 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
G-quadruplex DNA/iron cationic porphyrin catalyzes enantioselective carbon–silicon bond formation
Wenhui
Miao
ab,
Wenqin
Zhou
a,
Guoqing
Jia
*a and
Can
Li
 *ab
*ab
aState Key Laboratory of Catalysis, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, Liaoning, China. E-mail: canli@dicp.ac.cn
bUniversity of Chinese Academy of Sciences, Beijing 100049, China
First published on 11th November 2025
Abstract
Enantioselective carbon−silicon bond formation is achieved by using a DNA-based biocatalyst via self-assembly of a G-quadruplex DNA (2KYO) and iron cationic porphyrin (FeTMPyP4). Remarkably, single-site or multiple-site combined base mutations on the 2KYO sequence enable enantioselectivity modulation from 86% to −78% ee, delivering a neural network-like DNA catalyst development paradigm for stereocontrolled bioconversion.
DNA exhibits catalytic functions in vitro, constituting a class of molecules named DNAzymes.1,2 G-quadruplex DNA (G4), notable for its diverse and tunable 3D structures,3–10 can specifically interact with metal ions and other small molecules, and in turn facilitates catalytic transformations.11–19 Specifically, G4/hemin (Fe(III)-protoporphyrin IX) has been extended to catalyze the carbene transfer reaction, albeit with relatively low stereoselectivity.14 Previously, we engineered G4 complexed with cationic iron porphyrins such as Fe(III) meso-tetra (N-methyl-4-pyridyl) porphyrin (FeTMPyP4), achieving up to 90% enantiomeric excess (ee) in olefin cyclopropanation.15,20 These results confirmed that G4 scaffolds facilitate the formation of a reactive iron porphyrin carbene (IPC) intermediate and steer its subsequent transfer to substrate olefins during catalytic processes. Despite these successes, the types of chemical transformations accessible to G4/iron porphyrin catalysis remain limited. We now explore whether the G4/FeTMPyP4 system can be generalized to other IPC-mediated transformations.
Carbene-mediated carbon–silicon (C–Si) bond formation represents a powerful strategy for synthesizing organosilicon compounds. Synthetic chemists developed a series of chiral transition metal (Rh, Ir, and Cu) complexes catalyzing enantioselective C–Si bond formation.21–27 Distinct from these chemical methods, recent advance in protein engineering has unlocked a biological approach for this transformation.28
Herein, we report that the G4/FeTMPyP4 assembly catalyzes enantioselective C–Si bond formation, introducing DNA as a novel biological scaffold for this reaction. The 2KYO sequence, existing as a mixture of monomeric and dimeric G4, emerged as the optimal G4 scaffold. Upon base mutation screening cycles, we generated a diverse library of 2KYO scaffolds that complex with FeTMPyP4, enabling C–Si bond formation with tunable enantioselectivity ranging from 86% to −78% ee.
The benchmark reaction of C–Si bond formation uses dimethylphenylsilane (1a) as the silicon hydride substrate. The carbene source ethyl 2-diazopropanoate (2a) inserts into its Si–H bond. The G4/FeTMPyP4 is constructed via the non-covalent interaction between G4 and FeTMPyP4 (Scheme 1).
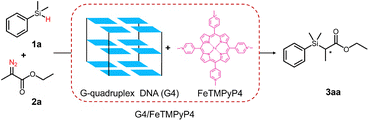 | ||
| Scheme 1 Schematic representation of the assembly formation of G4/FeTMPyP4 toward enantioselective C–Si bond formation. | ||
The catalytic ability of free FeTMPyP4 was first examined. FeTMPyP4 alone can catalyze the C–Si bond formation with a turnover number (TON) of 10, but gives the racemic product 3aa (Table 1, entry 1). Subsequently, a library of DNA sequences that can form G4 structures was selected from the Protein Data Bank (PDB; https://www.rcsb.org/) and complexed with FeTMPyP4 to construct G4/FeTMPyP4 (Table S1). Several G4/FeTMPyP4 promote the reaction in an enantioselective way, indicating that the enantioselectivity originates from the specific interactions within the G4/FeTMPyP4. However, no clear relationship between G4 secondary structure and enantioselectivity was observed (Fig. S1).
