Open Access Article
Open Access ArticleLigand-responsive ON–OFF anion transport using copper-caged acylhydrazone transporters
Umesh
Shivpuje
 ,
Manzoor
Ahmad†
,
Manzoor
Ahmad†
 ,
Naveen J.
Roy
,
Naveen J.
Roy
 and
Pinaki
Talukdar
and
Pinaki
Talukdar
 *
*
Department of Chemistry, Indian Institute of Science Education and Research Pune, Dr Homi Bhabha Road, Pashan, Pune 411008, Maharashtra, India. E-mail: ptalukdar@iiserpune.ac.in
First published on 6th November 2025
Abstract
Reversibly gated stimuli-responsive anion transport utilizing acylhydrazone-based transporters is reported. Acylhydrazone-copper complexation makes hydrazone protons unavailable for binding and transport, whilst decomplexation using Na2EDTA activates the ion transport.
Ion transport across bilayer membranes, facilitated by ion channels, carriers, and ion pumps, is vital for biological functions.1 Malfunctioning of these systems could result in severe diseases like cystic fibrosis (channelopathies).2,3 Artificial ion transport systems in the form of ion channels and ion carriers have emerged as an important class of compounds that promise therapeutic applications for treating channelopathies.4 Moreover, chloride transport by these systems is known to induce chloride-mediated apoptosis to the cancer cells, opening up a new avenue to combat cancer.5 However, controlled spatiotemporal activation is vital for target-specific applications, as classical transporters are known to affect healthy cells as well. Stimulus-responsive ion transport systems controlled by external stimuli, such as light, ligands, voltage, pH, enzymes, and redox switching, are rare.6 Examples include reversibly gated light-responsive systems based on azobenzene7 and stilbene,8 and irreversibly gated systems that provide effective ON–OFF control for ion transport. Responsive systems whose activity is controlled through the input of multiple stimuli remain rare. These provide better ON–OFF control compared to light-responsive ones, which exhibit background transport activity arising from incomplete photoconversion. Our lab has reported reversibly gated systems based on acylhydrazone9 and phenylhydrazone10 photoswitches. In 2020, a foldamer-based reversibly gated ion channel was reported with transmembrane ion transport controlled chemically using Cu(II) ion and EDTA.11 However, the use of complex foldamer molecules presents a challenge for the practical application of the system.
Herein, we report a similar reversibly gated ion transporter utilizing copper(II) and Na2EDTA as an external stimulus employing simpler acylhydrazone copper complexes. Acylhydrazone-based compounds are known to exhibit ion binding properties like heteroditopic ion pair binding. They also possess excellent ion transport properties. Acylhydrazone-based ligands, upon complexation with copper ion, are known to provide –N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–O–Cu linkage12 by removing hydrazone protons. We, therefore, envisaged that the complexation of carefully designed acylhydrazone-based ion transporters with copper would make hydrazone protons unavailable for anion binding and subsequent transport (Fig. 1). Decomplexation with Na2EDTA, which would remove the copper ions, would render anion binding and transmembrane ion transport activity by providing free hydrazone protons. Reversible control would be achieved using Na2EDTA and Cu(II) as an external stimulus. We chose isophthalic biscarboxamide hydrazone-based systems 1a–1d as active transporters, as similar molecules were found to be efficient ion transporters in our previous study.9 These transporters were expected to show an ion-pair transport because of an ion-pair binding in similar systems reported by Chmielewski and coworkers.13 However, only anion transport was observed, probably because the cation bound to the transporter would be exposed to the hydrophobic domains of the lipid membrane. Meanwhile, 2a was chosen as the protransporter. Incorporating different substituents to the amide moiety was expected to fine-tune the ion transport behavior of these systems by changing their lipophilicities.
