Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Chemical proteomics for a comprehensive understanding of functional activity and the interactome
Kostiantyn Kozoriz
and
Jun-Seok Lee
 *
*
Department of Pharmacology, Korea University College of Medicine, Seoul, Republic of Korea. E-mail: junseoklee@korea.ac.kr
First published on 19th May 2025
Abstract
Traditional mass spectrometry (MS)-based proteomics aims to detect and measure protein expression on a global scale and elucidate the link between protein function and phenotypic characteristics. Although advances in MS technology have significantly broadened the scope of detectable proteomes, these methodologies primarily provide data on protein abundance and offer limited insights into their functional activities. Phenotypic traits emerge from the interplay between protein abundance and functional activity, making the accurate measurement of activity a critical but challenging task, owing to the complexity of biological systems. Furthermore, the biological function of a protein is strongly linked to its interaction with other molecules within the cellular environment. Chemical proteomics offers a complementary approach that uses a toolkit developed in chemical biology to map the molecular interactome and provide initial insights into the activities of specific target proteins. However, the value of these techniques lies not in isolation, but as part of a broader experimental workflow that includes follow-up biological investigations to validate the findings and elucidate their functional relevance. This tutorial review highlights the design principles of chemical tools and examines their applications in two key areas: (i) functional activity profiling of biomolecules and (ii) molecular proximity profiling for interactome characterization. We also discuss the importance of the experimental context in shaping data interpretation and ensuring the practical adoption of these methods by biologists. Although chemical proteomics is not a standalone solution, it represents a promising step toward next-generation omics technologies and advances our understanding of biological functions at the molecular level.
Key learning points(1) Chemical proteomics was developed using activity-based protein profiling (ABPP).(2) The association of ABPP with reactive residue profiling is discussed, aiding in the identification of covalent drug targets and functional hot spots on proteins. (3) Advanced techniques for capturing new protein interactions in natural biological environments are discussed. (4) Unbiased drug-target profiling was compared between the labeled and label-free approaches. (5) The therapeutic potential of chemical proteomics was established using a discovery pipeline in biotechnology and pharmaceutical research, leading to the identification of several clinical candidates. |
1. Introduction
Biological processes are governed by the intricate actions of proteins, which serve as structural scaffolds and facilitate catalytic reactions. The traditional reductionist approach meticulously reveals the detailed molecular mechanisms of individual protein functions by investigating each protein on a case-by-case basis. However, the surge in omics research methods has prompted a shift toward system-wide exploration, providing broader insights into the complexities of biological systems. PCR amplification facilitates comprehensive exploration of the genome, delivers detailed sequence information, identifies sequence alterations, and assesses expression levels through copy numbers across the entire genomic spectrum. In contrast to genomics, where amplified copies of products are used for detection, proteomics involves the direct, non-amplified detection of peptides using mass spectrometry (MS) techniques.Given that a single gene can give rise to multiple proteoforms through enzymatic modifications and chemical processing, the scope of diversity that proteomics seeks to cover far exceeds the number of genes. Therefore, conventional proteomics is focused on identifying and differentiating proteins based on their expression levels across distinct samples.1,2 The current standards for both bottom-up and top-down proteomic analyses have been significantly influenced by advancements in electrospray ionization for MS analysis,3,4 nanoscale liquid chromatography,5 and database searching using MS/MS spectra (Fig. 1).6,7 In bottom-up techniques, proteins are digested using enzymes, such as trypsin, Lys-C, Lys-N, Glu-C, or Asp-N (Fig. 1a). The resulting peptides are typically analyzed using nano-liquid chromatography coupled with electrospray ionization or matrix-assisted laser desorption/ionization. The parent mass of each digested peptide is initially measured, followed by further fragmentation of the isolated peptides in the MS chamber by collision with gas. This process produces b-ions (charge is retained on the N-terminus as a primary ammonium ion) and y-ions (charge is retained at the C-terminus as an acylium ion), which correspond to peptide backbone cleavages at specific residues. Fragmentation tandem mass spectra are matched against a protein sequence database to determine protein identity and location (Fig. 1a). Although the bottom-up approach is suitable for protein identification, it is not optimized for single-point mutants, in-dels, alternative spliced variants, or intact modification studies. The top-down approach (Fig. 1b) measures intact protein ions without the need for enzymatic pre-digestion.8 The complete protein ion is introduced into the MS, where it underwent further fragmentation using gas collision. This analysis provides the primary structure and possible modifications. Although high-resolution tandem MS techniques have the theoretical capacity to analyze a protein completely, achieving extensive fragmentation of intact proteins, especially those with larger molecular weights, is challenging.
Substantial advancements in MS technology, resulting in enhanced resolution, sensitivity, and scan speed, have led to an increased capacity for protein detection. This progress has made it possible to conduct proteomic analyses at the single-cell level.9 However, several inherent technical challenges remain. First, in the absence of an amplification method similar to PCR, it becomes difficult to detect low-abundance proteins, which impedes comprehensive coverage of the entire proteomic landscape and is highly dependent on the dynamic range of the detection platform. Second, once translated, proteins present a level of complexity similar to that of post-translational modifications (PTMs) that surpasses their primary sequences. Given that peptides containing PTMs constitute only a small subset of the total peptide population, their quantity is often insufficient for proteomic identification. Furthermore, modifications can adversely affect the ionization efficiency of peptides, thereby presenting additional difficulties. Third, system-wide investigations of proteomic expression changes against phenotypic signatures have contributed valuable insights but are limited in their ability to predict functional outcomes, as protein activity is not solely determined by copy number. These encompass the state of catalytic activity and the presence of quaternary structures and their interaction partners. To address these challenges, chemical proteomics, which harnesses the power of chemical tools to enable analyses based on new principles for previously unmet requirements, has been established. In this review, we focus on the chemical biology strategies for exploring the proteomic landscape.
Systematic exploration of biology using small molecules started as a branch of genetics called chemical genetics.10 But these chemical genetics studies focused on perturbing a specific protein of interest,11 limiting the scope of proteins investigated. Chemical proteomics is the next conceptual paradigm in the field and aims to develop chemical probes that engage within a broader scope, allowing assessment of specific characteristics of all proteins targeted.
In this tutorial review, we summarize the recent progress in chemical proteomics, focusing on two key areas. We begin by introducing the concept of activity-based profiling for enzyme activity profiling and derivative investigation. Subsequently, chemical and enzymatic tags used in spatiotemporal mapping technologies to study protein–protein interactions (PPI) or drug interactome studies are discussed. Finally, we delve into future perspectives in this field and discuss the current challenges.
2. Functional activity profiling
Life processes involve complex signaling pathway networks. These biochemical events occur via a cascade of enzymatic reactions and their subsequent products. This implies that the physiological status of a living organism can be partially portrayed by the compiled information on proteins, including their sequences, PTMs, copy numbers, catalytic activities, compartmentalization, and molecular associations. Although conventional proteomic methods allow for the quantitative measurement of proteins, they fail to accurately determine the precise location and type of chemical modifications or the activity status of enzymes.A substantial advancement in overcoming these challenges was made with the development of bio-orthogonal click reactions, which was honored by the 2022 Nobel Prize in Chemistry.12 This advancement has made it feasible to comprehensively explore previously uncharted territories, enabling selective covalent bond formation at desired sites, even in complex biological environments. With a carefully designed chemical probe containing bio-orthogonal groups, subsequent functionalization can be performed for visualization or enrichment, regardless of the cellular location or associated proteins. The advent of bio-orthogonal reactions has led to the flourishing of chemoproteomics, with extensive investigations into the design of chemical probes, revealing new avenues for functional activity profiling at the individual protein level.
2.1. Activity-based protein profiling
Activity-based protein profiling (ABPP) uses enzyme-site-targeting chemical probes to investigate the functional states of enzyme families (Fig. 2a). This approach stems from the combination of mechanistic enzymology studies and the design of small-molecule inhibitors.13 In enzymology, fluorophosphonates (FPs) have been used as stable transition state analogs to covalently label the active-site serine (Ser-195)14 of chymotrypsin, mimicking the tetrahedral intermediate of catalysis (Fig. 2a). Additionally, labeling with tosyl-L-phenylalanine chloromethyl ketone (TPCK) helped identify His-57 as a critical residue at the active site of chymotrypsin. Notably, FP compounds are potent irreversible inhibitors of most serine hydrolases but are inert to other hydrolases, such as aspartyl, cysteine, and metallohydrolases.The Cravatt group recognized a key limitation of such chemical probes—namely, that they generate covalent bonds only with enzymes possessing specific catalytic mechanisms. Therefore, they proposed profiling entire enzyme superfamilies collectively, rather than investigating individual enzymes in isolation. Since the first demonstration of serine hydrolase activity profiling by 1D-PAGE using a biotinylated long-chain FP,15 researchers have demonstrated the immense potential of ABPP for the discovery of novel enzymes and characterization of the role of target enzymes in pathological events at the proteomic level (Fig. 2b). In subsequent studies, the architecture of the ABPP probe evolved into three components: (i) a reactive warhead, (ii) a spacer linker, and (iii) a reporter or bio-orthogonal handle (Fig. 2a). Depending on the assay platform, fluorescent tags were chosen for visualization using PAGE or live-cell imaging, whereas biotin tags were used for subfractionation and subsequent MS-based proteomic analysis (Fig. 2b). However, earlier findings have suggested that the presence of biotin or fluorophores can affect enzyme selectivity or cellular permeability. This has led to the frequent replacement of the reporter handle with a simpler bio-orthogonal motif to mitigate these effects. Enzyme targets are typically tethered by reactive warheads that mimic the intermediate state of enzymatic reactions. Electrophilic motifs have been incorporated into substrates for common enzyme classes such as caspases, cysteine proteases, deacylases,16 serine hydrolases, kinases, and phosphatases, which contain nucleophilic residues near their active sites. Decaging warheads into electrophiles depending on their activity or activating agent is another strategy for alkaline phosphatases,17 cytochrome P450s,18 deubiquitinating enzymes,19,20 and various oxidoreductases.21 Those specific case studies fall outside the scope of this review; however, numerous comprehensive reviews are available.22–25
The discovery of novel enzymes is a definite advantage of ABPP studies, which use the mechanisms of action of chemical probes. For instance, the FP group, among the most thoroughly studied ABPP probes, effectively revealed new serine hydrolases in human cell lines,26 animal tissues, and bacteria.27–30 The Bogyo group exploited ABPP using a series of bacteria.31 In their work on Bacteroides thetaiotaomicron,29 27 proteins were identified from chemoproteomic analysis using FP probes, whereas other bioinformatics prediction analyses showed more than triple the number of proteins as serine hydrolases. Although bioinformatics prediction offers sequence-based structural similarities, ABPP results in a combined effect of expression levels and intact enzymatic activity. Several classes of bacterial serine hydrolases that are absent in humans have been identified. Among the identified enzymes, two enzymes (BT4193 and BT3254) that are classified as homologs of human dipeptidyl peptidase 4 (hDPP4) were further analyzed to validate their biochemical functions, revealing several critical functions of BT4193 in the gut microbiota community. In a study on Staphylococcus aureus,30 chemical proteomic profiles led to the discovery of ten previously uncharacterized serine hydrolases (FphA-J) that do not have human homologs. Further functional examination indicated that FphB was localized to the cell surface and division septum. Perturbation of FphB expression affected bacterial infections in the heart and liver but not in the kidney, suggesting that FphB is crucial for initiating infections. They also demonstrated that a selective inhibitor of FphB reduced the infectivity. This example highlights the potential of the ABPP strategy for identifying new enzymes within the native environment, thus paving the way for the development of novel antibiotics.
In addition to identifying unannotated enzymes, ABPP offers the inherent advantage of evaluating the activity of multiple enzymes within a superfamily in a single assay, making it an ideal platform for assessing selectivity of inhibitors and other small molecules. By administering the drug candidate molecules together with ABPP probes, competitive ABPP facilitates the examination of the impact of these molecules on enzyme activity in an intact environment in the proteomic landscape.32,33 Notably, competitive ABPP can be implemented without the need for known substrates, recombinant expression, or purification processes and can be conducted using various techniques, including PAGE,33,34 fluorescence polarization,35 or MS-based proteomic profiling.36,37 Since enzyme activity correlates with the extent of probe labeling, competitive ABPP can detect both inhibitory and activating effects on target enzymes, although its primary focus has been on inhibitor discovery. This has been well demonstrated in competitive ABPP studies for lysophospholipase-like 1 (LYPLAL1).34,38 Previous genome-wide association studies have found that genetic variants in 19 loci near LYPLAL1 are associated with human metabolic diseases, such as type 2 diabetes and non-alcoholic fatty liver disease. However, these variant positions were not within the coding region; therefore, linking the gain or loss of activity was not straightforward. Although a selective covalent inhibitor of LYPLAL1 was previously identified in a competitive ABPP campaign,38 functional inhibition of LYPLAL1 was not beneficial for the treatment of metabolic diseases, prompting the search for an activator. Notably, the false discovery rate (FDR) for activators has been reported to be substantially higher than that for inhibitors, with only 5% of identified putative activators confirmed compared to 23% of inhibitors in follow-up gel-based ABPP assays.34 This highlights the inherent challenges in activator discovery and underscores the need for orthogonal validation assays. The successful identification of LYPLAL1 activators and their potential therapeutic benefits against metabolic disorders highlights the potential of ABPP as a tool for understanding enzyme function and as a platform for therapeutic development.
Although the majority of ABPP probes engage nucleophilic residues of the target because canonical amino acids lack reactive electrophilic functional groups, the recent emergence of reverse-polarity (RP)-ABPP offers a new perspective.21,39 In nature, enzymes acquire these reactive electrophiles through PTM or by binding to cofactors, creating an electron-deficient machinery. However, these targets have often been neglected because of the lack of suitable chemical tools. The RP-ABPP approach uses electron-rich hydrazine to investigate the biochemical effects of electrophilic enzyme cofactors or labile regulatory modifications. Although hydrazine probe labeling relies on the active-site functional state, it broadly interacts with multiple functional classes of enzymes, including monoamine oxidase (MAO), histone demethylase 1A (LSD1), ribosyldihydronichtinamide dehydrogenase (NQO2), nucleic acid dioxygenase (ALKBH1), aldehyde dehydrogenase (ALDH2), and copper chaperone cytochrome c oxidase (Cox17). This broad interaction is a characteristic of clinical hydrazine drugs. Notably, hydrazine probes revealed the N-terminal glyoxylyl (Glox) PTM in secerning-3 (SCRN3) for the first time, and subsequent investigations led to the discovery of naphthyl hydrazine as a potent and selective inhibitor of SCRN3. This observation is key to elucidating the regulation of Glox and SCRN3, and their physiological and pharmacological effects. The pursuit of expanding the repertoire of target enzymes by using rationally designed chemical probes is ongoing.
2.2. ABPP for reactive residue profiling
To broaden the scope of ABPP probes, early chemoproteomic studies exploited the effects of structural diversity in electrophilic scaffolds. For example, spiroepoxide40 or α-chloroacetamide41 groups can modulate target enzymes when incorporated into libraries of natural products or combinatorial ligands. However, these studies also revealed that probe labeling does not always occur at the enzyme active-site. Reactivity-dependent labeling often reflects the chemical environment and steric accessibility of the reactive side chains. Considering that numerous nucleophilic residues are essential for enzyme function and undergo various redox-related modifications or ligand-binding, it is crucial to catalog the functional residues that are more reactive than others, regardless of whether they are located at the active-site of the enzyme or at the PTM site. Therefore, cysteine and methionine are of particular importance because they are the only sulfur-containing amino acids essential for metal binding, redox regulation, and enzymatic catalysis.To identify hyperreactive cysteines, an isotopic tandem orthogonal proteolysis activity-based protein profiling (isoTOP-ABPP) strategy was proposed to quantify the labeling ratio from the MS signals of individual peptides (Fig. 3a).42 This approach differs from traditional proteomic methods that use an excess of iodoacetamide (IA) to alkylate all the cysteines. In contrast, isoTOP-ABPP gauges cysteine reactivity by comparing the labeling patterns under both low and high concentrations of the IA probe. Hyper-reactive cysteines are expected to be fully labeled, even at low IA probe doses. Although the vast majority of cysteines exhibited a continuous increase in the labeling ratio upon increasing the IA dose, <10% of the subset displayed a subtle difference in labeling, regardless of the IA dose, indicating hyperreactivity. Chemoproteomic reactivity profiling revealed distinct labeling ratios for cysteine residues in glutathione S-transferase (GSTO1) and acetyl-CoA acetyltransferase-1 (ACAT1). In both cases, only the active-site cysteines (GSTO1: C32; ACAT1: C126) exhibited minimal labeling changes upon incremental dosing, which correlated with the reported catalytic effect and hyperreactivity. Subsequently, cysteine reactivity profiling was performed during mitosis to examine the effects of PTMs on activity status.43 Differential labeling with an IA probe between lambda phosphatase-treated and untreated proteomes revealed phosphorylation-dependent perturbations in cysteine reactivity. Generally, proteins with a higher number of phosphorylation sites display substantial changes in reactivity, suggesting a cis-regulatory relationship. Furthermore, this study revealed that the reactivity of specific cysteine residues in key mitotic regulators, such as filamin A (FLNA) and SLAIN motif family member 2 (SLAIN2), increased in a phosphorylation-dependent manner during mitosis (FLNA: C1260, C1453; SLAIN2: C152). Beyond serving as a reliable predictor of cysteine functionality in enzymes, IA reactivity can also be used to identify metal-chelating cysteine residues,44 uncharacterized cysteine-containing peptides from short open reading frames,45 oxidation-sensitive cysteines in pathogenic bacteria,46 and covalent inhibitor discovery for mutant Kirsten rat sarcoma G12C and Bruton's tyrosine kinase.47 Ongoing efforts are underway to further optimize the probe for reduced cellular toxicity and improved temporal control of labeling, as demonstrated by the photo-caged IA probe.48 Efforts are being made to diversify electrophilic functional groups to expand the range of cysteines that can be targeted. This also encompasses the exploration of bioactive electrophiles as a small molecule for therapeutics, using several electrophiles, such as acrylamide,49 α-chloroacetamide,50 bromo- maleimide,51 1,3,5-triazines,52 and p-chloronitrobenzenes (Fig. 3b).53
The expansion of chemical proteomics to target amino acids other than cysteine has considerably broadened the landscape of ligandable residues. One notable advancement is the development of an amine-reactive sulfotetrafluorophenyl ester probe,54 which enabled the large-scale mapping of lysine reactivity and ligandability across over 9000 sites in the human proteome. This strategy facilitated the identification of functional lysines with the potential for covalent ligand interactions, including those involved in allosteric regulation and PPIs. Additionally, alternative lysine-targeting electrophiles, such as N-hydroxysuccinimide esters,55 activated pyridinium esters,56 dichlorotriazine,57 and α-acyloxyenamides58 have been used for ligandable site profiling. A more extensive lysine reactivity was later generated through a broad-spectrum study of approximately 180 compounds,59 representing over 30 aminophilic electrophiles. This approach quantifies the reactivity of more than 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 lysine residues in the human proteome.
000 lysine residues in the human proteome.
In contrast to the development of chemical probes targeting residues with inherent nucleophilicity, such as cysteine and lysine, the development of selective chemical probes for methionine remains in its infancy. This is largely because sulfur in methionine exhibits weak nucleophilicity under physiological conditions, making it less reactive toward electrophilic probes than more nucleophilic residues such as cysteine, lysine, serine, and tyrosine. The Chang group pioneered a redox-activated chemical tagging (ReACT) strategy using a strain-driven sulfur imidation with an oxaziridine scaffold (Fig. 3c).60 Although the oxidative sulfur imidation produced both the desired N-transfer product (sulfimide) and O-transfer side product (sulfoxide), they were able to optimize the reaction by introducing less electron-withdrawing groups into the oxaziridine structure. The application of the ReACT probe in chemoproteomic TOP-ABPP analysis represents a crucial step toward understanding proteome-wide hyperreactive methionines in HeLa cell lysates. The probe successfully labels various enzyme classes, with modified methionine residues predominantly localized on surface-exposed protein domains. While this represents an important step in mapping methionine reactivity, several considerations have been already noted.61 The observed labeling pattern may be influenced by steric accessibility rather than solely reflecting the intrinsic biological reactivity of methionine. Native oxidants, such as peroxides or hypochlorites, are unlikely to be restricted to surface-exposed residues, raising the possibility that the probe introduces a steric bias that differs from the native biological oxidation patterns. A broader challenge is to determine the extent to which synthetic probes mimic the native electrophilic species that target specific residues. In the case of ReACT, the oxaziridine scaffold drives bond formation with methionine; however, it remains unclear how well this system recapitulates the endogenous oxidative modifications. Furthermore, the compatibility of this probe with live-cell systems and intact specimens, rather than with lysates, requires further investigation. Another key factor is the selectivity of oxaziridine-mediated labeling, particularly whether oxygen transfer (rather than nitrogen-based nucleophilic addition) (Fig. 3c) could lead to additional, unintended modifications, as amine and thiolate nucleophiles may favor alternative reaction pathways. Despite these caveats, ReACT-based chemoproteomic profiling has identified hyperreactive methionines in both actin (M44 and M47) and enolase (M165, M169, and M244), which play vital roles in actin polymerization and functional redox regulation, respectively. A subsequent study by He et al. using a pancreatic organoid model further demonstrated the power of ReACT-based chemoproteomic profiling. This revealed that the oxidation state of methionine (M239) in pyruvate kinase M2 acts as a reversible redox switch controlling cancer metastasis.62 Beyond reactivity profiling, the ReACT probe has potential for position-specific labeling in antibody–drug conjugations, given that methionine is the second rarest amino acid in vertebrates.63 These findings highlight the intricate relationship between amino acid side chain properties and the specificity of chemical probes, offering new perspectives in both biochemistry and molecular pharmacology.
2.3. Intrinsic issues with ABPP
Despite its strengths, ABPP detects only a fraction of its intended target residues. For instance, among an estimated 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cysteines in the proteome, current ABPP approaches typically rank reactivity for approximately 1000–3000 specific cysteines, leaving the majority uncharacterized.64 Consequently, ABPP provides a high-resolution snapshot of a subset of the cysteome while overlooking the broader landscape of cysteine reactivity. Recent studies have highlighted the importance of addressing potential false positives and negatives to ensure analytical accuracy.65 For instance, proteins undergoing allosteric regulation, disulfide bond modifications, or transcriptional downregulation in response to reactive electrophilic species (RES) exposure may be mistakenly identified as direct RES targets, while proteins, such as Keap1, which harbor cysteines prone to conjugation with sp2-hybridized RES, such as 4-hydroxynonenal (HNE), may exhibit limited reactivity toward sp3-centered electrophilic probes, such as IA. Moreover, proteins susceptible to oxidation-induced aggregation or precipitation may degrade under sample-processing conditions, leading to their exclusion from subsequent MS analyses. Given that α,β-unsaturated RES modifications occur via time-dependent covalent labeling, precise control over their concentration, localization, and release timing is critical. Furthermore, MS sample preparation must be optimized, including the judicious selection of reducing agents to preserve sensor proteins prone to aggregation and to stabilize labile RES modifications, thereby ensuring robust and reproducible results.
000 cysteines in the proteome, current ABPP approaches typically rank reactivity for approximately 1000–3000 specific cysteines, leaving the majority uncharacterized.64 Consequently, ABPP provides a high-resolution snapshot of a subset of the cysteome while overlooking the broader landscape of cysteine reactivity. Recent studies have highlighted the importance of addressing potential false positives and negatives to ensure analytical accuracy.65 For instance, proteins undergoing allosteric regulation, disulfide bond modifications, or transcriptional downregulation in response to reactive electrophilic species (RES) exposure may be mistakenly identified as direct RES targets, while proteins, such as Keap1, which harbor cysteines prone to conjugation with sp2-hybridized RES, such as 4-hydroxynonenal (HNE), may exhibit limited reactivity toward sp3-centered electrophilic probes, such as IA. Moreover, proteins susceptible to oxidation-induced aggregation or precipitation may degrade under sample-processing conditions, leading to their exclusion from subsequent MS analyses. Given that α,β-unsaturated RES modifications occur via time-dependent covalent labeling, precise control over their concentration, localization, and release timing is critical. Furthermore, MS sample preparation must be optimized, including the judicious selection of reducing agents to preserve sensor proteins prone to aggregation and to stabilize labile RES modifications, thereby ensuring robust and reproducible results.
Hit identification in competitive ABPP, in which a small-molecule ligand of interest (LOI) competes with a proxy-electrophile probe, relies on an indirect readout. Proteins that lose proxy-electrophile engagement in cell lysates (from LOI-treated samples) compared to control specimens are identified as hits, with signal changes analyzed via an enrichment-based proteomics workflow.66 Nonetheless, this approach has been instrumental in mapping drug–target interactions, such as the ABPP-based identification of fumarate67 targets in cell-based HLRCC models, where physicochemical factors, such as pH, influence target reactivity.68 Despite advances in immune-relevant target profiling, ABPP-based approaches face inherent challenges. These include compartment-specific and low-occupancy target identification, as well as the indirect nature of the readout, as target capture occurs in-cell lysates rather than in live cells.66 These factors can complicate the interpretation of target engagement in complex biological systems.
The spatiotemporal nuances of redox signaling cannot be captured using ABPP methods in lysates, where proteins are exposed to a bolus of RES or other probes in a non-physiological manner.61 Although this approach allows for broad profiling of protein reactivity, it inherently lacks the ability to discriminate between specific localized cellular responses and bulk proteome-wide modifications. Bolus dosing of RES is a common experimental approach in ABPP studies that relies on externally applying excess RES to cells, tissues, or organisms for a defined period, followed by downstream pathway analysis.65 Although conceptually straightforward, this method introduces several confounding variables, including the potential for RES-induced membrane perturbation, generation of secondary reactive oxygen species (ROS), and widespread macromolecular modifications. Moreover, as multiple proteins are simultaneously modified under these conditions, the direct correlation between RES modifications and phenotypic outcomes remains unclear. The inability to selectively target a single protein under bulk treatment further complicates mechanistic interpretation, as downstream signaling effects are frequently not rescued by hypomorphic sensing mutants. Additionally, many identified RES-sensitive pathways exhibited hormetic responses, adding another layer of complexity to data interpretation. Another critical issue with bolus dosing is the potential overestimation of the reaction kinetics. Many putative RES sensors identified under such conditions exhibit significantly lower second-order reaction rates than well-established key protein sensors in physiologically relevant, RES-limited environments.69 This discrepancy raises concerns about the physiological relevance of protein hits obtained from ABPP-based screening using bolus conditions.
To address these limitations, alternative approaches, such as G-REX,70 Localis-REX,71 and T-REX72 have been developed. These techniques enable spatially and kinetically controlled profiling of protein interactions and modifications across multiple biological models, including cultured cells and whole organisms, such as worms and fish.66 The REX approach capitalizes on Halo proteins, which can be functionally expressed in a highly specific manner at the cellular, tissue, or organelle levels. After introducing bioinert, cell-permeable REX probes and allowing for washout, these probes irreversibly bind to Halo proteins with a precise 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry. Upon exposure to light at predefined times, these bound probes rapidly released electrophiles, typically within seconds (t1/2 < 1 min). This release mechanism emulates the natural generation of endogenous electrophiles, which are characterized by precise spatial and temporal resolution. Owing to their transient nature and limited availability, these electrophiles selectively engage with nearby native electrophile-sensing proteins located close to the Halo protein. However, it is important to note that REX technologies require genetic manipulation and are currently limited to studying reactive electrophilic species (RES), which restricts their scope. In contrast, ABPP is more broadly applicable across a wide range of reactive residues and protein classes.
1 stoichiometry. Upon exposure to light at predefined times, these bound probes rapidly released electrophiles, typically within seconds (t1/2 < 1 min). This release mechanism emulates the natural generation of endogenous electrophiles, which are characterized by precise spatial and temporal resolution. Owing to their transient nature and limited availability, these electrophiles selectively engage with nearby native electrophile-sensing proteins located close to the Halo protein. However, it is important to note that REX technologies require genetic manipulation and are currently limited to studying reactive electrophilic species (RES), which restricts their scope. In contrast, ABPP is more broadly applicable across a wide range of reactive residues and protein classes.
2.4. Glyco-activity profiling
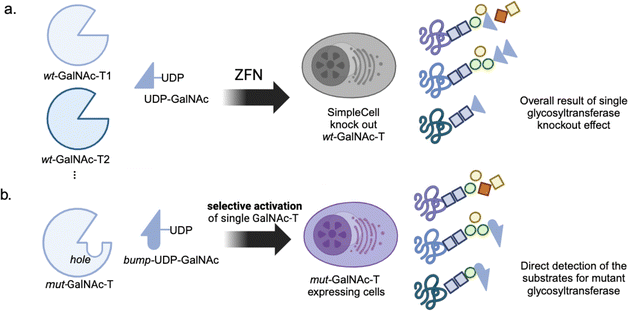
Glycosylation is a major PTM that profoundly influences protein function, stability, and cellular localization. The ultimate goal of glycosylation profiling is the system-wide identification of glycan structures and their specific modification sites on proteins. However, the heterogeneity of glycan structures poses a major obstacle to traditional proteomic techniques. Although the metabolic incorporation of azidosugars has substantially advanced the identification of target proteins and modification sites, monitoring glycosylation-related activities remains challenging. The enzymatic activities involved in glycosylation include the addition and removal of carbohydrates, mediated by glycosyltransferases and glycosidases, respectively. Most glycosidases have a conserved two-step catalytic mechanism mediated by both a nucleophilic residue and pair of acid/base residues; thus, ring-constrained epoxide (cyclophellitol probes) and aziridine (cyclophellitol aziridine probe) were incorporated into a carbohydrate motif to monitor activities of glycocerebrosidase (GBA),73 galactosylceramidase (GALC),74 heparinase (HPSE),75 and α-glucosidase.76Moreover, irreversible chemical probes for glycotransferases have not been developed yet. Instead, a single glycotransferase knockout model was used to monitor the system-wide glycosylation patterns by global glycoproteomic analysis (Fig. 4a).77 Nevertheless, glycosylation is a highly interdependent process; thus, a single loss-of-function model could substantially alter the overall glycosylation pattern. To selectively modulate the activity of glycosyltransferases, a bump-and-whole approach was applied to N-acetylgalactosaminyl transferases (GalNAc-Ts) (Fig. 4b). Double mutants of GalNAc-Ts (T1: I238A, L295A; T2: I253A, L310A; T10: I266A, L321A) were designed to contain a hole adjacent to the ligand-binding pocket, allowing the selective accommodation of a bulky UDP-GalNAc analog.78 This method was effective not only for GalNAc-T1 and -T2, which tend to use unglycosylated peptide substrates, but also for GalNAc-T10, which prefers glycosylated substrates. This approach was further applied in a live cell system, and chemoproteomic analysis revealed that the substrates of GalNAc depend on their activity.79 Although still in the early stages, such activity probes selective for a specific type of glycosyltransferase have the potential to provide a comprehensive map of substrate–enzyme relationships, thereby deepening our understanding of glycosylation events at the glycoproteomic level. For a detailed discussion of glycoproteomic approaches and opportunities for targeting O-GlcNAc, readers are encouraged to check recent reviews.80,81 Metabolic glycan labeling is a powerful technique that allows the incorporation of unnatural sugar analogs into cellular glycoproteins through the GalNAc salvage pathway.82–84
O-GlcNAc-labeled peptides or proteins can be further functionalized or conjugated through bio-orthogonal reactions, such as copper-catalyzed azide–alkyne cycloaddition (CuAAC) and strain-promoted azide–alkyne cycloaddition (SPAAC), facilitating the visualization, enrichment, quantification, and analysis of O-GlcNAc-mediated PPIs. Following the selective enrichment of O-GlcNAc-modified proteins or peptides, quantitative proteomic approaches have been used for in-depth characterization of O-GlcNAc modifications.
3. Interactome profiling
In addition to bioactivity assessed at the protein level, a considerable proportion of biomolecular functionality depends on interactions with other biomolecules. Interactions between biomolecules encompass a range of complexities, from relatively simple homomeric or heteromeric quaternary structure formations to multi-protein assemblies, such as the complex oxidative phosphorylation system in the mitochondria.85 In which the molecular proximity of intact proteins plays a pivotal role in various biochemical functions. Furthermore, in the context of drug molecules, the interactions between a drug and its on- and off-targets directly influence its bioactive phenotypic outcomes and potential side effects.Although most biomolecular function studies have focused on functional changes based on altering the abundance of a protein of interest (POI), molecular interactome profiling has emerged as a promising approach. In this field, highly reactive intermediates are generated in situ to capture spatiotemporal interactions using catalytic reactions or by activating inert probes through light or electricity, followed by enrichment and chemoproteomic analyses.
3.1. Endogenous physiological interactions
In the reductionist approach, molecular interactions are typically studied in vitro using methods, such as isothermal calorimetry or surface plasmon resonance, which measure the binding between two purified molecules. In more physiologically relevant settings, interactions have been examined in cell lysates using techniques, such as co-immunoprecipitation (Co-IP)86 or in genetic screening systems, such as the yeast-two-hybrid method, which can be applied to large-scale screening, allowing for the broader discovery of new interactors.87 However, these approaches have limitations: Co-IP often fails to capture weak or transient interactions, and the yeast two-hybrid method is conducted in a heterologous expression system, which may not fully recapitulate endogenous interactions. To address these issues, alternative methods, such as the proximity ligation assay (PLA)88 have been used to detect weak or transient PPIs in their native cellular context using species-orthogonal antibody pairs. Furthermore, protein complex analyses can be refined using blue native PAGE (BN-PAGE),89 which preserves protein interactions under non-denaturing conditions and allows the isolation of intact complexes. BN-PAGE can be followed by secondary separation techniques, such as tricine-SDS-PAGE or isoelectric focusing, to resolve subunit compositions. These electrophoretic methods provide additional insights into protein assembly without requiring prior knowledge of the interacting partners. For in-cell visualization, fluorescent-based strategies, such as Förster resonance energy transfer (FRET) and protein complementation assays, such as bimolecular fluorescent compartmentations (BiFC) or split-luciferase, have been developed for use in live cells.90 This involves expressing two recombinant POIs tagged with pairs of fluorescent proteins, tailored for either FRET or BiFC. While these techniques are commonly used to validate predefined interactions and comprehensively profile protein interactions, chemical proteomic approaches using covalent labeling strategies have gained traction. This is crucial, given that many biomolecular interactions have a dissociation constant in the range of tens to hundreds of micromolar91 and are easily disrupted during conventional lysis or Co-IP procedures.In addition to genetically introducing UAA in a site-specific manner, the stochastic incorporation of photoaffinity amino acids by endogenous translation machinery without using amber suppression is feasible because of their structural similarity to canonical amino acids. Photo-lysine and PTM-modified photo-lysine probes were incorporated into proteins by culturing cells in photo-lysine-containing medium, demonstrating that photo-lysine-based photoaffinity probes enable the identification of protein–protein interactions mediated by lysine post-translational modifications in vitro.102 A similar approach was applied for the alkyne-functionalized methionine surrogate, photo-ANA, in Salmonella typhimurium by expressing mutant methionyl-transfer RNA synthetase, and host factors playing roles during infections were revealed using photoaffinity labeling.103 As the bacterial proteins were solely incorporated into the photo-ANA, their interaction partners in the host cell were distinguishable and identified using chemoproteomic analysis. Glycan-mediated interactions are notably challenging to characterize compared with the above PPIs because of their weaker binding strength, sometimes in the micromolar to millimolar KD range. Furthermore, site-specific incorporation of unnatural carbohydrates into glycans remains elusive because it is not directed by genetic coding. Using metabolic azido sugar labeling,104 which has shown effective labeling from the cells105 to living mice,106 the chemoproteomic analysis of glycan interaction has been explored using bifunctional sugar analog containing photo-crosslinkers for sialic acids (Ac5-SiaDAz, 9AzSiaNAI, and 9AzSiaDAz),107,108 mannosamine (Ac3-ManNDAz),109 and N-acetylglucosamine (GlcNDAz).110,111 Despite of the low-binding affinity, GlcNDAz captured direct interaction between extracellular glycan binding proteins (galectin-1 and cholera toxin subunit B) and cell-surface N-linked glycoproteins111 as well as O-GlcNAc modified nucleoporins with nuclear transport factors.110 However, recent studies have highlighted the importance of meticulous placement of azido functional groups in azido sugar probes.112 Therefore, a thorough assessment of glycosylation processes for each probe is imperative, ensuring specific metabolic labeling substrates and thereby gaining a more nuanced understanding of glycan–protein interactions.
While investigating the enzymatic mechanism of BirA, the Roux group envisaged a potential for tethering biotinylation targets.117 BirA operates in a two-step process: the formation of biotinoyl-5′-AMP (bioAMP) from biotin and ATP; and substrate conjugation with activated bioAMP. A single amino acid mutation to BirA, (BirAR118G) has a weaker affinity for bioAMP than the wild type.118,119 This reduced affinity leads to promiscuous protein biotinylation because the bioAMP released from BirAR118G can spontaneously react with any neighboring primary amine (Fig. 5b). They named the enzyme tag proximity-dependent biotin identification (BioID) and coined the term “proximity labeling (PL)” for this technology. BioID (35 kDa) was further optimized to decrease its size and modulate the biotinylation radius, and the Roux group successfully presented an improved PL using a mutant biotin ligase of Aquifex aeolicus (R40G) called BioID2.120 Since these reports, both BioID and BioID2 have become invaluable tools for various PPI studies, including nuclear lamina,117 nuclear envelope protein,121 centrosome components,122,123 cell junction complexes,124,125 cytoskeletal structure,126 kinases,127–129 E3 ubiquitin ligases,130 and mitochondrial protease ClpP.131
The Ting group also recognized the unique advantages of genetically encoded enzyme tags for PL, using a distinct approach based on their high-resolution electron microscopy (EM) imaging studies.132 Conventionally, 3,3′-diaminobenzidine (DAB) staining is used to achieve clear EM image contrasts. This method relies on the polymerization of 3,3′-diaminobenzidine, triggered by hydrogen peroxide and horseradish peroxidase (HRP). Oxidized DABs are released from the HRP pocket, producing aggregate deposition in the vicinity. Observing HRP inefficiency in mammalian living conditions, they engineered a 28 kDa monomeric ascorbate peroxidase (APEX), which requires exogenous H2O2 for the oxidation of substrates. APEX oxidized the DAB as well as numerous phenol derivatives of phenoxyl radicals that are highly reactive to electron-rich amino acids with small crosslinking radius (Fig. 5c).133 Using the biotin-phenol probe as the APEX substrate for PL, they were able to specify protein localizations (Fig. 5d), even distinguishing sub-organelle regions, such as matrix and mitochondria intermembrane space.134 For enhanced sensitivity, APEX was further investigated using FACS selection of 106 yeast display libraries, leading to the discovery of APEX2, a variant containing a single amino acid mutation (A134P).135 The superior sensitivity of APEX2 elucidated intricate biological details, pinpointing the precise localization of Ca2+-binding protein (MICU1),135 revealing RNA granule trafficking machinery,136 and shedding light on transient interaction within the GPCR pathway137 or between enzymes and their substrate.138,139
Both types of PL tags have distinct advantages and drawbacks related to their labeling mechanisms. The strength of APEX2 lies in its rapid labeling duration (approximately 1 min). Furthermore, their reactivity can be tuned by altering their chemical probe structure. However, BioID is favored because it does not require an additional chemical probe (e.g., biotin-phenol) and avoids the use of toxic cofactors (e.g., hydrogen peroxide). Nonetheless, the labeling process is prolonged (>18 h), and regulating the passive diffusion of the activated ester form of biotin is challenging, leading to a broad labeling radius. The Ting group attempted to enhance BioID by combining the benefits of both the tags. Through yeast display screening using 107 libraries and after 29 rounds of selection, they found two random bioID mutants capable of 10 min PL in cells: TurboID (35 kDa) and miniTurbo (28 kDa).140 As TurboID improved the labeling speed compared to that with APEX2, both PL tags can be simultaneously applied in a single system and report a source and destination compartment within the cell using orthogonal enrichment handles.141 This technique, termed transitID, revealed subcellular trafficking between the cytosol-mitochondria, cytosol-nucleus, and cytosol-stress granules under oxidative stress. Recently, UltraID,142 an even smaller and faster biotin ligase, was developed to further enhance the PL efficiency.
These PL tags were further enhanced using the protein complementation method in BioID,143,144 TurboID,145 and peroxidase-based systems.146,147 The split forms of PL tags exhibit substantial spatial specificity based on a specific input, such as RNA–protein interactions during microRNA-mediated silencing pathways,144 organelle contacts,146 transient organelle formations,143 and neuronal synapses.147 In addition to biotin ligase and peroxidase, research on PL techniques has expanded to explore a diverse range of enzymatic reactions to broaden their application scope. Ubiquitination is a common PTM that involves the formation of covalent bonds at the lysine residue of a substrate via ubiquitin activation. Inspired by the high similarity between bioAMP-mediated proximal labeling and activated ubiquitin substrate labeling in the spatially restricted E3 ligase-binding mode, several approaches have been developed to functionally mimic ubiquitination. The first is the NEDDylator system,148,149 which performs the NEDDylation process using a chimeric protein that combines an NEDD8-conjugating enzyme with a bait protein. NEDD8 is a stable and rare ubiquitin homolog; therefore, the NEDD8-conjugating enzyme labels the interaction partner of the bait using NEDD8, which is particularly useful for identifying ubiquitin E3 ligase substrates. Another system is the PUP-IT system.150 Using the bacterial ubiquitin-like modifier Pup, a Pup ligase was developed as a PL tag, with a distinction between activated Pup that does not diffuse out of the enzyme. Substrate labeling occurs in the pup ligase, PUP-IT has a more restricted labeling radius and is used for membrane protein interaction discovery for CD28, a receptor for T lymphocyte activation. Notably, both NEDDylation and PUPylation attach large domains to target proteins, which can influence signaling pathways or protein functions independent of the intended experimental design. All engineered variants, including point mutants, should be assessed for potential disruptions to normal cellular functions, such as proliferation, signaling properties unrelated to the targeted process, canonical activity, and structural integrity.151 These evaluations should be conducted in relevant cell lines and, where necessary, further validated in knockout lines reconstituted with the fusion POI. In vitro assays provide valuable insights into protein folding and their activities. Beyond the ubiquitin analogs system, the transpeptidase sortase A (SrtA) has been adapted for PL to capture cell-to-cell interaction, especially between T and dendritic cells.152 This was achieved as SrtA transfers a “LPXTG” sequence motif into a nearby N-terminal polyglycine, and was later further engineered to target monoglycine since it is much more abundant on the cell surface than polyglycine.153
To address this gap, a recent study introduced organ-specific localized reactivity-based profiling (OS-Localis-REX),167 a method that enables the direct mapping of electrophile-responsive proteomes in live organisms. Using a photocaged HNE probe, this approach enables the controlled release of native electrophiles into specific tissues, enabling the identification of proteins with privileged reactivity under physiological conditions. Enrichment-based quantitative MS revealed that more than 70% of the identified targets exhibited localized responsiveness to electrophiles, independent of their tissue-specific abundance. This finding challenges the conventional assumption that locale-specific protein expression dictates the electrophilic reactivity. A comparison with existing datasets, such as tissue-specific single-cell RNA sequencing and PAXdb, confirmed that the electrophile responsivity was not strictly correlated with protein expression levels in the respective tissues.71,167 Additionally, OS-Localis-REX identified localized protein targets that were undetectable using tissue-specific biotinylation approaches, such as Ultra-ID, highlighting its unique capability to capture context-specific chemical actionability. Notably, OS-Localis-REX identified CYP-33E1, a protein that displays gut-specific electrophile responsivity in Caenorhabditis elegans, despite being expressed in multiple tissues. Functional analyses revealed that this localized response plays a crucial role in gut-driven lipid homeostasis, underscoring the potential of this method for identifying proteins involved in local metabolic regulation.
Despite its strengths, OS-Localis-REX has certain limitations. The approach is blind to total protein levels in specific tissues, making it necessary to integrate complementary techniques, such as OS-Ultra-ID, for a more comprehensive understanding. Moreover, because this method relies on a low-dose electrophile release, it selectively identifies the most responsive proteins, potentially missing lower-affinity interactors. Expanding the methodology through modified REX protocols or the inclusion of diverse photocaged electrophiles can enhance the depth and sensitivity of these methods.
3.2. Exogenous bioactive drug–target interactions
Bioactive molecules are invaluable pharmaceutical sources. Dramatic advances in molecular diversity generation168 and high-throughput screening have led to the identification of many bioactive molecules.169 However, determining their functional targets remains a complex exdeavour.170 Most bioactive compounds, including the majority of FDA-approved drugs, interact with their targets through non-covalent mechanisms—such as hydrogen bonds, ionic interactions, and hydrophobic forces—which are typically reversible and context-dependent. In contrast, covalent drugs form irreversible bonds with nucleophilic residues and require distinct profiling strategies. As we gather more detailed information about molecular perturbation networks at the genomic and proteomic levels, it is evident that the pharmacological effect of a bioactive molecule does not always stem from its single highest-affinity target.171 Instead, it is often influenced by its polypharmacological interactions. Regardless of whether bioactive compounds emerge from phenotype- or target-based approaches, their molecular interactions are not straightforward, particularly in complex cellular environments.172 Given that the conventional affinity pull-down method recovers only exceptionally high-affinity partners, comprehensive mapping of non-covalent small-molecule interactions is essential to fully understand their functions. Research in this area predominantly involves two methodologies: labeling and label-free target profiling.Although size remains a key consideration, it is not the only factor influencing drug–protein interactions. Therefore, the position of the tag and its potential for autoreactivity must be carefully evaluated. Any modification, even minimal, may profoundly affect the target engagement and selectivity. This is exemplified by the comparison of ibufenac and ibuprofen, two NSAIDs that differ only in their carboxyl groups. Despite their structural similarity, ibufenac was withdrawn from the market because of its hepatotoxicity,175 whereas ibuprofen remains one of the safest NSAIDs. Their acyl glucuronides exhibited only minor differences in reactivity; however, their CoA thioesters formed covalent adducts with proteins at significantly different rates, with ibufenac-CoA displaying an order-of-magnitude higher reactivity. This highlights the substantial biological consequences of seemingly small modifications.
The concept of minimalist tags has emerged to mitigate these challenges. This method was devised by integrating the smallest photoaffinity group, alkyl diazirine, with simple bio-orthogonal handles to facilitate subsequent visualization and enrichment (Fig. 7a). Due to their small size, the minimalist tag can effectively elucidate the target protein profiles of the BET inhibitor, JQ-1;176,177 the clinical diabetes medication, metformin;178 and nonsteroidal anti-inflammatory drugs.179 Notably, all three studies showed both on- and off-target effects. For example, the photoaffinity probe JQ-1 identified a novel off-targetapurinic/apyrimidinic endodeoxyribonuclease-1. This enzyme upregulates ROS levels upon JQ-1 binding, suggesting a synergistic anticancer effect in addition to its bromodomain-mediated epigenetic effect. The photoaffinity probe highlighted the mechanism by which metformin activates AMPK signaling. Upon binding to metformin, PEN2 forms a complex with ATP6AP1, which triggers AMPK activation regardless of cellular AMP levels. Despite the micromolar binding affinity between PEN2 and metformin, photoaffinity labeling effectively revealed this interaction. The use of photocatalysts is a unique photo-labeling method (Fig. 7b).180 Although the minimal tag captures the drug interactome, it exhibits a poor signal-to-noise ratio because a single label is produced from each photoaffinity molecule. In contrast, a drug molecule conjugated to a photocatalyst can generate multiple labels on targets in a spatially restricted manner via Dexter energy transfer. Admittedly, the large size of the photocatalyst may hinder drug–target interactions. However, the evident signal amplification highlights its potential for profiling the interactomes of various drugs, including JQ-1, paclitaxel, and dasatinib.
In addition to photo-crosslinking and photocatalytic labeling techniques, a small redox-active diazetidinone (DZE) was used to form covalent bonds through an alternative activation method (Fig. 7c).181 DZE can be easily produced from any carboxylic acid as a precursor and efficiently converted to ketene upon nitrogen extrusion using electrolysis. This electro-affinity labeling can engage various nucleophilic residues in lysates and living cells. To date, a series of drug interactions have been revealed using ligands for BRD4, ABL, AURKA, and MEK1.
Thermal proteome profiling (TPP) was used to evaluate alterations in the thermal stability of individual proteins upon ligand-binding at the proteomic level. Upon exposure to thermal stress, proteins unfold, and their hydrophobic domains are exposed. However, ligand-binding events alter this unfolding behavior, often stabilizing the protein owing to the binding energy of the interactions. TPP is particularly effective for high-affinity binders with sufficiently low dissociation constants (Kd) that induce measurable shifts in protein stability. While these stability shifts correlate with ligand affinity under saturated conditions, the ranking of binding affinities among multiple targets requires complementary approaches, such as isothermal dose–response (ITDR) profiling.182 ITDR refines affinity assessments by monitoring ligand-induced shifts at defined temperatures over a range of concentrations, enabling a finer resolution of binding interactions. Beyond affinity considerations, additional factors influence thermal stability shifts, including competitive binding events and disruption of PPIs, which can confound the TPP analyses. Using the superior resolution and rapid scanning capabilities of contemporary MS techniques, the simultaneous quantitation of more than 10 samples is achievable, streamlining the comparative assessment of protein thermal stabilities. TPP has expanded our understanding of the interactome of drugs for the pan-specificity of staurosporine,182 on- and off-target profiling of cancer drugs,183 and the stereoselectivity of (S)-crizotinib for MTH1 as a novel anticancer strategy.184 Although TPP offers an insightful target profile at the proteomic level, a subset of proteins that is either elusive or not quantifiable exists, delineating the current boundaries of the technique. The stability of proteins from rates of oxidation (SPROX) offers insights into protein–ligand interactions via oxidation of the methionine side chain.185 In particular, SPROX employs a denaturant influence on the oxidation rates of shielded methionine within proteins to calculate thermodynamic parameters, such as the folding free energy of the protein and the associated Kd values. Although SPROX has primarily been applied to cell lysates for small-molecule drugs186 or ubiquitous enzyme cofactors (NAD+),187 it can quantitatively measure the protein–ligand binding affinity. The last label-free target profiling method is drug affinity responsive target stability (DARTS).188 DARTS evaluates the resistance of a target to proteolysis, which increases upon ligand binding, because proteins often become more stabilized by drug binding. This method was initially demonstrated through the stabilization of mTOR and EF-1a upon treatment with rapamycin and didemnin B, respectively. DARTS has been applied to cell extracts and specific target proteins, such as alanine-serine-cysteine transporter 2 (ASCT2)189 and NOD-like receptor family pyrin domain-containing protein 3 (NLRP3),190 which demonstrated its versatility. However, DARTS relies on antibody-based monitoring of specific targets rather than MS-based proteomic profiling; its scope is limited, and does not offer a full proteomic-level analysis of off-targets. The intrinsic nature of bottom-up proteomic analysis, which involves piecing the detected peptide fragments together to infer proteins, complicates the straightforward integration of DARTS into conventional proteomics. One potential approach involves coupling 1D PAGE-like separation based on protein size with MS proteomics. This confirms the proteome-wide proteolytic resistance induced by drugs, as previously demonstrated by the PROTOMAP application.191,192 Nevertheless, these technological adaptations are still underdeveloped.
4. Conclusions and perspectives
The future of chemical proteomics is expected to advance substantially owing to its ability to bridge the gaps often overlooked in traditional proteomics and biochemistry. Recent advancements in MS techniques have expanded the scope of proteomics; however, they often fail to detect the so-called ‘dark proteome’193 and provide nuanced data on functional activity and molecular interactions. Chemoproteomics has addressed some of these gaps using chemical biological methods and tools.Proteomics experiments generate numerous candidate proteins that require rigorous validation to ensure their biological relevance. Ideally, some findings align with the existing literature, whereas novel candidates require further scrutiny. Selection criteria used for hit selection include confidence levels, pathway associations, and homology with known interactors. Although fold-change-based enrichment methods can be ambiguous and potential confounding factors must be considered, absolute occupancy techniques, such as ABPP, offer clearer insights. Ultimately, the goal is to link proteomic data to phenotypic outcomes to elucidate protein function or uncover mechanistic insights. Validation should be performed under biologically relevant conditions to reduce artifacts associated with in vitro systems or protein overexpression. Genetic perturbation models, including knockdowns and knockouts, provide strong evidence for functional roles. To validate PPIs, Co-IP remains a standard approach, though its success depends on antibody specificity and interaction stability post-lysis. Alternative approaches, such as PLAs and crosslinking techniques, enhance detection sensitivity. Point mutants and subcellular localization studies can further refine the mechanistic insights into protein function. Reagent validation is also essential. Antibody specificity should be confirmed using knockout or knockdown models with appropriate controls for imaging, western blotting, and immunoprecipitation. Ectopic protein fusion (e.g., APEX2, BioID, and REX technologies) should be assessed for functional perturbations. Given that commonly used cell lines may harbor genomic alterations, sequencing key genes or using multiple models may enhance reliability. For a comprehensive discussion of these validation strategies, including pitfalls and case studies, please refer to the relevant literature review.151
This review will serve as an ideal introduction to the topic of chemical proteomics for members of the chemical community, spark the interest of chemists from various fields to combine their expertise with new knowledge, and encourage them to contribute to the development of this relatively new field of research. Chemical proteomics has substantial implications in organic, inorganic, and medicinal chemistry. It is a great platform for chemists to showcase their creativity by developing new chemistries for covalent bond formation and improving the efficacy and performance of existing chemical warheads. Recent examples include halogenated enamine N-oxides, which can be reduced bio-orthogonally using diboron reagents to generate highly electrophilic α,β-unsaturated haloiminium ions that label a range of amino acid residues in proteins.194 This approach enables a deeper understanding of the metalloproteome and the interactions of metal-based drugs, such as platinum-, rhenium-, and bismuth-containing drugs, for cancer and bacterial infection treatment.195–197 Moreover, chemical proteomics is integral to inhibitor design, allowing early selective profiling of newly synthesized inhibitor candidates. Specifically, it is essential to identify and validate new therapeutic targets,198 and accelerate drug development by guiding the interactions of small molecules with disease-relevant proteins to impact human health.
Highly selective and promising chemical probes have unique practical applications for activity profiling. Further research is required to fine-tune the selectivity of these probes in both directions, allowing them to interact with a broader range of canonical amino acid residues. Moreover, there is a persistent need for enhanced spatiotemporal control of these probes to study biological systems practically. The targetable reactive electrophile and oxidant strategy is one example,199 demonstrating the potential of chemical probes for spatiotemporal control. Although exposure to specific wavelengths of light is the primary method of temporal control, pursuing alternative activation triggers may be worthwhile.
The scope of interactome profiling has expanded from studying PPIs in vitro to exploring interactions within subcellular organelles and cell-to-cell interactions. Numerous enzymatic and chemical catalysts have been exploited for the instant generation of reactive intermediates; however, multiplexing options remain limited. Given the intricate nature of biological signaling pathways, capturing interactome changes across multiple time points is essential. We foresee the development of next-generation methods that are specifically designed to monitor dynamic shifts in interactome profiles.
Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was supported by the Ministry of Science and ICT through the National Research Foundation of Korea (RS-2025-00523442, RS-2024-00396224, RS-2023-00217701) and a National Research Council of Science and Technology (NST) grant (CAP23011-400).Notes and references
- K. Suhre, M. I. McCarthy and J. M. Schwenk, Nat. Rev. Genet., 2021, 22, 19–37 CrossRef CAS PubMed.
- B. Zhang, J. R. Whiteaker, A. N. Hoofnagle, G. S. Baird, K. D. Rodland and A. G. Paulovich, Nat. Rev. Clin. Oncol., 2019, 16, 256–268 CrossRef PubMed.
- J. B. Fenn, M. Mann, C. K. Meng, S. F. Wong and C. M. Whitehouse, Science, 1989, 246, 64–71 CrossRef CAS PubMed.
- M. Wilm, A. Shevchenko, T. Houthaeve, S. Breit, L. Schweigerer, T. Fotsis and M. Mann, Nature, 1996, 379, 466–469 CrossRef CAS PubMed.
- D. F. Hunt, R. A. Henderson, J. Shabanowitz, K. Sakaguchi, H. Michel, N. Sevilir, A. L. Cox, E. Appella and V. H. Engelhard, Science, 1992, 255, 1261–1263 CrossRef CAS PubMed.
- J. K. Eng, A. L. McCormack and J. R. Yates, J. Am. Soc. Mass Spectrom., 1994, 5, 976–989 CrossRef CAS PubMed.
- M. Mann and M. Wilm, Anal. Chem., 1994, 66, 4390–4399 CrossRef CAS PubMed.
- B. T. Ruotolo, J. L. Benesch, A. M. Sandercock, S. J. Hyung and C. V. Robinson, Nat. Protoc., 2008, 3, 1139–1152 CrossRef CAS PubMed.
- M. Labib and S. O. Kelley, Nat. Rev. Chem., 2020, 4, 143–158 CrossRef PubMed.
- D. P. Walsh and Y. T. Chang, Chem. Rev., 2006, 106, 2476–2530 CrossRef CAS PubMed.
- S. L. Schreiber, Chem. Eng. News, 2003, 81, 51–61 CrossRef.
- J. Zaia, Anal. Bioanal. Chem., 2023, 415, 527–532 CrossRef CAS PubMed.
- C. Walsh, Enzymatic reaction mechanisms, W. H. Freeman, San Francisco, 1979 Search PubMed.
- G. H. Dixon, S. Go and H. Neurath, Biochim. Biophys. Acta, 1956, 19, 193–195 CrossRef CAS PubMed.
- Y. Liu, M. P. Patricelli and B. F. Cravatt, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 14694–14699 CrossRef CAS PubMed.
- J. E. Bolding, P. Martin-Gago, N. Rajabi, L. F. Gamon, T. N. Hansen, C. R. O. Bartling, K. Stromgaard, M. J. Davies and C. A. Olsen, Angew. Chem., Int. Ed., 2022, 61, e202204565 CrossRef CAS PubMed.
- H. Song, Y. Li, Y. Chen, C. Xue and H. Xie, Chemistry, 2019, 25, 13994–14002 CrossRef CAS PubMed.
- A. T. Wright, J. D. Song and B. F. Cravatt, J. Am. Chem. Soc., 2009, 131, 10692–10700 CrossRef CAS PubMed.
- U. Rolen, V. Kobzeva, N. Gasparjan, H. Ovaa, G. Winberg, F. Kisseljov and M. G. Masucci, Mol. Carcinog., 2006, 45, 260–269 CrossRef CAS PubMed.
- N. C. Taylor, G. Hessman, H. B. Kramer and J. F. McGouran, Chem. Sci., 2020, 11, 2967–2972 RSC.
- Z. Lin, X. Wang, K. A. Bustin, K. Shishikura, N. R. McKnight, L. He, R. M. Suciu, K. Hu, X. Han, M. Ahmadi, E. J. Olson, W. H. Parsons and M. L. Matthews, ACS Cent. Sci., 2021, 7, 1524–1534 CrossRef CAS PubMed.
- D. K. Nomura, M. M. Dix and B. F. Cravatt, Nat. Rev. Cancer, 2010, 10, 630–638 CrossRef CAS PubMed.
- L. Krammer and R. Breinbauer, Isr. J. Chem., 2023, 63, e202200086 CrossRef CAS.
- L. Wu, Z. Armstrong, S. P. Schroder, C. de Boer, M. Artola, J. M. Aerts, H. S. Overkleeft and G. J. Davies, Curr. Opin. Chem. Biol., 2019, 53, 25–36 CrossRef CAS PubMed.
- B. F. Cravatt, A. T. Wright and J. W. Kozarich, Annu. Rev. Biochem., 2008, 77, 383–414 CrossRef CAS PubMed.
- N. Jessani, Y. Liu, M. Humphrey and B. F. Cravatt, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 10335–10340 CrossRef CAS PubMed.
- S. K. Hatzios, S. Abel, J. Martell, T. Hubbard, J. Sasabe, D. Munera, L. Clark, D. A. Bachovchin, F. Qadri, E. T. Ryan, B. M. Davis, E. Weerapana and M. K. Waldor, Nat. Chem. Biol., 2016, 12, 268–274 CrossRef CAS PubMed.
- S. Zweerink, V. Kallnik, S. Ninck, S. Nickel, J. Verheyen, M. Blum, A. Wagner, I. Feldmann, A. Sickmann, S. V. Albers, C. Brasen, F. Kaschani, B. Siebers and M. Kaiser, Nat. Commun., 2017, 8, 15352 CrossRef CAS PubMed.
- L. J. Keller, T. H. Nguyen, L. J. Liu, B. M. Hurysz, M. Lakemeyer, M. Guerra, D. J. Gelsinger, R. Chanin, N. Ngo, K. M. Lum, F. Faucher, P. Ipock, M. J. Niphakis, A. S. Bhatt, A. J. O’Donoghue, K. C. Huang and M. Bogyo, Nat. Chem. Biol., 2023, 19, 1469–1479 CrossRef CAS PubMed.
- C. S. Lentz, J. R. Sheldon, L. A. Crawford, R. Cooper, M. Garland, M. R. Amieva, E. Weerapana, E. P. Skaar and M. Bogyo, Nat. Chem. Biol., 2018, 14, 609–617 CrossRef CAS PubMed.
- L. J. Keller, B. M. Babin, M. Lakemeyer and M. Bogyo, Curr. Opin. Chem. Biol., 2020, 54, 45–53 CrossRef CAS PubMed.
- W. Li, J. L. Blankman and B. F. Cravatt, J. Am. Chem. Soc., 2007, 129, 9594–9595 CrossRef CAS PubMed.
- D. Leung, C. Hardouin, D. L. Boger and B. F. Cravatt, Nat. Biotechnol., 2003, 21, 687–691 CrossRef CAS PubMed.
- B. P. Kok, S. Ghimire, W. Kim, S. Chatterjee, T. Johns, S. Kitamura, J. Eberhardt, D. Ogasawara, J. Xu, A. Sukiasyan, S. M. Kim, C. Godio, J. M. Bittencourt, M. Cameron, A. Galmozzi, S. Forli, D. W. Wolan, B. F. Cravatt, D. L. Boger and E. Saez, Nat. Chem. Biol., 2020, 16, 997–1005 CrossRef CAS PubMed.
- D. A. Bachovchin, S. J. Brown, H. Rosen and B. F. Cravatt, Nat. Biotechnol., 2009, 27, 387–394 CrossRef CAS PubMed.
- B. R. Lanning, L. R. Whitby, M. M. Dix, J. Douhan, A. M. Gilbert, E. C. Hett, T. O. Johnson, C. Joslyn, J. C. Kath, S. Niessen, L. R. Roberts, M. E. Schnute, C. Wang, J. J. Hulce, B. Wei, L. O. Whiteley, M. M. Hayward and B. F. Cravatt, Nat. Chem. Biol., 2014, 10, 760–767 CrossRef CAS PubMed.
- A. M. Zuhl, J. T. Mohr, D. A. Bachovchin, S. Niessen, K. L. Hsu, J. M. Berlin, M. Dochnahl, M. P. Lopez-Alberca, G. C. Fu and B. F. Cravatt, J. Am. Chem. Soc., 2012, 134, 5068–5071 CrossRef CAS PubMed.
- K. Ahn, M. Boehm, M. F. Brown, J. Calloway, Y. Che, J. Chen, K. F. Fennell, K. F. Geoghegan, A. M. Gilbert, J. A. Gutierrez, A. S. Kalgutkar, A. Lanba, C. Limberakis, T. V. Magee, I. O'Doherty, R. Oliver, B. Pabst, J. Pandit, K. Parris, J. A. Pfefferkorn, T. P. Rolph, R. Patel, B. Schuff, V. Shanmugasundaram, J. T. Starr, A. H. Varghese, N. B. Vera, C. Vernochet and J. Yan, ACS Chem. Biol., 2016, 11, 2529–2540 CrossRef CAS PubMed.
- X. Wang, Z. Lin, K. A. Bustin, N. R. McKnight, W. H. Parsons and M. L. Matthews, J. Am. Chem. Soc., 2022, 144, 5377–5388 CrossRef CAS PubMed.
- M. J. Evans, A. Saghatelian, E. J. Sorensen and B. F. Cravatt, Nat. Biotechnol., 2005, 23, 1303–1307 CrossRef CAS PubMed.
- K. T. Barglow and B. F. Cravatt, Chem. Biol., 2004, 11, 1523–1531 CrossRef CAS PubMed.
- E. Weerapana, C. Wang, G. M. Simon, F. Richter, S. Khare, M. B. Dillon, D. A. Bachovchin, K. Mowen, D. Baker and B. F. Cravatt, Nature, 2010, 468, 790–795 CrossRef CAS PubMed.
- E. K. Kemper, Y. Zhang, M. M. Dix and B. F. Cravatt, Nat. Methods, 2022, 19, 341–352 CrossRef CAS PubMed.
- N. J. Pace and E. Weerapana, ACS Chem. Biol., 2014, 9, 258–265 CrossRef CAS PubMed.
- A. G. Schwaid, D. A. Shannon, J. Ma, S. A. Slavoff, J. Z. Levin, E. Weerapana and A. Saghatelian, J. Am. Chem. Soc., 2013, 135, 16750–16753 CrossRef CAS PubMed.
- X. Deng, E. Weerapana, O. Ulanovskaya, F. Sun, H. Liang, Q. Ji, Y. Ye, Y. Fu, L. Zhou, J. Li, H. Zhang, C. Wang, S. Alvarez, L. M. Hicks, L. Lan, M. Wu, B. F. Cravatt and C. He, Cell Host Microbe, 2013, 13, 358–370 CrossRef CAS PubMed.
- M. Kuljanin, D. C. Mitchell, D. K. Schweppe, A. S. Gikandi, D. P. Nusinow, N. J. Bulloch, E. V. Vinogradova, D. L. Wilson, E. T. Kool, J. D. Mancias, B. F. Cravatt and S. P. Gygi, Nat. Biotechnol., 2021, 39, 630–641 CrossRef CAS PubMed.
- M. Abo and E. Weerapana, J. Am. Chem. Soc., 2015, 137, 7087–7090 CrossRef CAS PubMed.
- E. V. Vinogradova, X. Zhang, D. Remillard, D. C. Lazar, R. M. Suciu, Y. Wang, G. Bianco, Y. Yamashita, V. M. Crowley, M. A. Schafroth, M. Yokoyama, D. B. Konrad, K. M. Lum, G. M. Simon, E. K. Kemper, M. R. Lazear, S. Yin, M. M. Blewett, M. M. Dix, N. Nguyen, M. N. Shokhirev, E. N. Chin, L. L. Lairson, B. Melillo, S. L. Schreiber, S. Forli, J. R. Teijaro and B. F. Cravatt, Cell, 2020, 182(1009–1026), e1029 Search PubMed.
- K. M. Backus, B. E. Correia, K. M. Lum, S. Forli, B. D. Horning, G. E. Gonzalez-Paez, S. Chatterjee, B. R. Lanning, J. R. Teijaro, A. J. Olson, D. W. Wolan and B. F. Cravatt, Nature, 2016, 534, 570–574 CrossRef CAS PubMed.
- M. E. Smith, F. F. Schumacher, C. P. Ryan, L. M. Tedaldi, D. Papaioannou, G. Waksman, S. Caddick and J. R. Baker, J. Am. Chem. Soc., 2010, 132, 1960–1965 CrossRef CAS PubMed.
- R. Banerjee, N. J. Pace, D. R. Brown and E. Weerapana, J. Am. Chem. Soc., 2013, 135, 2497–2500 CrossRef CAS PubMed.
- D. A. Shannon, R. Banerjee, E. R. Webster, D. W. Bak, C. Wang and E. Weerapana, J. Am. Chem. Soc., 2014, 136, 3330–3333 CrossRef CAS PubMed.
- S. M. Hacker, K. M. Backus, M. R. Lazear, S. Forli, B. E. Correia and B. F. Cravatt, Nat. Chem., 2017, 9, 1181–1190 CrossRef CAS PubMed.
- C. C. Ward, J. I. Kleinman and D. K. Nomura, ACS Chem. Biol., 2017, 12, 1478–1483 CrossRef CAS PubMed.
- C. Wan, D. Y. Yang, C. L. Song, M. C. Liang, Y. H. An, C. S. Lian, C. Dai, Y. X. Ye, F. Yin, R. Wang and Z. G. Li, Chem. Sci., 2024, 15, 5340–5348 RSC.
- D. A. Shannon, R. Banerjee, E. R. Webster, D. W. Bak, C. Wang and E. Weerapana, J. Am. Chem. Soc., 2014, 136, 3330–3333 CrossRef CAS PubMed.
- Y. Y. Zhao, K. Duan, Y. L. Fan, S. R. Li, L. Y. Huang, Z. C. Tu, H. Y. Sun, G. M. Cook, J. Yang, P. H. Sun, Y. Tan, K. Ding and Z. Q. Li, Commun. Chem., 2024, 7, 31 CrossRef CAS PubMed.
- M. E. Abbasov, M. E. Kavanagh, T. A. Ichu, M. R. Lazear, Y. F. Tao, V. M. Crowley, C. W. A. Ende, S. M. Hacker, J. R. Ho, M. M. Dix, R. Suciu, M. M. Hayward, L. L. Kiessling and B. F. Cravatt, Nat. Chem., 2021, 13, 1151 CrossRef CAS PubMed.
- S. Lin, X. Yang, S. Jia, A. M. Weeks, M. Hornsby, P. S. Lee, R. V. Nichiporuk, A. T. Iavarone, J. A. Wells, F. D. Toste and C. J. Chang, Science, 2017, 355, 597–602 CrossRef CAS PubMed.
- S. Parvez, M. J. C. Long, J. R. Poganik and Y. Aye, Chem. Rev., 2018, 118, 8798–8888 CrossRef CAS PubMed.
- D. He, H. Feng, B. Sundberg, J. Yang, J. Powers, A. H. Christian, J. E. Wilkinson, C. Monnin, D. Avizonis, C. J. Thomas, R. A. Friedman, M. D. Kluger, M. A. Hollingsworth, P. M. Grandgenett, K. A. Klute, F. D. Toste, C. J. Chang and I. I. C. Chio, Mol. Cell, 2022, 82(3045–3060), e3011 Search PubMed.
- S. K. Elledge, H. L. Tran, A. H. Christian, V. Steri, B. Hann, F. D. Toste, C. J. Chang and J. A. Wells, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 5733–5740 CrossRef CAS PubMed.
- M. J. C. Long and Y. Aye, Cell Chem. Biol., 2017, 24, 787–800 CrossRef CAS PubMed.
- X. Y. Liu, M. J. C. Long and Y. Aye, Trends Biochem. Sci., 2019, 44, 75–89 CrossRef CAS PubMed.
- K. T. Huang and Y. Aye, Commun. Chem., 2024, 7, 195 CrossRef CAS PubMed.
- M. M. Blewett, J. J. Xie, B. W. Zaro, K. M. Backus, A. Altman, J. R. Teijaro and B. F. Cravatt, Sci. Signal, 2016, 9(445), rs10 Search PubMed.
- R. A. Kulkarni, D. W. Bak, D. Wei, S. E. Bergholtz, C. A. Briney, J. H. Shrimp, A. Alpsoy, A. L. Thorpe, A. E. Bavari, D. R. Crooks, M. Levy, L. Florens, M. P. Washburn, N. Frizzell, E. C. Dykhuizen, E. Weerapana, W. M. Linehan and J. L. Meier, Nat. Chem. Biol., 2019, 15, 391–400 CrossRef CAS PubMed.
- S. L. Surya, M. J. C. Long, D. A. Urul, Y. Zhao, E. J. Mercer, I. M. Elsaid, T. Evans and Y. Aye, ACS Chem. Biol., 2018, 13, 1824–1831 CrossRef CAS PubMed.
- Y. Zhao, M. J. C. Long, Y. R. Wang, S. Zhang and Y. Aye, ACS Central Sci., 2018, 4, 246–259 CrossRef CAS PubMed.
- Y. Zhao, P. A. M. Herrera, D. L. Chang, R. Hamelin, M. J. C. Long and Y. Aye, Proc. Natl. Acad. Sci. U. S. A., 2022, 119(5), e2120687119 CrossRef CAS PubMed.
- S. Parvez, M. J. C. Long, H. Y. Lin, Y. Zhao, J. A. Haegele, V. N. Pham, D. K. Lee and Y. Aye, Nat. Protoc., 2016, 11, 2328–2356 CrossRef CAS PubMed.
- M. D. Witte, W. W. Kallemeijn, J. Aten, K. Y. Li, A. Strijland, W. E. Donker-Koopman, A. M. van den Nieuwendijk, B. Bleijlevens, G. Kramer, B. I. Florea, B. Hooibrink, C. E. Hollak, R. Ottenhoff, R. G. Boot, G. A. van der Marel, H. S. Overkleeft and J. M. Aerts, Nat. Chem. Biol., 2010, 6, 907–913 CrossRef CAS PubMed.
- A. R. Marques, L. I. Willems, D. Herrera Moro, B. I. Florea, S. Scheij, R. Ottenhoff, C. P. van Roomen, M. Verhoek, J. K. Nelson, W. W. Kallemeijn, A. Biela-Banas, O. R. Martin, M. B. Cachon-Gonzalez, N. N. Kim, T. M. Cox, R. G. Boot, H. S. Overkleeft and J. M. Aerts, ChemBioChem, 2017, 18, 402–412 CrossRef CAS PubMed.
- L. Wu, J. Jiang, Y. Jin, W. W. Kallemeijn, C. L. Kuo, M. Artola, W. Dai, C. van Elk, M. van Eijk, G. A. van der Marel, J. D. C. Codee, B. I. Florea, J. Aerts, H. S. Overkleeft and G. J. Davies, Nat. Chem. Biol., 2017, 13, 867–873 CrossRef CAS PubMed.
- J. Jiang, C. L. Kuo, L. Wu, C. Franke, W. W. Kallemeijn, B. I. Florea, E. van Meel, G. A. van der Marel, J. D. Codee, R. G. Boot, G. J. Davies, H. S. Overkleeft and J. M. Aerts, ACS Cent. Sci., 2016, 2, 351–358 CrossRef CAS PubMed.
- C. Steentoft, S. Y. Vakhrushev, M. B. Vester-Christensen, K. T. Schjoldager, Y. Kong, E. P. Bennett, U. Mandel, H. Wandall, S. B. Levery and H. Clausen, Nat. Methods, 2011, 8, 977–982 CrossRef CAS PubMed.
- J. Choi, L. J. S. Wagner, S. Timmermans, S. A. Malaker, B. Schumann, M. A. Gray, M. F. Debets, M. Takashima, J. Gehring and C. R. Bertozzi, J. Am. Chem. Soc., 2019, 141, 13442–13453 CrossRef CAS PubMed.
- B. Schumann, S. A. Malaker, S. P. Wisnovsky, M. F. Debets, A. J. Agbay, D. Fernandez, L. J. S. Wagner, L. Lin, Z. Li, J. Choi, D. M. Fox, J. Peh, M. A. Gray, K. Pedram, J. J. Kohler, M. Mrksich and C. R. Bertozzi, Mol. Cell, 2020, 78(824–834), e815 Search PubMed.
- S. V. Cheng, A. C. Mody and C. M. Woo, Chem. Rev., 2024, 124, 12918–13019 CrossRef CAS PubMed.
- I. Bagdonaite, S. A. Malaker, D. A. Polasky, N. M. Riley, K. Schjoldager, S. Y. Vakhrushev, A. Halim, K. F. Aoki-Kinoshita, A. I. Nesvizhskii, C. R. Bertozzi, H. H. Wandall, B. L. Parker, M. Thaysen-Andersen and N. E. Scott, Nat. Rev. Method Prime, 2022, 2, 48 CrossRef CAS.
- N. Darabedian, B. Yang, R. Ding, G. Cutolo, B. W. Zaro, C. M. Woo and M. R. Pratt, Front. Chem., 2020, 8, 318 CrossRef CAS PubMed.
- N. Darabedian, J. X. Gao, K. N. Chuh, C. M. Woo and M. R. Pratt, J. Am. Chem. Soc., 2018, 140, 7092–7100 CrossRef CAS PubMed.
- C. M. Woo, P. J. Lund, A. C. Huang, M. M. Davis, C. R. Bertozzi and S. J. Pitteri, Mol. Cell. Proteomics, 2018, 17, 764–775 CrossRef CAS PubMed.
- J. Smeitink, L. van den Heuvel and S. DiMauro, Nat. Rev. Genet., 2001, 2, 342–352 CrossRef CAS PubMed.
- E. Golemis, Protein-protein interactions: a molecular cloning manual, Cold Spring Harbor Laboratory Press, NY, 2002 Search PubMed.
- S. Fields and R. Sternglanz, Trends Genet., 1994, 10, 286–292 CrossRef CAS PubMed.
- S. Fredriksson, M. Gullberg, J. Jarvius, C. Olsson, K. Pietras, S. M. Gústafsdóttir, A. Östman and U. Landegren, Nat. Biotechnol., 2002, 20, 473–477 CrossRef CAS PubMed.
- I. Wittig, H. P. Braun and H. Schägger, Nat. Protoc., 2006, 1, 418–428 CrossRef CAS PubMed.
- T. K. Kerppola, Nat. Methods, 2006, 3, 969–971 CrossRef CAS PubMed.
- O. Vinogradova and J. Qin, Top. Curr. Chem., 2012, 326, 35–45 CrossRef CAS PubMed.
- B. Yang, S. Tang, C. Ma, S. T. Li, G. C. Shao, B. Dang, W. F. DeGrado, M. Q. Dong, P. G. Wang, S. Ding and L. Wang, Nat. Commun., 2017, 8, 2240 CrossRef PubMed.
- N. Hino, Y. Okazaki, T. Kobayashi, A. Hayashi, K. Sakamoto and S. Yokoyama, Nat. Methods, 2005, 2, 201–206 CrossRef CAS PubMed.
- D. P. Murale, S. C. Hong, M. M. Haque and J. S. Lee, Proteome Sci., 2016, 15, 14 CrossRef PubMed.
- G. W. Preston and A. J. Wilson, Chem. Soc. Rev., 2013, 42, 3289–3301 RSC.
- L. Davis and J. W. Chin, Nat. Rev. Mol. Cell Biol., 2012, 13, 168–182 CrossRef CAS PubMed.
- Y. Yang, H. Song and P. R. Chen, IUBMB Life, 2016, 68, 879–886 CrossRef CAS PubMed.
- M. Zhang, S. Lin, X. Song, J. Liu, Y. Fu, X. Ge, X. Fu, Z. Chang and P. R. Chen, Nat. Chem. Biol., 2011, 7, 671–677 CrossRef CAS PubMed.
- I. Coin, V. Katritch, T. Sun, Z. Xiang, F. Y. Siu, M. Beyermann, R. C. Stevens and L. Wang, Cell, 2013, 155, 1258–1269 CrossRef CAS PubMed.
- S. Lin, D. He, T. Long, S. Zhang, R. Meng and P. R. Chen, J. Am. Chem. Soc., 2014, 136, 11860–11863 CrossRef CAS PubMed.
- Y. Yang, H. Song, D. He, S. Zhang, S. Dai, S. Lin, R. Meng, C. Wang and P. R. Chen, Nat. Commun., 2016, 7, 12299 CrossRef CAS PubMed.
- T. Yang, X. M. Li, X. Bao, Y. M. Fung and X. D. Li, Nat. Chem. Biol., 2016, 12, 70–72 CrossRef CAS PubMed.
- X. M. Li, S. Huang and X. D. Li, Nat. Chem. Biol., 2023, 19, 614–623 CrossRef CAS PubMed.
- B. Cheng, Q. Tang, C. Zhang and X. Chen, Annu. Rev. Anal. Chem., 2021, 14, 363–387 CrossRef CAS PubMed.
- E. Saxon and C. R. Bertozzi, Science, 2000, 287, 2007–2010 CrossRef CAS PubMed.
- A. Hashimoto, K. Suenaga, A. Gloter, K. Urita and S. Iijima, Nature, 2004, 430, 870–873 CrossRef CAS PubMed.
- S. Han, B. E. Collins, P. Bengtson and J. C. Paulson, Nat. Chem. Biol., 2005, 1, 93–97 CrossRef CAS PubMed.
- L. Feng, S. Hong, J. Rong, Q. You, P. Dai, R. Huang, Y. Tan, W. Hong, C. Xie, J. Zhao and X. Chen, J. Am. Chem. Soc., 2013, 135, 9244–9247 CrossRef CAS PubMed.
- Y. Tanaka and J. J. Kohler, J. Am. Chem. Soc., 2008, 130, 3278–3279 CrossRef CAS PubMed.
- S. H. Yu, M. Boyce, A. M. Wands, M. R. Bond, C. R. Bertozzi and J. J. Kohler, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 4834–4839 CrossRef CAS PubMed.
- H. Wu, A. Shajahan, J. Y. Yang, E. Capota, A. M. Wands, C. M. Arthur, S. R. Stowell, K. W. Moremen, P. Azadi and J. J. Kohler, Cell Chem. Biol., 2022, 29(84–97), e88 Search PubMed.
- F. Liu, H. M. Chen, Z. Armstrong and S. G. Withers, ACS Cent. Sci., 2022, 8, 656–662 CrossRef CAS PubMed.
- S. A. Slavoff, I. Chen, Y. A. Choi and A. Y. Ting, J. Am. Chem. Soc., 2008, 130, 1160–1162 CrossRef CAS PubMed.
- I. Chen, M. Howarth, W. Lin and A. Y. Ting, Nat. Methods, 2005, 2, 99–104 CrossRef CAS PubMed.
- C. W. Lin and A. Y. Ting, J. Am. Chem. Soc., 2006, 128, 4542–4543 CrossRef CAS PubMed.
- M. Fernandez-Suarez, T. S. Chen and A. Y. Ting, J. Am. Chem. Soc., 2008, 130, 9251–9253 CrossRef CAS PubMed.
- K. J. Roux, D. I. Kim, M. Raida and B. Burke, J. Cell Biol., 2012, 196, 801–810 CrossRef CAS PubMed.
- K. Kwon and D. Beckett, Protein Sci., 2000, 9, 1530–1539 CrossRef CAS PubMed.
- K. Kwon, E. D. Streaker, S. Ruparelia and D. Beckett, J. Mol. Biol., 2000, 304, 821–833 CrossRef CAS PubMed.
- D. I. Kim, S. C. Jensen, K. A. Noble, B. Kc, K. H. Roux, K. Motamedchaboki and K. J. Roux, Mol. Biol. Cell, 2016, 27, 1188–1196 CrossRef CAS PubMed.
- D. I. Kim, K. C. Birendra, W. Zhu, K. Motamedchaboki, V. Doye and K. J. Roux, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, E2453–E2461 CrossRef CAS PubMed.
- E. N. Firat-Karalar, N. Rauniyar, J. R. Yates, 3rd and T. Stearns, Curr. Biol., 2014, 24, 664–670 CrossRef CAS PubMed.
- D. Comartin, G. D. Gupta, E. Fussner, E. Coyaud, M. Hasegan, M. Archinti, S. W. Cheung, D. Pinchev, S. Lawo, B. Raught, D. P. Bazett-Jones, J. Luders and L. Pelletier, Curr. Biol., 2013, 23, 1360–1366 CrossRef CAS PubMed.
- C. M. Van Itallie, A. J. Tietgens, A. Aponte, K. Fredriksson, A. S. Fanning, M. Gucek and J. M. Anderson, J. Cell Sci., 2014, 127, 885–895 CAS.
- C. M. Van Itallie, A. Aponte, A. J. Tietgens, M. Gucek, K. Fredriksson and J. M. Anderson, J. Biol. Chem., 2013, 288, 13775–13788 CrossRef CAS PubMed.
- B. Morriswood, K. Havlicek, L. Demmel, S. Yavuz, M. Sealey-Cardona, K. Vidilaseris, D. Anrather, J. Kostan, K. Djinovic-Carugo, K. J. Roux and G. Warren, Eukaryot. Cell, 2013, 12, 356–367 CrossRef CAS PubMed.
- A. L. Couzens, J. D. Knight, M. J. Kean, G. Teo, A. Weiss, W. H. Dunham, Z. Y. Lin, R. D. Bagshaw, F. Sicheri, T. Pawson, J. L. Wrana, H. Choi and A. C. Gingras, Sci. Signal, 2013, 6, rs15 Search PubMed.
- E. Prikas, A. Poljak and A. Ittner, Protein Sci., 2020, 29, 1196–1210 CrossRef CAS PubMed.
- J. A. Cutler, R. Tahir, S. K. Sreenivasamurthy, C. Mitchell, S. Renuse, R. S. Nirujogi, A. H. Patil, M. Heydarian, X. Wong, X. Wu, T. C. Huang, M. S. Kim, K. L. Reddy and A. Pandey, Leukemia, 2017, 31, 1513–1524 CrossRef CAS PubMed.
- E. Coyaud, M. Mis, E. M. Laurent, W. H. Dunham, A. L. Couzens, M. Robitaille, A. C. Gingras, S. Angers and B. Raught, Mol. Cell. Proteomics, 2015, 14, 1781–1795 CrossRef CAS PubMed.
- A. Cole, Z. Wang, E. Coyaud, V. Voisin, M. Gronda, Y. Jitkova, R. Mattson, R. Hurren, S. Babovic, N. Maclean, I. Restall, X. Wang, D. V. Jeyaraju, M. A. Sukhai, S. Prabha, S. Bashir, A. Ramakrishnan, E. Leung, Y. H. Qia, N. Zhang, K. R. Combes, T. Ketela, F. Lin, W. A. Houry, A. Aman, R. Al-Awar, W. Zheng, E. Wienholds, C. J. Xu, J. Dick, J. C. Wang, J. Moffat, M. D. Minden, C. J. Eaves, G. D. Bader, Z. Hao, S. M. Kornblau, B. Raught and A. D. Schimmer, Cancer Cell, 2015, 27, 864–876 CrossRef CAS PubMed.
- J. D. Martell, T. J. Deerinck, Y. Sancak, T. L. Poulos, V. K. Mootha, G. E. Sosinsky, M. H. Ellisman and A. Y. Ting, Nat. Biotechnol., 2012, 30, 1143–1148 CrossRef CAS PubMed.
- H. W. Rhee, P. Zou, N. D. Udeshi, J. D. Martell, V. K. Mootha, S. A. Carr and A. Y. Ting, Science, 2013, 339, 1328–1331 CrossRef CAS PubMed.
- V. Hung, P. Zou, H. W. Rhee, N. D. Udeshi, V. Cracan, T. Svinkina, S. A. Carr, V. K. Mootha and A. Y. Ting, Mol. Cell, 2014, 55, 332–341 CrossRef CAS PubMed.
- S. S. Lam, J. D. Martell, K. J. Kamer, T. J. Deerinck, M. H. Ellisman, V. K. Mootha and A. Y. Ting, Nat. Methods, 2015, 12, 51–54 CrossRef CAS PubMed.
- Y. C. Liao, M. S. Fernandopulle, G. Wang, H. Choi, L. Hao, C. M. Drerup, R. Patel, S. Qamar, J. Nixon-Abell, Y. Shen, W. Meadows, M. Vendruscolo, T. P. J. Knowles, M. Nelson, M. A. Czekalska, G. Musteikyte, M. A. Gachechiladze, C. A. Stephens, H. A. Pasolli, L. R. Forrest, P. George-Hyslop, J. Lippincott-Schwartz and M. E. Ward, Cell, 2019, 179(147–164), e120 Search PubMed.
- B. T. Lobingier, R. Huttenhain, K. Eichel, K. B. Miller, A. Y. Ting, M. von Zastrow and N. J. Krogan, Cell, 2017, 169(350–360), e312 Search PubMed.
- A. A. Dumont, L. Dumont, J. Berthiaume and M. Auger-Messier, Biochim. Biophys. Acta, Mol. Cell Res., 2019, 1866, 118557 CrossRef CAS PubMed.
- K. M. Mannix, R. M. Starble, R. S. Kaufman and L. Cooley, Development, 2019, 146, 14 CrossRef PubMed.
- T. C. Branon, J. A. Bosch, A. D. Sanchez, N. D. Udeshi, T. Svinkina, S. A. Carr, J. L. Feldman, N. Perrimon and A. Y. Ting, Nat. Biotechnol., 2018, 36, 880–887 CrossRef CAS PubMed.
- W. Qin, J. S. Cheah, C. Xu, J. Messing, B. D. Freibaum, S. Boeynaems, J. P. Taylor, N. D. Udeshi, S. A. Carr and A. Y. Ting, Cell, 2023, 186(3307–3324), e3330 Search PubMed.
- L. Kubitz, S. Bitsch, X. Y. Zhao, K. Schmitt, L. Deweid, A. Roehrig, E. C. Barazzone, O. Valerius, H. Kolmar and J. Béthune, Commun. Biol., 2022, 5, 657 CrossRef CAS PubMed.
- C. Kwak, S. Shin, J. S. Park, M. Jung, T. T. M. Nhung, M. G. Kang, C. Lee, T. H. Kwon, S. K. Park, J. Y. Mun, J. S. Kim and H. W. Rhee, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 12109–12120 CrossRef CAS PubMed.
- I. M. Schopp, C. C. Amaya Ramirez, J. Debeljak, E. Kreibich, M. Skribbe, K. Wild and J. Bethune, Nat. Commun., 2017, 8, 15690 CrossRef CAS PubMed.
- K. F. Cho, T. C. Branon, S. Rajeev, T. Svinkina, N. D. Udeshi, T. Thoudam, C. Kwak, H. W. Rhee, I. K. Lee, S. A. Carr and A. Y. Ting, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 12143–12154 CrossRef CAS PubMed.
- Y. Han, T. C. Branon, J. D. Martell, D. Boassa, D. Shechner, M. H. Ellisman and A. Ting, ACS Chem. Biol., 2019, 14, 619–635 CrossRef CAS PubMed.
- J. D. Martell, M. Yamagata, T. J. Deerinck, S. Phan, C. G. Kwa, M. H. Ellisman, J. R. Sanes and A. Y. Ting, Nat. Biotechnol., 2016, 34, 774–780 CrossRef CAS PubMed.
- Z. B. Hill, S. B. Pollock, M. Zhuang and J. A. Wells, J. Am. Chem. Soc., 2016, 138, 13123–13126 CrossRef CAS PubMed.
- M. Zhuang, S. Guan, H. Wang, A. L. Burlingame and J. A. Wells, Mol. Cell, 2013, 49, 273–282 CrossRef CAS PubMed.
- Q. Liu, J. Zheng, W. Sun, Y. Huo, L. Zhang, P. Hao, H. Wang and M. Zhuang, Nat. Methods, 2018, 15, 715–722 CrossRef CAS PubMed.
- M. J. C. Long, J. M. Liu and Y. Aye, RSC Chem. Biol., 2023, 4, 110–120 RSC.
- G. Pasqual, A. Chudnovskiy, J. M. J. Tas, M. Agudelo, L. D. Schweitzer, A. Cui, N. Hacohen and G. D. Victora, Nature, 2018, 553, 496–500 CrossRef CAS PubMed.
- Y. Ge, L. Chen, S. Liu, J. Zhao, H. Zhang and P. R. Chen, J. Am. Chem. Soc., 2019, 141, 1833–1837 CrossRef CAS PubMed.
- C. P. Seath, A. J. Burton, X. Sun, G. Lee, R. E. Kleiner, D. W. C. MacMillan and T. W. Muir, Nature, 2023, 616, 574–580 Search PubMed.
- J. B. Geri, J. V. Oakley, T. Reyes-Robles, T. Wang, S. J. McCarver, C. H. White, F. P. Rodriguez-Rivera, D. L. Parker, Jr., E. C. Hett, O. O. Fadeyi, R. C. Oslund and D. W. C. MacMillan, Science, 2020, 367, 1091–1097 CrossRef CAS PubMed.
- K. Kozoriz, O. Shkel, K. T. Hong, D. H. Kim, Y. K. Kim and J. S. Lee, Acc. Chem. Res., 2023, 56, 25–36 CrossRef CAS PubMed.
- Z. Huang, Z. Liu, X. Xie, R. Zeng, Z. Chen, L. Kong, X. Fan and P. R. Chen, J. Am. Chem. Soc., 2021, 143, 18714–18720 CrossRef CAS PubMed.
- H. Wang, Y. Zhang, K. Zeng, J. Qiang, Y. Cao, Y. Li, Y. Fang, Y. Zhang and Y. Chen, JACS Au, 2021, 1, 1066–1075 CrossRef CAS PubMed.
- B. F. Buksh, S. D. Knutson, J. V. Oakley, N. B. Bissonnette, D. G. Oblinsky, M. P. Schwoerer, C. P. Seath, J. B. Geri, F. P. Rodriguez-Rivera, D. L. Parker, G. D. Scholes, A. Ploss and D. W. C. MacMillan, J. Am. Chem. Soc., 2022, 144, 6154–6162 CrossRef CAS PubMed.
- N. E. S. Tay, K. A. Ryu, J. L. Weber, A. K. Olow, D. C. Cabanero, D. R. Reichman, R. C. Oslund, O. O. Fadeyi and T. Rovis, Nat. Chem., 2023, 15, 101–109 CrossRef CAS PubMed.
- R. C. Oslund, T. Reyes-Robles, C. H. White, J. H. Tomlinson, K. A. Crotty, E. P. Bowman, D. Chang, V. M. Peterson, L. Li, S. Frutos, M. Vila-Perello, D. Vlerick, K. Cromie, D. H. Perlman, S. Ingale, S. D. O. Hara, L. R. Roberts, G. Piizzi, E. C. Hett, D. J. Hazuda and O. O. Fadeyi, Nat. Chem. Biol., 2022, 18, 850–858 CrossRef CAS PubMed.
- S. D. Knutson, B. F. Buksh, S. W. Huth, D. C. Morgan and D. W. C. MacMillan, Cell Chem. Biol., 2024, 31, 1145–1161 CrossRef CAS PubMed.
- T. J. Bechtel, T. Reyes-Robles, O. O. Fadeyi and R. C. Oslund, Nat. Chem. Biol., 2021, 17, 641–652 CrossRef CAS PubMed.
- J. V. Oakley, B. F. Buksh, D. F. Fernández, D. G. Oblinsky, C. P. Seath, J. B. Geri, G. D. Scholes and D. W. C. MacMillan, Proc. Natl. Acad. Sci. U. S. A., 2022, 119(34), e2208077119 CrossRef PubMed.
- B. Alvarez-Castelao, C. T. Schanzenbächer, C. Hanus, C. Glock, S. T. Dieck, A. R. Dörrbaum, I. Bartnik, B. Nassim-Assir, E. Ciirdaeva, A. Mueller, D. C. Dieterich, D. A. Tirrell, J. D. Langer and E. M. Schuman, Nat. Biotechnol., 2017, 35, 1196–1201 CrossRef CAS PubMed.
- J. R. Poganik, K. T. Huang, S. Parvez, Y. Zhao, S. Raja, M. J. C. Long and Y. Aye, Nat. Commun., 2021, 12 Search PubMed.
- J. M. Liu, A. Kulkarni, Y. Q. Gao, D. A. Urul, R. Hamelin, B. A. Novotny, M. J. C. Long and Y. Aye, Cell, 2024, 187(26), 7450–7469 CrossRef CAS PubMed.
- C. J. Gerry and S. L. Schreiber, Nat. Rev. Drug Discovery, 2018, 17, 333–352 CrossRef CAS PubMed.
- B. J. Leslie and P. J. Hergenrother, Chem. Soc. Rev., 2008, 37, 1347–1360 RSC.
- G. C. Terstappen, C. Schlupen, R. Raggiaschi and G. Gaviraghi, Nat. Rev. Drug Discovery, 2007, 6, 891–903 CrossRef CAS PubMed.
- A. L. Hopkins, Nat. Chem. Biol., 2008, 4, 682–690 CrossRef CAS PubMed.
- M. Schenone, V. Dancik, B. K. Wagner and P. A. Clemons, Nat. Chem. Biol., 2013, 9, 232–240 CrossRef CAS PubMed.
- L. P. Conway, A. M. Jadhav, R. A. Homan, W. Li, J. S. Rubiano, R. Hawkins, R. M. Lawrence and C. G. Parker, Chem. Sci., 2021, 12, 7839–7847 RSC.
- J. Park, M. Koh, J. Y. Koo, S. Lee and S. B. Park, ACS Chem. Biol., 2016, 11, 44–52 CrossRef CAS PubMed.
- C. Limban, Toxicol. Rep., 2021, 8, 28 CrossRef PubMed.
- S. Pan, S. Y. Jang, D. Wang, S. S. Liew, Z. Li, J. S. Lee and S. Q. Yao, Angew. Chem., Int. Ed., 2017, 56, 11816–11821 CrossRef CAS PubMed.
- Z. Li, D. Wang, L. Li, S. Pan, Z. Na, C. Y. Tan and S. Q. Yao, J. Am. Chem. Soc., 2014, 136, 9990–9998 CrossRef CAS PubMed.
- T. Ma, X. Tian, B. Zhang, M. Li, Y. Wang, C. Yang, J. Wu, X. Wei, Q. Qu, Y. Yu, S. Long, J. W. Feng, C. Li, C. Zhang, C. Xie, Y. Wu, Z. Xu, J. Chen, Y. Yu, X. Huang, Y. He, L. Yao, L. Zhang, M. Zhu, W. Wang, Z. C. Wang, M. Zhang, Y. Bao, W. Jia, S. Y. Lin, Z. Ye, H. L. Piao, X. Deng, C. S. Zhang and S. C. Lin, Nature, 2022, 603, 159–165 CrossRef CAS PubMed.
- J. Gao, A. Mfuh, Y. Amako and C. M. Woo, J. Am. Chem. Soc., 2018, 140, 4259–4268 CrossRef CAS PubMed.
- A. D. Trowbridge, C. P. Seath, F. P. Rodriguez-Rivera, B. X. Li, B. E. Dul, A. G. Schwaid, B. F. Buksh, J. B. Geri, J. V. Oakley, O. O. Fadeyi, R. C. Oslund, K. A. Ryu, C. White, T. Reyes-Robles, P. Tawa, D. L. Parker, Jr. and D. W. C. MacMillan, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2208077119 CrossRef CAS PubMed.
- Y. Kawamata, K. A. Ryu, G. N. Hermann, A. Sandahl, J. C. Vantourout, A. K. Olow, L. A. Adams, E. Rivera-Chao, L. R. Roberts, S. Gnaim, M. Nassir, R. C. Oslund, O. O. Fadeyi and P. S. Baran, Nat. Chem., 2023, 15, 1267–1275 CrossRef CAS PubMed.
- M. M. Savitski, F. B. Reinhard, H. Franken, T. Werner, M. F. Savitski, D. Eberhard, D. Martinez Molina, R. Jafari, R. B. Dovega, S. Klaeger, B. Kuster, P. Nordlund, M. Bantscheff and G. Drewes, Science, 2014, 346, 1255784 CrossRef PubMed.
- D. Martinez Molina, R. Jafari, M. Ignatushchenko, T. Seki, E. A. Larsson, C. Dan, L. Sreekumar, Y. Cao and P. Nordlund, Science, 2013, 341, 84–87 CrossRef PubMed.
- K. V. Huber, E. Salah, B. Radic, M. Gridling, J. M. Elkins, A. Stukalov, A. S. Jemth, C. Gokturk, K. Sanjiv, K. Stromberg, T. Pham, U. W. Berglund, J. Colinge, K. L. Bennett, J. I. Loizou, T. Helleday, S. Knapp and G. Superti-Furga, Nature, 2014, 508, 222–227 CrossRef CAS PubMed.
- E. C. Strickland, M. A. Geer, D. T. Tran, J. Adhikari, G. M. West, P. D. DeArmond, Y. Xu and M. C. Fitzgerald, Nat. Protoc., 2013, 8, 148–161 CrossRef CAS PubMed.
- G. M. West, C. L. Tucker, T. Xu, S. K. Park, X. Han, J. R. Yates, 3rd and M. C. Fitzgerald, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 9078–9082 CrossRef CAS PubMed.
- P. D. Dearmond, Y. Xu, E. C. Strickland, K. G. Daniels and M. C. Fitzgerald, J. Proteome Res., 2011, 10, 4948–4958 CrossRef CAS PubMed.
- B. Lomenick, R. Hao, N. Jonai, R. M. Chin, M. Aghajan, S. Warburton, J. Wang, R. P. Wu, F. Gomez, J. A. Loo, J. A. Wohlschlegel, T. M. Vondriska, J. Pelletier, H. R. Herschman, J. Clardy, C. F. Clarke and J. Huang, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 21984–21989 CrossRef CAS PubMed.
- M. L. Schulte, A. Fu, P. Zhao, J. Li, L. Geng, S. T. Smith, J. Kondo, R. J. Coffey, M. O. Johnson, J. C. Rathmell, J. T. Sharick, M. C. Skala, J. A. Smith, J. Berlin, M. K. Washington, M. L. Nickels and H. C. Manning, Nat. Med., 2018, 24, 194–202 CrossRef CAS PubMed.
- R. C. Coll, J. R. Hill, C. J. Day, A. Zamoshnikova, D. Boucher, N. L. Massey, J. L. Chitty, J. A. Fraser, M. P. Jennings, A. A. B. Robertson and K. Schroder, Nat. Chem. Biol., 2019, 15, 556–559 CrossRef CAS PubMed.
- M. M. Dix, G. M. Simon and B. F. Cravatt, Cell, 2008, 134, 679–691 CrossRef CAS PubMed.
- M. M. Dix, G. M. Simon, C. Wang, E. Okerberg, M. P. Patricelli and B. F. Cravatt, Cell, 2012, 150, 426–440 CrossRef CAS PubMed.
- B. W. Wright, Z. Yi, J. S. Weissman and J. Chen, Trends Cell Biol., 2022, 32, 243–258 CrossRef CAS PubMed.
- S. Siriwongsup, A. M. Schmoker, S. B. Ficarro, J. A. Marto and J. Kim, Chem, 2024, 10, 1306–1315 CAS.
- H. Wang, Y. Zhou, X. Xu, H. Li and H. Sun, Curr. Opin. Chem. Biol., 2020, 55, 171–179 CrossRef CAS PubMed.
- H. Li, R. Wang and H. Sun, Acc. Chem. Res., 2019, 52, 216–227 CrossRef CAS PubMed.
- B. Neuditschko, A. P. King, Z. Huang, L. Janker, A. Bileck, Y. Borutzki, S. C. Marker, C. Gerner, J. J. Wilson and S. M. Meier-Menches, Angew. Chem., Int. Ed., 2022, 61, e202209136 CrossRef CAS PubMed.
- V. W. Rebecca, M. C. Nicastri, C. Fennelly, C. I. Chude, J. S. Barber-Rotenberg, A. Ronghe, Q. McAfee, N. P. McLaughlin, G. Zhang, A. R. Goldman, R. Ojha, S. Piao, E. Noguera-Ortega, A. Martorella, G. M. Alicea, J. J. Lee, L. M. Schuchter, X. Xu, M. Herlyn, R. Marmorstein, P. A. Gimotty, D. W. Speicher, J. D. Winkler and R. K. Amaravadi, Cancer Discovery, 2019, 9, 220–229 CrossRef CAS PubMed.
- M. J. C. Long, C. Rogg and Y. Aye, Acc. Chem. Res., 2021, 54, 618–631 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |