Open Access Article
Open Access ArticleDigitalisation of catalytic processes for sustainable production of biobased chemicals and exploration of wider chemical space
Firdaus
Parveen
* and
Anna G.
Slater

Department of Chemistry and Materials Innovation Factory, School of Environmental Sciences, University of Liverpool, Liverpool, UK. E-mail: fparveen@liverpool.ac.uk
First published on 29th January 2025
Abstract
Global warming and the depletion of petroleum resources require immediate and focused attention, and there is a pressing need to accelerate progress. Digital approaches can be leveraged in these efforts, for example in exploring effective replacements for petrochemicals or effectively identifying molecules with better performance. One such potential replacement is lignocellulosic biomass: a sustainable feedstock for producing chemicals and fuels that does not compete with essential food supply. However, the inherent complexity of lignocellulosic biomass and the technical challenges in its transformation pose significant obstacles that require data-driven approaches to solve. Here, we use the catalytic transformation of lignocellulose to value added chemicals as a case study highlighting the critical role of digital technologies, including improved data integration, process optimization, and system-level decision-making in catalyst design, synthesis, and characterization. Data-driven approaches work hand-in-hand with technology: the integration of machine learning (ML) and artificial intelligence (AI) allows for efficient molecule design and optimization; coupling ML/AI with the use of flow chemistry and high-throughput synthesis techniques enhances scalability and sustainability. Together, these innovations can facilitate a more resilient and sustainable chemical industry, reducing dependency on fossil fuels and mitigating environmental impact.
Introduction
The climate emergency demands immediate solutions to reduce the use of petroleum resources, e.g., via the development of alternative chemicals and fuels.1 However, it is of critical importance that any proposed solution does not use land that is needed for food production. Lignocellulosic biomass, which does not require agricultural land, has promising potential to meet the demand for non-renewable feedstocks. Lignocellulose is a renewable source of carbon which is produced from CO2 through the process of photosynthesis,2–4 and is abundant in nature: >170 billion metric tonnes are produced per year. However, only 5% of the available lignocellulosic biomass is used to produce chemicals and fuels and the remaining 95% is treated as waste: the complex nature and varied functionality of lignocellulosic biomass makes its transformation to commodity products challenging and time consuming, limiting its use.3Lignocellulosic biomass mainly consists of lignin (10–20%), cellulose (30–50%) and hemicellulose (20–40%).3,5 Lignin is a complex cross-linked polymer of aromatic rings, such as coumaryl, coniferyl and sinapyl alcohols. Cellulose is a homopolymer of hexoses (β-D-glucopyranose units) linked together by β-glycosidic bonds making a cellulose microfibril. Hemicellulose is a branched polymer of pentoses and hexoses.6 Each can be transformed to various platform chemicals such as 5-hydroxymethyl furfural, levulinic acid, furfural, xylitol and protocatechuic acid7 (Fig. 1).
Lignocellulosic biomass will not be used as a petrochemical replacement without cost effective, fast, selective and atom efficient routes to its transformation into commodity products, but the structure of lignocellulose and its component constituents, lignin, cellulose, and hemicellulose, poses several difficulties. The presence of intra- and intermolecular hydrogen bonding between cellulose microfibrils make them recalcitrant towards dissolution in any organic solvent, meaning harsh reaction conditions are required, e.g., high temperature (320 °C) and pressure (25 MPa).8 Lignin is a heterogeneous polymer comprised of a complex mixture of phenolic and non-phenolic compounds that are difficult to separate and characterize.9 Unlike cellulose, hemicellulose is relatively easy to depolymerize; its amorphous and highly branched structure improves solubility. However, the composition of hemicellulose varies depending on the source (e.g., hard wood vs. soft wood), meaning reaction conditions vary considerably. Furthermore, the chemicals obtained after depolymerization possess varied oxygen containing functional groups which makes the transformations non-selective and atom inefficient.10
Despite these challenges, progress has been made in chemical transformations of biomass: catalysis has revolutionized this field by lowering the activation energy of the processes while improving selectivity and reaction kinetics.11 Various studies have been conducted on lignocellulosic transformations to fuels and chemicals using diverse catalysts, such as ionic liquids,12,13 zeolites,14 metal supported catalysts,15 metal organic frameworks,16 and single atom catalysts.17,18 Typical catalytic reactions of lignocellulose include the depolymerization of C–O bonds in the polymeric chain of cellulose, formation/ rearrangement of C–C bonds in intermediates, and hydrodeoxygenation (HDO) reactions to remove of oxygen-containing functional groups and yield platform chemicals.19–22
Despite these advances, and due to the complexity of the system under study, there are limits to progress. Catalyst selection is still typically based on a trial-and-error approach; detailed structure–activity relationships are missing; optimisation to find robust and economical catalysts that can offer better selectivity, repeatability, and durability is in its early stages. Catalysis has inherent major challenges in terms of reproducibility, recoverability and durability to deliver sustainable and scalable processes.23,24 The complex nature of the biomass feedstock makes it difficult to decide which pathway to follow: difficulties in understanding catalyst–substrate binding mechanisms, the nature of active sites, and active site–support interaction25 typically result in poor selectivity and challenges in scale up.26–28 Thus, despite the availability of sophisticated tools such as high throughput testing systems, in situ catalyst characterization techniques, and powerful theoretical tools to predict structure activity relationships and compute energy landscapes, industries are still relying on petrochemical-based feedstocks.29
To solve these challenges and deliver sustainable and efficient production of chemicals from biomass will require a combined approach, including a) computational modelling; b) data-driven catalyst design; c) process optimization leveraging artificial intelligence (AI) and machine learning (ML) tools; and d) synthesis technology, e.g., high-throughput experimentation and flow self-optimized systems, to efficiently explore chemical space.30,31 The community is building such capabilities: for example, the Nachwuchs Reaktionstechnik (NaWuReT)32 and Young German Catalysis Society (YounGeCatS)33 summer schools emphasize collaborative efforts between engineers and chemists to develop sustainable and economically viable technologies focussing on defossilization, carbon capture and utilization, fostering a circular economy through cooperation, communication and digitalization.34
In this article we will highlight the state of art in digital catalysis, particularly focusing on strategies that can be implemented for catalytic biomass transformation. By employing essential digital frameworks, adopting data-driven catalyst design and optimization methods, and using AI/ML models to optimize the process and rationally design synthetic pathways, we anticipate the transition to biomass-based feedstocks will be accelerated.
Data frameworks for digital catalysis
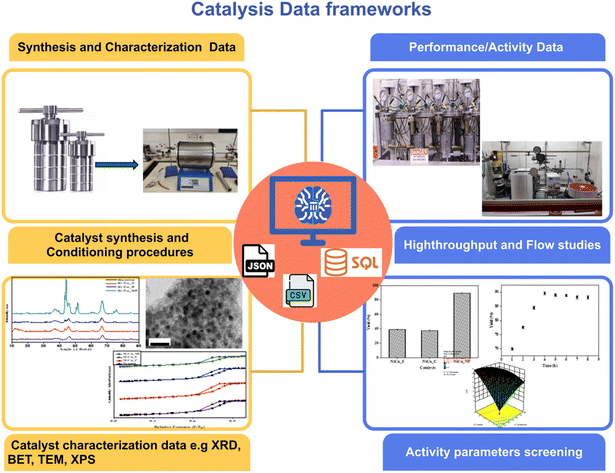
Data structure is a fundamental first consideration for any data-driven scientific field. Data standardization in catalysis research is crucial for creating datasets that are truly useful, reproducible, and shareable. Standardized protocols should be adopted to record the data, including negative results. These protocols should be regularly reviewed to integrate emerging best practices, community standards, and technological advancements in catalysis research.Catalysis is interdisciplinary in nature, including inorganic, organic, analytical, physical, computational chemistry, engineering and chemical physics; each of these disciplines involves different techniques and methods, generating data in a range of formats. Catalysis data can be broadly classified into two types: catalyst synthesis and characterization data (catalyst/material centric) and reaction data (reaction/experiment centric) as depicted in Fig. 2. Capturing catalyst production data is particularly important: catalyst properties such as surface area, metal dispersion, and oxidation states of metal changes with minute variations from batch to batch, contributing to reproducibility challenges. Furthermore, the active form of catalyst is generally achieved only under reaction conditions, making it difficult to understand the complex relationship between catalyst properties and catalyst activity. Hence, integrating in operando characterization data is critical. The German Catalytic Society, GeCATS, reported the five pillars of data frameworks for meaningful description of catalytic processes: data exchange with theory, performance data, synthesis data, characterization data and operando data.35,36
The diverse data formats across various areas of catalysis characterization and performance data and metadata create significant challenges in comprehensively recording and managing all the information. For instance, synthesis data such as details of glassware, reactors and furnace used, lot number of chemicals, order of addition of reagents, aging time, pretreatment conditions (such as flow rate of gases and ramp rate in the furnace) are often ignored in the literature, yet influence catalyst activity, causing irreproducibility from batch to batch.37 The nature of the metadata to be recorded is a key consideration in database design, optimization, governance, and integration, ensuring the database structure is the right fit for the desired application. Winther et al. recorded the data and metadata for catalytic surface reactions using the “ARRAY” data type, generating an open repository including atomic positions and numbers determining the chemical composition of the catalytic surface and minimum adsorption energies based on density functional theory (DFT) calculations: ‘https://www.Catalysis-Hub.org’. Structured query language (SQL), used to manage and manipulate relational databases, was implemented to store the data in ordered tables, meaning that property selection (e.g., reactions involving CO2, or surfaces containing Ni) can be used to recall a subset of column and rows from the tables.38
Digital frameworks are required to record the data with metadata in a structured manner with the adoption of principles of digital catalysis, using FAIR (findable, accessible, interoperable, usable) data principles39 as developed by Wilkinson et al., a diverse group of stakeholders from academia, industry, funding agencies, and scholarly publishers.39 FAIR principles prioritize enabling machines to autonomously locate, access, and use data, while still supporting human users. Ensuring data are easy to find in standardized formats is a key step in integrating them with automated workflows for better reproducibility.40 One benefit of FAIR data is that it promotes cross-disciplinary research by establishing common standards, allowing data from one field to be applied to new contexts, such as leveraging semiconductor studies41 for catalysis research.42 Whether recording data that has been generated by the user, or collating information from third-party sources such as the scientific literature, data curation is essential to ensure that data is accurate, reliable, well-documented and accessible for future use while adhering to ethical and legal standards. Data curation includes the collection of data from diverse sources, data cleaning to remove inconsistencies and enriching it with metadata such as catalyst chemical composition, reaction conditions, characterization data and performance metrics. It is critical for advancing catalytic science by fostering collaboration, improving data transparency, and accelerating the design of most effective catalytic systems.43,44
Marshall et al. discussed the current status of data infrastructure and future directions of data management with FAIR data principles for the catalysis community.45 Automated solutions and standard operating procedures, incorporating benchmarks, play a crucial role in improving data management and laying the groundwork for autonomous catalyst discovery, a goal that remains distant but achievable. In their viewpoint, these advancements can be initiated in individual laboratories, the broader responsibilities lie with the scientific community to establish overarching repositories that respect access rights and intellectual property concerns. Progress depends on the active participation of all researchers—enhancing IT literacy, launching local initiatives, appointing data stewards to mediate between researchers and IT specialists, and mentoring younger scientists.45
Considering the quantity of parameters that should be recorded, datasets can quickly reach very large sizes. Research data management (RDM) is essential especially when the complexity and size of the required datasets is vast. Despite its importance, many laboratories still rely on paper notebooks, and data is frequently stored in proprietary or obsolete formats, lacking proper experimental context. This practice limits the use of data beyond being reported in supplementary information (SI) of research publications. Electronic Lab Notebooks (ELNs) and Laboratory Information Management Systems (LIMS) offer solutions for more effective data management, simplifying both research processes and publication. Researchers can also benefit from approaches developed within the logistics and financial industries, where large and complex datasets are commonplace, and solutions have been developed to answer these challenges. For example, cloud storage frameworks such as “data marts”, “data warehouses” and “data lake” architectures can be used to store structured and unstructured data. A “data warehouse” is an organization-wide repository that integrates structured data from multiple sources, offering a centralized platform for analytics and decision-making. A “data mart” is a subset of a data warehouse, used for specific projects to store structured data for fast querying and reporting. “Data lakes” can be used to store raw, semi structured and unstructured data.46 However, these large-scale architectures require specific IT infrastructure and may be out of reach for many academic groups: it is important that the chosen data framework fits the needs of the data and the application, and that the energy and resource use inherent in data storage and handling are considered and carefully justified.
To ensure consistency in any data framework, the adoption of minimum information standards for data handling is crucial.40 For example, AC/Cat Lab launched in 2003 and has been continuously developing as ELN to record the findings in catalysis.47 For data collection, platforms that run alongside or work with equipment-specific software are being developed. For example, Adacta is a research data management platform developed for catalysis that creates a digital twin of the testing environment and stores time-accurate data to measure catalyst performance, with options to store generated data in ELNs or databases.48 Other available data frameworks and platforms for catalysis include: Nomad (advanced in the field of computational chemistry with FAIR principles and unified data storage),49 Catalysis Hub (database of surface reaction generated by DFT),50 Catalyst Acquisition by Data Science (CADS),51 the Cambridge Structural Database,52 Swiss CAT+,53 Zenodo,54 the Material Project,55 the Material Cloud,56 and the Nationale Forschungsdateninfrastruktur für die Chemie (NDFI4cat).57 Although not currently focused on biomass, each of these can be adopted to record the catalysis data for biomass transformation.
To ensure robust and comparable datasets, worldwide standardized operating procedures should be used by laboratories, enabling the benchmarking of catalytic processes.45 It is important to standardize catalyst data collection with high quality, consistent, and complete data, and to include negative results to understand the boundaries between positive and negative outcomes and to enable the effective training of AI models. Research data management, integrating feedback loops at every stage of the data collection chain, can enhance the information and knowledge gained and influence the next set of experiments. Iterative reaction design further helps in building quantitative models based on AI/ML to predict other regions of interest both in catalyst discovery and chemical space for the processes.36 In the specific case of bio-based transformation, feedstock source and life cycle assessment data should also be recorded and included in catalytic data that helps in decision making towards sustainable transitioning to biobased industry. Ensuring that high-quality data and advanced digital frameworks are available is also critical to feed into effective and/or autonomous catalyst discovery.
Data driven catalyst design and optimization
Access to comprehensive catalytic data is particularly beneficial for catalyst design. However, catalyst design is challenging due to the complexity of catalyst behaviour under different conditions, and this is further increased when dealing with the complex nature of biomass. Catalyst informatics58 can be a potential solution to enable informed design. Catalysis informatics is based on three concepts: catalyst data, ‘data to knowledge’, and catalyst platform, all operating simultaneously. In this way, experimental, computational, and literature data is used to transform raw data into actionable knowledge using data science techniques, extracting insights and driving advancements in the field. A catalyst platform serves as a centralized hub, integrating databases and data science tools to support this process.59First an informatics environment is set up, using Python, Linux, and suitable available tools (e.g., scikit-learn,60 pandas,61 matplotlib62). A workflow of catalyst informatics (Fig. 3) then typically uses the following steps: data collection; setting the objective variable; data pre-processing; statistical analysis and data visualization; machine learning and inverse analysis.63 Tailored data collection is carried out to target the objectives, such as yield and selectivity. Often, the data collected have inconsistencies in units and formats which must be harmonised, and data in text format needs to be converted into numerical values for machine readability and visualization: this process is known as data pre-processing or data cleansing. Data pre-processing is also important to identify outliers and treat them appropriately.64 Data visualization is used to identify the pattern and trends for multidimensional data, using techniques such as parallel coordinates and RadViz; these plots later guide machine learning models to predict the descriptor variables to achieve the objective defined earlier.65 Inverse analysis, also referred to as inverse design, is where existing catalysis data is used to predict and design a new catalyst that would have desired properties, rather than starting from a library of known catalysts and modelling or testing whether any meet the requirements. Here, data science plays an important role in linking catalyst design with catalyst data, and, through machine learning, identifying trends and rules that can be used to suggest new catalysts that meet the desired criteria.
The success of catalysis informatics depends on the quality and structure of data. Difficulties arise from poor data uniformity which can arise from data loss via media conversion, exclusion of metadata, communication barriers, and lack of field-wide standardization. To avoid such issues, data ontology can be employed to structure the data and define information. Ontology is a structured system that defines a domain, its objects, and the relationships between these objects.66 While it shares surface similarities with traditional database structures, ontology fundamentally differs due to its reliance on description logic and formal semantics. These features, enabled by technologies such as web ontology languages (OWL), allow ontologies to define data vocabularies and their relationships in a manner that facilitates intelligent machine navigation and reasoning.67 Ontologies can integrate vast datasets including metadata, annotations, and observations in a layered approach by using logically consistent ontological rules that connect the datasets with each other. Additionally, ontologies can enhance data retrieval by enabling semantic querying based on definition and restrictions. The inferential capabilities of such structures allow autonomous reclassification and reorganization into new subclasses, which can reveal new information and unconventional solutions to the query. Ontologies enable the continuous addition and refinement of definition, which can be particularly beneficial for complex problems such as catalytic biomass transformation.67–69
Behr et al. investigated the landscape of ontologies for catalysis data by exploring the systematic collection of ontology metadata.68 A code-based workflow was adapted to convert metadata to easy-to-read markdown files that automatically mapped the classes between the ontologies pairs of catalysis metadata and could be reused or easily adapted by other ontologies. These codes were made accessible via Github.68 Github integration provided a visual representation of metadata which is then easier to understand by humans while preserving machine readability. Later, they integrated the ontology learning with ‘named entity recognition’ (NER) to automate the extraction of key scientific data from publications, then organized this implicit knowledge into a machine and user readable knowledge graph with the help of a pretrained model, CatalysisIE. This model was fine-tuned with the addition of new datasets resulting in improved precision and recall of the model with regard to the added dataset.70
Tools to record, visualise, and interrogate catalytic data are becoming available to the community. For example, CatApp and Catalyst Hub71 are web-based catalytic platforms developed for data recording and visualisation, although do not include data analysis tools. Later, Fujima et al. added the feature of data analysis and prediction in their open source platform, Catalyst Acquisition by Data Science (CADS)51a,b for catalysis informatics. It can be used for data repository, collaboration, and publishing, as an analytic workspace for visual analysis and for catalyst property prediction with pretrained machine learning models.
Use of such platforms has been demonstrated for catalysis design. For example, oxidative coupling of methane (OCM) is an industrially important method to produce ethylene, offering an alternative to naphtha cracking routes.72 There is a 40-year history of catalytic studies of OCM with conventional methods, but as yet a cost-effective route is missing.73 CADS was used as a means to reveal the underlying patterns and trends in the data sets for OCM and to feed into design of new catalysts.74,75
Later, Nishimura et al. implemented supervised machine learning using support vector regression (SVR) and Bayesian optimization (BO) based on expected improvement index on published literature data, coupling this with systemic high throughput screening (HTS) experiments. SVR was first used to identify potential catalysts that could produce more than 15% C2 (ethylene and ethane). However, when more data including experimental and validation data sets were added for a second trial, the method could not further improve results because the new data did not include standout discoveries. Bayesian optimization (BO), on the other hand, gradually improved predictions over three rounds by adding experimental data after each validation. The results frequently predicted La2O3-based materials as potential catalysts with a C2 yield maximum of 16% under the same test conditions. The limitation of BO was spatial shrinkage during prediction, which limits the room to explore diverse options and reduces the chance of serendipitous discoveries.
The choice of exploration and exploitation strategy should be guided by the context, e.g., the dimensionality and size of chemical space to be explored, the objective to be met, and the availability and quality of existing knowledge that can guide the search. As more tools are developed, workflows will evolve to include combined use of each tool at the point at which it is of most use: for example, starting with DoE approaches to explore a wide space, then the use of BO or ML approaches when the necessary datasets are available. The development of new algorithms to explore wide chemical space and the combination of ML with human intuition could make the search for better catalysts more effective by balancing data-driven predictions with creative insights.76
Machine learning models with inverse analysis (where catalysts can be suggested from desired properties, rather than starting from a known catalyst and predicting its behaviour) can be used to suggest new catalysts with desired activity by uncovering underlying trends and patterns within the data of published reports.77 Various studies have been published on material informatics but research on inverse analysis of heterogeneous catalysis78 is still in its infancy.
Smith et al. used ML frameworks to explore the predictability limit of catalytic activity based on 27 experimental descriptors that collectively represent catalyst formulations and reaction conditions for water–gas shift reactions (WGSR). The framework included principal component analysis (PCA), which reduces the dimensionality of the descriptor data while retaining the maximum information, artificial neural network (ANN), which summarized the data from PCA and predicted catalytic activity, and constrained-PCA to predict new catalyst formulation in unexplored information space. The framework was applied to 2228 experimental datasets of WGSR, which systematically guided the design of experiments and descriptor selection and predicted new catalyst formulations that reduced cost but retained activity. They trained the model on catalyst formulation data such as primary metal, promoter and support, and logarithmic reaction rate as ‘activity data’ from the literature. The model was validated using data from reported literature that wasn't the part of the training data set. They suggested predictability can be improved by adding more descriptors such as stability of active site, centre of mass of unoccupied orbital and d-band centre value, by integrating ML techniques with the experimental data, and by using first principles data collection for descriptor from density functional theory (DFT).79
Suvarna et al. used transformer models, a deep learning encoder–decoder architecture designed to handle sequential data such as text,80 to extract synthesis protocols from literature reports and transform them into structured action sequences for heterogeneous Fe-based single atom catalysts. By converting synthesis protocols into structured action sequences, the model facilitated statistical analysis of synthesis trends, helping to streamline literature review and support predictive modelling to accelerate synthesis planning. The model demonstrated adaptability across various catalyst types, showcasing its potential use for diverse applications in heterogeneous catalysis, not just single atom catalysis. However, inconsistent reporting standards in protocol documentation still hindered machine readability. To address this, they proposed guidelines for standardizing protocol reports to enhance machine-readability and support digital advancements in the field.81
Later, the same group accentuated the importance of data science in the field of catalysis. They reviewed 240 publications from the last decade and categorized them into two types of study: deductive (that is, going from general principles to specific conclusions) and inductive (that is, using observation to form hypotheses), specifically mapping out structure–property–performance relationships. Based on this classification they identified the challenges and their data driven solutions in the field of catalysis, in terms of catalyst task, data sources and representation, and choice of algorithm. They suggested the adoption of data science in catalysis research with the incorporation of “descriptive, predictive, causal and prescriptive” strategies would accelerate innovation.82
Such strategies clearly have relevance for biobased transformations, but thus far have rarely been used for biomass catalysis. In 2022, Uusitalo et al. demonstrated the application of such tools for bio-based transformations for the first time. They used the systematic approach of mathematical modelling and machine learning. Focussing on variable selection using regularization algorithms83 that minimize overfitting, to explain and predict the catalyst performance of bimetallic catalysts towards the hydrogenation of 5-ethoxymethyl furfural. They adopted various ML methods including support vector regression (SVM), Gaussian process regression (GPR), and decision tree models to estimate outcomes. The model showed strong correlation (0.9–0.98) in estimating the conversion, selectivity and yield. Although, the model outcome was good, the variable selection methods relied entirely on data-driven approaches, leaving the physical interpretation of many variables unclear. Also, some values in the descriptor datasets were derived from lists of both experimental and simulated studies, potentially leading to inaccuracies. Furthermore, the lasso algorithm84 had limitations when handling highly correlated variables, which were prioritised at the expense of others, potentially missing significant variables in the process. They predicted that expanding the descriptor dataset, and investigating the exploration capabilities of models with the addition of relevant descriptors, for instance, d-band centre value,85 would be fruitful directions for predicting the optimum catalyst for biomass-based transformations.86
Data driven optimization of catalytic processes and exploration of chemical space for biomass derived molecules
Process optimization
The optimal catalyst is only part of the picture: process optimisation is key in any catalytic process. In the case of catalytic transformation of bio-based molecules, this is particularly challenging due to the complex structure of biomass, hence, in addition to finding the highly active, selective, cost-effective and sustainable catalyst, focus should be placed on making process optimisation faster, more selective, and cost effective. Data driven and ML/AI based approaches can be adopted along with high throughput experimentation, flow technologies, and real time analysis to enhance the process performance by facilitating rapid decision-making and supporting synthetic methods.87 This can help in translating lab-scale research to industrial-scale production, facilitating a shift toward a biomass-based economy. Although ML/AI with high throughput, flow, and real time analysis has already been adopted for pharmaceutical chemistry88,89 and catalysis,90,91 very few examples of its use have been reported for bio-based transformations.92Eyke et al. highlighted the importance of synergies of ML and high throughput techniques towards rapid chemical space exploration and optimization, using experimental and analytical data to iteratively improve ML algorithm performance in a feedback loop. They suggested the merging of traditional statistical methods like design of experiment (DoE) with ML models to deliver optimal experiment design with high dimensional chemical reaction space, taking advantage of both methods. To reduce the cost of the process dimensionality, reduction algorithms like principal component analysis (PCA) can be employed. Bayesian neural networks can be used to construct probabilistic surrogate models, and ‘traditional’ algorithms such as neural networks (NN) and random forests (RF) can be used as surrogate models to describe and explore the high dimensionality space that results when many parameters must be optimised.93
Choosing the most time- and resource-efficient optimization method can be challenging, but examples of their use in catalysis offer compelling reasons to try. Install et al. recently integrated a statistical DoE approach with a high throughput platform to optimize the solvent composition for maximum conversion of glucose to methyl lactate with SnCl4·5H2O. Using this strategy, optimal reaction conditions (75.9% yield using 7.5% water in methanol) were determined in just 58 runs.94
Yang et al. adopted machine learning frameworks for catalyst screening and process optimization for indirect hydrogenation of CO2 to methanol and ethylene glycol. Datasets based on catalyst descriptors, i.e. preparation conditions, operational parameters, and feed conditions were initially analysed by PCA, then further improved with additional catalyst descriptor datasets. Among three machine learning models trialled (RF, NN, and SVR), NN with two hidden neural layers was found to have the highest prediction accuracy after optimizing the hyperparameter for each model with minimum mean square error (MSE), mean absolute error (MAE), and highest determination coefficient (R2). Feature engineering was used to remove redundant features from the model with minimal loss of data and improved prediction accuracy of the model. Shapley additive explanation (SHAP) was used to interpret the improved machine model and predict that space velocity and hydrogen/ester ratio are the most important factors that impact the conversion and product yield. ML models with genetic algorithms were used to maximizes the yield of products from indirect CO2 hydrogenation system. The results proposed xMoOx–Cu/SiO2 as the candidate with the best catalytic activity as compared to other catalytic systems. However, experimental validation is essential prior to their industrial application.95 A similar methodology was adopted by Liu et al. for the hydrogenation of biomass-derived levulinic acid to γ-valerolactone. ML model analysis with SHAP predicted that temperature was an important factor for the hydrogenation of levulinic acid, and genetic algorithms with multiobjective optimization identified Ru/N@CNTs as a promising catalyst.96
Wang et al. developed a trained ML model for the prediction and optimization of catalytic steam reforming of biomass tar using a database of 584 data points from the published literature. The RF algorithm predicted the reaction temperature as the most important factor to influence the conversion rate of toluene as major component of tar, followed by support, additive, Ni loading and calcination temperature. The proposed model was empirically validated with experimental trials using Ni–Co supported on γ-Al2O3 as catalyst, and predictions were found to be in good agreement with the experimental data. The optimal ranges for the key parameters in the catalytic process were reaction temperature of 600–700 °C, Ni loading of 5–15 wt%, and calcination temperature of 500–650 °C, which maximizes toluene conversion rates. Additionally, they highlighted the importance of suitable supports and additives which significantly enhance catalytic performance by providing more active sites and promoting Ni dispersion, resulting in improved activity and stability of the catalyst.97
Reproducible process control, e.g., the reliable maintenance and data logging of mixing, temperature profile, addition rates, etc., is as important as reproducibility in catalyst synthesis and formulation; both underpin meaningful optimization. In this space, digitalization and industry 4.0 (ref. 98) are poised to significantly transform chemicals and materials discovery and development. By integrating various technologies—such as flow synthesis, automation, analytics, and real-time reaction control—the industry is moving toward highly efficient, data-driven discovery and synthesis protocols.99–103
Flow chemistry enhances control over parameters like flow rates, temperature, and pressure, resulting in improved efficiency of the process and sustainability through waste minimization.104,105 Additionally, flow chemistry supports integration with downstream processing and enables in situ process monitoring by capturing large amounts of process and product data.106–108 Kaisin et al. reported the challenges in transformation of biomass derived chemicals to pharmaceutical ingredients in terms of chemical, process, supply chain and regulatory aspect. In their perspective they highlighted the benefit of flow in synthesizing the chemicals in a safer, scalable manner with reduced environmental impact and improved process efficiency. Incorporation of downstream PAT analytical techniques can provide the real time data and control the quality of the product during the production campaign. However, the varied impurity profiles of biomass sources and their resultant by-products is still a major concern.109
Flow chemistry is also finding use in the transformation of bioderived chemicals into commodity products. Muzyka et al. used a flow process to produce biobased glycerol carbonate at large scale with a space time yield of 2.7 kg h−1 L−1 and an environmental factor (E factor)110 as low as 4.7.111 Sivo et al. developed and optimized a continuous-flow process for producing glycidol from glycerol, addressing challenges such as long reaction times, harsh conditions, and unstable intermediates. The optimized process demonstrated higher yields, improved reaction mass intensity, and improved sustainability compared to batch methods. Further exploration enabled integrated preparation of glycidol derivatives, showcasing protocols for aminolysis, polymerization, and tosylation reactions, highlighting the scalability and versatility of the continuous-flow approach. Techno-economic and life cycle assessments confirmed its superiority in cost, efficiency, and environmental impact.112 Continuous flow has been used in multiple studies upgrading biomass-derived glycerol to fine chemicals and pharmaceuticals.113–120 As yet, routes to upgrade other platform chemicals to value added chemicals and fuels under continuous flow conditions are rare, with limited studies using heterogenous catalysts.121–123
Flow optimisation using downstream PAT tools and ML algorithms can autonomously adjust reaction conditions like temperature, pressure, flow rates, and reagent concentrations in real-time. Such self-optimizing synthesis platforms minimize human intervention and can accelerate the identification of optimal reaction parameters, improving yield and selectivity, and reducing waste. Various examples have been reported for the automated synthesis of organic molecules,102,124–128 pharmaceuticals,129 and nanoparticles130–132 enabling selective, cost effective and scalable synthesis of molecules with the desired properties.
Recently, workflows has been developed using a hybrid approach of active machine learning with ‘human in the loop’ to generate informative datasets.133 Kuddusi et al. adopted this methodology to evaluate Ni- and Co-based catalysts supported on Al2O3 for the thermo-catalytic conversion of CO2 to CH4. Researchers conducted 48 catalytic activity tests within a design space exceeding 50 million potential experiments, using an automated reactor system to ensure controlled conditions. Key experimental variables included temperature, pressure, catalyst composition, and synthesis conditions such as calcination and reduction temperatures. The dataset trained three regression algorithms—Gaussian processes, RF, and extreme gradient boosting—to predict CO2 conversion, methane selectivity, and methane space–time yield. Feature importance analysis highlighted temperature, Ni load, and calcination temperature as critical factors for catalyst activity. Experimental validation identified an optimal calcination temperature range (673–723 K), beyond which catalyst activity diminished due to structural changes in the material. This approach, leveraging a modest dataset, achieved a 50% improvement in methane space–time yield compared to the training set's maximum. The study demonstrates the potential of combining active machine learning with experimental workflows to optimize chemical reactions and suggests broad applicability to other reactions with diverse design spaces.134
Chemical space exploration
When datasets are rigorously recorded, discovery and exploration of new chemical products from catalytic reactions dovetails with catalyst and process optimisation. AI and ML tools can be used to design new biomass-derived replacements for petrochemicals by navigating multidimensional input and output relationships, e.g., candidate structures from desired properties.135 ML algorithms can analyse vast datasets of biomass-derived compounds, predict their properties, and suggest novel molecular structures tailored for specific applications, such as biofuels, bioplastics, or pharmaceuticals.136 AI-driven techniques like generative models (e.g., generative adversarial networks (GANs) or variational autoencoders) and reinforcement learning enable the exploration of complex chemical spaces, facilitating the design of molecules from sustainable feedstocks.137 This approach accelerates discovery, reduces reliance on trial-and-error experimentation, and promotes a circular bioeconomy by optimizing the valorisation of renewable biomass resources.Batchu et al. highlighted the areas to focus on to explore and accelerate the manufacturing of high-performance biomass-based molecules that have no analog in traditional refineries, advocating the use of retrosynthetic approaches, text mining, natural language processing and modern machine learning models to identify opportunities. Automated laboratory and simulation data, enhanced through active learning methods, enable the efficient generation of thermochemistry and kinetics data, crucial for developing detailed and validated process models, understanding product structure–property relationships, and establishing correlations between catalyst and solvent descriptors with their performance.92
Chang et al. used such methods to identify bioderived replacements for aviation fuel and their catalytic synthetic routes, mostly based on furanics derived from hemicellulosic feedstock. Automated network generation and semi-empirical thermochemistry calculations predicted more than 100 potential sustainable aviation fuel candidates (C8–C16 alkanes and cycloalkanes) across 300 synthesis routes. 2-Methyl heptane, ethyl cyclohexane, and propyl cyclohexane were found to be the most promising candidates, but all require multiple synthetic steps, including energy intensive hydrogenation and oxygen removal steps. Process intensification with multifunctional catalyst systems was suggested as a means to overcome these challenges.138
Singh et al. recently showed the potential of machine learning models for reaction discovery with relatively small and sparsely labelled datasets. RF methods reliably predicted catalytic reaction yields and enantioselectivity for asymmetric hydrogenation of imines. It is difficult to derive molecular features from experimental data, hence quantum mechanically derived molecular descriptors (i.e., charge, frequency, intensity, HOMO, LUMO, and NMR shifts) of reactants, solvents, catalyst etc. served as input vectors for feature engineering. The feature learning techniques using SMILES-based molecular representations and customized natural language processing (NLP) techniques proven to be a promising strategy for yield and enantioselectivity predictions. A transfer learning approach was adopted, where model was trained on a large data set (105–106 molecules) to explore latent chemical space, then fine-tuned for targeted reaction library (102–103 reactions). Additionally, the exploration of latent space within deep neural networks offered a promising generative strategy for identifying new and useful substrates tailored to specific reactions. These approaches highlighted the potential of molecular ML to accelerate reaction discovery and optimization.139
ML has been used to improve the synthesis and design of new biobased polymers for the sustainable energy and fuel sectors. A review by Abu Sofian et al. reported the state of the art of ML based biopolymers and highlighted scope for future development via modification of algorithms or exploring deep learning models to enhance thermal stability and mechanical strength and reduce degradation rates.140
In a similar vein, Akinpelu et al. highlighted the application of machine learning in pyrolysis: from biorefinery to end-of-life product management. ML methods, particularly artificial neural networks (ANN), are widely used to study pyrolysis due to their ability to model a ‘highly nonlinear’ input–output relationship. They highlighted ML's potential to accelerate research, development, and scalability in biomass pyrolysis, and recommended its further use in life cycle assessment (LCA) and technoeconomic analysis.141
It is important to state that LCA and sustainability metrics are equally important for biomass derived alternative molecules as for their petrochemical counterparts. LCA is a methodology used to evaluate the environmental impacts of a process, system, or product throughout its entire life cycle, from raw material extraction to disposal.142 The primary goal of LCA is to provide decision-makers with data to choose sustainable technology options that meet societal needs.143 Sustainable reaction identification is a complex interdisciplinary challenge. Weber et al. addressed different methods for automated discovery and assessment of sustainable reaction routes for chemicals derived from renewables and waste feedstocks. These methods explored the opportunity for circular economy with the help of chemical data intelligence with focus on data, evaluation metrics and decision making.144 The major bottleneck for LCA and sustainability evaluations was found to be incomplete datasets that hinder mass balance calculations, and difficulty in linking various data sources such as regional waste stream composition, pretreatment method and end of life use. To overcome this, a roadmap for systematic reaction pathway planning through digitalized chemical data, sustainability evaluation metrics and decision making has been suggested.144
Conclusions and future perspectives
Defossilization and moving away from a petro-based industry can be achieved using alternative molecules derived from renewable lignocellulosic feedstock—but will require interdisciplinary collaboration and investment in data-driven approaches. The catalytic transformation of lignocellulosic biomass to value added chemicals and fuel precursors is challenging because of the complex nature of biomass and their derived molecules. This is further complicated when using heterogeneous catalysts due to inherent issues with reproducibility, stability and durability.Digitalization of the catalytic process is a potential solution to solve this multidimensional problem. Recording, sharing, curating, analysing, and using data in advanced optimization and discovery workflows will impact each step, from catalyst development and process optimization to the exploration of alternative bio-based molecules.
In this perspective, we focussed on the state of art in digital catalysis, considering how these methods can be adopted for catalytic biomass transformation. Data frameworks are required to record both catalyst-focussed data (synthesis and characterization) and reaction-focussed data (reaction performance). Various frameworks have been suggested that are being used for heterogeneous catalyst and material synthesis, and these can be adopted for catalysis for biomass. To ensure widespread use and progress in the field, such frameworks should use FAIR principles, ensure metadata is recorded in both machine and human readable formats, and be curated to remove inconsistencies. Ontologies have been used to structure vast datasets in a layer approach connecting them with each other and making them searchable; this will be especially important for the complex reaction processes in biomass catalysis. In this way, reported literature data can be used for catalyst design and development, leveraging catalyst informatics and ML models to discover the optimum catalyst for a given transformation, and increasing the chances that biomass will become part of the chemical supply chain.
The multistep and complex nature of biomass transformation demands advanced solutions but also provides challenges that will stimulate advances in digital catalysis methods and reactor technologies alike. The integration of AI/ML with high throughput experimentation, flow reactors, and real time analysis can speed up process optimization and the exploration of chemical space to discover new molecules. AI/ML models alongside with DOE and PCA analysis reduce the cost of the process with the exploration of wider chemical reaction space. Validating and improving these models with experimental data is an important next step for the growing community using such methods in catalysis.
A major challenge in achieving the digitalization of catalytic biomass transformation is the lack of available structured data and metadata. Future research should focus on recording metadata on available web-based platforms, and development of data frameworks to record catalyst- and reaction-centric data with the integration of AI/ML workflows for process optimization. Additionally, data on LCA and sustainability metrics is important to translate lab-based research to the industrial scale and achieve the desired circular economy. Ultimately, solving this challenge will require international and interdisciplinary collaboration between chemists, chemical engineers, computer and data scientists; the methods developed in recent years offer the strongest chance that the 95% of unused lignocellulose feedstock will form the basis of a biofuel-derived economy.
Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Author contributions
Firdaus Parveen: conceptualization, visualization, writing – original draft, writing – review & editing. Anna G. Slater: writing – review & editing, supervision, funding acquisition.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
AGS thanks the Royal Society for a University Research Fellowship (URF\R1\201168) that supported this work.Notes and references
- G. W. Huber, S. Iborra and A. Corma, Chem. Rev., 2006, 106, 4044–4098 CrossRef CAS PubMed.
- Sushma, S. Chamoli, S. Upadhyayula and F. Parveen, in Handbook of Biomass Valorization for Industrial Applications, 2022, pp. 41–53, DOI:10.1002/9781119818816.ch3.
- A. Corma, S. Iborra and A. Velty, Chem. Rev., 2007, 107, 2411–2502 CrossRef CAS PubMed.
- W. Deng, Y. Feng, J. Fu, H. Guo, Y. Guo, B. Han, Z. Jiang, L. Kong, C. Li, H. Liu, P. T. T. Nguyen, P. Ren, F. Wang, S. Wang, Y. Wang, Y. Wang, S. S. Wong, K. Yan, N. Yan, X. Yang, Y. Zhang, Z. Zhang, X. Zeng and H. Zhou, Green Energy Environ., 2023, 8, 10–114 Search PubMed.
- L. T. Mika, E. Cséfalvay and Á. Németh, Chem. Rev., 2018, 118, 505–613 CrossRef CAS PubMed.
- F. Parveen, K. Ahmad and S. Upadhyayula, in Integrating Green Chemistry and Sustainable Engineering, 2019, pp. 113–163, DOI:10.1002/9781119509868.ch5.
- P. P. Upare, R. E. Clarence, H. Shin and B. G. Park, Processes, 2023, 11, 2912–2926 CrossRef CAS.
- M. V. Rodionova, A. M. Bozieva, S. K. Zharmukhamedov, Y. K. Leong, J. Chi-Wei Lan, A. Veziroglu, T. N. Veziroglu, T. Tomo, J.-S. Chang and S. I. Allakhverdiev, Int. J. Hydrogen Energy, 2022, 47, 1481–1498 CrossRef CAS.
- W. Boerjan, J. Ralph and M. Baucher, Annu. Rev. Plant Biol., 2003, 54, 519–546 Search PubMed.
- Z. Usmani, M. Sharma, A. K. Awasthi, T. Lukk, M. G. Tuohy, L. Gong, P. Nguyen-Tri, A. D. Goddard, R. M. Bill, S. C. Nayak and V. K. Gupta, Renewable Sustainable Energy Rev., 2021, 148, 111258 CrossRef CAS.
- J. Cai, L. Wei, J. Wang, N. Lin, Y. Li, F. Li, X. Zha and W. Li, Catalysts, 2024, 14(8), 499 Search PubMed.
- K. Kumar, F. Parveen, T. Patra and S. Upadhyayula, New J. Chem., 2018, 42, 228–236 Search PubMed.
- F. Parveen, T. Patra and S. Upadhyayula, Carbohydr. Polym., 2016, 135, 280–284 CrossRef CAS PubMed.
- Y. Wang, X. Yuan, J. Liu and X. Jia, ChemPlusChem, 2024, 89, e202300399 CrossRef CAS PubMed.
- K. Tomishige, Y. Nakagawa and M. Tamura, Curr. Opin. Green Sustainable Chem., 2020, 22, 13–21 CrossRef.
- V. Srivastava, K. Lappalainen, A. Rusanen, G. Morales and U. Lassi, ChemPlusChem, 2023, 88, e202300309 CrossRef CAS PubMed.
- S. De, A. S. Burange and R. Luque, Green Chem., 2022, 24, 2267–2286 RSC.
- J.-Y. Chen, Y. Xiao, F.-S. Guo, K.-M. Li, Y.-B. Huang and Q. Lu, ACS Catal., 2024, 14, 5198–5226 CrossRef CAS.
- J. B. Binder and R. T. Raines, J. Am. Chem. Soc., 2009, 131, 1979–1985 CrossRef CAS PubMed.
- S. Wang, A. Cheng, F. Liu, J. Zhang, T. Xia, X. Zeng, W. Fan and Y. Zhang, Ind. Chem. Mater., 2023, 1, 188–206 RSC.
- H. Wang, B. Yang, Q. Zhang and W. Zhu, Renewable Sustainable Energy Rev., 2020, 120, 109612 CrossRef CAS.
- A. Shivhare, A. Kumar and R. Srivastava, Green Chem., 2021, 23, 3818–3841 RSC.
- F. Zaera, Catal. Lett., 2012, 142, 501–516 CrossRef CAS.
- K. F. Kalz, R. Kraehnert, M. Dvoyashkin, R. Dittmeyer, R. Gläser, U. Krewer, K. Reuter and J.-D. Grunwaldt, ChemCatChem, 2017, 9, 17–29 CrossRef CAS.
- T. W. Walker, A. H. Motagamwala, J. A. Dumesic and G. W. Huber, J. Catal., 2019, 369, 518–525 CrossRef CAS.
- G. M. Preethi, G. Kumar, O. P. Karthikeyan, S. Varjani and R. B. J, Environ. Technol. Innovation, 2021, 24, 102080 CrossRef.
- Y. Jing, Y. Guo, Q. Xia, X. Liu and Y. Wang, Chem, 2019, 5, 2520–2546 CAS.
- S. P. S. Chundawat, G. T. Beckham, M. E. Himmel and B. E. Dale, Annu. Rev. Chem. Biomol. Eng., 2011, 2, 121–145 CrossRef CAS PubMed.
- Q. Wu, M. Pan, S. Zhang, D. Sun, Y. Yang, D. Chen, D. Weitz and X. Gao, Energies, 2022, 15, 6666 CrossRef CAS.
- R. Schlögl, ChemCatChem, 2017, 9, 533–541 CrossRef.
- A. Toniato, A. C. Vaucher and T. Laino, Catal. Today, 2022, 387, 140–142 CrossRef CAS.
- J. Friedland, M. Börnhorst, B. Kreitz, E. Moioli and G. Wehinger, Chem. Ing. Tech., 2022, 94, 629–633 CrossRef CAS.
- YounGeCatS, ChemCatChem, 2023, 15, e202201420 CrossRef CAS.
- P. Naliwajko, J. Friedland and M. Börnhorst, ChemCatChem, 2023, 15, e202201548 CrossRef CAS.
- O. Deutschmann, D. Demtröder, B. Eck, R. Franke, R. Gläser, L. Goosen, J. D. Grunwaldt, R. Krähnert, D. G. f. Katalyse and G. f. C. T. u. B. DECHEMA, The Digitalization of Catalysis-related Sciences: Whitepaper, German Catalysis Society (GeCatS), 2019 Search PubMed.
- C. Wulf, M. Beller, T. Boenisch, O. Deutschmann, S. Hanf, N. Kockmann, R. Kraehnert, M. Oezaslan, S. Palkovits, S. Schimmler, S. A. Schunk, K. Wagemann and D. Linke, ChemCatChem, 2021, 13, 3223–3236 CrossRef CAS.
- S. L. Scott, T. B. Gunnoe, P. Fornasiero and C. M. Crudden, ACS Catal., 2022, 12, 3644–3650 CrossRef CAS.
- K. T. Winther, M. J. Hoffmann, J. R. Boes, O. Mamun, M. Bajdich and T. Bligaard, Sci. Data, 2019, 6, 75 CrossRef PubMed.
- M. D. Wilkinson, M. Dumontier, I. J. Aalbersberg, G. Appleton, M. Axton, A. Baak, N. Blomberg, J.-W. Boiten, L. B. da Silva Santos, P. E. Bourne, J. Bouwman, A. J. Brookes, T. Clark, M. Crosas, I. Dillo, O. Dumon, S. Edmunds, C. T. Evelo, R. Finkers, A. Gonzalez-Beltran, A. J. G. Gray, P. Groth, C. Goble, J. S. Grethe, J. Heringa, P. A. C. 't Hoen, R. Hooft, T. Kuhn, R. Kok, J. Kok, S. J. Lusher, M. E. Martone, A. Mons, A. L. Packer, B. Persson, P. Rocca-Serra, M. Roos, R. van Schaik, S.-A. Sansone, E. Schultes, T. Sengstag, T. Slater, G. Strawn, M. A. Swertz, M. Thompson, J. van der Lei, E. van Mulligen, J. Velterop, A. Waagmeester, P. Wittenburg, K. Wolstencroft, J. Zhao and B. Mons, Sci. Data, 2016, 3, 160018 CrossRef PubMed.
- S. Herres-Pawlis, F. Bach, I. J. Bruno, S. J. Chalk, N. Jung, J. C. Liermann, L. R. McEwen, S. Neumann, C. Steinbeck, M. Razum and O. Koepler, Angew. Chem., Int. Ed., 2022, 61, e202203038 CrossRef CAS PubMed.
- T. Hisatomi and K. Domen, Nat. Catal., 2019, 2, 387–399 CrossRef CAS.
- H. Tao, T. Wu, M. Aldeghi, T. C. Wu, A. Aspuru-Guzik and E. Kumacheva, Nat. Rev. Mater., 2021, 6, 701–716 CrossRef.
- P. S. F. Mendes, S. Siradze, L. Pirro and J. W. Thybaut, ChemCatChem, 2021, 13, 836–850 CrossRef CAS.
- A. H. Poole, Arch. Sci., 2015, 15, 101–139 CrossRef.
- C. P. Marshall, J. Schumann and A. Trunschke, Angew. Chem., Int. Ed., 2023, 62, e202302971 CrossRef CAS PubMed.
- https://www.aws.amazon.com/compare/the-difference-between-a-data-warehouse-data-lake-and-data-mart/ .
- https://www.github.com/fhimpg/archive .
- H. Gossler, J. Riedel, E. Daymo, R. Chacko, S. Angeli and O. Deutschmann, Chem. Ing. Tech., 2022, 94, 1798–1807 CrossRef CAS.
- https://www.nomad-lab.eu/, 2014.
- https://www.catalysis-hub.org/ .
- (a) https://www.cads.eng.hokudai.ac.jp/ ; (b) J. Fujima, React. Chem. Eng., 2020, 5, 903–911 RSC.
- https://www.ccdc.cam.ac.uk/structures/ .
- https://www.swisscatplus.ch/ .
- https://www.zenodo.org/ .
- https://www.materialsproject.org/catalysis .
- https://www.materialscloud.org .
- https://www.nfdi4cat.org/ .
