CRISPR-coupled triple cascade amplification for simultaneous lateral flow detection of Mycoplasma pneumoniae and H1N1
Ruyue
Wei†
a,
Shuqi
Wang†
a,
Yingbo
Pan
a,
Wei
Pan
 a,
Na
Li
a,
Na
Li
 *a and
Bo
Tang
*a and
Bo
Tang
 ab
ab
aCollege of Chemistry, Chemical Engineering and Materials Science, Key Laboratory of Molecular and Nano Probes, Ministry of Education, Collaborative Innovation Center of Functionalized Probes for Chemical Imaging in Universities of Shandong, Institute of Molecular and Nano Science, Shandong Normal University, Jinan 250014, P. R. China. E-mail: lina@sdnu.edu.cn
bLaoshan Laboratory, Qingdao 266237, P. R. China
First published on 4th November 2025
Abstract
We developed a CRISPR-coupled triple cascade system integrating recombinase polymerase amplification (RPA), CRISPR/Cas12a, and catalytic hairpin assembly (CHA) for simultaneous lateral flow detection of Mycoplasma pneumoniae and H1N1 in saliva samples, achieving a LOD of 10 aM for H1N1 RNA and 25 aM for MP DNA on a single LFA.
Respiratory pathogens pose a persistent public health challenge, with influenza A virus being responsible for seasonal flu outbreaks and Mycoplasma pneumoniae (MP) serving as a leading cause of community-acquired pneumonia.1–3 Notably, these two pathogens often co-circulate during outbreaks and it is difficult to distinguish infections caused by different pathogens based solely on clinical manifestations. Consequently, a sensitive and specific diagnosis approach is essential for preventing their spread and guiding appropriate medications. Conventional immunological methods have the advantages of being rapid and convenient, but more sensitive detection methods are needed for early detection of the pathogen in infection.4–7 In contrast, real-time quantitative polymerase chain reaction (qPCR)-based nucleic acid detection is considered the gold standard due to its high sensitivity and specificity.8,9 However, the reliance on specialized equipment, thermal cycling, and highly trained personnel limits its application for on-site analysis. There is a growing need for alternative detection methods with high sensitivity for on-site analysis of the nucleic acids of respiratory pathogens, promoting mass screening and guided medication to impede disease transmission.
Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) enzymes, a kind of natural adaptive immune system of microorganisms, have been repurposed as a potential tool for in vitro diagnosis.10–12 Cas12a, as one of the important members of the Cas family, can be programmed and activated with CRISPR RNA (crRNA) for non-specific degradation of single-stranded nucleic acids (trans-cleavage).13–15 The high specificity and enzyme activity enable the CRISPR/Cas12a system to be a promising platform for biosensing applications.16,17 However, the trans-cleavage amplification effect of CRISPR/Cas12a alone cannot achieve the desired sensitivity, and this inherent limitation necessitates integration with auxiliary nucleic acid amplification methods to enhance detection performance.18–20 Isothermal nucleic acid amplification technologies, such as recombinase polymerase amplification (RPA),21 strand displacement amplification (SDA),22 catalytic hairpin assembly (CHA),23 and loop-mediated isothermal amplification (LAMP),24 require only simple thermal control, making them ideal for portable diagnostic applications. Consequently, the synergistic integration of CRISPR/Cas12a with isothermal amplification enables the development of sensitive and user-friendly diagnostic technologies.