C–O–Cu linkage12 by removing hydrazone protons. We, therefore, envisaged that the complexation of carefully designed acylhydrazone-based ion transporters with copper would make hydrazone protons unavailable for anion binding and subsequent transport (Fig. 1). Decomplexation with Na2EDTA, which would remove the copper ions, would render anion binding and transmembrane ion transport activity by providing free hydrazone protons. Reversible control would be achieved using Na2EDTA and Cu(II) as an external stimulus. We chose isophthalic biscarboxamide hydrazone-based systems 1a–1d as active transporters, as similar molecules were found to be efficient ion transporters in our previous study.9 These transporters were expected to show an ion-pair transport because of an ion-pair binding in similar systems reported by Chmielewski and coworkers.13 However, only anion transport was observed, probably because the cation bound to the transporter would be exposed to the hydrophobic domains of the lipid membrane. Meanwhile, 2a was chosen as the protransporter. Incorporating different substituents to the amide moiety was expected to fine-tune the ion transport behavior of these systems by changing their lipophilicities.
 | ||
| Fig. 1 Working principle of ligand-responsive ion transport (A). Molecular structures of transporters 1a–1d and their clogP values (B) and protransporter 2a, respectively (C). | ||
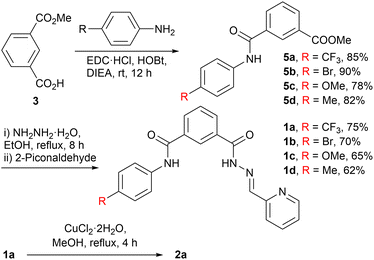
The active transporters 1a–1d were synthesized from mono-hydrolyzed ester 3, prepared from the reported literature.9 Subsequently, 3 was coupled with aromatic amines 4a–4d using EDC·HCl/HOBt to yield the amide derivatives 5a–5d. The reaction of 5a–5d with hydrazine hydrate, followed by coupling with 2-piconaldehyde, furnished the active transporters 1a–1d (Scheme 1). Further, compound 1b was recrystallized from methanol. The single crystal structure data are provided in the SI (Fig. S23 and S24).
Anion-binding affinities of 1a were evaluated through 1H NMR studies. Titration of 1a with tetrabutylammonium chloride (TBACl), bromide (TBABr), iodide (TBAI) and nitrate (TBANO3) salts in acetonitrile-d3 led to significant downfield chemical shifts of the protons H1, H2, H3, and H4, indicating their involvement in anion binding through amide-NH1⋯A−, ArC-H2⋯A−, hydrazone-NH3⋯A−, imine-CH4⋯A− hydrogen bonding interactions.
BindFit14 analysis furnished a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (receptor: anion) binding stoichiometry with association constant values (Ka(1:1)/Cl−) of (9.1 ± 0.7) × 104 M−1, Ka(1:1)/Br− = (3.47 ± 0.1) × 103 M−1, and Ka(1:1)/I− = 76.68 ± 4.9 M−1 and Ka(1:1)/NO3− = 95.13 ± 0.41 M−1, respectively (Fig. S3–S10). Further proof of chloride binding was obtained by high-resolution mass spectrometry (HRMS). Peaks at m/z = 447.0842 and 449.0818 corresponding to the [1a + Cl−] complex were obtained in the solution state (Fig. S11).
1 (receptor: anion) binding stoichiometry with association constant values (Ka(1:1)/Cl−) of (9.1 ± 0.7) × 104 M−1, Ka(1:1)/Br− = (3.47 ± 0.1) × 103 M−1, and Ka(1:1)/I− = 76.68 ± 4.9 M−1 and Ka(1:1)/NO3− = 95.13 ± 0.41 M−1, respectively (Fig. S3–S10). Further proof of chloride binding was obtained by high-resolution mass spectrometry (HRMS). Peaks at m/z = 447.0842 and 449.0818 corresponding to the [1a + Cl−] complex were obtained in the solution state (Fig. S11).
