), and the B band is submerged into the region of the E2 bands (Fig. 2, panel c). ChatGPT 3.5 picked the longest wavelength E2 band (374 nm, ε = 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300), and the value is in an acceptable range. ChatGPT 4o denoted the positions of the E1 (252 nm) and E2 (350 nm) bands correctly; however, the ε values were completely unreasonable. All other chatbots properly employed the data from literature values.47
300), and the value is in an acceptable range. ChatGPT 4o denoted the positions of the E1 (252 nm) and E2 (350 nm) bands correctly; however, the ε values were completely unreasonable. All other chatbots properly employed the data from literature values.47
9,10-DPA exhibits four absorption peaks at wavelengths greater than 300 nm: 338 nm (ε = 4000), 354 nm (ε = 8740), 373 nm (ε = 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), and 392 nm (ε = 9200) (Fig. 2, panel d). These values are based on literature data,47 whereas the data shown in the figure are different by as much as 20%. ChatGPT 3.5 denoted the first peak (333 nm) whereas ChatGPT 4o picked the second peak (354 nm); however, the ε values were overestimated. All other chatbots properly employed data from literature values.47
000), and 392 nm (ε = 9200) (Fig. 2, panel d). These values are based on literature data,47 whereas the data shown in the figure are different by as much as 20%. ChatGPT 3.5 denoted the first peak (333 nm) whereas ChatGPT 4o picked the second peak (354 nm); however, the ε values were overestimated. All other chatbots properly employed data from literature values.47
(ii) All chatbots responded quite well to the questions for the long-established fluorophores quinine, fluorescein, and rhodamine 6G.
(iii) The absorption spectrum of acridine orange is drastically altered by protonation/deprotonation70 and is concentration-dependent due to monomeric and dimeric forms.68,70,75 As determined here, the absorption maximum of acridine orange in ethanol (490 nm, ε = 48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600) is shifted hypsochromically and hypochromically in basic ethanol (431 nm, ε = 21
600) is shifted hypsochromically and hypochromically in basic ethanol (431 nm, ε = 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900) (Fig. 3).
900) (Fig. 3).
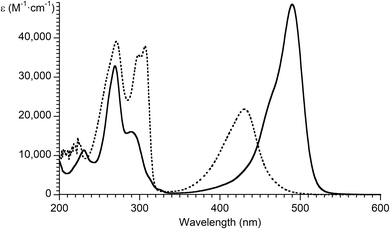 | ||
| Fig. 3 Absorption spectrum of acridine orange in ethanol (solid line) and basic ethanol (dotted line). | ||
The ε values reported in the literature67–76 for acridine orange are summarized together with the data measured herein, as shown in Table 4.
| λ max (nm) | ε (M−1 cm−1) | Solvent | Reference |
|---|---|---|---|
| 490 | 82![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 300 |
Ethanol | 67 |
| 490 | 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
Ethanol | 68 |
| 492 | 63![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 800 |
0.001 M HCl aq | 75 |
| 491.5 | 58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 500 |
Ethanol | 69 |
| 492 | 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
Ethanol | 76 |
| 493 | 53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 560 560 |
Ethanol with H2O and CO2 gas | 70 |
| 490 | 48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 600 |
Ethanol | This work |
| 492 | 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
Aqueous solution below pH 2 | 72 |
| 489 | 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
Aqueous solution | 73 |
| 496 | 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
SDS buffer | 71 |
| 420 | 59![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
Aqueous solution pH 7 | 74 |
| 432 | 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 600 |
Basic ethanol | 70 |
| 431 | 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 900 |
Basic ethanol | This work |
The responses from ChatGPT 3.5 and 4o were based on data in ethanol solution and are in a reasonable range (∼495 nm, ε = 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 or 70
000 or 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000). The responses from Copilot and Meta AI were values in basic ethanol that originate from literature data (431 nm, ε = 27
000). The responses from Copilot and Meta AI were values in basic ethanol that originate from literature data (431 nm, ε = 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000).47 The response from Gemini advanced was affected by the propagation of misplaced values from the commercial vendor's data,50 which are composed of the molar absorption coefficient in basic ethanol (ε = 27
000).47 The response from Gemini advanced was affected by the propagation of misplaced values from the commercial vendor's data,50 which are composed of the molar absorption coefficient in basic ethanol (ε = 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) and the wavelength maxima (492 nm) in ethanol.
000) and the wavelength maxima (492 nm) in ethanol.
(iv) The ε value of coumarin 1 from ChatGPT 3.5 (ε = 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) and 4o (ε = 29
000) and 4o (ε = 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) was close to the value from PhotochemCAD (ε = 23
000) was close to the value from PhotochemCAD (ε = 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500); however, the corresponding absorption wavelength (λmax = 350 nm) is different from the value from PhotochemCAD (λmax = 375 nm). On the other hand, all other chatbots provided the values from PhotochemCAD. The ChatGPTs were found to tend to estimate and give approximate values in the current study, which may stem from the characteristic features of ChatGPTs.
500); however, the corresponding absorption wavelength (λmax = 350 nm) is different from the value from PhotochemCAD (λmax = 375 nm). On the other hand, all other chatbots provided the values from PhotochemCAD. The ChatGPTs were found to tend to estimate and give approximate values in the current study, which may stem from the characteristic features of ChatGPTs.
(v) The absorption spectra of chlorophylls a, b, d, and f are displayed in Fig. 4 to visually guide the comparison described below. The spectra are drawn from a comprehensive database of chlorophyll spectra6 that have been included in PhotochemCAD.
 | ||
| Fig. 4 Absorption spectra of chlorophylls a (green, in diethyl ether), b (black, in diethyl ether), d (blue, in methanol), and f (red, in methanol).6 | ||
The ε value of chlorophyll a from ChatGPT 3.5 was 0.25–0.5 times that of the values from PhotochemCAD, whereas the values from ChatGPT 4o were highly likely taken directly from PhotochemCAD data. The ε values of chlorophyll b from ChatGPT 3.5, ChatGPT 4o and Gemini advanced were also far less than the widely accepted value.