Lateral flow assays (LFAs) are particularly attractive for point-of-care biomarker detection due to their simplicity, affordability, and equipment-free operation.25–27 The combination of LFAs with CRISPR-isothermal nucleic acid amplification technologies offers a sensitive and low-cost nucleic acid detection strategy. Currently, CRISPR/Cas12a-based LFAs utilize ssDNA probes as reporters, with the test line (T line) located above the control line (C line), which increases the difficulty of result interpretation and only one target can be analysed via a single test strip. In this study, we developed a lateral flow assay (LFA) platform integrated triple cascade amplification system (LFA-TCA), incorporating recombinase polymerase amplification (RPA), CRISPR/Cas12a, and a catalytic hairpin assembly (CHA) circuit, for sensitive and accurate detection of Mycoplasma pneumoniae DNA (MP DNA) and influenza A virus subtype H1N1 RNA. The LFA-TCA platform can simultaneously detect two pathogen nucleic acids on a single LFA with high sensitivity. As shown in Scheme 1, in the presence of specific targets, MP DNA and H1N1 RNA are exponentially amplified via RPA to generate numerous amplicons, which can induce the trans-cleavage activity of CRISPR/Cas12a. Activated Cas12a can cleave blocked strands (BS), blocking their hybridization with the initiator strands (IS) and allowing IS to trigger subsequent CHA reactions. For H1N1 RNA detection, free IS-1 catalyzes the cross-hybridization of FAM-labelled H1 (FAM-H1) and biotin-labelled H2 (Bio-H2) via a CHA-1 circuit to produce numerous hybrid duplexes (FAM-H1/Bio-H2), which bind to the streptavidin-modified gold nanoparticles (Au NPs) pre-packaged in the conjugate pad and are further captured by anti-FAM antibodies on test line 1 (TL1), resulting in a visible red band. For MP DNA detection, its presence will release dissociative IS-2 to initiate the CHA-2 circuit to obtain hybrid duplexes of TAMRA-labelled H3 (TAMRA-H3) and digoxin-labelled H4 (Dig-H4) (TAMRA-H3/Dig-H4). TAMRA-H3/Dig-H4 acts as a bridging element to bind to anti-TAMRA antibody-modified Au NPs, leading to the aggregation of the Au NPs on test line 2 (TL2) to generate a red band. Conversely, the lack of targets inhibits the trans-cleavage activity of CRISPR-Cas12a and preserves the integrity of BS. Then, BS hybridized with IS to prevent the CHA circuits from generating a signal, and only the blue control line (CL) can be observed. LFA-TCA successfully detects H1N1 RNA and MP DNA on a single LFA with high sensitivity and robustness via multiple isothermal amplification, facilitating a field-deployable solution for pathogen testing.
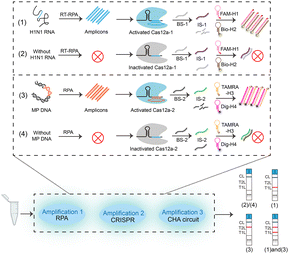 | ||
| Scheme 1 Schematic of the LFA platform based on a triple cascade amplification system (LFA-TCA) for sensitive detection of MP DNA and H1N1 RNA. | ||
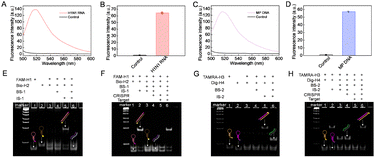
The feasibility of RPA-mediated primary amplification was investigated. According to the nucleic acid sequences of H1N1 RNA and MP DNA, the specific primers for RPA were designed (Table S1). As shown in Fig. S1, the bright amplified bands of H1N1 RNA and MP DNA appeared, suggesting that the designed primers of RPA and RT-RPA were suitable for target detection. The fluorescence experiment further showed that the amplicons successfully activated the trans-cleavage activity of Cas12a (Fig. 1A–D). The polyacrylamide gel electrophoresis (PAGE) image revealed the destruction of the BS by Cas12a (Fig. S2). Subsequently, the BS blocked CHA circuit was verified. For the CHA-1 circuit, as shown in Fig. 1E, both FAM-H1 and Bio-H2 maintained a stable hairpin conformation without unexpected cross reaction when BS-1 was present, indicating that the CHA-1 circuit was successfully prevented. Upon introduction of the H1N1 RNA amplicons and CRISPR/Cas12a-1, a significant FAM-H1/Bio-H2 band was found in lane 6, which was attributed to the H1N1 RNA amplicons activating Cas12a-1 for breaking this blockade and promoting the occurrence of tertiary amplification (Fig. 1F). For MP DNA detection, we also designed an exclusive CHA-2 circuit, and as shown in Fig. 1G and H, a bright TAMRA-H3/Dig-H4 hybrid band appeared on the addition of MP DNA amplicons and relevant CRISPR/Cas12a-2, manifesting that target MP DNA successfully triggered the corresponding cascade reaction. These experimental results confirmed the successful establishment of the TCA system based on multiple isothermal amplifications.