The ion transport activity of the compounds 1a–1d was evaluated across large unilamellar vesicles (LUVs).5 8-Hydroxy pyrene-1,3,6-trisulfonic acid trisodium salt (HPTS, 1 mM) was entrapped within the vesicles containing 100 mM of NaCl and 10 mM of HEPES buffer. A pH gradient (pHin = 7.0 and pHout = 7.8) was created across the lipid membrane by the addition of 0.5 M NaOH (20 µL) to the extravesicular solution. The subsequent addition of compounds 1a–1d resulted in the collapse of the pH gradient, monitored by recording the change in fluorescence intensity at λem = 510 nm (λex = 450 nm). The activity comparison yielded a transport activity sequence of 1a > 1b > 1c > 1d (Fig. 2A). Hill analysis of dose-dependent ion transport data furnished the EC50 values of 5.36 µM, 6.15 µM, and 46.31 µM for compounds 1a, 1b, and 1c, respectively (Fig. S13 and S14). Hill coefficients (n) of ∼1 indicated that ion transport across the lipid bilayer is mediated by a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 receptor
1 receptor![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) anion interaction, and this result corroborates 1H NMR titration studies. Hill analysis of 1d could not be done due to its precipitation at higher concentrations.
anion interaction, and this result corroborates 1H NMR titration studies. Hill analysis of 1d could not be done due to its precipitation at higher concentrations.
Subsequently, Cl− transport across EYPC–LUVs⊃lucigenin was monitored for the most active transporter 1a. Vesicles entrapping lucigenin dye were prepared in a 200 mM NaNO3 solution, and then a Cl−/NO3−gradient was created by adding NaCl (33.3 µL, 2.0 M) in the extravesicular buffer. Chloride influx was evaluated by monitoring the change in the fluorescence intensity (λex = 455 nm and λem = 535 nm) after the addition of 1a. The dose-dependent Cl− influx by 1a is shown in Fig. S17. Hill analysis furnished an EC50 value of 9.34 µM with a Hill coefficient (n) of ∼1 as obtained in the above-mentioned HPTS studies.
Mechanistically, ion transport in the lucigenin assay could occur through H+/Cl− symport, M+/Cl− symport, or Cl−/OH− antiport modes. However, varying anions in the external buffer using different NaX salts (X = Cl−, Br−, OAc−, NO3−, and ClO4−) strongly affect the ion transport (Fig. 2B), and variation of extravesicular MCl salts (M = Li+, Na+, K+, Rb+, and Cs+) in lucigenin-based studies did not change the ion transport (Fig. S19). These observations ruled out M+/Cl− symport and M+/H+ antiport, which indicated that ion transport could occur either through H+/Cl− symport or Cl−/OH− antiport mode. To get further insights, chloride efflux using 1a was monitored using a chloride ion selective electrode (ISE) in the presence and absence of either valinomycin (a highly selective K+ transporter) or monensin (an H+/K+ antiporter) with intravesicular KCl (300 mM) and extravesicular potassium gluconate (300 mM) solutions.15 A significant enhancement in the Cl− efflux rate was observed for 1a in the presence of valinomycin, while it remained nearly unchanged with monensin. This enhanced Cl− efflux in the presence of valinomycin provided evidence of transporter-mediated electrogenic transport. Selective K+ transport by valinomycin complements anion transport by 1a, suggesting an antiport mechanism of transport (i.e., Cl−/OH−). However, the lack of Cl− efflux enhancement with monensin rules out an H+/Cl− symport mechanism for 1a (Fig. 2C, D and Fig. S21).
The necessary evidence for a mobile carrier mechanism for ion transport was explored through experiments conducted in the liquid gel phase of dipalmitoylphosphatidylcholine (DPPC) large unilamellar vesicles (LUVs).16 Inactivity at 25 °C, and restoration of activity at 45 °C, which is above the gel–liquid phase transition temperature for DPPC (Tm = 41 °C), is indicative of a mobile carrier process rather than transport mediated by self-assembly into an ion channel, that activity of which would be typically expected to be independent of the lipid phase (Fig. S18). Based on the experimentally determined Hill coefficient value of n ∼ 1 the geometry-optimized structure of [1a + Cl−] complex was obtained first by generating the most probable conformation by using the CONFLEX 8 program17 (Fig. S25). Subsequently, the geometry optimization of the generated conformation was done by the Gaussian 09 program (see SI) using the B3LYP functional and 6-31G(d,p)18 basis set. The geometry-optimized structure confirmed that the receptor participates in the anion recognition through amide-NH1⋯Cl− (H⋯Cl− = 2.20 Å), hydrazone NH3⋯Cl (H⋯Cl− = 2.30 Å), imine CH4⋯Cl− (H⋯Cl− = 2.61 Å), and Ar-CH5⋯Cl− (H⋯Cl− = 2.86 Å) hydrogen bonding interactions (Fig. 2E and Fig. S27). The computed binding energy was found to be −45.02 kcal mol−1.