(vi) The ε value of chlorophyll d from ChatGPT 4o is given for the peak position for the Q band (662 nm); however, 662 nm is not a peak maximum. The correct value is 697 nm, and the given molar absorption coefficient (21 000) is 0.3 times that of the accepted value (63 680). Discrepancies of 35 nm in peak position and 3-fold in peak intensity are profound errors in the context of function in a photosynthetic apparatus as well as in many other systems. Gemini advanced returned a source journal reference for chlorophyll d and f wherein the title, journal name, year, and volume were correct; the list of authors was only partially correct (key author Blankenship was excluded whereas the estimable chlorophyll scientist Scheer was erroneously included); and the journal page number was wrong. Such errors were easy to spot. Such errors in this field are referred to as fabrication.
(vii) The ε value of chlorophyll f from ChatGPT 4o fabricated an additional peak (740 nm) that is non-existent. The values for chlorophyll f from Gemini (ε = 57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 at 705 nm) and Gemini advanced (ε = 71
500 at 705 nm) and Gemini advanced (ε = 71![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 at 437 nm and ε = 56
000 at 437 nm and ε = 56![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 at 706 nm) also were fabricated, even though the values were close to those reported in the specified reference by Gemini and Gemini advanced.79 On top of that, the absorption maximum for the near-ultraviolet absorption band (termed the B band) from Gemini advanced (437 nm) was completely incorrect; the correct value is 406.5 nm.
800 at 706 nm) also were fabricated, even though the values were close to those reported in the specified reference by Gemini and Gemini advanced.79 On top of that, the absorption maximum for the near-ultraviolet absorption band (termed the B band) from Gemini advanced (437 nm) was completely incorrect; the correct value is 406.5 nm.
(viii) The ε value of TPP from both Copilot and Meta AI was incorrect (ε = 4450 at 532 nm). The sources were from different web-based homework helpers for students: Bartleby for Copilot and Chegg for Meta AI. The original text material displayed in Bartleby59 was identical with that in Chegg60 as shown in Fig. 5 (also see the ESI† for screenshots of the web site).
 | ||
| Fig. 5 Image of (erroneous) parameter values in a source material59,60 cited by a chatbot. | ||
The absorption spectrum of TPP is comprised of a strong band in the blue region (denoted as B) and a set of comparatively weaker bands in the green-red region (denoted as Q); however, no peak exists at 532 nm. The absorption spectrum of TPP is shown in Fig. 6.8 The source of the material shown in Fig. 5, presumably drawn from a textbook, could not be located regardless of further internet searches or examination of diverse printed materials. The origin of the inaccurate data (peak at 532 nm) is unknown. This subtle but non-negligible incident exemplifies how the propagation on the internet of pernicious errors concerning properties of even the most common benchmark materials can damage scientific research and corrode understanding.
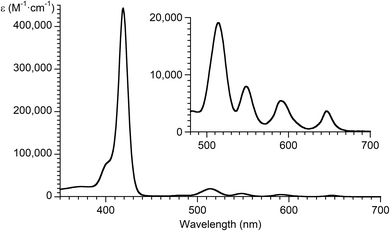 | ||
| Fig. 6 Absorption spectrum of TPP in toluene at room temperature.8 | ||
(ix) The ε value of ICG from Gemini advanced (ε = 78![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) was provided without any sources and was dissimilar to the values from other retrieved data or known from other sources.
000) was provided without any sources and was dissimilar to the values from other retrieved data or known from other sources.
(x) The ε value of Alexa 488 from Gemini advanced (ε = 91![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400) was slightly different from other retrieved data or known from other sources. The disparity appears to be due to the different environment (the value was for Alexa 488 conjugated to secondary antibodies, but no sources were provided).
400) was slightly different from other retrieved data or known from other sources. The disparity appears to be due to the different environment (the value was for Alexa 488 conjugated to secondary antibodies, but no sources were provided).
It is noteworthy that all chatbots provided the wavelength (nm) together with the ε value even though in most cases the question requested only the latter parameter. The ε value varies depending on the wavelength and is senseless without a specified wavelength. An absorption spectrum typically consists of multiple peaks due to the presence of distinct electronic levels often each accompanied by a manifold of vibrational energy levels; therefore, it is not easy to describe a spectrum solely by tabulated numbers without the spectral trace, as shown by the material in this section. The availability of spectra is an essential matter in the photosciences.12,15–17,19
3.3 Retrieval of the fluorescence quantum yield (Φf)
The results of data retrieval for Φf values by chatbots are summarized in Table 5 together with data from reliable sources. The data retrieval for the Φf from chatbots was straightforward and successful for the most part. The Φf values for naphthalene, anthracene, 9,10-DPA, quinine, fluorescein, rhodamine 6G and Alexa Fluor 488 were retrieved accurately by all chatbots.| ChatGPT 3.5 | ChatGPT 4o | Copilot | Gemini | Gemini advanced | Meta AI | Data from reliable sourcesb | |
|---|---|---|---|---|---|---|---|
| a Reported values are significantly different from the authentic, generally accepted data. b Data from PhotochemCAD are curated, and original literature sources are provided therein. | |||||||
| Naphthalene | 0.25 | 0.23 | 0.2346 | 0.2346 | 0.2346 | 0.2346 | 0.2347 |
| 1,8-ANS | 0.28 to 0.33 | 0.001 in H2O | 0.2 to 0.380 | Low | 0.004 in H2Oa![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 81 81 |
0.003 in H2Oa | 0.2447 |
0.154 in ethylene glycola![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 81 81 |
0.004 in H2O82 | ||||||
| Anthracene | 0.28 | 0.27 | 0.3646 | 0.3646 | 0.3646 | 0.3646 | 0.3647 |
| 9,10-DPA | 0.98 | 0.90 to 0.95 | 146 | 0.8–146 | 146 | 146 | 147 |
| Quinine | 0.54 to 0.58 | 0.54 | 0.54646 | 0.54646 | 0.54646 | 0.54646 | 0.54647 |
| Acridine orange | 0.7 to 0.85a | 0.3 to 0.4 | 0.246 | 0.246 | 0.246 | 0.246 | 0.247 |
| Coumarin 1 | 0.15 to 0.40a | 0.73 | 0.5, 0.7346 | 0.5, 0.7346 | 0.7346 | NA | 0.547 |
| Fluorescein | 0.85 to 0.90 | 0.92 | 0.7954 | 0.92583 | 0.92583 | 0.9746 | 0.9747 |
| Rhodamine 6G | 0.95 to 0.99 | 0.95 | 0.9546 | 0.9583 | 0.9583 | 0.9546 | 0.9547 |
| Chlorophyll a | 0.001 to 0.01a | 0.3 | 0.2584 | 0.3246 | 0.3246 | 0.01 to 0.06 deep water88![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a a |
0.3247 |
| Chlorophyll b | 0.003 to 0.01a | 0.16 | 0.11746 | 0.06 to 0.1184 | 0.11746 | 0.11746 | 0.11747 |
| Chlorophyll d | 0.001 to 0.003a | 0.1 | NA | NA | NA | NA | 0.3686 |
| Chlorophyll f | 0.001 to 0.003a | 0.1 | 0.1685 | NA | NA | NA | 0.3986 |
| TPP | 0.15 to 0.25a | 0.11 | 0.1146 | 0.03a to 0.1146 | 0.1146 | 0.1146 | 0.1147 |
| ICG | 0.13 to 0.16a | 0.02 | 0.02587 | 0.0957 | 0.0957 | 0.0452 | 0.0547 |
| Alexa 488 | 0.92 | 0.92 | 0.9256 | 0.9256 | 0.9256 | 0.9256 | 0.9256 |
The responses from ChatGPT 3.5 and 4o are distinctive: dramatic improvements in the accuracy of data retrieval were observed for ChatGPT 4o compared to ChatGPT 3.5. The responses from ChatGPT 3.5 were unreliable; indeed, the responses for the Φf values of chlorophylls indicated each was a weakly fluorescent compound. A major drawback of the ChatGPT family is the absence of reported data sources (e.g., web links or research articles). ChatGPT 4o performed considerably well for the 16 compounds listed here, but of course that does not imply that ChatGPT 4 will afford reliable results for the Φf value of other compounds. Indeed, for a non-expert, the absence of annotation of sources presents a situation where the retrieved values must be taken on faith.