| Entry | G4 | Cofactor | ee%b | TON | TOF (h−1) |
|---|---|---|---|---|---|
| a Reaction conditions: 1a (20 mM), 2a (10 mM), G4 (50 µM), FeTMPyP4 (40 µM), Na2S2O4 (10 mM) and DMSO (5% V/V) in 50 mM potassium phosphate buffer (pH 7.0, [K+] = 100 mM) at 20 °C. TON and ee values are based on the areas of HPLC peaks as compared to those of ethyl 3-propionate as an internal standard. b (major)–(minor). | |||||
| 1 | — | FeTMPyP4 | 0 | 10 ± 5 | 0.4 ± 0.2 |
| 2 | 2KYO | FeTMPyP4 | 53 ± 7 | 28 ± 10 | 1.2 ± 0.5 |
| 3 | 2KYO-18 | FeTMPyP4 | −57 ± 1 | 23 ± 7 | 1.0 ± 0.3 |
| 4 | 2KYO-30 | FeTMPyP4 | 63 ± 5 | 68 ± 13 | 2.8 ± 0.6 |
| 5 | 2KYO-30-9 | FeTMPyP4 | 74 ± 6 | 35 ± 8 | 1.5 ± 0.4 |
2KYO (5′-CGGGCGGGCGCGAGGGAGGGT-3′), a modified DNA of wild-type c-kit2 (5′-CGGGCGGGCGCGAGGGAGGGG-3′),29 stands out from other sequences. The resulting 2KYO/FeTMPyP4 shows the highest ee (53%) (Fig. S1 and Table 1, entry 2). Therefore, 2KYO/FeTMPyP4 was selected as the platform for developing a G4-based C−Si bond formation catalyst.
2KYO/FeTMPyP4 was optimized via iterative DNA mutagenesis (Fig. S2), with non-G4 plane regions of 2KYO divided into five segments: 5′-flanking bases (FB5), 3′-flanking bases (FB3), the first loop (L1), the second loop (L2), and the Linker region. In the initial mutagenesis cycle (Fig. 1 and Table S2), adding a G at FB5 of 2KYO produced a mutant 2KYO-30, raising the enantioselectivity from 53% to 63% ee, and TON from 28 to 68 (Table 1, entry 4). Substituting the G at position 12 with C in the Linker generated mutant 2KYO-18 (C9-G10-C11-G12-A13 → C9-G10-C11-C12-A13), inducing complete ee reversal from 53% to −57% (Table 1, entry 3). In the second cycle (Table S3), substitution of C with T in L1 (2KYO-30-9) elevated the enantioselectivity from 63% to 74% ee (Table 1, entry 5). Further mutations on 2KYO-18 failed to enhance enantioselectivity (Table S4). The trajectory from 2KYO → 2KYO-30 → 2KYO-30-9 demonstrated progressive enantioselectivity enhancement (53% → 63% → 74% ee), while the 2KYO → 2KYO-18 achieved enantioselectivity reversal (53% → −57% ee). Additionally, G4 improved the catalytic efficiency by 2.3–6.8-fold vs. FeTMPyP4 alone.
Circular dichroism spectroscopy reveals a parallel G4 structure for 2KYO (Fig. 2a). Native polyacrylamide gel electrophoresis demonstrates the structural polymorphism of 2KYO (Fig. 2b and Fig. S3). These findings were further corroborated by imino proton NMR analysis (Fig. 2c), which further confirms the presence of a mixture of monomer and dimer, and this is in line with previous reports by Patel et al.29 By analyzing the peak areas of the characteristic peaks for the monomer (G2m and G14m) and dimer (G2d), we discovered that the content of the dimer is nearly 1.8 times that of the monomer. Thus, a mixture of monomeric and dimeric G4 models are proposed for 2KYO (Fig. 2d). Notably, the structure of 2KYO and the dimer/monomer ratio can vary with the conditions (Fig. S4–S6).
The multiple binding sites of 2KYO to FeTMPyP4 were observed from UV-visible titration and Job plot (Fig. S7 and S8). Therefore, as shown in Fig. 1, we explored the enantioselectivity-determining FeTMPyP4 binding sites by correlating 2KYO mutagenesis sites with the enantioselectivity of 3aa. Mutations in the FB5, FB3, L1, and L2 regions showed no significant effects on enantioselectivity. In contrast, specific mutations within the Linker region profoundly impacted enantioselectivity, even inducing inversion of the product configuration (Fig. 1 and Table S2). Notably, mutation of dA13 in the Linker region (2KYO-19, 2KYO-20, and 2KYO-21) nearly abolished the enantioselective control of 2KYO/FeTMPyP4, demonstrating the critical role of dA13 in enantiocontrol during catalysis. Mutational analyses identified the primary binding site of FeTMPyP4 within the Linker regions of the monomer and dimer 2KYO, forming the specific catalytic microenvironment.
However, it remains to be elucidated whether enantioselective control is mediated primarily by the monomeric or the dimeric form of 2KYO. Time-evolution imino NMR spectroscopy revealed a progressive structural reorganization of 2KYO from the monomer/dimer mixture to a dimer over 98 h, as evidenced by the diminishment of the monomer signatures (G6/G15 peaks) (Fig. 3a). Intriguingly, the ee value exhibited non-monotonic changes during this transition, decreasing initially before rising as dimerization completed (Fig. 3b). This unexpected correlation between structural transition and catalytic behavior remains under investigation.
 | ||
| Fig. 3 (a) Time-evolution imino NMR spectroscopy of 2KYO; (b) the relationship between the enantioselectivity of 3aa and incubation time. | ||
We then investigated the substrate scope of C–Si bond formation catalyzed by 2KYO-30-9/FeTMPyP4 and 2KYO-18/FeTMPyP4 (Table S5). We found that both 2KYO-30-9/FeTMPyP4 and 2KYO-18/FeTMPyP4 are highly sensitive to minor substrate modifications. 2KYO-30-9/FeTMPyP4 achieves peak enantioselectivity (86% ee) for Bn-modified aryl silanes, whereas 2KYO-18/FeTMPyP4 attains optimal performance (−78% ee) with t-Bu-modified diazoesters.
Besides optimizing highly enantioselective G4/FeTMPyP4 for C–Si bond formation, the above mutation process also delivers a neural network-like DNA catalyst development paradigm. Combinatorial base mutagenesis in five regions of 2KYO (FB5, FB3, L1, L2, and Linker) enables programmable enantioselectivity tuning through four distinct mutation pair patterns (Fig. 4): (1) synergistic enhancement (e.g., case 1): combining mutant 2KYO-30 (63% ee) and 2KYO-5 (32% ee) significantly improved ee to 74% (2KYO-30-9). (2) Antagonistic suppression (e.g., case 2): combining 2KYO-30 (63% ee) and 2KYO-14 (43% ee) resulted in lower ee (38%, 2KYO-30-12). (3) Additive interpolation: more commonly, combined ee fell between those of the single mutants. For example, combining 2KYO-30 (63% ee) and 2KYO-4 (46% ee) results in intermediate enantioselectivity of 55% ee (2KYO-30-8). (4) Enantio-inversion (e.g., case 7): the combination of two mutation sites with inverse enantioselectivity, 2KYO-7 (−11% ee) and 2KYO-11 (−15% ee), results in positive enantioselectivity of 16% ee. These combinatorial principles persist in triple mutants (Fig. S9). Expanding this mutational library may diversify enantioselectivity profiles, enabling neural networks to predict and control stereoselectivity via targeted mutations.
 | ||
| Fig. 4 A neural network-like DNA catalyst development paradigm, symbolizing the intricate relationship between the input 2-site combined mutation mutations of the DNA sequence and the resultant ee. | ||
In summary, this work achieves enantioselective C–Si bond formation using a DNA-based biocatalyst by non-covalent assembly of FeTMPyP4 with a mixture of monomer/dimer 2KYO. We demonstrate that enantiocontrol originates from the G4 scaffold, particularly bases in the Linker region. Through systematic optimization of 2KYO mutants and screening of several substrates, the enantioselectivities reached 86% and −78% ee. This establishes G4 topologies as programmable platforms for stereodivergent biocatalysis, and delivers a neural network-like DNA catalyst development paradigm.
Conflicts of interest
There are no conflicts to declare.Data availability
Experimental procedures and data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5cc04968g.Notes and references
- C. R. Geyer and D. Sen, Chem. Biol., 1997, 4, 579–593 CrossRef CAS.
- M. Yang, Y. Xie, L. Zhu, X. Li and W. Xu, ACS Catal., 2024, 14, 16392–16422 CrossRef CAS.
- M. L. Bochman, K. Paeschke and V. A. Zakian, Nat. Rev. Genet., 2012, 13, 770–780 CrossRef CAS.
- H. L. Lightfoot, T. Hagen, N. J. Tatum and J. Hall, FEBS Lett., 2019, 593, 2083–2102 CrossRef CAS.