The copper complexed protransporter 2a was synthesized by refluxing the best active transporter 1a with copper chloride dihydrate in methanol (Scheme 1). The broad peaks observed in the 1H NMR spectra of compound 2a indicate paramagnetic characteristics, which were further confirmed by EPR spectra, revealing the presence of an unpaired electron consistent with a 3d9 configuration (Fig. S2). The lack of acyl hydrazone-NH3 proton in the 1H NMR spectrum of 2a, along with the (Cu2+) 3d9 configuration confirmation through EPR, suggests the bonding structure of 2a is –N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–O–Cu. Furthermore, to assess the rigidity of [Cu(1a)2], variable-temperature (VT) EPR measurements were conducted. The results showed no difference in the g value, indicating that [Cu(1a)2] remains stable from 100 K to 300 K.11 The decomplexation of copper–hydrazone complex 2a to form 1a using Na2EDTA was initially analyzed by 1H NMR spectroscopy. Na2EDTA solution in D2O was added to a sample of 2a in DMSO-d6, and the NMR spectrum was recorded. 1H NMR of the copper complex was observed to be broad due to the paramagnetic characteristics of the Cu-d9 system. However, the addition of Na2EDTA led to the appearance of NMR peaks, such as the hydrazine N–H3 at 12.25 ppm and the Ar C–H5 proton at 7.5 ppm. All these changes indicate the formation of active transporter 1a (Fig. S22). The decomplexation of compound 2a was additionally confirmed through UV-Vis absorption analyses. The absorbance observed at 370 nm for compound 2a (20 µM) in MeOH
C–O–Cu. Furthermore, to assess the rigidity of [Cu(1a)2], variable-temperature (VT) EPR measurements were conducted. The results showed no difference in the g value, indicating that [Cu(1a)2] remains stable from 100 K to 300 K.11 The decomplexation of copper–hydrazone complex 2a to form 1a using Na2EDTA was initially analyzed by 1H NMR spectroscopy. Na2EDTA solution in D2O was added to a sample of 2a in DMSO-d6, and the NMR spectrum was recorded. 1H NMR of the copper complex was observed to be broad due to the paramagnetic characteristics of the Cu-d9 system. However, the addition of Na2EDTA led to the appearance of NMR peaks, such as the hydrazine N–H3 at 12.25 ppm and the Ar C–H5 proton at 7.5 ppm. All these changes indicate the formation of active transporter 1a (Fig. S22). The decomplexation of compound 2a was additionally confirmed through UV-Vis absorption analyses. The absorbance observed at 370 nm for compound 2a (20 µM) in MeOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (9
H2O (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) solvent may be due to a charge transfer (CT) transition.19 The stepwise addition of Na2EDTA to 2a led to a hypochromic shift at 370 nm and a hyperchromic shift seen at 297 nm, indicative of the formation of 1a from 2a which was validated by the absorbance measurement of compound 1a in the same solvent at a concentration of 37 µM (Fig. 3A). Approximately 1.4 equivalents of Na2EDTA were required to fully decomplex compound 2a during this titration. This analysis also revealed a rapid decoupling of the copper hydrazone complex upon addition of Na2EDTA. The quick disappearance of the 370 nm peak associated with copper ligand interaction confirmed the efficacy of the ligand exchange process. After analyzing the decomplexation of 2a in the solution phase, responsive ion transport studies were performed in HPTS-based vesicular experiments, as mentioned earlier. Compound 2a at 10 µM concentration did not show any ion transport activity, while transporter 1a at the same concentration displayed maximal activity. The significant improvement in fluorescent intensity after decomplexation of 2a with Na2EDTA, suggests the formation of 1a (Fig. 3B). Additionally, reversible ON–OFF ion transport assay was conducted by utilizing Na2EDTA and copper as external triggers (Fig. 3C). When Na2EDTA (1.4 eq.) was added to the solution of 2a (20 µM in DMSO) to chelate Cu(II), it resulted in a marked enhancement in ion transport, while the addition of copper chloride dihydrate (1.1 eq.) to the activated solution halted the ion transport activity. This ON–OFF ion transport behavior using two distinct stimuli was successfully maintained across multiple cycles with minimal loss in performance (Fig. 3D).