The following are notable points.
(i) The Φf value of 1,8-ANS exhibits a strong solvent dependence and ranges from 0.004 in water to 0.63 in n-octanol.82 The Φf value of 1,8-ANS retrieved from Gemini advanced and Meta AI was actually for 1,8-ANS derivatives – not 1,8-ANS itself – and although those values were very close by coincidence,81 the data retrieval by Gemini advanced and Meta AI are judged as unsuccessful.
(ii) The web links of PubMed are often embedded as sources (especially by Gemini), and in most of the cases, the links are valid; however, an invalid (fabricated) PubMed ID was provided for the Φf value of acridine orange.
(iii) The initial response for chlorophyll a from Meta AI was the Φf value of oceanic phytoplankton, not that of the molecule chlorophyll a. While phytoplankton likely contain chlorophyll a, the former is a living organism whereas the latter is a molecule; the chatbot mixup is non-trivial. A more specific question “What is the fluorescence quantum yield of the chlorophyll a molecule?” to Meta AI generated reasonable answers (0.32 and 0.25).
(iv) The response for TPP from Gemini reflects the lethal problem of the incapability of distinguishing chemical derivatives by generative AI. The response included not only the Φf value of TPP (0.11) but also that of the zinc chelate of TPP, namely Zn-TPP (0.03). This is a common problem for not only all chatbots but also the results from search engines. The responses for TPP also reflect a longstanding problem perhaps appreciated only by the photosciences aficionado – that values of Φf depend on a number of experimental conditions, including whether the solution is aerated or deaerated, and even if aeration is controlled and specified, the reported values can span a distressingly large range.13 The recent consensus values for Φf of TPP are 0.090 in deaerated toluene versus 0.070 in toluene in air,13 replacing a longstanding reliance on the generic value of 0.11 for the Φf of TPP in toluene. The passage of time – and perhaps the advent of more powerful chatbots – may be required for the new values to supplant the old.
(v) To our knowledge, there is only one reported value for the Φf of chlorophyll d [0.36 in benzene],86 and only two values for chlorophyll f [0.39 in benzene86 and 0.16 in pyridine85]. The latter two values were recorded by different research groups and could reflect different experimental methods or true solvent effects. Thus, it is quite understandable that most chatbots have trouble retrieving the data. Note that ChatGPT 4o quoted the Φf value of both chlorophyll d and f as 0.1, which must originate by estimations from other related compounds.
(vi) For chlorophyll f, ChatGPT 3.5 gave two results: one was the fabricated value of 0.001 to 0.003, whereas the other was ‘there is no widely accepted value’.
3.4 Questions about fictive dyes and fluorophores
A question concerning a fictive compound comprises a good test of the reliability of the chatbot. Six fictive compounds were conceived and used for questions with each of the six chatbots examined herein. The responses for non-existing fictive compounds from chatbots are summarized in Table 6. In general, ChatGPT 3.5 and 4o were susceptible to the fictive compounds, Gemini and Gemini advanced were reasonably careful, whereas Copilot and Meta AI were cautious. The questions (Q1–Q6) are listed below followed by additional information concerning the results. The responses can be categorized in one of several ways: (1) numbers are provided that do not exist in the context that is queried, which is referred to by the established term “fabricated”; (2) the responses indicate mis-identification of the fictive compound but no data are returned, which is referred to here as “fooled”; and (3) the responses are as if the fictive compound is real but no data are provided, which is referred to here as “feigned” and is tantamount to a confidence game where there is a superficial appearance of knowledge but nothing beneath, in other words the trickery is only by half; and (4) the responses are that the compound is not known or is not available, which is an undeceived report and here is referred to as “faithful”.| ChatGPT 3.5 | ChatGPT 4o | Copilot | Gemini | Gemini advanced | Meta AI | |
|---|---|---|---|---|---|---|
| Q1 | Fabricated | Fabricated | Faithful | Fabricated | Faithful | Faithful |
| Q2 | Fabricated | Feigned | Faithful | Faithful | Feigned | Faithful |
| Q3 | Feigned | Faithful | Faithful | Faithful | Faithful | Faithful |
| Q4 | Fabricated | Faithful | Feigned | Faithful | Fooled | Fooled |
| Q5 | Fabricated | Fabricated | Faithful | Fabricated | Faithful | Faithful |
| Q6 | Faithful | Fabricated | Faithful | Faithful | Faithful | Faithful |
(Q1) 10,10-Diphenylanthracene: Gemini regarded 10,10-diphenylanthracene as equal to 9,10-diphenylanthracene. Copilot, Gemini advanced, and Meta AI apprised that data for 10,10-diphenylanthracene were not readily available. No chatbots pointed out that the 10,10-diphenyl substitution is chemically wrong and that 10,10-diphenylanthracene is a non-existent compound.