- C. Wang, G. Xu, X. Liu, L. Jiang, X. Zhou, M. Liu and C. Li, J. Am. Chem. Soc., 2024, 146, 4741–4751 CrossRef CAS PubMed.
- M. Nishio, K. Tsukakoshi and K. Ikebukuro, Biosens. Bioelectron., 2021, 178, 113030 CrossRef CAS.
- L. Chen, J. Dickerhoff, S. Sakai and D. Yang, Acc. Chem. Res., 2022, 55, 2628–2646 CrossRef CAS PubMed.
- J. Dong, M. P. O'Hagan and I. Willner, Chem. Soc. Rev., 2022, 51, 7631–7661 RSC.
- J. T. Grun and H. Schwalbe, Biopolymers, 2022, 113, e23477 CrossRef.
- Y. Hashimoto, S. Shil, M. Tsuruta, K. Kawauchi and D. Miyoshi, RSC Chem. Biol., 2025, 6, 466–491 RSC.
- E. Golub, R. Freeman and I. Willner, Angew. Chem., Int. Ed., 2011, 50, 11710–11714 CrossRef CAS.
- C. H. Wang, G. Q. Jia, J. Zhou, Y. H. Li, Y. Liu, S. M. Lu and C. Li, Angew. Chem., Int. Ed., 2012, 51, 9352–9355 CrossRef CAS.
- C. H. Wang, Y. H. Li, G. Q. Jia, Y. Liu, S. M. Lu and C. Li, Chem. Commun., 2012, 48, 6232–6234 RSC.
- H. Ibrahim, P. Mulyk and D. Sen, ACS Omega, 2019, 4, 15280–15288 CrossRef CAS PubMed.
- J. Y. Hao, W. H. Miao, Y. Cheng, S. M. Lu, G. Q. Jia and C. Li, ACS Catal., 2020, 10, 6561–6567 CrossRef CAS.
- L. Hadian-Dehkordi, Z. Abdi, A. Zarei, H. Aghahosseini, Z. Mohammadi, A. Farokhi, T. M. Yazdely, S. H. Nouri, M. Hosseini, A. Ramazani, H. Zheng, K. H. Qadir, M. A. Hamad and A. Rezaei, Coord. Chem. Rev., 2024, 505, 215635 CrossRef CAS.
- Z. Wang, X. Dong, Y. Chen and C. Wang, ChemBioChem, 2025, 26, e202400909 CrossRef CAS.
- H. Yang, Y. Zhou and J. Liu, Inorg. Chem. Front., 2021, 8, 2183–2199 RSC.
- J. H. Yum, H. Sugiyama and S. Park, Chem. Rec., 2022, 22, e202100333 CrossRef CAS.
- J. Y. Hao, W. H. Miao, S. M. Lu, Y. Cheng, G. Q. Jia and C. Li, Chem. Sci., 2021, 12, 7918–7923 RSC.
- D. Chen, D.-X. Zhu and M.-H. Xu, J. Am. Chem. Soc., 2016, 138, 1498–1501 CrossRef CAS PubMed.
- S. Hyde, J. Veliks, B. Liégault, D. Grassi, M. Taillefer and V. Gouverneur, Angew. Chem., Int. Ed., 2016, 55, 3785–3789 CrossRef CAS PubMed.
- J. Wu and J. S. Panek, J. Org. Chem., 2011, 76, 9900–9918 CrossRef CAS PubMed.
- Y. Yasutomi, H. Suematsu and T. Katsuki, J. Am. Chem. Soc., 2010, 132, 4510–4511 CrossRef CAS PubMed.
- Y. Z. Zhang, S. F. Zhu, L. X. Wang and Q. L. Zhou, Angew. Chem., Int. Ed., 2008, 47, 8496–8498 CrossRef CAS PubMed.
- B. Su and J. F. Hartwig, Angew. Chem., Int. Ed., 2022, 61, e202113343 CrossRef CAS.
- R. J. Albadr, W. M. Taher, M. Alwan, M. J. Jawad, H. Mushtaq, B. M. Saadi, A. Smerat, M. Kazemi and R. Javahershenas, J. Inorg. Organomet. Polym. Mater., 2025, 35, 3197–3221 CrossRef CAS.
- S. B. J. Kan, R. D. Lewis, K. Chen and F. H. Arnold, Science, 2016, 354, 1048–1051 CrossRef CAS.
- V. Kuryavyi, A. T. Phan and D. J. Patel, Nucleic Acids Res., 2010, 38, 6757–6773 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2025 |