1) solvent may be due to a charge transfer (CT) transition.19 The stepwise addition of Na2EDTA to 2a led to a hypochromic shift at 370 nm and a hyperchromic shift seen at 297 nm, indicative of the formation of 1a from 2a which was validated by the absorbance measurement of compound 1a in the same solvent at a concentration of 37 µM (Fig. 3A). Approximately 1.4 equivalents of Na2EDTA were required to fully decomplex compound 2a during this titration. This analysis also revealed a rapid decoupling of the copper hydrazone complex upon addition of Na2EDTA. The quick disappearance of the 370 nm peak associated with copper ligand interaction confirmed the efficacy of the ligand exchange process. After analyzing the decomplexation of 2a in the solution phase, responsive ion transport studies were performed in HPTS-based vesicular experiments, as mentioned earlier. Compound 2a at 10 µM concentration did not show any ion transport activity, while transporter 1a at the same concentration displayed maximal activity. The significant improvement in fluorescent intensity after decomplexation of 2a with Na2EDTA, suggests the formation of 1a (Fig. 3B). Additionally, reversible ON–OFF ion transport assay was conducted by utilizing Na2EDTA and copper as external triggers (Fig. 3C). When Na2EDTA (1.4 eq.) was added to the solution of 2a (20 µM in DMSO) to chelate Cu(II), it resulted in a marked enhancement in ion transport, while the addition of copper chloride dihydrate (1.1 eq.) to the activated solution halted the ion transport activity. This ON–OFF ion transport behavior using two distinct stimuli was successfully maintained across multiple cycles with minimal loss in performance (Fig. 3D).
In conclusion, we have developed reversibly-gated stimuli-responsive anion transporters based on acylhydrazones frameworks, exemplified by compound 1a. Coordination of 1a with Cu(II) ions affords a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometric complex 2a, which remains inactive towards anion transport owing to the sequestration of the hydrazone proton essential for anion recognition. Subsequent decomplexation with Na2EDTA restores the free hydrazone form 1a, thereby reinstating its anion transport activity. Reversible ON–OFF switching of ion transport was reproducibly achieved over multiple cycles by the alternate addition of Cu(II) ions and Na2EDTA as external chemical stimuli. This study highlights the utility of metal–ligand coordination as a dynamic regulatory element in the design of stimuli-responsive molecular systems capable of controlled chloride transport. The present strategy is anticipated to be transferable to congeners 1b–1d and other appropriately designed ligands, providing a versatile platform for the development of bioinspired materials with potential therapeutic relevance.
1 stoichiometric complex 2a, which remains inactive towards anion transport owing to the sequestration of the hydrazone proton essential for anion recognition. Subsequent decomplexation with Na2EDTA restores the free hydrazone form 1a, thereby reinstating its anion transport activity. Reversible ON–OFF switching of ion transport was reproducibly achieved over multiple cycles by the alternate addition of Cu(II) ions and Na2EDTA as external chemical stimuli. This study highlights the utility of metal–ligand coordination as a dynamic regulatory element in the design of stimuli-responsive molecular systems capable of controlled chloride transport. The present strategy is anticipated to be transferable to congeners 1b–1d and other appropriately designed ligands, providing a versatile platform for the development of bioinspired materials with potential therapeutic relevance.
PT acknowledges the financial support from the Anusandhan National Research Foundation (ANRF) (Project No. CRG/2022/001640) and the Indian Institute of Science Education and Research (IISER), Pune. US thanks the University Grants Commission (UGC), Govt of India, for a fellowship (UGC-NFOBC). MA thanks the University Grants Commission (UGC), Govt of India, for a fellowship. NR acknowledges the Council of Scientific and Industrial Research (CSIR), Govt of India, for a research fellowship.
Conflicts of interest
There are no conflicts to declare.Data availability
All supporting data for this article are provided in the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5cc04105h.CCDC 2473653 (1b) contains the supplementary crystallographic data for this paper.20
Notes and references
- T. J. Jentsch, C. A. Hübner and J. C. Fuhrmann, Nat. Cell Biol., 2004, 6, 1039–1047 CrossRef CAS PubMed.
- D. C. Gadsby, P. Vergani and L. Csanády, Nature, 2006, 440, 477–483 CrossRef CAS PubMed.
- G. Bernard and M. I. Shevell, Pediatr. Neurol., 2008, 38, 73–85 CrossRef PubMed.
- R. Planells-Cases and T. J. Jentsch, Biochim. Biophys. Acta, Mol. Basis Dis., 2009, 1792, 173–189 CrossRef CAS PubMed.
- T. Saha, M. S. Hossain, D. Saha, M. Lahiri and P. Talukdar, J. Am. Chem. Soc., 2016, 138, 7558–7567 CrossRef CAS PubMed.
- S. Chattopadhayay and P. Talukdar, Curr. Opin. Chem. Biol., 2025, 85, 102582 CrossRef CAS PubMed.
- W. Szymański, J. M. Beierle, H. A. V. Kistemaker, W. A. Velema and B. L. Feringa, Chem. Rev., 2013, 113, 6114–6178 CrossRef.
- S. J. Wezenberg, L.-J. Chen, J. E. Bos, B. L. Feringa, E. N. W. Howe, X. Wu, M. A. Siegler and P. A. Gale, J. Am. Chem. Soc., 2022, 144, 331–338 CrossRef CAS PubMed.
- M. Ahmad, S. Chattopadhayay, D. Mondal, T. Vijayakanth and P. Talukdar, Org. Lett., 2021, 23, 7319–7324 CrossRef CAS PubMed.
- M. Ahmad, D. Mondal, N. J. Roy, T. Vijayakanth and P. Talukdar, ChemPhotoChem, 2022, 6, e202200002 CrossRef CAS.
- A. D. Peters, S. Borsley, F. della Sala, D. F. Cairns-Gibson, M. Leonidou, J. Clayden, G. F. S. Whitehead, I. J. Vitórica-Yrezábal, E. Takano, J. Burthem, S. L. Cockroft and S. J. Webb, Chem. Sci., 2020, 11, 7023–7030 RSC.
- S. Mondal, S. Naskar, A. K. Dey, E. Sinn, C. Eribal, S. R. Herron and S. K. Chattopadhyay, Inorg. Chim. Acta, 2013, 398, 98–105 CrossRef CAS.
- Z. Kokan and M. J. Chmielewski, J. Am. Chem. Soc., 2018, 140, 16010–16014 CrossRef CAS PubMed.
- P. Thordarson, Chem. Soc. Rev., 2011, 40, 1305–1323 RSC.
- L. Rose and A. T. A. Jenkins, Bioelectrochemistry, 2007, 70, 387–393 CrossRef CAS.
- A. Kerckhoffs and M. J. Langton, Chem. Sci., 2020, 11, 6325–6331 RSC.
- (a) H. Goto and E. Osawa, J. Am. Chem. Soc., 1889, 111, 8950–8951 CrossRef; (b) H. Goto, S. Obata, N. Nakayama and K. Ohta, CONFLEX 8, CONFLEX Corporation, Tokyo, Japan, 2017 Search PubMed.
- A. D. Becke, J. Chem. Phys., 1993, 98, 5648–5652 CrossRef CAS.
- A. K. Patel, R. N. Jadeja, H. Roy, R. N. Patel, S. K. Patel, R. J. Butcher, M. Cortijo and S. Herrero, Polyhedron, 2020, 186, 114624 CrossRef CAS.
- CCDC 2473653: Experimental Crystal Structure Determination, 2022 DOI:10.5517/ccdc.csd.cc2p1182.
Footnote |
| † Present address: Chemistry Research Laboratory, Mansfield Road, Oxford, OX1 3TA, UK. |
| This journal is © The Royal Society of Chemistry 2025 |