(Q2) Coumarin 808: ChatGPT 3.5 and 4o presumed that coumarin 808 is a known coumarin derivative (which is a typical behavior of ChatGPT) and provided values similar to those of other coumarin derivatives. Gemini advanced reported that “coumarin 808 absorbs light in the near-infrared range,” most likely due to the beguiling number of 808. Such a labeling scheme is common, as exemplified by commercial dyes (e.g., DyLight 800).
(Q3) Chlorophyll k: ChatGPT 3.5 defined chlorophyll k as a recently discovered pigment; otherwise, all other chatbots skipped this booby trap.
(Q4) Lucifer Red: ChatGPT 4o declared that Lucifer Red is not readily available. Copilot regarded Lucifer Red as a compound similar to that of Lucifer Yellow, but did not fabricate any data. Gemini stated that “There isn't a well-established fluorophore called “Lucifer Red”.” On the other hand, Gemini advanced concluded that Lucifer Red is a red-emitting luciferin analog used in bioluminescence imaging. Meta AI deduced that Lucifer Red is a derivative of rhodamine (Lucifer Yellow is an amino-naphthalimide derivative).
(Q5) Rhodamine 7G: ChatGPT 3.5, ChatGPT 4o, and Gemini regarded rhodamine 7G as a synonym of other rhodamine derivatives. Conversely, Copilot, Gemini advanced, and Meta AI recognized that rhodamine 7G is a non-existing fluorophore.
(Q6) Alexa Fluor 850: all chatbots other than ChatGPT 4o clearly discerned that Alexa Fluor 850 is not a valid dye and does not exist.
Again, the intention for giving these tricky questions was not meant to cheat or depreciate the value of chatbots, but rather to show the consequences and capabilities even with zero-shot prompts. By adding (i) appropriate additional prompts (e.g., if you cannot find the relevant information, please say “I don't know”) or (ii) using few-shot prompts (first ask if the titled compounds exist or not, then provide additional questions), chatbots should be able to respond honestly.
3.5 Retrieval of absorption and fluorescence spectral traces
The capability of chatbots for retrieval of absorption and fluorescence spectral traces was examined next. Questions Q7–Q9 concerned the spectra for beta-carotene, tetraphenylporphyrin (TPP), and chlorophyll a, respectively. The results are provided in Table 7.| ChatGPT 3.5 | ChatGPT 4o | Copilot | Gemini | Gemini advanced | Meta AI | |
|---|---|---|---|---|---|---|
| Q7 | ASCII art | Gaussian | — | Spectrum | Spectrum | Tabulated |
| Q8 | ASCII art | Gaussian | — | Spectrum | Spectrum | Tabulated |
| Q9 | ASCII art | Gaussian | — | Spectrum | Spectrum | Tabulated |
| Q10 | — | Gaussian | — | — | Spectra (false) | Tabulated |
ChatGPT 3.5 tried to display absorption and fluorescence spectra by ASCII art, yet the generated graphics were nonsensical and unsatisfactory. ChatGPT 4o created spectral traces by applying a Gaussian distribution, which was a good upgrade from that of ChatGPT 3.5. Nonetheless, the spectra were of limited use due to lack of information such as the full-width-at-half-maximum (fwhm), the intensity ratio of each peak, and the inclusion of multiple peaks. Copilot and Meta AI lack capabilities for drawing spectra in graphics. Gemini and Gemini advanced pulled out the corresponding spectra in graphical form together with web links of sources, a function equivalent to that of a Google image search.
Question 10 requested the spectral overlap integral derived from the absorption spectrum of Nile Blue and the fluorescence spectrum of fluorescein. Such an overlap entails the unusual reverse FRET – in other words, uphill energy transfer from fluorescein to Nile Blue. The question is demanding by requiring immensely specific data that may be neither published nor available. No chatbot could provide a suitable response. The attempt by Gemini advanced displayed the homo overlap of the absorption and fluorescence spectra of rhodamine 6G, and hence was a failure.
3.6 Perspective on methods and results
The work described herein represents the evaluation of six chatbots for performance in response to granular questions in the photosciences. As the development of AI is a rapidly evolving field, clarity and perspective are warranted concerning the methods employed for this particular study. The following points are germane.(1) It is known that Gemini, Meta AI, and Copilot leverage both search engine results and LLMs (thereby accessing a wide range of literature), whereas GPT 3.5 and GPT 4o rely solely on their training data. An alternative means of comparison could rely on use of an application programming interface (API), which enables local operation of the chatbots independent from servers.89 Accessing chatbots via an API can automate tasks by batch processing, which is an efficient approach compared to that of a web user interface (WUI), which is time consuming given the reliance on manual input.90,91 Similar accuracy rates were typically obtained by both WUI and API approaches for GPT-4V,92 yet the processing time for data extraction from scientific graphs via an API can be 30 times faster than that of a WUI.92 With speed comes cost, however: an API can cost ∼6 times more ($125 per month) than a WUI ($20 per month).92 The tasks examined herein were relatively simple given that limited numbers of photophysical parameters were examined; thus, WUI-based chatbots were employed. Data extraction in bulk upon submitting questions concerning photophysical parameters may enjoy benefits from use of an API in the future.
(2) Chatbots are known to generate different answers when the identical question is repeated.93,94 An identical question was fed to GPT 4.1 for three subjects five times (see the ESI†). For those photophysical parameter retrieval tasks that are relatively straightforward, almost identical responses were obtained. Thus, each question was asked only once thereafter.
(3) Chatbots are known to be sensitive to how a prompt is formulated.94 A prompt can be engineered to improve the quality of the responses from chatbots without tedious fine-tuning of the training data.95 For example, repeatedly modifying the questions can engender responses in a desired format.96 Examples in this domain include adding conditions, pinpointing solvents, indicating the environment (pH, bound to protein), limiting the phase (solid or liquid or gas), specifying the composition (e.g., molecules), and so forth.
(4) During the preparation and review of this manuscript, a new version of ChatGPT (GPT 4.1) was released. Are the results presented herein already out of date? Some domain specific tasks may enjoy greater benefits due to the features of GPT 4.1; however, a foray using GPT 4.1 revealed little improvement for the retrieval of values for ε and Φf. The results are summarized in the ESI (Tables S1 and S2†). GPT 4.1 is not immune to questions involving fictive dyes and fluorophores. GPT 4.1 does exhibit notable improvements for the graphical display of spectra, albeit utilizing Gaussian distributions.
4. Outlook
Searching the scientific literature for specific granular data as well as spectra has been a surprisingly difficult task. Said differently, it is very hard to glean from the immense scientific literature – estimated at >108 publications12 – those specific papers wherein a spectrum or quantitative parameter is located. The difficulty has severely crimped the ability to assemble curated databases of spectra, as in PhotochemCAD, with which we have had direct experience for nearly 40 years. The advent of conversational AI chatbots, made possible by transformer-based large language models, offers potential advances in finding such information. While the potential impact of chatbots on creativity in science remains unclear, merely improved information acquisition is expected to constitute a substantial benefit.It is clear that the present chatbots are of limited reliability for tasks that require broad processing, such as unguided education in the photosciences.97 The results shown in Fig. 5 herein and in the accompanying text substantiate this conclusion. On the other hand, extant chatbots are of significant benefit already for punctate and singular tasks such as identification of the values of photophysical parameters that otherwise are often buried deeply in the vast scientific literature.
What is the significant difference between chatbots and extant search engines? Chatbots provide clear-cut definitive answers (a double-edged sword), which can save time and accelerate acquisition of desired information; in this regard, chatbots surpass present search engines in terms of efficiency. Chatbots do not generate novel data from scratch, but create sentences by simply adding the most suited words by following natural language theories utilizing trained data. The knowledge of chatbots acquired through training processes relies heavily on internet resources, causing the knowledge of chatbots to overlap heavily with web resources. Thus, the information accessible to chatbots and search engines has partial commonality. The well-deserved criticisms leveled at chatbots may reflect shortcomings that are not entirely idiosyncratic to chatbots, but rather arise from the nature of the source materials, particularly internet web resources. As an example, an erroneous value of a parameter in a textbook (due to author error or publication production error) could enter a website or scientific publication, and from the book or the latter sources be accessed by a chatbot. The thread-like lineage of such values often is frayed if not clipped. In other words, the propagation of mistakes and typographical errors across the internet may arise from decades-old (if not centuries-old), traditional printed materials, not chatbots.
Chatbots may not always provide accurate information, thus the domain-specific expert needs to inspect the answers carefully. With the spectral databases of PhotochemCAD in hand, evaluations of ε and Φf are straightforward tasks for compounds represented therein. An outcome of the present study indicates the importance of using multiple chatbots to elicit results followed by evaluation by the domain-specific expert. Multiple chatbots are readily accessible, and others are under development. The possibility exists of course that all chatbots elicit identical, incorrect responses; however, the current study already demonstrates the diversity of responses from chatbots at least in the photosciences field. Even inaccurate or incorrect results often have subtleties that may warrant further scrutiny. In summary, the chatbots examined here are quite effective (but not universally so) for retrieval of granular data (ε and Φf) of considerable importance in the photosciences, are only marginally effective for finding spectral traces, and can be susceptible to inquiries concerning (intentionally or inadvertently) fictive compounds. Molecular design in the photosciences can make use of information beyond absorption and fluorescence spectra; for example, a database of the yield of intersystem crossing, phosphorescence spectrum, and triplet state lifetime would enable design of molecules for diverse photoprocesses.98–100 In sum, chatbots would appear to be in their infancy, yet if judiciously applied, already offer a valuable means for searching the scientific literature, for which new strategies are urgently required.
Data availability
All data used herein are contained in the body of the paper and the companion ESI.†Conflicts of interest
The authors declare no conflicts of interestAcknowledgements
This work was supported by a grant from the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy (DE-SC0025317), and by NC State University. We thank the reviewers for constructive suggestions.References
- H. Du, R.-C. A. Fuh, J. Li, L. A. Corkan and J. S. Lindsey, Photochem. Photobiol., 1998, 68, 141–142, DOI:10.1111/j.1751-1097.1998.tb02480.x.
- J. M. Dixon, M. Taniguchi and J. S. Lindsey, Photochem. Photobiol., 2005, 81, 212–213, DOI:10.1111/j.1751-1097.2005.tb01544.x.
- M. Taniguchi, H. Du and J. S. Lindsey, Photochem. Photobiol., 2018, 94, 277–289, DOI:10.1111/php.12862.
- M. Taniguchi and J. S. Lindsey, Photochem. Photobiol., 2018, 94, 290–327, DOI:10.1111/php.12860.
- A. R. M. Soares, Y. Thanaiah, M. Taniguchi and J. S. Lindsey, New J. Chem., 2013, 37, 1087–1097, 10.1039/C3NJ41042K.
- M. Taniguchi and J. S. Lindsey, Photochem. Photobiol., 2021, 97, 136–165, DOI:10.1111/php.13319.
- T. J. O'Donnell, J. R. Gurr, J. Dai, M. Taniguchi, P. G. Williams and J. S. Lindsey, New J. Chem., 2021, 45, 11481–11494, 10.1039/D1NJ02108G.
- M. Taniguchi, D. F. Bocian, D. Holten and J. S. Lindsey, J. Photochem. Photobiol., C, 2022, 52, 100513, DOI:10.1016/j.jphotochemrev.2022.100513.
- C. A. Karg, M. Taniguchi, J. S. Lindsey and S. Moser, Planta Med., 2023, 89, 637–662, DOI:10.1055/a-1955-4624.
- M. Taniguchi, C. A. LaRocca, J. D. Bernat and J. S. Lindsey, J. Nat. Prod., 86, 1087–1119, DOI:10.1021/acs.jnatprod.2c00720.
- M. Taniguchi and J. S. Lindsey, J. Photochem. Photobiol., C, 2023, 55, 100585, DOI:10.1016/j.jphotochemrev.2023.100585.
- M. Taniguchi and J. S. Lindsey, Proc. SPIE, 2020, 11256, 112560J, DOI:10.1117/12.2542859.
