Open Access Article
Open Access ArticleA highly active and regioselective cannabigerolic acid synthase engineered from a promiscuous prenyltransferase NphB†
Ye Seop
Park‡
 *a,
Minju
Kim‡
a,
Chae Yeong
Na
a,
Hyeon Woo
Ham
a,
Jun-Young
Cho
b,
Boyoung
Park
bc,
Cheulhee
Jung
*a,
Minju
Kim‡
a,
Chae Yeong
Na
a,
Hyeon Woo
Ham
a,
Jun-Young
Cho
b,
Boyoung
Park
bc,
Cheulhee
Jung
 d,
Daechan
Park
ae and
Tae Hyeon
Yoo
d,
Daechan
Park
ae and
Tae Hyeon
Yoo
 *ae
*ae
aDepartment of Molecular Science and Technology, Ajou University, Suwon 16499, Korea. E-mail: wizard309@ajou.ac.kr; taehyeonyoo@ajou.ac.kr
bDepartment of Fundamental Pharmaceutical Science, College of Pharmacy, Kyung Hee University, Seoul 02447, Korea
cDepartment of Biomedical and Pharmaceutical Science, College of Pharmacy, Kyung Hee University, Seoul 02447, Korea
dDepartment of Biotechnology, College of Life Sciences and Biotechnology, Korea University, Seoul 02841, Korea
eAdvanced College of Bio-Convergence Engineering, Ajou University, Suwon 16499, Korea
First published on 1st May 2025
Abstract
Cannabinoids are a class of natural products originally isolated from the plant Cannabis sativa, and the demand for cannabinoid-derived compounds has continuously grown. Recombinant microorganisms harboring metabolic pathways for synthesizing these complex molecules have drawn considerable attention as alternatives to purification from the plant. NphB is a soluble aromatic prenyltransferase from Streptomyces sp. that has been shown to prenylate diverse aromatic substrates and can condense geranyl pyrophosphate (GPP) with olivetolic acid (OA) to form cannabigerolic acid (CBGA), a crucial precursor of various cannabinoids. However, the low activity and lack of regioselectivity of NphB have been a hurdle to developing efficient biological processes. Several engineered variants have been reported, but their catalytic properties still need further improvement for application in commercial production. We identified a critical residue (S214) for interaction with OA by characterizing a small library of NphB. The modeled structure docked with OA suggested two additional positions (A232 and Y288) for further engineering of the binding pocket of OA. Combined with a previously reported mutation of V49W, a variant having four changes (V49W/S214H/A232S/Y288P) showed the highest kcat/Km value with OA (275.89 ± 38.248 min−1 mM−1) at least to our knowledge, which was 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold higher than that of the wild-type enzyme (0.0052 ± 0.00184 min−1 mM−1). The structural analyses using the molecular dynamics simulation indicated that the remodeled binding pocket had favorable non-covalent interactions with OA, contributing to the decreased Km value for the substrate. The purified NphB variant synthesized CBGA using OA and GPP faster than other reported enzymes, which was more evident with low concentrations of the substrates. These findings suggested that the CBGA synthase engineered in this study holds promise for application in the production of various cannabinoids in microbial cell factories.
000-fold higher than that of the wild-type enzyme (0.0052 ± 0.00184 min−1 mM−1). The structural analyses using the molecular dynamics simulation indicated that the remodeled binding pocket had favorable non-covalent interactions with OA, contributing to the decreased Km value for the substrate. The purified NphB variant synthesized CBGA using OA and GPP faster than other reported enzymes, which was more evident with low concentrations of the substrates. These findings suggested that the CBGA synthase engineered in this study holds promise for application in the production of various cannabinoids in microbial cell factories.
Introduction
Cannabinoids are biologically active compounds naturally produced by the plant Cannabis sativa. To date, more than 150 cannabinoids have been identified. They interact with the endocannabinoid system in the human body, which plays a crucial role in regulating various physiological processes, including mood, appetite, pain sensation, memory, and immune response.1–3 In 2018, the cannabinoid-derived compound cannabidiol was first approved as Epidiolex by the U.S. Food and Drug Administration.4,5 The drug is prescribed for the treatment of seizures associated with Lennox–Gastaut syndrome or Dravet syndrome, two rare and severe forms of epilepsy.4,5 The success has motivated the development of cannabinoids as therapeutics.6,7 Even though some of them have been reported to exhibit undesirable psychotropic effects, attempts have been made to minimize these side effects, which are expected to expand cannabinoid-related therapeutics.7,8Purification from plants is the main route to obtain cannabinoids, but these methods suffer from the constrained supply of plants and the low process yield.9,10 In particular, it is often difficult to isolate one type of cannabinoid from the many derivatives present in raw materials.10,11 Methods based on chemical synthesis have been developed; however, the complex structures of cannabinoids require complicated chemical steps.12,13 Recently, the production of cannabinoids in microbial cell factories has attracted attention as an alternative method due to their fast growth rates and the feasibility of introducing exogenous metabolic pathways.10,14–20 Various cannabinoids have been biosynthesized via engineered heterologous pathways in diverse microbes including Saccharomyces cerevisiae, Pichia pastoris, Escherichia coli, Nicotiana benthamiana, and Candida viswanathii.10,14–19
The prenylation reaction of olivetolic acid (OA) into cannabigerolic acid (CBGA) (Scheme S1†) has been known as the rate-limiting step in the production of cannabinoids through the heterologous synthetic pathways.21–23 Aromatic prenyltransferases catalyze the transfer of a prenyl group from a prenyl-pyrophosphate donor (e.g., geranyl pyrophosphate; GPP) to an acceptor.24–26 Among these, CsPT4 from C. sativa was utilized for the prenylation reaction.14 However, because the enzyme is membrane-integrated, it is challenging to express it functionally in various microbial chassis.22 In contrast, NphB from Streptomyces sp., a soluble enzyme, has been shown to prenylate several aromatic substrates including OA, and has been utilized to biologically synthesize CBGA.21,22,24,27,28 However, the enzyme has two critical limitations for applying to the CBGA synthesis: a relatively low activity toward OA and the generation of the side product, 2-geranyl-olivetolic acid (2-O-GOA)21,22 (Scheme 1). Rational approaches were previously tried to engineer the binding pocket of aromatic acceptors to address the issues, and the resulting variants exhibited higher reaction rates and improved regioselectivity toward CBGA.21,22 However, some variants designed based on the modeled complex structure of NphB and OA did not exhibit the expected catalytic properties, which motivated a different approach to NphB engineering. In this study, we used a computational tool to generate a small library in that active variants can be enriched, and characterization of them revealed that S412 plays a critical role in the interaction with OA. Variants with the amino acid change in the residue showed improved catalytic properties toward OA, particularly Km values, and the best one (V49W/S214H/A232S/Y288P) exhibited more than 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 times higher kcat/Km than the wild-type enzyme without any detectable production of the side product of 2-O-GOA.