The LFA consists of a sample pad, a conjugate pad, an absorbent pad, test lines (TL1 and TL2), and a control line (CL), of which the conjugate pad was pre-packaged with anti-TAMRA antibody-modified Au NPs and streptavidin-modified Au NPs. The anti-FAM and anti-digoxin antibodies were pre-decorated at TL1 and TL2, respectively. The products, carrying FAM and biotin, served as a linker that can absorb streptavidin-modified Au NPs and detain them on TL1 by binding to anti-FAM antibodies, producing a red TL1 band visible to the naked eye. In addition, the presence of products with TAMRA and digoxin will generate a red TL2 band by fixing anti-TAMRA antibody-modified Au NPs on TL2. Then we combine the kind of LFA with the developed TCA system and explore the detection of H1N1 RNA and MP DNA (Fig. 2A and B). As shown in Fig. 2C, red TL1 and TL2 occurred when the H1N1 RNA and MP DNA were present, respectively, because the targets triggered the TCA system to generate abundant FAM-H1/Bio-H2 and TAMRA-H3/Dig-H4 hybrid products. In addition, the coexistence of H1N1 RNA and MP DNA resulted in two test lines on the LFA, while only CL was found when there was no target nucleic acid in the sample (Fig. 2C and D). The results verified that the TCA system could be applied for the simultaneous detection of H1N1 RNA and MP DNA on a single LFA.
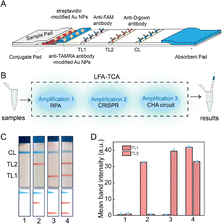 | ||
| Fig. 2 (A) The construction of the LFA. (B) The detection process of the LFA-TCA. (C) The LFA images and (D) grayscale analysis of TL1 and TL2 for different samples. TL1: H1N1 RNA, TL2: MP DNA. | ||
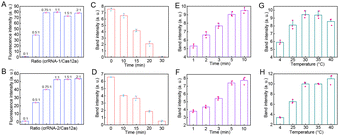
To achieve ideal detection results, it is necessary to explore the optimal reaction conditions. The ratio of primers (IS) to hairpins was first optimized (Fig. S3). The optimal reaction ratios for the CHA-1 and CHA-2 circuits were 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 and 1
5 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, respectively. Additionally, hairpins with different stem lengths were designed to optimize the CHA reaction. As expected, hairpins with shorter stems showed a higher background signal and longer stems resulted in low reactivity. H1 (12)–H2 (12) was selected for the CHA-1 circuit reaction and H3 (10)–H4 (11) was selected for the CHA-2 circuit reaction (Fig. S4 and S5). Fig. 3A and B display that the fluorescence signal reached a maximal state when the ratios of crRNA to Cas12a were 0.75
4, respectively. Additionally, hairpins with different stem lengths were designed to optimize the CHA reaction. As expected, hairpins with shorter stems showed a higher background signal and longer stems resulted in low reactivity. H1 (12)–H2 (12) was selected for the CHA-1 circuit reaction and H3 (10)–H4 (11) was selected for the CHA-2 circuit reaction (Fig. S4 and S5). Fig. 3A and B display that the fluorescence signal reached a maximal state when the ratios of crRNA to Cas12a were 0.75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, respectively. And the cleavage efficacy of Cas12a at different times was assessed. As illustrated in Fig. 3C, D and Fig. S6A, B, with the increase of the reaction time, the BS-1 and BS-2 bands gradually weakened and completely disappeared at 30 min, which was exploited for the subsequent assay. The duration and temperature of the CHA circuits used in the tertiary amplification process were optimized to improve the detection performance. FAM-H1/Bio-H2 (Fig. 3E and Fig. S6C) and TAMRA-H3/Dig-H4 (Fig. 3F and Fig. S6D) were found to initially increase with the incubation time and then they stabilized at 5 min. Moreover, the bands of FAM-H1/Bio-H2 and TAMRA-H3/Dig-H4 showed minimal difference when incubated at 30–40 °C. To retain the isothermal reaction, the CHA circuits were carried out at 37 °C for 5 min (Fig. 3G, H and Fig. S6E, F).