- M. Taniguchi, J. S. Lindsey, D. F. Bocian and D. Holten, J. Photochem. Photobiol., C, 2021, 46, 100401, DOI:10.1016/j.jphotochemrev.2020.100401.
- M. Taniguchi and J. S. Lindsey, Proc. SPIE, 2024, 12862, 128620B, DOI:10.1117/12.3000407.
- M. Taniguchi, Z. Wu, C. Sterling and J. S. Lindsey, Proc. SPIE, 2023, 12398, 1239806, DOI:10.1117/12.2651694.
- Y. Guo, Z. Xu, A. E. Norcross, M. Taniguchi and J. S. Lindsey, Proc. SPIE, 2019, 10893, 108930O, DOI:10.1117/12.2508077.
- Z. Wu, A. Kittinger, A. E. Norcross, M. Taniguchi and J. S. Lindsey, Proc. SPIE, 2021, 116600, 116600I, DOI:10.1117/12.2577840.
- M. Taniguchi, G. Hu, R. Liu, H. Du and J. S. Lindsey, Proc. SPIE, 2018, 10508, 1050806, DOI:10.1117/12.2302709.
- Q. Qi, M. Taniguchi and J. S. Lindsey, J. Chem. Inf. Model., 2019, 59, 652–667, DOI:10.1021/acs.jcim.8b00753.
- J. S. Lindsey, M. Taniguchi, D. F. Bocian and D. Holten, Chem. Phys. Rev., 2021, 2, 011302, DOI:10.1063/5.0041132.
- A. Dhara, T. Sadhukhan, E. G. Sheetz, A. H. Olsson, K. Raghavachari and A. H. Flood, J. Am. Chem. Soc., 2020, 142, 12167–12180, DOI:10.1021/jacs.0c02450.
- B. C. Freitas-Dörr, C. O. Machado, A. C. Pinheiro, A. B. Fernandes, F. A. Dörr, E. Pinto, M. Lopes-Ferreira, M. Abdellah, J. Sá, L. C. Russo, F. L. Forti, L. C. P. Gonçalves and E. L. Bastos, Sci. Adv., 2020, 6, eaaz0421, DOI:10.1126/sciadv.aaz0421.
- E. J. Beard, G. Sivaraman, Á. Vázquez-Mayagoitia, V. Vishwanath and J. M. Cole, Sci. Data., 2019, 6, 307, DOI:10.1038/s41597-019-0306-0.
- J. F. Joung, M. Han, M. Jeong and S. Park, Sci. Data., 2022, 7, 295, DOI:10.1038/s41597-020-00634-8.
- R. S. Da Silva, L. F. Marins, D. V. Almeida, K. Dos Santos Machado and A. V. Werhli, Comput. Biol. Chem., 2019, 83, 107089, DOI:10.1016/j.compbiolchem.2019.107089.
- Z.-R. Ye, I.-S. Huang, Y.-T. Chan, Z.-J. Li, C.-C. Liao, H.-R. Tsai, M.-C. Hsieh, C.-C. Chang and M.-K. Tsai, RSC Adv., 2020, 10, 23834–23841, 10.1039/D0RA05014H.
- C.-W. Ju, H. Bai, B. Li and R. Liu, J. Chem. Inf. Model., 2021, 61, 1053–1065, DOI:10.1021/acs.jcim.0c01203.
- J. F. Joung, M. Han, J. Hwang, M. Jeong, D. H. Choi and S. Park, JACS Au, 2021, 1, 427–438, DOI:10.1021/jacsau.1c00035.
- J. F. Joung, M. Han, M. Jeong and S. Park, J. Chem. Inf. Model., 2022, 62, 2933–2942, DOI:10.1021/acs.jcim.2c00173.
- J. F. Jeong, M. Joung, J. Hwang, M. Han, C. W. Koh, D. H. Choi and S. Park, npj Comput. Mater., 2022, 8, 147, DOI:10.1038/s41524-022-00834-3.
- A. A. Ksenofontov, M. M. Lukanov and P. S. Bocharov, Spectrochim. Acta, Part A, 2022, 279, 121442, DOI:10.1016/j.saa.2022.121442.
- J. Wang, J. Jin, Y. Geng, S. Sun, H. Xu, Y. Lu and Z. Su, J. Comput. Chem., 2013, 34, 566–575, DOI:10.1002/jcc.23168.
- K. P. Greenman, W. H. Green and R. Gómez-Bombarelli, Chem. Sci., 2022, 13, 1152–1162, 10.1039/D1SC05677H.
- A. D. McNaughton, R. P. Joshi, C. R. Knutson, A. Fnu, K. J. Luebke, J. P. Malerich, P. B. Madrid and N. Kumar, J. Chem. Inf. Model., 2023, 63, 1462–1471, DOI:10.1021/acs.jcim.2c01662.
- G. Montavon, M. Rupp, V. Gobre, A. Vazquez-Mayagoitia, K. Hansen, A. Tkatchenko, K.-R. Müller and O. Anatole Von Lilienfeld, New J. Phys., 2013, 15, 095003, DOI:10.1088/1367-2630/15/9/095003.
- R. Ramakrishnan, M. Hartmann, E. Tapavicza and O. A. Von Lilienfeld, J. Chem. Phys., 2015, 143, 084111, DOI:10.1063/1.4928757.
- M. Nakata and T. Shimazaki, J. Chem. Inf. Model., 2017, 57, 1300–1308, DOI:10.1021/acs.jcim.7b00083.
- K. Ghosh, A. Stuke, M. Todorović, P. B. Jørgensen, M. N. Schmidt, A. Vehtari and P. Rinke, Adv. Sci., 2019, 6, 1801367, DOI:10.1002/advs.201801367.
- B. Kang, C. Seok and J. Lee, J. Chem. Inf. Model., 2020, 60, 5984–5994, DOI:10.1021/acs.jcim.0c00698.
- J. Westermayr and P. Marquetand, J. Chem. Phys., 2020, 153, 154112, DOI:10.1063/5.0021915.
- K. Singh, J. Münchmeyer, L. Weber, U. Leser and A. Bande, J. Chem. Theory Comput., 2022, 18, 4408–4417, DOI:10.1021/acs.jctc.2c00255.
- A. Smith, V. Bhat, Q. Ai and C. Risko, Chem. Mater., 2022, 34, 4821–4827, DOI:10.1021/acs.chemmater.2c00445.