000 times higher kcat/Km than the wild-type enzyme without any detectable production of the side product of 2-O-GOA.
Materials and methods
Plasmid construction
The synthetic gene of NphB24 (Table S1†) was cloned into pET28a using the NdeI and XhoI sites, resulting in pSPEL1066. The plasmids expressing V49W/Y288P22 and G286S/Y288A21 were constructed by introducing the two mutations into the wild-type NphB gene (pSPEL1067 and pSPEL1318). The ten synthetic genes of the NphB variants from FuncLib29 (Table S1†) were cloned into pET28a using the NdeI and XhoI sites, resulting in pSPEL1132–1141. The NphB variant genes in Table S2,† other than the FuncLib variants, were constructed by assembly PCR. The synthetic genes were provided by Twist Bioscience (USA). All the primers used in this study were synthesized by Macrogen (Seoul, Korea).Protein production
Each plasmid encoding one of NphB variants was transformed into Escherichia coli BL21(DE3). The recombinant strains were cultured in 250 mL of 2× YT containing 35 μg mL−1 of kanamycin until OD600 reached 0.5. The expression of NphB variants was induced with 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG; Bioshop, Canada) at 20 °C for 16 h. The cells were harvested by centrifugation (9300 × g for 15 min) and stored at −20 °C until purification. The cell pellets were resuspended in 10 mL of lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 10 mM imidazole). After resuspension, cell lysates were lysed by sonication and centrifuged to remove cellular debris, and the supernatant was collected. Proteins with an N-terminal His6-tag were purified using Ni-NTA resin (Qiagen, Germany) according to the manufacturer's protocol. The purified protein was eluted using an elution buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 250 mM imidazole). The protein solutions were buffer-exchanged with lysis buffer using a centrifugal filter unit (MWCO: 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000; Merck Millipore, USA) and stored with 30% (v/v) glycerol (Samchun, Korea).
000; Merck Millipore, USA) and stored with 30% (v/v) glycerol (Samchun, Korea).
Enzyme reaction conditions and analyses of the products
Enzyme reactions of NphB variants were conducted in the solution of 100 mM Tris-HCl (pH 7.5) containing 10 mM MgCl2. For the results shown in Fig. 1C, 2, 4 and 6A and S3,† 2 mM olivetolic acid (OA; Cayman Chemical Company, USA) and 2 mM geranyl pyrophosphate (GPP; Sigma-Aldrich, USA) were incubated with 5 μM of each purified enzyme at 30 °C for 18 h. The results of Fig. 6B were obtained by reacting 0.2 mM of substrates (OA and GPP) with 0.5 μM of enzyme at 30 °C. The concentrations of CBGA and 2-O-GOA were analyzed using an HPLC system (Agilent 1260 Infinity II, USA) equipped with the YMC-Triart Bio C18 column (250 × 4.6 mm) (YMC, Japan). Methyl-4-hydroxybenzoate (Sigma-Aldrich, USA) was used as an internal standard to reduce errors involved in the extraction process. The internal standard dissolved in 0.1 N HCl (5 μL) was added to each reaction sample. Then, the products were extracted by methanol (v![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v = 1
v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The samples were separated via a gradient of acetonitrile with 0.1% trifluoroacetic acid from 50% to 100% for 10 min. The peak areas were used to calculate the fold change of CBGA and 2-O-GOA.
1). The samples were separated via a gradient of acetonitrile with 0.1% trifluoroacetic acid from 50% to 100% for 10 min. The peak areas were used to calculate the fold change of CBGA and 2-O-GOA.