1, respectively. And the cleavage efficacy of Cas12a at different times was assessed. As illustrated in Fig. 3C, D and Fig. S6A, B, with the increase of the reaction time, the BS-1 and BS-2 bands gradually weakened and completely disappeared at 30 min, which was exploited for the subsequent assay. The duration and temperature of the CHA circuits used in the tertiary amplification process were optimized to improve the detection performance. FAM-H1/Bio-H2 (Fig. 3E and Fig. S6C) and TAMRA-H3/Dig-H4 (Fig. 3F and Fig. S6D) were found to initially increase with the incubation time and then they stabilized at 5 min. Moreover, the bands of FAM-H1/Bio-H2 and TAMRA-H3/Dig-H4 showed minimal difference when incubated at 30–40 °C. To retain the isothermal reaction, the CHA circuits were carried out at 37 °C for 5 min (Fig. 3G, H and Fig. S6E, F).
Under optimal conditions, the detection performance of LFA-TCA was explored. As shown in Fig. S7A–D, the color intensity of TL1 and TL2 progressively intensified with the addition of H1N1 RNA and MP DNA concentration, respectively, indicating that LFA-TCA could sense the change in target concentration and detect different kinds of targets independently (Fig. S7E and F). Subsequently, the specificity of the proposed LFA-TCA was further assessed. Fig. S7G and H show that only the H1N1 RNA and MP DNA led to bright red bands on TL1 and TL2, respectively, indicating the excellent specificity of LFA-TCA.
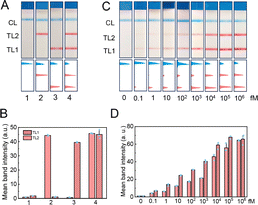
To validate the anti-interference capability of LFA-TCA, the saliva-adulterated target nucleic acid was detected. The visualized detection results showed that the saliva did not influence the detection performance of LFA-TCA (Fig. 4A and B). TL1 and TL2 gradually increased with the increase of the corresponding target concentrations, consistent with the assay in PBS buffer, revealing that LFA-TCA performed well under the change of target concentrations (Fig. 4C and D). There was a linear correlation between the TL signal and target concentrations raising from 0.1 to 1000 fM. According to the linear equation y = 8.0409![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lg
lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C + 14.8490 (H1N1 RNA) and y = 4.6803
C + 14.8490 (H1N1 RNA) and y = 4.6803![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lg
lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C + 7.3407 (MP DNA), the limit of detection (LOD) for H1N1 RNA and MP DNA was 10 aM and 25 aM, respectively (Fig. S8). Compared with other strategies for analyzing nucleic acids, our LFA-TCA exhibited a comparable detection performance and portable on-site deployment (Table S2). Additionally, the simulated pathogen samples were added to saliva solutions to further assess the practical analytical capabilities of LFA-TCA. As the concentration of the pathogen increased, the signals of TL1 and TL2 gradually strengthened (Fig. S9). The visual detection ability for Mycoplasma pneumoniae and H1N1 was below 50 copies per µL. Then, we employed the LFA-TCA strategy as well as the common PCR method to detect pathogen nucleic acids. The detection results of LFA-TCA were consistent with those of the PCR method, indicating the reliability of the detection system (Fig. S10). Given the limited availability of clinical samples, this study provided a preliminary verification of the LFA-TCA's clinical application potential. Subsequent large-scale validation with diverse samples will be essential to thoroughly evaluate the generalizability of this strategy.
C + 7.3407 (MP DNA), the limit of detection (LOD) for H1N1 RNA and MP DNA was 10 aM and 25 aM, respectively (Fig. S8). Compared with other strategies for analyzing nucleic acids, our LFA-TCA exhibited a comparable detection performance and portable on-site deployment (Table S2). Additionally, the simulated pathogen samples were added to saliva solutions to further assess the practical analytical capabilities of LFA-TCA. As the concentration of the pathogen increased, the signals of TL1 and TL2 gradually strengthened (Fig. S9). The visual detection ability for Mycoplasma pneumoniae and H1N1 was below 50 copies per µL. Then, we employed the LFA-TCA strategy as well as the common PCR method to detect pathogen nucleic acids. The detection results of LFA-TCA were consistent with those of the PCR method, indicating the reliability of the detection system (Fig. S10). Given the limited availability of clinical samples, this study provided a preliminary verification of the LFA-TCA's clinical application potential. Subsequent large-scale validation with diverse samples will be essential to thoroughly evaluate the generalizability of this strategy.
In summary, we constructed an LFA platform based on a triple cascade amplified detection system (LFA-TCA), which integrates recombinase polymerase amplification (RPA), CRISPR-Cas12a and catalytic hairpin assembly (CHA) for the detection of Mycoplasma pneumoniae DNA (MP DNA) and H1N1 influenza virus RNA (H1N1 RNA). LFA-TCA could analyze two pathogen nucleic acids simultaneously through a single LFA with the advantages of high sensitivity (the LOD for H1N1 RNA and MP DNA is 10 aM and 25 aM, respectively) and satisfactory specificity. This study demonstrates the potential of multiplex amplification for enhanced sensitivity and multiple detections of LFA. In future work, expanding the multiplexing capacity could be achieved by modifying RPA primers and crRNA sequences, which would allow the design of customized detection modules targeting different nucleic acids. Additionally, this strategy showed high specificity but potential limitations remain when extending the platform to diverse pathogen classes. The genetic diversity within pathogen species, such as mutations, may necessitate further optimization of the primers and crRNA sequences. Besides, extensive validation with different pathogens and complex clinical samples will be required to ensure robustness across various pathogens.
This work was supported by the National Natural Science Foundation of China (22374092) and the Project of Shandong Provincial Center for Fundamental Science Research (YDZX2024150).
Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information: experimental conditions and procedures, and additional characterization data. See DOI: https://doi.org/10.1039/d5cc04993h.Notes and references
- Z. Zhao, C. Huang, Z. Huang, F. Lin, Q. He, D. Tao, N. Jaffrezic-Renault and Z. Guo, TrAC, Trends Anal. Chem., 2021, 139, 116253 CrossRef CAS.
- Y. Li, X. Fu, J. Ma, J. Zhang, Y. Hu, W. Dong, Z. Wan, Q. Li, Y. Q. Kuang, K. Lan, X. Jin, J. H. Wang and C. Zhang, Nat. Commun., 2019, 10, 2288 CrossRef.
- N. Zhang, L. Wang, X. Deng, R. Liang, M. Su, C. He, L. Hu, Y. Su, J. Ren, F. Yu, L. Du and S. Jiang, J. Med. Virol., 2020, 92, 408–417 CrossRef CAS PubMed.
- K. Chan, K. Chan, Y. Ho, Y. Lam, H. Tong, L. Poon, B. Cowling and J. Peiris, J. Virol. Methods, 2012, 186, 184–188 CrossRef CAS PubMed.
- N. Ishiguro, H. Kikuta, M. Konno, R. Sato and A. Manabe, J. Lab. Med., 2021, 45, 189–192 CrossRef CAS.
- N. Miyashita, Y. Kawai, T. Tanaka, H. Akaike, H. Teranishi, T. Wakabayashi, T. Nakano, K. Ouchi and N. Okimoto, J. Infect. Chemother., 2015, 21, 473–475 CrossRef CAS.
- O. Puhach, B. Meyer and I. Eckerle, Nat. Rev. Microbiol., 2023, 21, 147–161 CAS.
- D. Kong, X. Wang, C. Gu, M. Guo, Y. Wang, Z. Ai, S. Zhang, Y. Chen, W. Liu, Y. Wu, C. Dai, Q. Guo, D. Qu, Z. Zhu, Y. Xie, Y. Liu and D. Wei, J. Am. Chem. Soc., 2021, 143, 17004–17014 CrossRef CAS PubMed.
- C. Shi, D. Yang, X. Ma, L. Pan, Y. Shao, G. Arya, Y. Ke, C. Zhang, F. Wang, X. Zuo, M. Li and P. Wang, Angew. Chem., Int. Ed., 2024, 63, e202320179 CrossRef CAS PubMed.
- C. Diez-Villasenor and F. Rodriguez-Valera, Nat. Commun., 2019, 10, 294 CrossRef PubMed.
- C. Zhang, X. Zhao, X. Chen, X. Lin, Z. Huang, J. Hu, R. Liu and Y. Lv, Chem. Commun., 2024, 60, 13259–13262 RSC.
- H. Yan, Y. Wen, Z. Tian, N. Hart, S. Han, S. J. Hughes and Y. Zeng, Nat. Biomed. Eng., 2023, 7, 1583–1601 CrossRef CAS PubMed.
- H. Liu, J. T. Dong, Z. J. Duan, F. Xia, I. Willner and F. J. Huang, Sci. Adv., 2024, 10, eadp6166 CrossRef CAS PubMed.
- S. R. Rananaware, E. K. Vesco, G. M. Shoemaker, S. S. Anekar, L. S. W. Sandoval, K. S. Meister, N. C. Macaluso, L. T. Nguyen and P. K. Jain, Nat. Commun., 2023, 14, 5409 CrossRef CAS PubMed.
- X. Ding, K. Yin, Z. Li, R. V. Lalla, E. Ballesteros, M. M. Sfeir and C. Liu, Nat. Commun., 2020, 11, 4711 CrossRef CAS PubMed.
- S. Gong, K. Song, W. Pan, N. Li and B. Tang, Anal. Chem., 2024, 96, 15789–15796 CrossRef CAS.
- S. Peng, Z. Tan, S. Chen, C. Lei and Z. Nie, Chem. Sci., 2020, 11, 7362–7368 RSC.
- J. M. Lesinski, T. Moragues, P. Mathur, Y. Shen, C. Paganini, L. Bezinge, B. Verberckmoes, B. Van Eenooghe, S. Stavrakis, A. J. deMello and D. A. Richards, Anal. Chem., 2024, 96, 10443–10450 CrossRef CAS PubMed.
- R. Deng, J. Sheng, Z. Xie, H. Yang, S. Yang, S. Xie, X. Tang, S. Zhao, H. Dong, M. Chen and K. Chang, Anal. Chem., 2025, 97, 799–810 CrossRef CAS PubMed.
- M. A. Ahamed, M. A. U. Khalid, M. Dong, A. J. Politza, Z. Zhang, A. Kshirsagar, T. Liu and W. Guan, Biosens. Bioelectron., 2024, 246, 115866 CrossRef CAS PubMed.
- Z. Yao, K. He, H. Wang, S. Feng, X. Ding, Y. Xu, Q. Wang, X. Xu, Q. Wu and L. Wang, ACS Sens., 2024, 9, 3511–3519 CrossRef CAS PubMed.
- Y. Wu, B. Lv, X. Ni, S. Zhu and D. Li, ACS Sens., 2025, 10, 965–976 CrossRef CAS PubMed.
- P. Chen, L. Wang, P. Qin, B. C. Yin and B. C. Ye, Biosens. Bioelectron., 2022, 207, 114152 CrossRef CAS PubMed.
- C. Chen, S. Chen, Y. Fu, Y. Wei, L. Xie and M. Chen, Biosens. Bioelectron., 2024, 263, 116635 CrossRef CAS PubMed.
- D. Lee, T. Ozkaya-Ahmadov and A. F. Sarioglu, Small, 2023, 19, e2208035 CrossRef PubMed.
- S. Wang, Y. Zhu, Z. Zhou, Y. Luo, Y. Huang, Y. Liu and T. Xu, Adv. Sci., 2024, 11, e2406196 CrossRef.
- R. Wei, D. Wang, P. Zhou, Y. Pan, X. Wan, W. Pan, N. Li and B. Tang, Chem. Commun., 2024, 60, 7491–7494 RSC.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2025 |