- K. Takahashi and L. Takahashi, in Materials Informatics and Catalysts Informatics: An Introduction, ed. K. Takahashi and L. Takahashi, Springer Nature Singapore, Singapore, 2024, pp. 113–142, DOI:10.1007/978-981-97-0217-6_5.
- K. Takahashi, J. Ohyama, S. Nishimura, J. Fujima, L. Takahashi, T. Uno and T. Taniike, Chem. Commun., 2023, 59, 2222–2238 RSC.
- F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, P. Prettenhofer, R. Weiss, V. Dubourg, J. VanderPlas, A. Passos, D. Cournapeau, M. Brucher, M. Perrot and E. Duchesnay, J. Mach. Learn. Res., 2011, 12, 2825–2830 Search PubMed.
- W. McKinney, Python High Performance Science Computer, 2011 Search PubMed.
- J. D. Hunter, Comput. Sci. Eng., 2007, 9, 90–95 Search PubMed.
- K. Takahashi, L. Takahashi, I. Miyazato, J. Fujima, Y. Tanaka, T. Uno, H. Satoh, K. Ohno, M. Nishida, K. Hirai, J. Ohyama, T. N. Nguyen, S. Nishimura and T. Taniike, ChemCatChem, 2019, 11, 1146–1152 CrossRef CAS.
- A. J. Medford, M. R. Kunz, S. M. Ewing, T. Borders and R. Fushimi, ACS Catal., 2018, 8, 7403–7429 CrossRef CAS.
- K. Takahashi, L. Takahashi, S. Nishimura, J. Fujima and J. Ohyama, in Crystalline Metal Oxide Catalysts, ed. W. Ueda, Springer Nature Singapore, Singapore, 2022, pp. 349–371, DOI:10.1007/978-981-19-5013-1_12.
- T. R. Gruber, Int. J. Hum. Comput. Stud., 1995, 43, 907–928 CrossRef.
- L. Takahashi and K. Takahashi, J. Phys. Chem. Lett., 2019, 10, 7482–7491 CrossRef CAS PubMed.
- A. S. Behr, H. Borgelt and N. Kockmann, J. Cheminf., 2024, 16, 16 Search PubMed.
- A. S. Behr, M. Völkenrath and N. Kockmann, Knowl. Inf. Syst., 2023, 65, 5503–5522 CrossRef.
- A. S. Behr, D. Chernenko, D. Koßmann, A. Neyyathala, S. Hanf, S. A. Schunk and N. Kockmann, Catal. Sci. Technol., 2024, 14, 5699–5713 RSC.
- J. S. Hummelshøj, F. Abild-Pedersen, F. Studt, T. Bligaard and J. K. Nørskov, Angew. Chem., Int. Ed., 2012, 51, 272–274 CrossRef.
- Y. Gao, L. Neal, D. Ding, W. Wu, C. Baroi, A. M. Gaffney and F. Li, ACS Catal., 2019, 9, 8592–8621 CrossRef CAS.
- G. Keller and M. Bhasin, J. Catal., 1982, 73, 9–19 CrossRef CAS.
- J. Fujima, Y. Tanaka, I. Miyazato, L. Takahashi and K. Takahashi, React. Chem. Eng., 2020, 5, 903–911 RSC.
- K. Suzuki, T. Toyao, Z. Maeno, S. Takakusagi, K.-I. Shimizu and I. Takigawa, ChemCatChem, 2019, 11, 4537–4547 CrossRef CAS.
- S. Nishimura, X. Li, J. Ohyama and K. Takahashi, Catal. Sci. Technol., 2023, 13, 4646–4655 RSC.
- J. Benavides-Hernández and F. Dumeignil, ACS Catal., 2024, 14, 11749–11779 CrossRef.
- Y. Guan, D. Chaffart, G. Liu, Z. Tan, D. Zhang, Y. Wang, J. Li and L. Ricardez-Sandoval, Chem. Eng. Sci., 2022, 248, 117224 CrossRef CAS.
- A. Smith, A. Keane, J. A. Dumesic, G. W. Huber and V. M. Zavala, Appl. Catal., B, 2020, 263, 118257 CrossRef CAS.
- A. Vaswani, Advances in Neural Information Processing Systems, 2017 Search PubMed.
- M. Suvarna, A. C. Vaucher, S. Mitchell, T. Laino and J. Pérez-Ramírez, Nat. Commun., 2023, 14, 7964 CrossRef CAS PubMed.
- M. Suvarna and J. Pérez-Ramírez, Nat. Catal., 2024, 7, 624–635 CrossRef.
- J. H. Friedman, T. Hastie and R. Tibshirani, J. Stat. Softw., 2010, 33, 1–22 Search PubMed.
- J. O. Ogutu, T. Schulz-Streeck and H.-P. Piepho, BMC Proc., 2012, 6, S10 CrossRef PubMed.
- S. Bhattacharjee, U. V. Waghmare and S.-C. Lee, Sci. Rep., 2016, 6, 35916 CrossRef CAS PubMed.
- P. Uusitalo, A. Sorsa, F. R. Abegão, M. Ohenoja and M. Ruusunen, Ind. Eng. Chem. Res., 2022, 61, 4752–4762 CrossRef CAS PubMed.
- Y. Su, X. Wang, Y. Ye, Y. Xie, Y. Xu, Y. Jiang and C. Wang, Chem. Sci., 2024, 15, 12200–12233 Search PubMed.
- S. M. Mennen, C. Alhambra, C. L. Allen, M. Barberis, S. Berritt, T. A. Brandt, A. D. Campbell, J. Castañón, A. H. Cherney, M. Christensen, D. B. Damon, J. Eugenio de Diego, S. García-Cerrada, P. García-Losada, R. Haro, J. Janey, D. C. Leitch, L. Li, F. Liu, P. C. Lobben, D. W. C. MacMillan, J. Magano, E. McInturff, S. Monfette, R. J. Post, D. Schultz, B. J. Sitter, J. M. Stevens, I. I. Strambeanu, J. Twilton, K. Wang and M. A. Zajac, Org. Process Res. Dev., 2019, 23, 1213–1242 CrossRef CAS.
- A. D. Clayton, E. O. Pyzer-Knapp, M. Purdie, M. F. Jones, A. Barthelme, J. Pavey, N. Kapur, T. W. Chamberlain, A. J. Blacker and R. A. Bourne, Angew. Chem., Int. Ed., 2023, 62, e202214511 CrossRef CAS PubMed.
- D. E. Fitzpatrick, C. Battilocchio and S. V. Ley, Org. Process Res. Dev., 2016, 20, 386–394 CrossRef CAS.
- E. S. Isbrandt, R. J. Sullivan and S. G. Newman, Angew. Chem., Int. Ed., 2019, 58, 7180–7191 Search PubMed.
- S. P. Batchu, B. Hernandez, A. Malhotra, H. Fang, M. Ierapetritou and D. G. Vlachos, React. Chem. Eng., 2022, 7, 813–832 Search PubMed.
- N. S. Eyke, B. A. Koscher and K. F. Jensen, Trends Chem., 2021, 3, 120–132 Search PubMed.
- J. Install, R. Zhang, J. Hietala and T. Repo, RSC Adv., 2024, 14, 35578–35584 RSC.
- Q. Yang, Y. Fan, J. Zhou, L. Zhao, Y. Dong, J. Yu and D. Zhang, Green Chem., 2023, 25, 7216–7233 RSC.
- D. Liu, Z. Jia, L. Shen, W. Liu, R. Pang, S. Yu, S. Liu, L. Li, Y. Liu and L. Yu, ACS Sustainable Chem. Eng., 2024, 12, 16340–16353 CrossRef CAS.
- N. Wang, H. He, Y. Wang, B. Xu, J. Harding, X. Yin and X. Tu, Energy Convers. Manage., 2024, 300, 117879 CrossRef CAS.
- L. S. Dalenogare, G. B. Benitez, N. F. Ayala and A. G. Frank, Int. J. Prod. Econ., 2018, 204, 383–394 CrossRef.
- C. A. Hone and C. O. Kappe, Chem.:Methods, 2021, 1, 454–467 CAS.
- O. J. Kershaw, A. D. Clayton, J. A. Manson, A. Barthelme, J. Pavey, P. Peach, J. Mustakis, R. M. Howard, T. W. Chamberlain, N. J. Warren and R. A. Bourne, Chem. Eng. J., 2023, 451, 138443 CrossRef CAS.
- M. B. Plutschack, B. Pieber, K. Gilmore and P. H. Seeberger, Chem. Rev., 2017, 117, 11796–11893 CrossRef CAS PubMed.
- A. M. Schweidtmann, A. D. Clayton, N. Holmes, E. Bradford, R. A. Bourne and A. A. Lapkin, Chem. Eng. J., 2018, 352, 277–282 CrossRef CAS.
- A. Echtermeyer, Y. Amar, J. Zakrzewski and A. Lapkin, Beilstein J. Org. Chem., 2017, 13, 150–163 CrossRef CAS PubMed.
- L. Capaldo, Z. Wen and T. Noël, Chem. Sci., 2023, 14, 4230–4247 RSC.
- F. Parveen, N. Watson, A. M. Scholes and A. G. Slater, Curr. Opin. Green Sustainable Chem., 2024, 100935 CrossRef.
- F. Parveen, H. J. Morris, H. West and A. G. Slater, J. Flow Chem., 2024, 14, 23–31 CrossRef CAS.
- V. Sans, L. Porwol, V. Dragone and L. Cronin, Chem. Sci., 2015, 6, 1258–1264 RSC.
- T. H. Rehm, C. Hofmann, D. Reinhard, H.-J. Kost, P. Löb, M. Besold, K. Welzel, J. Barten, A. Didenko and D. V. Sevenard, React. Chem. Eng., 2017, 2, 315–323 RSC.
- G. Kaisin, L. Bovy, Y. Joyard, N. Maindron, V. Tadino and J.-C. M. Monbaliu, J. Flow Chem., 2023, 13, 77–90 CrossRef PubMed.
- D. J. C. Constable, A. D. Curzons and V. L. Cunningham, Green Chem., 2002, 4, 521–527 RSC.
- C. Muzyka, S. Renson, B. Grignard, C. Detrembleur and J.-C. M. Monbaliu, Angew. Chem., Int. Ed., 2024, 63, e202319060 CrossRef CAS PubMed.
- A. Sivo, I. Montanari, M. C. Ince and G. Vilé, Green Chem., 2024, 26, 7911–7918 RSC.
- S. Guidi, M. Noè, P. Riello, A. Perosa and M. Selva, Molecules, 2016, 21, 657 CrossRef PubMed.
- N. Ozbay, N. Oktar, G. Dogu and T. Dogu, Top. Catal., 2013, 56, 1790–1803 CrossRef CAS.
- V. Domínguez-Barroso, C. Herrera, M. Á. Larrubia, R. González-Gil, M. Cortés-Reyes and L. J. Alemany, Catalysts, 2019, 9, 609 CrossRef.
- C. Len, F. Delbecq, C. C. Corpas and E. R. Ramos, Synthesis, 2018, 50, 723–741 CrossRef CAS.
- C. Len and R. Luque, Sustainable Chem. Processes, 2014, 2, 1–10 CrossRef.
- A. Kostyniuk, D. Bajec, P. Djinović and B. Likozar, Chem. Eng. J., 2020, 394, 124945 CrossRef CAS.
- M. R. Nanda, Z. Yuan, W. Qin, H. S. Ghaziaskar, M.-A. Poirier and C. C. Xu, Fuel, 2014, 128, 113–119 CrossRef CAS.
- M. R. Nanda, Z. Yuan, W. Qin, H. S. Ghaziaskar, M.-A. Poirier and C. C. Xu, Appl. Energy, 2014, 123, 75–81 CrossRef CAS.
- R. Gérardy, D. P. Debecker, J. Estager, P. Luis and J.-C. M. Monbaliu, Chem. Rev., 2020, 120, 7219–7347 CrossRef PubMed.
- Y. Zhang, H. Xue, M. Cheng, X. Yang, Z. Zhang, X. Zhao, A. Rezayan, D. Han, D. Wu and C. Xu, ACS Catal., 2024, 14, 10009–10021 CrossRef CAS.
- X. Zhu, X. Feng, C. Yao, W. Sun, J. Ma, F. Zhong, J. Zeng, X. Ge, W. Chen, G. Qian, X. Duan, Y. Cao, Z. Liu, X.-G. Zhou and J. Zhang, Ind. Eng. Chem. Res., 2024, 63, 8175–8186 CrossRef CAS.
- D. Cortés-Borda, E. Wimmer, B. Gouilleux, E. Barré, N. Oger, L. Goulamaly, L. Peault, B. Charrier, C. Truchet, P. Giraudeau, M. Rodriguez-Zubiri, E. Le Grognec and F.-X. Felpin, J. Org. Chem., 2018, 83, 14286–14299 CrossRef PubMed.
- M. I. Jeraal, S. Sung and A. A. Lapkin, Chem.:Methods, 2021, 1, 71–77 CAS.
- C. J. Taylor, A. Pomberger, K. C. Felton, R. Grainger, M. Barecka, T. W. Chamberlain, R. A. Bourne, C. N. Johnson and A. A. Lapkin, Chem. Rev., 2023, 123, 3089–3126 CrossRef CAS PubMed.
- L. Schrecker, J. Dickhaut, C. Holtze, P. Staehle, M. Vranceanu, K. Hellgardt and K. K. Hii, React. Chem. Eng., 2023, 8, 41–46 RSC.
- M. J. Takle, B. J. Deadman, K. Hellgardt, J. Dickhaut, A. Wieja and K. K. M. Hii, ACS Catal., 2023, 13, 10541–10546 CrossRef CAS.
- C. J. Taylor, A. Baker, M. R. Chapman, W. R. Reynolds, K. E. Jolley, G. Clemens, G. E. Smith, A. J. Blacker, T. W. Chamberlain, S. D. R. Christie, B. A. Taylor and R. A. Bourne, J. Flow Chem., 2021, 11, 75–86 CrossRef CAS.
- N. Munyebvu, Z. Akhmetbayeva, S. Dunn and P. D. Howes, Nanoscale Adv., 2025, 7, 495–505 RSC.
- J. Park, Y. M. Kim, S. Hong, B. Han, K. T. Nam and Y. Jung, Matter, 2023, 6, 677–690 CrossRef CAS.
- C. P. Breen, A. M. K. Nambiar, T. F. Jamison and K. F. Jensen, Trends Chem., 2021, 3, 373–386 CrossRef.
- E. Mosqueira-Rey, E. Hernández-Pereira, D. Alonso-Ríos, J. Bobes-Bascarán and Á. Fernández-Leal, Artif. Intell. Rev., 2023, 56, 3005–3054 CrossRef.
- Y. Kuddusi, M. R. Dobbelaere, K. M. Van Geem and A. Züttel, Catal. Sci. Technol., 2024, 14, 6307–6320 RSC.
- R. P. Joshi and N. Kumar, Molecules, 2021, 26, 6761–6781 CrossRef CAS PubMed.
- P. Fantke, C. Cinquemani, P. Yaseneva, J. De Mello, H. Schwabe, B. Ebeling and A. A. Lapkin, Chem, 2021, 7, 2866–2882 CAS.
- B. Sanchez-Lengeling and A. Aspuru-Guzik, Science, 2018, 361, 360–365 CrossRef CAS PubMed.
- C.-F. Chang, K. Paragian, S. Sadula, S. Rangarajan and D. G. Vlachos, ACS Sustainable Chem. Eng., 2024, 12, 12927–12937 CrossRef CAS PubMed.
- S. Singh and R. B. Sunoj, Acc. Chem. Res., 2023, 56, 402–412 CrossRef CAS PubMed.
- A. D. A. B. Abu Sofian, X. Sun, V. K. Gupta, A. Berenjian, A. Xia, Z. Ma and P. L. Show, Energy Fuels, 2024, 38, 1593–1617 CrossRef.
- D. A. Akinpelu, O. A. Adekoya, P. O. Oladoye, C. C. Ogbaga and J. A. Okolie, Digit. Chem. Eng., 2023, 8, 100103 CrossRef.
- M. A. Curran, Curr. Opin. Chem. Eng., 2013, 2, 273–277 CrossRef.
- J. Dong, Y. Tang, A. Nzihou, Y. Chi, E. Weiss-Hortala and M. Ni, Sci. Total Environ., 2018, 626, 744–753 CrossRef CAS PubMed.
- J. M. Weber, Z. Guo, C. Zhang, A. M. Schweidtmann and A. A. Lapkin, Chem. Soc. Rev., 2021, 50, 12013–12036 RSC.
| This journal is © The Royal Society of Chemistry 2025 |