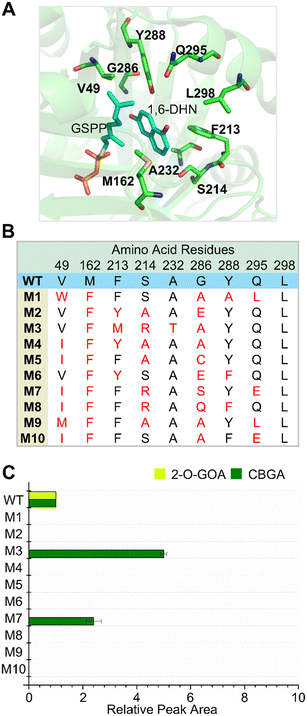 | ||
| Fig. 1 NphB library generated by FuncLib. (A) Nine residues in the binding pocket of aromatic acceptors were selected as ‘designable residues’ for FuncLib, based on the structure of NphB complexed with 1,6-dihydroxynaphthalene (1,6-DHN) and geranyl S-thiolodiphosphate (GSPP) (PDB: 1ZB6). (B) Amino acid sequences of NphB variants generated by FuncLib. (C) Production of CBGA and 2-O-GOA by each NphB variant. 2 mM of OA and GPP were incubated with 5 μM of each enzyme for 18 h at 30 °C, and the concentrations of CBGA and 2-O-GOA were shown as the peak areas of chromatograms relative to those of the wild-type enzyme. The experiments were repeated at least three times, and the error bars indicate the standard deviations. | ||
Determination of the enzymatic kinetic parameters (kcat and Km)
Kinetic assays of NphB and the four NphB variants (V49W/Y288P, G286S/Y288A, V49W/S214H/Y288P, and V49W/S214H/A232S/Y288P) were conducted under the following conditions: 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, and varied concentrations of OA and GPP. The kinetic parameters toward OA were determined using 0.001–1 mM of OA and 2 mM of GPP. For those for GPP, 0.005–1 mM of GPP and 2 mM of OA were used. The concentration of each enzyme was adjusted ranging from 1 μM to 15 μM depending on their catalytic activities. The reaction products were analyzed using the method described above. The initial velocities were fitted to the Michaelis–Menten equation through the non-linear regression of OriginPro 8.5 (Northampton, MA, USA).Molecular dynamics (MD) simulation of the NphB quadruple variant
The modeled structure of the quadruple variant without ligands was constructed using AlphaFold2.30 The substrate-free structure was subjected to the molecular dynamics (MD) simulation for 150 ns. The simulation was performed using the CHARMM36 force field31 with the water model TIP3P on a GPU cluster within GROMACS 2022.1.32 The constructed system was first minimized for 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps using the steepest descent method. After 100 ps of NVT equilibration, a 100 ps production simulation was performed at 300 K and 1 bar in the NPT ensemble. The particle mesh Ewald (PME) method was used to treat all electrostatic interactions beyond a cutoff of 12 Å. The LINCS algorithm was used to record the lengths of the bonds involving hydrogen during the simulation with an integration time step of 2 fs. The production runs lasted 150 ns with no atom restrictions and a time step of 2 fs. The coordinates were extracted from steered MD trajectories. The root mean square deviation (RMSD) during the simulation were calculated using the rmsd module. The GROMACS default analysis commands, visual molecular dynamics (VMD),33 PyMOL (https://pymol.org), and XMGRACE34 were used for data processing and visualization. The RMSD of backbone atoms was stable after 75 ns, and the structure at 100 ns was used to build the complex structure with the ligands. Then, to the modeled structure of the quadruple variant, GSPP and OA were manually introduced using UCSF Chimera.35 GSPP was introduced into the modeled structure at the same place as 1ZB6. OA was placed in the modeled structure as follows: 1) the phenyl ring of OA was superimposed on that of 1,6-DHN, and 2) the C3 carbon of OA was located at the C5 carbon of 1,6-DHN, which is prenylated by NphB. The generated complex structure was subject to the MD simulation. The parameters for the docked substrates were generated using CGenFF.36 The distances between specific atoms in the ligands or mutated residues during the simulation were calculated using the gmx distance module.
000 steps using the steepest descent method. After 100 ps of NVT equilibration, a 100 ps production simulation was performed at 300 K and 1 bar in the NPT ensemble. The particle mesh Ewald (PME) method was used to treat all electrostatic interactions beyond a cutoff of 12 Å. The LINCS algorithm was used to record the lengths of the bonds involving hydrogen during the simulation with an integration time step of 2 fs. The production runs lasted 150 ns with no atom restrictions and a time step of 2 fs. The coordinates were extracted from steered MD trajectories. The root mean square deviation (RMSD) during the simulation were calculated using the rmsd module. The GROMACS default analysis commands, visual molecular dynamics (VMD),33 PyMOL (https://pymol.org), and XMGRACE34 were used for data processing and visualization. The RMSD of backbone atoms was stable after 75 ns, and the structure at 100 ns was used to build the complex structure with the ligands. Then, to the modeled structure of the quadruple variant, GSPP and OA were manually introduced using UCSF Chimera.35 GSPP was introduced into the modeled structure at the same place as 1ZB6. OA was placed in the modeled structure as follows: 1) the phenyl ring of OA was superimposed on that of 1,6-DHN, and 2) the C3 carbon of OA was located at the C5 carbon of 1,6-DHN, which is prenylated by NphB. The generated complex structure was subject to the MD simulation. The parameters for the docked substrates were generated using CGenFF.36 The distances between specific atoms in the ligands or mutated residues during the simulation were calculated using the gmx distance module.