- D. Hu, B. Liu, X. Zhu, X. Lu and N. Wu, Int. J. Med. Inf., 2024, 183, 105321, DOI:10.1016/j.ijmedinf.2023.105321.
- X. Wei, X. Cui, N. Cheng, X. Wang, X. Zhang, S. Huang, P. Xie, J. Xu, Y. Chen, M. Zhang, Y. Jiang and W. Han, arXiv, 2024, Preprint, arXiv:2302.10205v2, DOI:10.48550/arXiv.2302.10205.
- A. Azaria1, R. Azoulay and S. Reches, Data Intell., 2024, 6, 240–296, DOI:10.1162/dint_a_00235.
- Oregon Medical Laser Center, PhotoChemCAD Chemicals, https://omlc.org/spectra/PhotochemCAD/index.html, accessed 2024-07-10.
- PhotoChemCAD, Common Compounds Spectra Database, https://www.photochemcad.com/databases/common-compounds, accessed 2024-07-10.
- MilliporeSigma, 8-Anilino-1-naphthalenesulfonic acid ammonium salt, https://www.sigmaaldrich.com/deepweb/assets/sigmaaldrich/product/documents/362/696/a3125pis.pdf, accessed 2024-07-10.
- MPbio, 8-Anilino-1-Naphthalene Sulfonic Acid,https://www.mpbio.com/us/8-anilino-1-naphthalene-sulfonic-acid, accessed 2024-07-10.
- AAT Bioquest, Acridine orange,https://www.aatbio.com/products/acridine-orange-10-mg-ml-solution-in-water?unit=17503, accessed 2024-07-10.
- AAT Bioquest, Fluoresceins,https://www.aatbio.com/catalog/fluoresceins, accessed 2024-07-10.
- AAT Bioquest, Indocyanine Green,https://www.aatbio.com/catalog/indocyanine-green, accessed 2024-07-10.
- AAT Bioquest, Extinction Coefficient Alexa Fluor,https://www.aatbio.com/resources/extinction-coefficient/alexa_fluor_488, accessed 2024-07-10.
- AAT Bioquest, What is the quantum yield of fluorescein,https://www.aatbio.com/resources/faq-frequently-asked-questions/What-is-the-quantum-yield-of-fluorescein, accessed 2024-07-10.
- Thermo Fisher Scientific, The Alexa Fluor Dye Series—Note 1.1,https://www.thermofisher.com/us/en/home/references/molecular-probes-the-handbook/technical-notes-and-product-highlights/the-alexa-fluor-dye-series.html, accessed 2024-07-10.
- Thermo Fisher Scientific, Fluorescence quantum yields (QY) and lifetimes (τ) for Alexa Fluor dyes—Table 1.5,https://www.thermofisher.com/us/en/home/references/molecular-probes-the-handbook/tables/fluorescence-quantum-yields-and-lifetimes-for-alexa-fluor-dyes.html, accessed 2024-07-10.
- Lumiprobe, Indocyanine Green (ICG), https://www.lumiprobe.com/p/icg-3599-32-4, accessed 2024-07-10.
- Plants in Action, 1.2.2 - Chlorophyll absorption and photosynthetic action spectra,https://rseco.org/content/122-chlorophyll-absorption-and-photosynthetic-action-spectra.html, accessed 2024-07-10.
- Bartleby.com Answered iv.Tetraphenylporphyrin (TPP) has a molar absorption coefficient of 4450 M-1cm-1 at 532 nm , https://www.bartleby.com/questions-and-answers/iv.-tetraphenylporphyrin-tpp-has-a-molar-absorption-coefficient-of-4450-m-cm-at-532-nm.-tpp-has-a-fl/41c9058f-5229-4426-afda-56df52a0732e, accessed 2024-07-10.
- Chegg.com, Solved iv. Tetraphenylporphyrin (TPP) has a molar absorption,https://www.chegg.com/homework-help/questions-and-answers/iv-tetraphenylporphyrin-tpp-molar-absorption-coefficient-4450-mathrm-m-1-mathrm-%7Ecm-1-532--q104927229, accessed 2024-07-10.
- R. M. Silverstein, G. C. Bassler and T. C. Morrill, Spectrometric identification of organic compounds, John Wiley & Sons, New York, 5th edn, 1991, pp. 289–315 Search PubMed.
- I. B. Berlman, Handbook of Fluorescence Spectra of Aromatic Molecules, Academic Press, New York, 2nd edn, 1971 Search PubMed.
- B. A. Braude, Annu. Rep. Prog. Chem., 1945, 42, 105–130 Search PubMed.
- H. B. Klevens and J. R. Platt, J. Chem. Phys., 1949, 17, 470–481, DOI:10.1063/1.1747291.
- W. V. Mayneord and E. M. F. Roe, Proc. R. Soc. London, Ser. A, 1935, 52, 299–324, DOI:10.1098/rspa.1935.0193.
- A. Azzi, Methods Enzymol., 1974, 32, 234–246, DOI:10.1016/0076-6879(74)32024-1.
- E. J. Ngen, P. Rajaputra and Y. You, Bioorg. Med. Chem., 2009, 17, 6631–6640, DOI:10.1016/j.bmc.2009.07.074.
- N. Mataga, Bull. Chem. Soc. Jpn., 1957, 30, 375–379, DOI:10.1246/bcsj.30.375.
- E.-Y. Cho, J.-M. Gu, I.-H. Choi, W.-S. Kim, Y.-K. Hwang, S. Huh, S.-J. Kim and Y. Kim, Cryst. Growth Des., 2014, 14, 5026–5033, DOI:10.1021/cg5005837.
- J. Ferguson and A. W. H. Mau, Chem. Phys. Lett., 1972, 17, 543–546, DOI:10.1016/0009-2614(72)85101-7.
- M. N. Berberan-Santos, M. J. E. Prieto and A. G. Szabo, J. Chem. Soc., Faraday Trans., 1992, 88, 255–261, 10.1039/FT9928800255.
- P. Sawunyama and S. B. Jonnalagadda, J. Phys. Org. Chem., 1995, 8, 175–185, DOI:10.1002/poc.610080308.
- D. A. Makarov, N. A. Kuznetsova and O. L. Kaliya, Russ. J. Phys. Chem., 2006, 80, 268–274, DOI:10.1134/S0036024406020270.
- J. K. Ghosh, A. K. Mandal and M. K. Pal, Spectrochim. Acta, Part A, 1999, 55, 1877–1886, DOI:10.1016/S1386-1425(99)00046-3.
- L. Costantlno, G. Guarino, O. Ortona and V. Vttagllano, J. Chem. Eng. Data, 1984, 29, 62–66, DOI:10.1021/je00035a021.
- R. Caramazza, L. Costantino and V. Vitagliano, Ric. Sci., Parte 2 Sez. A, 1964, 34, 67–73 Search PubMed.
- R. J. Porra, W. A. Thompson and P. E. Kriedmann, Biochim. Biophys. Acta, 1989, 975, 384–394, DOI:10.1016/S0005-2728(89)80347-0.
- S. W. Jeffrey, R. F. C. Mantoura and T. Bjørnland, in Phytoplankton Pigments in Oceanography: Guidelines to Modern Methods, ed. S. W. Jeffrey, R. F. C. Mantoura and S. W. Wright, UNESCO Publishing, Paris, 1997, pp. 449–559 Search PubMed.
- Y. Li, N. Scales, R. E. Blankenship, R. D. Willows and M. Chen, Biochim. Biophys. Acta, 2012, 1817, 1292–1298, DOI:10.1016/j.bbabio.2012.02.026.
- D. H. Haynes and H. Staerk, J. Membr. Biol., 1974, 17, 313–340, DOI:10.1007/BF01870190.
- N. Wang, E. B. Faber and G. I. Georg, ACS Omega, 2019, 4, 18472–18477, DOI:10.1021/acsomega.9b03002.
- L. Stryer, J. Mol. Biol., 1965, 13, 482–495, DOI:10.1016/S0022-2836(65)80111-5.
- D. Magde, R. Wong and P. G. Seybold, Photochem. Photobiol., 2002, 75, 327–334, DOI:10.1562/0031-8655(2002)0750327FQYATR2.0.CO2.
- L. S. Forster and R. Livingston, J. Chem. Phys., 1952, 20, 1315–1320, DOI:10.1063/1.1700727.
- D. M. Niedzwiedzki, H. Liu, M. Chen and R. E. Blankenship, Photosynth. Res., 2014, 121, 25–34, DOI:10.1007/s11120-014-9981-z.
- M. Kobayashi, Y. Sorimachi, D. Fukayama, H. Komatsu, T. Kanjoh, K. Wada, M. Kawachi, H. Miyashita, M. Ohnishi-Kameyama and H. Ono, in Handbook of Photosynthesis, ed. M. Pessarakli, CRC Press, Florida, 2016, pp. 95–147 Search PubMed.
- T. Jin, S. Tsuboi, A. Komatsuzaki, Y. Imamura, Y. Muranaka, T. Sakata and H. Yasuda, Med. Chem. Commun., 2016, 7, 623–631, 10.1039/C5MD00580A.
- S. Maritorena, A. Morel and B. Gentili, Appl. Opt., 2000, 39, 6725–6737, DOI:10.1364/AO.39.006725.
- M. P. Polak, S. Modi, A. Latosinska, J. Zhang, C.-W. Wang, S. Wang, A. D. Hazra and D. Morgan, Digital Discovery, 2024, 3, 1221–1235, 10.1039/d4dd00016a.
- Z. Zheng, O. Zhang, C. Borgs, J. T. Chayes and O. M. Yaghi, J. Am. Chem. Soc., 2023, 145, 18048–18062, DOI:10.1021/jacs.3c05819.
- K. G. Yager, Digital Discovery, 2023, 2, 1850–1861, 10.1039/d3dd00112a.
- Z. Zheng, Z. He, O. Khattab, N. Rampal, M. A. Zaharia, C. Borgs, J. T. Chayes and O. M. Yaghi, Digital Discovery, 2024, 3, 491–501, 10.1039/d3dd00239j.
- T. Guo, K. Guo, B. Nan, Z. Liang, Z. Guo, N. V. Chawla, O. Wiest and X. Zhang, arXiv, 2023, Preprint, arXiv:2305.18365v3, DOI: DOI:10.48550/arXiv.2305.18365.
- L. S. Balhorn, J. M. Weber, S. Buijsman, J. R. Hildebrandt, M. Ziefle and A. M. Schweidtmann, Sci. Rep., 2024, 14, 4998, DOI:10.1038/s41598-024-54936-7.
- M. P. Polak and D. Morgan, Nat. Commun., 2024, 15, 1569, DOI:10.1038/s41467-024-45914-8.
- W. Zhang, Q. Wang, X. Kong, J. Xiong, S. Ni, D. Cao, B. Niu, M. Chen, Y. Li, R. Zhang, Y. Wang, L. Zhang, X. Li, Z. Xiong, Q. Shi, Z. Huang, Z. Fu and M. Zheng, Chem. Sci., 2024, 15, 10600–10611, 10.1039/D4SC00924J.
- M. Taniguchi and J. S. Lindsey, Photochem. Photobiol., 2024 DOI:10.1111/php.14037.
- T. V. Esipova, M. J. P. Barrett, E. Erlebach, A. E. Masunov, B. Weber and S. A. Vinogradov, Cell Metab., 2019, 29, 736–744, DOI:10.1016/j.cmet.2018.12.022.
- W. Li, P. Chasing, P. Nalaoh, T. Chawanpunyawat, N. Chantanop, C. Sukpattanacharoen, N. Kungwan, P. Wongkaew, T. Sudyoadsuka and V. Promarak, J. Mater. Chem. C, 2022, 10, 9968–9979, 10.1039/D2TC01406H.
- D. Kim, M. C. Rosko, F. N. Castellano, T. G. Gray and T. S. Teets, J. Am. Chem. Soc., 2024, 146, 19193–19204, DOI:10.1021/jacs.4c04288.
Footnote |
| † Electronic supplementary information (ESI) available: Retrieval of the molar absorption coefficient (ε) and the fluorescence quantum yield (Φf). Questions involving fictive dyes and fluorophores. Retrieval of absorption and fluorescence spectral traces. Comparison of results from ChatGPT 4o and GPT 4.1. See DOI: https://doi.org/10.1039/d4dd00255e |
| This journal is © The Royal Society of Chemistry 2025 |