Results and discussion
Two variants from a FuncLib library exhibited higher productivity and regioselectivity for CBGA synthesis than the wild-type enzyme
Rational strategies were previously tried to engineer the substrate binding site of NphB.21,22 In these studies, the crystal structure of NphB24 was docked with OA, and the resulting structure was used to evaluate mutations in the active site computationally. These approaches led to the discovery of NphB variants with improved catalytic activities and regioselective modification of OA. However, all the calculation results were not consistent with the experimental characterizations, which motivated us to try a different approach to engineer NphB. Algorithms have been reported to make smart libraries in that active variants are enriched, which can dramatically reduce the library size to examine.29,37,38 We turned to one of such algorithms named FuncLib.29 Substitutions at target positions are decided based on the phylogenetic analyses, and the variants having multiple mutations are evaluated for their stability using Rosetta. When residues in the active site of an enzyme are targeted, the top-ranked candidates are expected to fold into stable structures with pre-organized active site pockets.29,39,40The crystal structure in which NphB is complexed with its substrates of 1,6-dihydroxynaphthalene (1,6-DHN) and geranyl S-thiolodiphosphate (GSPP) (PDB: 1ZB6 (ref. 24)) was used to decide the residues in the binding pocket of aromatic acceptors for mutation. Six residues (M162, F213, S214, Y288, Q295, and L298) located nearby the aromatic substrate-binding pocket were selected based on the structure (Fig. 1A) as the targets for FuncLib. In addition, three residues (V49, A232, and G286),21,22 which had been reported to positively affect the regioselectivity and the activity of the enzyme toward OA, were included. The total nine residues were decided as ‘designable residues’ for FuncLib, and the algorithm proposed 1000 variants (Data file S1†). Among them, the 10 variants with the highest stability scores (designated as M1-M10; Fig. 1B) were characterized for the synthesis of CBGA using OA and GPP. The enzymes were expressed with the N-terminal His6-tag in Escherichia coli and then purified through Ni-NTA affinity chromatography. The purified enzymes were incubated with OA and GPP for 18 h at 30 °C, and the products were analyzed using RP-HPLC. The peak for CBGA (∼9.6 min, Fig. S1†) was identified using the chemically synthesized one (Fig. S2 and Table S3†). The wild-type enzyme has been reported to produce CBGA and 2-O-GOA, and the other peak (∼10.2 min, Fig. S1†) was assigned to 2-O-GOA. The peak areas of CBGA and 2-O-GOA for each variant were compared to those of the wild-type enzyme (Fig. 1C). Eight enzymes besides M3 and M7 did not produce either CBGA or 2-O-GOA. Interestingly, only the CBGA peak was detected for M3 and M7, which suggested the regioselectivity of these two variants toward CBGA. In particular, M3 produced CBGA 5-fold more than that of the wild-type enzyme.
Mutations in the S214 position played a critical role in the improvement of the CBGA productivity
Among the five amino acid changes in M3 (Fig. 1B), we focused on F213M, S214R, and A232T, which were unique (F213M and A232T) or found also in M7 (S214R). Three variants having each mutation were characterized for CBGA synthesis (Fig. 2). The F213M and A232T variants produced both CBGA and 2-O-GOA, and their CBGA concentrations were lower than that of the wild-type enzyme. Interestingly, the S214R variant produced CBGA exclusively, and its concentration was higher than that of M3. Motivated by these results, we changed the S214 position to ten different amino acids which were selected based on the physical properties of their side chains (positively charged: His or Lys; negatively charged: Glu; hydrophilic: Cys, Thr, or Asn; hydrophobic: Phe, Ala, Gly, or Val). One variant (S214H) converted OA into CBGA without any detectable synthesis of 2-O-GOA, and the concentration of CBGA was higher than that of both M3 and S214R (Fig. 2).The S214H structure was modeled based on the crystal structure of NphB complexed with 1,6-DHN and GSPP (PDB: 1ZB6) using AlphaFold2.30 There was no change in the binding pocket for GPP, and GSPP was manually introduced into the modeled structure of S214H at the same place as 1ZB6. Then, OA was docked into the variant structure in place of 1,6-DHN using AutoDock.41 We checked the top-ranked ten poses carefully, but none of them gave a clue as to how the substitution at position 214 improved the catalytic properties (data not shown). Instead, we manually placed OA in the modeled structure as follows: 1) the phenyl ring of OA was superimposed on that of 1,6-DHN, and 2) the C3 carbon of OA was located at the C5 carbon of 1,6-DHN, which is prenylated by NphB (Fig. 3A). The manually docked structure showed that the carboxy group of OA was in proximity to the imidazole ring of S214H (Fig. 3B). The favorable electrostatic interaction between them might play a role in positioning OA at the active site as the C3 carbon is prenylated preferentially. Based on this observation, we decided to use the two variants in which S214 was mutated into His or Arg as templates for further engineering. In contrast, although the side chain of Lys is also positively charged, the S214K variant produced less CBGA than the other two variants, so we did not pursue the change further.
Additional amino acid substitutions in the OA binding pocket synergized with the S214 mutations
The modeled structure of S214H docked with OA predicted a spatial clash between the alkyl (pentanyl) chain of OA and the phenol group of Y288 (Fig. 3B). The Y288 residue was mutated into a small hydrophobic residue (Ala, Val, Gly, or Pro) for both S214H and S214R. Total eight double mutants were reacted with OA and GPP for CBGA synthesis (Fig. 4). The changes into Ala or Val resulted in higher CBGA concentrations than the templates of S214H and S214R. However, the substitution into a smaller and flexible Gly abolished (S214R) or decreased (S214H) the CBGA synthesis activity. Interestingly, the Y288P mutation caused the opposite effect for the two template variants. While S214R/Y288P barely produced CBGA, S214H/Y288P enzyme showed a 15.3-fold higher CBGA concentration than that of the wild-type enzyme.Our results suggested that the interaction between the carboxyl group of OA and the positively charged residue at position 214 played a crucial role in positioning the substrate for the regioselective prenylation. We hypothesized that a hydrogen bond with the hydroxyl group at the C2 carbon would strengthen the interactions for preferential substrate orientation in the active site. In the modeled structure (Fig. 3B), the side chain of A232 was located close to the hydroxyl group, and substitution into one of three polar amino acids (Ser, Thr, or Asn) was made for the S214H/Y288P variant. Only the S214H/A232S/Y288P variant showed an improved CBGA titer compared to the S214H/Y288P (Fig. 4). Among several mutations previously reported to improve the catalytic activity, we focused on the V49W mutation since the synergetic effect had been observed for the combination of V49W and Y288P. The additional mutation improved the CBGA synthesis activities22 (V49W/S214H/Y288P and V49W/S214H/A232S/Y288P) (Fig. 4). The quadruple variant showed the best performance for CBGA synthesis, with a 20.4-fold higher concentration than that of the wild-type (Fig. 4).
The quadruple variant showed a 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) 000-fold enhanced catalytic efficiency toward OA compared to the wild-type enzyme
000-fold enhanced catalytic efficiency toward OA compared to the wild-type enzyme
The wild-type and the two variant enzymes (V49W/S214H/Y288P and V49W/S214H/A232S/Y288P) showing the highest CBGA concentrations in Fig. 4 were characterized for their catalytic parameters (kcat and Km) for the two substrates of OA and GPP (Table 1, Fig. 5 and S3†). Two previously engineered NphB variants (G286S/Y288A21 and V49W/Y288P22) were also characterized for comparison. Consistent with the reported results, the two variants (G286S/Y288A and V49W/Y288P) showed much higher catalytic activities than the wild-type enzyme, and V49W/Y288P had a higher kcat/Km value for OA than G286S/Y288A. The triple variant (V49W/S214H/Y288P) had an additional change (S214H) compared with the V49W/Y288P, and the comparison of the kcat and Km values of the two enzymes suggested the role of the S214H mutation in interacting with the substrate, lowering the Km value with OA from 0.13 ± 0.024 mM (V49W/Y288P) to 0.006 ± 0.0021 mM (V49W/S214H/Y288P). The quadruple variant (V49W/S214H/A232S/Y288P) showed the highest kcat/Km value with OA (275.89 ± 38.248 min−1 mM−1), which was 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold higher than that of the wild-type enzyme (0.0052 ± 0.00184 min−1 mM−1) and 28-fold higher than that of V49W/Y288P (9.83 ± 0.939 min−1 mM−1). Interestingly, even though the engineering efforts, including the two previous variants, were made toward OA, the variants showed a wide range of kcat/Km values for GPP (Table 1). The G286S/Y288A enzyme showed the highest catalytic activity toward GPP. Even though V49W/Y288P was better with OA than G286S/Y288A, the latter showed around a 5-fold higher kcat/Km value for GPP (51.34 ± 13.367 min−1 mM−1) than the former one (9.96 ± 1.265 min−1 mM−1). The quadruple variant exhibited a catalytic activity with GPP comparable to that of G286/Y288A. The concentration of OA (2 mM) used to determine the kinetic parameters for GPP was not high enough to expect the saturation with the substrate for some variants. Therefore, we tried a three-dimensional Michaelis–Menten equation42 (see Table S3† for the equation) by fitting all the data for each enzyme. The resulting kinetic parameters (Table S3†) were similar to those shown in Table 1, except for G286/Y288A, whose values showed relatively large standard errors.
000-fold higher than that of the wild-type enzyme (0.0052 ± 0.00184 min−1 mM−1) and 28-fold higher than that of V49W/Y288P (9.83 ± 0.939 min−1 mM−1). Interestingly, even though the engineering efforts, including the two previous variants, were made toward OA, the variants showed a wide range of kcat/Km values for GPP (Table 1). The G286S/Y288A enzyme showed the highest catalytic activity toward GPP. Even though V49W/Y288P was better with OA than G286S/Y288A, the latter showed around a 5-fold higher kcat/Km value for GPP (51.34 ± 13.367 min−1 mM−1) than the former one (9.96 ± 1.265 min−1 mM−1). The quadruple variant exhibited a catalytic activity with GPP comparable to that of G286/Y288A. The concentration of OA (2 mM) used to determine the kinetic parameters for GPP was not high enough to expect the saturation with the substrate for some variants. Therefore, we tried a three-dimensional Michaelis–Menten equation42 (see Table S3† for the equation) by fitting all the data for each enzyme. The resulting kinetic parameters (Table S3†) were similar to those shown in Table 1, except for G286/Y288A, whose values showed relatively large standard errors.
| OAa | GPPb | |||||
|---|---|---|---|---|---|---|
| k cat (min−1) | K m (mM) | k cat/Km (min−1 mM−1) | k cat (min−1) | K m (mM) | k cat/Km (min−1 mM−1) | |
| a k cat and Km for OA were determined using 2 mM GPP. b k cat and Km values of GPP were determined using 2 mM OA. The experiments were repeated at least three times. Values are presented as mean ± standard deviation. | ||||||
| Wild-type | 0.00243 ± 0.000304 | 0.49 ± 0.113 | 0.00525 ± 0.001841 | 0.00246 ± 0.000484 | 0.030 ± 0.0122 | 0.0856 ± 0.01864 |
| G286S/Y288A | 1.50 ± 0.204 | 0.38 ± 0.103 | 4.08 ± 0.753 | 2.11 ± 0.232 | 0.042 ± 0.0065 | 51.3 ± 13.37 |
| V49W/Y288P | 1.27 ± 0.183 | 0.13 ± 0.024 | 9.83 ± 0.939 | 1.61 ± 0.306 | 0.16 ± 0.020 | 9.96 ± 1.265 |
| V49W/S214H/Y288P | 0.402 ± 0.1139 | 0.0062 ± 0.00211 | 68.1 ± 19.53 | 0.500 ± 0.0886 | 0.029 ± 0.0026 | 17.3 ± 3.58 |
| V49W/S214H/A232S/Y288P | 2.87 ± 0.183 | 0.010 ± 0.0009 | 276 ± 38.2 | 3.35 ± 0.389 | 0.083 ± 0.0160 | 40.8 ± 5.73 |
 | ||
| Fig. 5 Catalytic activities of NphB enzymes toward OA. The kcat/Km values with OA from Table 1 were shown on a log scale for comparison. | ||
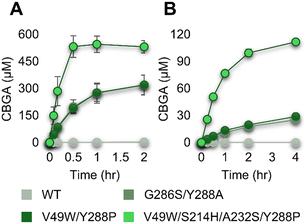
While the finally engineered variant (V49W/S214H/A232S/Y288P) showed a much higher catalytic activity than the other enzymes (Table 1 and Fig. 5), the long incubation yielded relatively small differences in the CBGA production among the variants (Fig. 4). Thus, we investigated the time-dependent CBGA concentration changes with 2 mM of OA and GPP at 30 °C for 5 μM of each enzyme, which was the same condition as those of Fig. 4 (Fig. 6A and S4†). The quadruple variant produced around 500 μM CBGA in 30 min, and the CBGA concentration was maintained after the time point. In contrast, the CBGA concentration increased for 2 h for the two other variants (G286S/Y288A and V49W/Y288P), and the steady-state one was around 300 μM. In the case of the wild-type enzyme, the production of CBGA continued throughout the entire period. Interestingly, the steady-state concentrations of CBGA differed among the enzymes, and the quadruple variant resulted in a higher concentration than other variants. We hypothesized that the degradation of CBGA43–45 was correlated with the observed steady-state concentrations. The mathematical analysis described in the ESI† shows that the steady-state CBGA concentration ([CBGA]) increases as the rate constant (k1) for the reaction of OA and GPP to CBGA increases. Consistent with the Km values (Table 1), the difference in the CBGA production rate was more obvious when the concentrations of OA and GPP were decreased 10-fold (Fig. 6B).
The modeled structure of the quadruple variant suggested the favorable interactions of OA in the substrate-binding pocket
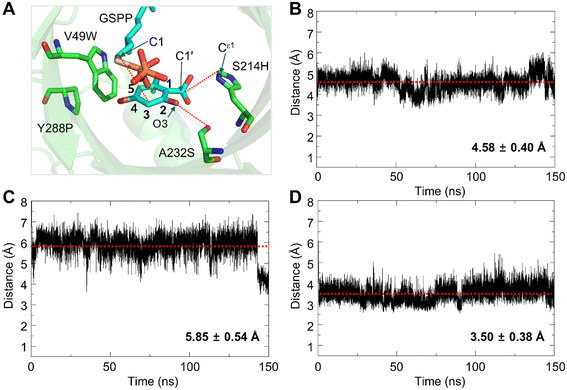
Interactions between OA and the substrate-binding pocket of V49W/S214H/A232S/Y288P were investigated using a modeled structure complexed with OA and GSPP. Initially, the substrate-free structure modeled using AlphaFold2 (ref. 30) was subjected to the molecular dynamics (MD) simulation for 150 ns. The root mean square deviation (RMSD) of backbone atoms was stable after 75 ns (Fig. S5†), and the structure at 100 ns was used to build the complex structure with the ligands. GSPP and OA were manually introduced to the modeled structure following the same procedure as done for the S214H variant. The complex structure was subjected to the MD simulation for another 150 ns (Fig. S6†). OA remained stable in the active site throughout the simulation period (Fig. 7). The average distance between the C3 carbon of OA and the C1 carbon of GSPP during the simulation was 4.58 ± 0.40 Å (Fig. 7B), which is comparable to the distance (4.2 Å) between the C5 of 1,6-DHN (Fig. 3A) and the C1 of GSPP in the reported NphB structure (PDB: 1ZB6 (ref. 24)). An electrostatic interaction was detected between the side chain of S214H and the carboxylate group of OA (Fig. 7C). In addition, the side chain of A232S formed a hydrogen bond with the hydroxyl group at C2 of OA (Fig. 7D). The structural analyses indicated that the remodeled binding pocket of the quadruple variant could facilitate additional favorable non-covalent interactions with OA compared to the wild-type enzyme, which resulted in the improvement in the catalytic efficiency, particularly the decrease of the Km value for the substrate.Conclusion
The condensation reaction of GPP with OA into CBGA, catalyzed by aromatic prenyltransferases, has been known as the rate-limiting step in the production of cannabinoids through heterologous synthetic pathways. NphB from Streptomyces sp. has shown the potential for the application in the biological synthesis of CBGA. However, the enzyme has suffered from a low activity toward OA and the side reaction to produce 2-O-GOA. In this study, we engineered the binding pocket of aromatic acceptors (OA) of NphB prenyltransferase to improve its catalytic efficiency and regioselectivity. Screening of a small library generated by a computational algorithm (FuncLib) and then a focused library led to the identification of residue S214 playing a critical role in the interaction with OA. The docking of OA in the binding pocket proposed Y288 and A232 as targets for substitution to make space for the alkyl chain and introduce a favorable interaction with the hydroxyl group of C2, respectively. The finally engineered variant (V49W/S214H/A232S/Y288P), which had the previously reported mutation (V49W) in addition to the changes in the three positions (S214H/A232S/Y288P), showed the highest kcat/Km value toward OA among the reported enzymes, without any detectable synthesis of 2-O-GOA.The structural analyses based on the molecular dynamics simulation suggested favorable interactions between OA and the engineered binding pocket: 1) the electrostatic interaction between the imidazole ring of S214H and the carboxylate group of OA and 2) the hydrogen bonding between the side chain of A232S and the hydroxyl group at C2 of OA. In particular, the strong charge–charge interaction seems to play a crucial role in lowering the Km value for OA. The two studies reporting G286S/Y288A21 and V49W/Y288P22 provided the simulated structures complexed with OA. The relative orientations of the C3 carbon of OA to the C1 carbon of GSPP in their structures were similar to ours, but the interactions of OA in the binding pocket seemed different based on the description in the manuscript. The structures complexed with OA need to be determined to understand the roles of the mutations, which might provide clues for further engineering. The results of the in vitro CBGA synthesis using the engineered NphB variants (Fig. 6) could estimate the ability of the NphB variants in the complex cellular environments. The microorganisms harboring the metabolic pathways to synthesize OA have been reported to produce the metabolite in the range of 50 to 150 mg L−1 (0.22 to 0.67 mM),14,20,46 which would be the maximum concentration without further conversion into other metabolites. Thus, it is likely that the OA concentration is even lower under the condition where it serves as a precursor for CBGA. The quadruple variant synthesized CBGA faster than the other variants, and the improvement was more evident at a low concentration of OA (0.2 mM). The NphB variants need to be evaluated in recombinant hosts for application to the biological production of cannabinoids using microbial cell factories, which is planned for further study.
Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
Ye Seop Park: conceptualization, data curation, methodology, investigation, formal analysis, writing – original draft, writing – review & editing. Minju Kim: methodology, data curation, investigation, formal analysis, writing – original draft. Chae Yeong Na: data curation, formal analysis. Hyeon Woo Ham: data curation, formal analysis. Jun-Young Cho: methodology, resources. Boyoung Park: methodology, resources. Cheulhee Jung: conceptualization, resources. Daechan Park: conceptualization, supervision. Tae Hyeon Yoo: conceptualization, data curation, methodology, investigation, supervision, funding acquisition, writing – original draft, writing – review & editing.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This research was supported by the Bio&Medical Technology Development Program of the National Research Foundation funded by the Korean government (2022M3A9B6082670 and RS-2024-00439931).References
- R. Tanasescu and C. S. Constantinescu, Immunobiology, 2010, 215, 588–597 CrossRef CAS PubMed.
- T. W. Klein, C. Newton, K. Larsen, L. Lu, I. Perkins, L. Nong and H. Friedman, J. Leukocyte Biol., 2003, 74, 486–496 CrossRef CAS PubMed.
- P. G. Fine and M. J. Rosenfeld, Rambam Maimonides Med. J., 2013, 4, e0022 Search PubMed.
- J. M. Nichols and B. L. F. Kaplan, Cannabis Cannabinoid Res., 2020, 5, 12–31 CrossRef CAS PubMed.
- R. Abu-Sawwa, B. Scutt and Y. Park, J. Pediatr. Pharmacol. Ther., 2020, 25, 485–499 Search PubMed.
- T. W. Klein, Nat. Rev. Immunol., 2005, 5, 400–411 CrossRef CAS PubMed.
- C. Pagano, G. Navarra, L. Coppola, G. Avilia, M. Bifulco and C. Laezza, Int. J. Mol. Sci., 2022, 23, 3344 CrossRef CAS PubMed.
- S. Gottschling, O. Ayonrinde, A. Bhaskar, M. Blockman, O. D'Agnone, D. Schecter, L. D. Suarez Rodriguez, S. Yafai and C. Cyr, Int. J. Gen. Med., 2020, 13, 1317–1333 CrossRef CAS PubMed.
- A. Hazekamp, R. Simons, A. Peltenburg-Looman, M. Sengers, R. van Zweden and R. Verpoorte, J. Liq. Chromatogr. Relat. Technol., 2009, 27, 2421–2439 CrossRef.
- M. Hesami, M. Pepe, A. Baiton and A. M. P. Jones, Biotechnol. Adv., 2023, 62, 108074 CrossRef CAS PubMed.
- F. Bucar, A. Wube and M. Schmid, Nat. Prod. Rep., 2013, 30, 525–545 RSC.
- D. H. Dethe, R. D. Erande, S. Mahapatra, S. Das and B. V. Kumar, Chem. Commun., 2015, 51, 2871–2873 RSC.
- Z. P. Shultz, G. A. Lawrence, J. M. Jacobson, E. J. Cruz and J. W. Leahy, Org. Lett., 2018, 20, 381–384 CrossRef CAS PubMed.
- X. Luo, M. A. Reiter, L. d'Espaux, J. Wong, C. M. Denby, A. Lechner, Y. Zhang, A. T. Grzybowski, S. Harth, W. Lin, H. Lee, C. Yu, J. Shin, K. Deng, V. T. Benites, G. Wang, E. E. K. Baidoo, Y. Chen, I. Dev, C. J. Petzold and J. D. Keasling, Nature, 2019, 567, 123–126 CrossRef CAS PubMed.
- G. R. Favero, G. V. de Melo Pereira, J. C. de Carvalho, D. P. de Carvalho Neto and C. R. Soccol, Fermentation, 2022, 8, 84 CrossRef CAS.
- A. Carvalho, E. H. Hansen, O. Kayser, S. Carlsen and F. Stehle, FEMS Yeast Res., 2017, 17, 4 CrossRef PubMed.
- K. Blatt-Janmaat and Y. Qu, Int. J. Mol. Sci., 2021, 22, 2454 CrossRef CAS PubMed.
- D. Wiles, B. K. Shanbhag, M. O'Brien, M. S. Doblin, A. Bacic and T. Beddoe, Phytochemistry, 2022, 203, 113380 CrossRef CAS PubMed.
- B. Zirpel, F. Stehle and O. Kayser, Biotechnol. Lett., 2015, 37, 1869–1875 CrossRef CAS PubMed.
- Z. Tan, J. M. Clomburg and R. Gonzalez, ACS Synth. Biol., 2018, 7, 1886–1896 CrossRef CAS PubMed.
- M. A. Valliere, T. P. Korman, N. B. Woodall, G. A. Khitrov, R. E. Taylor, D. Baker and J. U. Bowie, Nat. Commun., 2019, 10, 565 CrossRef CAS PubMed.
- K. J. H. Lim, Y. D. Hartono, B. Xue, M. K. Go, H. Fan and W. S. Yew, ACS Catal., 2022, 12, 4628–4639 CrossRef CAS.
- F. Thomas, C. Schmidt and O. Kayser, Appl. Microbiol. Biotechnol., 2020, 104, 9551–9563 CrossRef CAS PubMed.
- T. Kuzuyama, J. P. Noel and S. B. Richard, Nature, 2005, 435, 983–987 CrossRef CAS PubMed.
- U. Metzger, S. Keller, C. E. Stevenson, L. Heide and D. M. Lawson, J. Mol. Biol., 2010, 404, 611–626 CrossRef CAS PubMed.
- R. Chen, B. Gao, X. Liu, F. Ruan, Y. Zhang, J. Lou, K. Feng, C. Wunsch, S. M. Li, J. Dai and F. Sun, Nat. Chem. Biol., 2017, 13, 226–234 CrossRef CAS PubMed.
- C. Schmidt, M. Aras and O. Kayser, Biotechnol. J., 2024, 19, e2300507 CrossRef PubMed.
- S. Spitzer, J. Wloka, J. Pietruszka and O. Kayser, ChemBioChem, 2023, 24, e202300441 CrossRef CAS PubMed.
- O. Khersonsky, R. Lipsh, Z. Avizemer, Y. Ashani, M. Goldsmith, H. Leader, O. Dym, S. Rogotner, D. L. Trudeau, J. Prilusky, P. Amengual-Rigo, V. Guallar, D. S. Tawfik and S. J. Fleishman, Mol. Cell, 2018, 72(178–186), e175 Search PubMed.
- J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Zidek, A. Potapenko, A. Bridgland, C. Meyer, S. A. A. Kohl, A. J. Ballard, A. Cowie, B. Romera-Paredes, S. Nikolov, R. Jain, J. Adler, T. Back, S. Petersen, D. Reiman, E. Clancy, M. Zielinski, M. Steinegger, M. Pacholska, T. Berghammer, S. Bodenstein, D. Silver, O. Vinyals, A. W. Senior, K. Kavukcuoglu, P. Kohli and D. Hassabis, Nature, 2021, 596, 583–589 CrossRef CAS PubMed.
- J. Huang and A. D. MacKerell Jr., J. Comput. Chem., 2013, 34, 2135–2145 CrossRef CAS PubMed.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess and E. Lindahl, SoftwareX, 2015, 1-2, 19–25 CrossRef.
- A. D. William Humphrey and K. Schulten, J. Mol. Graphics, 1996, 14, 33–38 CrossRef PubMed.
- P. Turner, Center for Coastal and Land-Margin Research, Oregon Graduate Institute of Science and Technology, Beaverton, OR, 2005, vol. 2 Search PubMed.
- E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng and T. E. Ferrin, J. Comput. Chem., 2004, 25, 1605–1612 CrossRef CAS PubMed.
- K. Vanommeslaeghe, E. Hatcher, C. Acharya, S. Kundu, S. Zhong, J. Shim, E. Darian, O. Guvench, P. Lopes, I. Vorobyov and A. D. Mackerell Jr., J. Comput. Chem., 2010, 31, 671–690 CrossRef CAS PubMed.
- L. Sumbalova, J. Stourac, T. Martinek, D. Bednar and J. Damborsky, Nucleic Acids Res., 2018, 46, W356–W362 CrossRef CAS PubMed.
- R. Shroff, A. W. Cole, D. J. Diaz, B. R. Morrow, I. Donnell, A. Annapareddy, J. Gollihar, A. D. Ellington and R. Thyer, ACS Synth. Biol., 2020, 9, 2927–2935 CrossRef CAS PubMed.
- P. Gomez de Santos, I. Mateljak, M. D. Hoang, S. J. Fleishman, F. Hollmann and M. Alcalde, J. Am. Chem. Soc., 2023, 145, 3443–3453 CrossRef CAS PubMed.
- S. Barber-Zucker, I. Mateljak, M. Goldsmith, M. Kupervaser, M. Alcalde and S. J. Fleishman, ACS Catal., 2022, 12, 13164–13173 CrossRef CAS PubMed.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, J. Comput. Chem., 2009, 30, 2785–2791 CrossRef CAS PubMed.
- A. V. Fejzagic, S. Myllek, F. Hogenkamp, J. Greb, J. Pietruszka and T. Classen, ChemistryOpen, 2020, 9, 959–966 CrossRef CAS PubMed.
- M. Wang, Y. H. Wang, B. Avula, M. M. Radwan, A. S. Wanas, J. van Antwerp, J. F. Parcher, M. A. ElSohly and I. A. Khan, Cannabis Cannabinoid Res., 2016, 1, 262–271 CrossRef CAS PubMed.
- F. Taura, S. Morimoto and Y. Shoyama, J. Biol. Chem., 1996, 271, 17411–17416 CrossRef CAS PubMed.
- M. T. Garcia-Valverde, C. Sanchez-Carnerero Callado, M. C. Diaz-Linan, V. Sanchez de Medina, J. Hidalgo-Garcia, X. Nadal, L. Hanus and C. Ferreiro-Vera, Front. Chem., 2022, 10, 1038729 CrossRef CAS PubMed.
- Y. Zhang, J. Guo, P. Gao, W. Yan, J. Shen, X. Luo and J. D. Keasling, Metab. Eng., 2023, 80, 232–240 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cy01367k |
| ‡ Ye Seop Park and Minju Kim contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |