 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Towards a hydrolysis stable artificial base pair with C-glycosides†
Robert
Dörrenhaus
 ,
Philip K.
Wagner
and
Stephanie
Kath-Schorr
,
Philip K.
Wagner
and
Stephanie
Kath-Schorr
 *
*
Department of Chemistry and Biochemistry, Institute of Organic Chemistry, 50939 Cologne, Germany. E-mail: skathsch@uni-koeln.de
First published on 13th June 2025
Abstract
Artificial base-pairs expand the genetic alphabet and allow for site-specific labeling, for example with fluorophores or spin labels, thereby enhancing the capabilities of biomolecular sensing. We developed CTPT3, a stable C-nucleoside pairing with NaM to form a hydrophobic base pair, and demonstrate its enzymatic incorporation into DNA/RNA and facile modification for labeling.
Interest in expanding the genetic alphabet has grown since its initial proposal in 1962.1 In the 1990s, the first unnatural base pairs (UBPs) were developed and successfully incorporated into DNA and RNA by polymerases.2 Beyond modified natural nucleobases, entirely novel nucleosides have attracted attention, as with different shape and different attraction forces, compared to the natural hydrogen bonding (H-bonding) moiety, a more selective incorporation of these artificial nucleobases was achieved and it was demonstrated, that H-bonds are no requirement for replication.2f,3 In hydrophobic base pairs, neighbouring interactions and π–π stacking play key roles,4 further stabilized by electrostatic effects, van der Waals forces and solvophobic effects.5 The TPT3–NaM base pair developed by the Romesberg group presents a well-studied hydrophobic UBP.6 Its optimization enabled the development of semisynthetic organisms with an expanded genetic alphabet.7 TPT3 has been functionalized with fluorophores and spin labels, facilitating site-specific labeling and structural analysis of long RNAs.8 Aside from high replication fidelity, chemical stability is essential for UBPs. While canonical nucleosides form N-glycosidic bonds, some modified nucleosides, such as pseudouridine (Ψ) or bacterial Formicin A, feature more stable C-glycosidic linkages.9 Artificial nucleosides are usually incorporated into DNA or RNA in few copies, making enhanced stability particularly important. As an N-glycoside, TPT3 can be susceptible to acid- or enzyme-catalyzed cleavage during synthesis or purification.10 Although C-glycosidic bonds offer significantly enhanced hydrolytic stability, their formation remains synthetically challenging due to low yields and limited substrate scope.11 A common, yet low-yielding and laborious method involves aryl lithium addition to sugar lactones. Here, protected lactones form hemiketals with C-nucleophiles and are reduced by silanes and Lewis acids via hydride transfer.12 This method has been applied to the synthesis of dNaM and its derivatives, such as PTMO.7,8 More recently, efforts have shifted toward more versatile strategies, including classical C–C cross-coupling (CC) reactions, to improve yields and expand substrate scope.11,13
Recent studies have explored a wide range of metal ions, C1′ modifications, and reductive pathways for C1′ activation, often via the C1′ glycal radical and using bench-stable glycal donors. Accordingly, we investigated such CC reactions to synthesize the novel C-glycosides nucleobase CTPT3 and its ribonucleoside rCTPT3 (Fig. 1).
To assess whether the design of CTPT3 can replicate the electrostatic characteristics of TPT3, we computed the electrostatic surface potentials of both TPT3 and CTPT3. N → C and C → N substitutions in CTPT3 shift the electrostatic potential compared to TPT3. However, these changes occur on the face of the nucleobase oriented away from NaM, unlikely to impact base-pairing interactions significantly. On the NaM-facing side, the carbonyl oxygen of CTPT3 exhibits a higher surface electrostatic potential compared to the thiocarbonyl sulfur in TPT3, with this elevated potential also influencing adjacent carbon atoms. Overall, their electrostatic profiles are similar, suggesting, that CTPT3 binds to NaM similarly to TPT3.
This study optimized Ni-catalyzed CCs to enhance rNaM (Scheme 1 VI) and enable rCTPT3 synthesis (Scheme 1 II), applied a Heck-type reaction for dCTPT3 (Scheme 1 III) and evaluated both in enzymatic triphosphate (TP) synthesis and polymerase assays. We thus present a hydrolysis-resistant C-glycoside complementary to dNaM to expand nucleic acid chemical tools.
CTPT3 nucleobase 3a was first synthesized from thieno[2,3-b]pyridin-7-ol by acid catalysed bromination with NBS (Scheme 1 I). The intermediate was either N-methylated using iodomethane or functionalized with a (CH2)4NBoc linker via its iodinated substrate, yielding 3a and 3b in near-quantitative yields. N-Functionalisation stabilizes the ketone in the keto-tautomeric form. Moreover, as demonstrated in our experiments, the following cross-coupling reactions do not proceed effectively without N-methylation. Coupling of the CTPT3 nucleobase to ribose was challenging as classical methods, such as using aryl lithium with protected lactones, did not yield the desired product, regardless of whether the methylated or non-methylated nucleobase analogue was used. Building on our previous progress with Ni-catalysed CC14 with the rAc ribofuranose15 for rNaM synthesis (VI), we tested this approach with the iodinated CTPT3 nucleobase, but only obtained side product formation induced by Zn.16
The route was changed to diastereoselective photo-reductive Ni-catalysed CC with the rDHP sugar moiety (Scheme 1 II).17 The rDHP sugar was synthesized from ribose by acid catalysed acetonide protection of the 2′ and 3′ OH, followed by benzoyl protection of the 5′ OH. Dihydropyridine was synthesized from ethyl-3-aminocrotonate and glyoxylic acid and coupled to the ribose 1′-OH by amide coupling with N,N′-diisopropylcarbodiimide (see ESI,† Section 2). The Ni-catalysed CC follows a photoreductive mechanism, where the glycosyl ester is oxidized by an activated photosensitizer (4CzIPN), forming a radical after deprotonation. Through a series of single-electron transfers, a Hantzsch pyridine and an alcoxycarbonyl radical are generated. An unstable C1′ radical forms via CO2 loss and participates in a Ni-catalysed CC with a Ni(II) species formed by oxidative addition of the Br-CTPT3 nucleobase 3a,17 producing the protected β-rCTPT3 nucleoside 4a in 17% yield. Deprotection using p-toluenesulfonic acid to remove the acetonide group, followed by nucleophilic cleavage of the benzoyl group with NaOMe, gave the free nucleoside in 95% yield over two steps. The overall yield from thieno[2,3-b]pyridin-7-ol to the rCTPT3 nucleoside was 14% with the CC step being yield-limiting. Compared to rTPT3, which has an overall nucleoside yield of ∼3%,6 this represents a significant improvement, making the route promising for further research and application. Further optimization of the CC step may improve yields. To assess enzymatic compatibility, the nucleoside was subjected to TP synthesis using enzymes typically applied to natural or modified nucleosides, aiming to simplify the triphosphorylation process and avoid low-yielding chemical routes. With support by BioNukleo (Berlin), an enzymatic synthesis of rCTPT3 TP 5 was achieved.
The synthesis of DNA nucleotide dTPT3 TP 7 (Scheme 1 III) originated from the Br-CTPT3 nucleobase 3a used for rCTPT3 and a TMS-protected glycal (Scheme 1 III). The dgly was synthesized in one step from thymidine by silylation of the hydroxy groups and nucleobase elimination in 85% yield.18 A subsequent Heck-reaction19 and selective C3-ketone reduction with with NaBH(OAc)320 afforded the β-nucleoside in 6% yield over two steps. The dCTPT3 nucleoside was enzymatically converted to TP 7 in 7.5% yield. Thus, both rCTPT3 and dCTPT3 are compatible with enzymatic TP synthesis—an encouraging result, as enzymatic acceptance offers here a more efficient and higher-yielding alternative to traditional chemical methods.
rCTPT3 was incorporated into RNA via in vitro transcription (Fig. 2a and c) using T7 RNA polymerase and dNaM-containing DNA template (DNA1_dNaM). The resulting RNA was purified and analysed by LC-MS and denaturing PAGE confirming the presence of full-length products (see ESI,† Section S7 and Fig. 2a). Full-length RNA with rCTPT3 incorporated opposite dNaM with an additional non-templated rCTPT3 at the end of the sequence (sodium adduct: 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 321 g mol−1; calculated 10
321 g mol−1; calculated 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 321 g mol−1; double sodium adduct 10
321 g mol−1; double sodium adduct 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 343 g mol−1; calculated 10
343 g mol−1; calculated 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 344 g mol−1) was observed. The addition of non-templated nucleotides by T7 RNA polymerase is well-documented,21 and was also found in control samples with unmodified templates and classical rTPT3 (ESI,† Section S7). With rTPT3 also a tendency of adding non-templated rTPT3 at the 3′ end of the product has been observed previously.22In vitro transcriptions of Spinach_dNaM DNA template were conducted to produce a functional Spinach RNA aptamer (ESI,† Section S7.3) with one rCTPT3 modification. In gel staining with DFHBI confirms correct folding of the full length rCTPT3 modified Spinach aptamer (ESI,† S7.3).
344 g mol−1) was observed. The addition of non-templated nucleotides by T7 RNA polymerase is well-documented,21 and was also found in control samples with unmodified templates and classical rTPT3 (ESI,† Section S7). With rTPT3 also a tendency of adding non-templated rTPT3 at the 3′ end of the product has been observed previously.22In vitro transcriptions of Spinach_dNaM DNA template were conducted to produce a functional Spinach RNA aptamer (ESI,† Section S7.3) with one rCTPT3 modification. In gel staining with DFHBI confirms correct folding of the full length rCTPT3 modified Spinach aptamer (ESI,† S7.3).
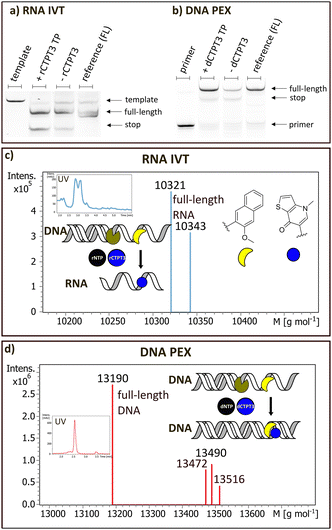 | ||
Fig. 2 Enzymatic incorporation of rCTPT3 TP and dCTPT3 TP. (a) Denaturing PAGE of in vitro transcription by T7 RNA Pol with rCTPT3-TP. Lanes left to right: template; NaM template + rCTPT3-TP; NaM template without rCTPT3-TP; natural template + NTPs. (b) Denaturing PAGE of DNA primer extension by KlenTaq. Lanes left to right: DNA primer; NaM template + dCTPT3-TP; NaM template without dCTPT3-TP; (4) unmodified template + dNTPs. (c) Deconvoluted ESI mass spectrum of purified in IVT using DNA1_dNaM template and T7 RNA polymerase giving rCTPT3-full length product with additional non-templated rCTPT3 addition at the 3′-end (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 321 g mol−1; calculated 10 321 g mol−1; calculated 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 321 g mol−1) and the 2× sodium adduct of this sequence (10 321 g mol−1) and the 2× sodium adduct of this sequence (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 343 g mol−1; calculated 10 343 g mol−1; calculated 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 344 g mol−1). (d) Deconvoluted ESI mass spectrum of purified primer extension assay using template DNA2_dNaM and KlenTaq DNA polymerase resulting in dCTPT3-full length product (13 344 g mol−1). (d) Deconvoluted ESI mass spectrum of purified primer extension assay using template DNA2_dNaM and KlenTaq DNA polymerase resulting in dCTPT3-full length product (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 190 g mol−1; calculated 13 190 g mol−1; calculated 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 191 g mol−1). Further, mismatches of dNaM/dA plus non-templated dA (13 191 g mol−1). Further, mismatches of dNaM/dA plus non-templated dA (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 472 g mol−1; calculated 13 472 g mol−1; calculated 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 474 g mol−1), dNaM/dA plus non-templated dG (13 474 g mol−1), dNaM/dA plus non-templated dG (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 490 g mol−1; calculated 13 490 g mol−1; calculated 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 490) and the sodium adduct of full-length product with non-templated dT (13 490) and the sodium adduct of full-length product with non-templated dT (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 516 g mol−1; calculated 13 516 g mol−1; calculated 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 518 g mol−1) are observed. Raw data and supplementary data are available in the ESI† (Section S6). 518 g mol−1) are observed. Raw data and supplementary data are available in the ESI† (Section S6). | ||
We also investigated DNA primer extension with dCTPT3 TP, dNaM-containing template (DNA2_dNaM) and KlenTaq DNA polymerase (Fig. 2b and d). Equivalent primer extensions were additionally performed with classical dTPT3 TP (see ESI† Section S7.2). Analysis in PAGE showed full length products as well as minor amounts of termination products (Fig. 2b). LC-MS analysis confirmed the full-length product with incorporated dCTPT3 (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 190 g mol−1; calculated 13
190 g mol−1; calculated 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 191 g mol−1) and its sodium adduct with a non-templated dT (13
191 g mol−1) and its sodium adduct with a non-templated dT (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 516 g mol−1; calculated 13
516 g mol−1; calculated 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 518 g mol−1). Additional products included dNaM extensions with natural nucleosides dA mismatches and non-templated dA (13
518 g mol−1). Additional products included dNaM extensions with natural nucleosides dA mismatches and non-templated dA (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 472 g mol−1; calculated 13
472 g mol−1; calculated 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 474 g mol−1), as well as full-length fragments of mismatch dNaM/dA plus non-templated dG (13
474 g mol−1), as well as full-length fragments of mismatch dNaM/dA plus non-templated dG (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 490 g mol−1; calculated 13
490 g mol−1; calculated 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 490) were found (see ESI† Section S7.1). Similar mismatches between natural nucleobases and unnatural bases have been previously reported.23 These findings align with the in vitro transcription data and confirm that standard DNA polymerases can incorporate CTPT3 TPs into full-length DNA strands (DNA template, primer and promoter sequences: ESI† Section S5).
490) were found (see ESI† Section S7.1). Similar mismatches between natural nucleobases and unnatural bases have been previously reported.23 These findings align with the in vitro transcription data and confirm that standard DNA polymerases can incorporate CTPT3 TPs into full-length DNA strands (DNA template, primer and promoter sequences: ESI† Section S5).
To evaluate the enhanced acid stability conferred by the C-glycosidic bond, nucleotide rCTPT3 TP was compared with rTPT3 TP under acidic conditions (pH 5, 80 °C, 5 h) (Fig. 3). Prior studies by us have shown that TPT3 derivatives are prone to base loss during dPAGE purification and are susceptible to acid cleavage due to their N-glycosidic linkage.10a–e
At pH 5, both TPs hydrolysed to the monophosphate as expected (Fig. 3). However, for rTPT3 nucleobase fragments were found, indicating cleavage of the N-glycosidic bond. These fragments are not observed in the MS fragmentation pattern of the triphosphate prior to acidic incubation, which confirms that they are generated by degradation under acidic conditions (see ESI† Section S7.4). Quantitative analysis of the UV chromatogram revealed that 11% of TPT3 underwent hydrolysis under the applied conditions (see ESI† Section S7.4). In contrast, rCTPT3 remained intact, confirming its superior stability due to the C-glycosidic linkage.
In summary, we developed efficient synthetic routes to the stable, non-hydrolysable C-glycosides dCTPT3 and rCTPT3 via Heck and Ni-catalysed CC reactions. CTPT3 nucleobase was functionalized and demonstrated compatibility with CC conditions. Both nucleosides were successfully converted into their TPs via enzymatic synthesis, offering moderate to high yields and a simplified alternative to traditional chemical methods. The TPs were enzymatically incorporated into full-length products using in vitro transcription (rCTPT3) and primer extension (dCTPT3) with dNaM-containing templates, confirming polymerase acceptance. While the C-glycosidic linkage provides enhanced stability and modifiability, challenges such as the achievement of a nature-like fidelity for enzymatic amplification of the UBP remain. Our work expands the nucleic acid chemistry toolbox by introducing a robust and versatile C-glycosidic artificial nucleobase as part of the unnatural base pair CTPT3-NaM.
The authors would like to thank Dr Christina Wartmann, Dr Niklas Hermes and Lukas Neu for fruitful discussions. Further, the authors want to thank Sawar Aziz, Moritz Steiner and Jessica Kubis for experimental support. A special thanks to Dr Hannah Depmeier for advice in biochemical methods.
Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the ESI.†Notes and references
- A. Rich, Horizons in biochemistry, Academic Press, New York, 1962 Search PubMed.
- (a) C. Switzer, S. E. Moroney and S. A. Benner, J. Am. Chem. Soc., 1989, 111(21), 8322–8323 CrossRef CAS; (b) J. Horlacher, M. Hottiger, V. N. Podust, U. Hübscher and S. A. Benner, Proc. Natl. Acad. Sci. U. S. A., 1995, 92(14), 6329–6333 CrossRef CAS PubMed; (c) T. Mitsui, A. Kitamura, M. Kimoto, T. To, A. Sato, I. Hirao and S. Yokoyama, J. Am. Chem. Soc., 2003, 125(18), 5298–5307 CrossRef CAS PubMed; (d) T. Ohtsuki, M. Kimoto, M. Ishikawa, T. Mitsui, I. Hirao and S. Yokoyama, Proc. Natl. Acad. Sci. U. S. A., 2001, 98(9), 4922–4925 CrossRef CAS PubMed; (e) I. Hirao, Y. Harada, M. Kimoto, T. Mitsui, T. Fujiwara and S. Yokoyama, J. Am. Chem. Soc., 2004, 126(41), 13298–13305 CrossRef CAS PubMed; (f) T. J. Matray and E. T. Kool, J. Am. Chem. Soc., 1998, 120(24), 6191–6192 CrossRef CAS PubMed; (g) J. C. Morales and E. T. Kool, J. Am. Chem. Soc., 1999, 121(10), 2323 CrossRef CAS PubMed; (h) I. Hirao, T. Mitsui, M. Kimoto, R. Kawai, A. Sato and S. Yokoyama, Nucleic Acids Symp. Ser., 2005, 49, 33–34 CrossRef PubMed; (i) J. A. Piccirilli, S. A. Benner, T. Krauch, S. E. Moroney and S. A. Benner, Nature, 1990, 343(6253), 33–37 CrossRef CAS PubMed.
- (a) B. A. Schweitzer and E. T. Kool, J. Am. Chem. Soc., 1995, 117(7), 1863–1872 CrossRef CAS PubMed; (b) A. M. Leconte, G. T. Hwang, S. Matsuda, P. Capek, Y. Hari and F. E. Romesberg, J. Am. Chem. Soc., 2008, 130(7), 2336–2343 CrossRef CAS PubMed; (c) Y. J. Seo, G. T. Hwang, P. Ordoukhanian and F. E. Romesberg, J. Am. Chem. Soc., 2009, 131(9), 3246–3252 CrossRef CAS PubMed; (d) D. L. McMinn, A. K. Ogawa, Y. Wu, J. Liu, P. G. Schultz and F. E. Romesberg, J. Am. Chem. Soc., 1999, 121(49), 11585–11586 CrossRef CAS; (e) I. Hirao, M. Kimoto, T. Mitsui, T. Fujiwara, R. Kawai, A. Sato, Y. Harada and S. Yokoyama, Nat. Methods, 2006, 3(9), 729–735 CrossRef CAS PubMed; (f) I. Hirao, T. Mitsui, M. Kimoto and S. Yokoyama, J. Am. Chem. Soc., 2007, 129(50), 15549–15555 CrossRef CAS PubMed.
- (a) K. J. Breslauer, R. Frank, H. Blöcker and L. A. Marky, Proc. Natl. Acad. Sci. U. S. A., 1986, 83(11), 3746–3750 CrossRef CAS PubMed; (b) C. A. Hunter and J. K. M. Sanders, J. Am. Chem. Soc., 1990, 112(14), 5525–5534 CrossRef CAS; (c) M. Petersheim and D. H. Turner, Biochemicals, 1983, 22(2), 256–263 CAS.
- (a) V. R. Cooper, T. Thonhauser and D. C. Langreth, J. Chem. Phys., 2008, 128(20), 204102 CrossRef PubMed; (b) C. D. M. Churchill and S. D. Wetmore, J. Phys. Chem. B, 2009, 113(49), 16046–16058 CrossRef CAS PubMed; (c) C. A. Hunter, J. Mol. Biol., 1993, 230(3), 1025–1054 CrossRef CAS PubMed; (d) J. w Hwang, P. Li and K. D. Shimizu, Org. Biomol. Chem., 2017, 15(7), 1554–1564 RSC; (e) S. Cui, J. Yu, F. Kühner, K. Schulten and H. E. Gaub, J. Am. Chem. Soc., 2007, 129(47), 14710–14716 CrossRef CAS PubMed; (f) K. M. Guckian, B. A. Schweitzer, R. X. F. Ren, C. J. Sheils, D. C. Tahmassebi and E. T. Kool, J. Am. Chem. Soc., 2000, 122(10), 2213–2222 CrossRef CAS PubMed.
- L. Li, M. Degardin, T. Lavergne, D. A. Malyshev, K. Dhami, P. Ordoukhanian and F. E. Romesberg, J. Am. Chem. Soc., 2014, 136(3), 826–829 CrossRef CAS PubMed.
- V. T. Dien, M. Holcomb, A. W. Feldman, E. C. Fischer, T. J. Dwyer and F. E. Romesberg, J. Am. Chem. Soc., 2018, 140(47), 16115–16123 CrossRef CAS PubMed.
- (a) T. Lavergne, R. Lamichhane, D. A. Malyshev, Z. Li, L. Li, E. Sperling, J. R. Williamson, D. P. Millar and F. E. Romesberg, ACS Chem. Biol., 2016, 11(5), 1347–1353 CrossRef CAS PubMed; (b) F. Eggert, K. Kulikov, C. Domnick, P. Leifels and S. Kath-Schorr, Methods, 2017, 120, 17–27 CrossRef CAS PubMed; (c) C. Domnick, F. Eggert, C. Wuebben, L. Bornewasser, G. Hagelueken, O. Schiemann and S. Kath-Schorr, Angew. Chem., Int. Ed., 2020, 59(20), 7891–7896 CrossRef CAS PubMed; (d) Y. Wang, Y. Chen, Y. Hu and X. Fang, Proc. Natl. Acad. Sci. U. S. A., 2020, 117(37), 22823–22832 CrossRef CAS PubMed; (e) Y. Hu, Y. Wang, J. Singh, R. Sun, L. Xu, X. Niu, K. Huang, G. Bai, G. Liu, X. Zuo, C. Chen, P. Z. Qin and X. Fang, ACS Chem. Biol., 2022, 17(9), 2448–2460 CrossRef CAS PubMed; (f) Y. Wang, V. Kathiresan, Y. Chen, Y. Hu, W. Jiang, G. Bai, G. Liu, P. Z. Qin and X. Fang, Chem. Sci., 2020, 11(35), 9655–9664 RSC; (g) M. Kimoto and I. Hirao, Front. Mol. Biosci., 2022, 9, 851656 Search PubMed.
- (a) D. R. Davis, Nucleic Acids Res., 1995, 23(24), 5020–5026 CrossRef CAS PubMed; (b) F. W. J. Collins, P. M. O’Connor, O. Sullivan, M. C. Rea, C. Hill and R. P. Ross, Microbiology, 2016, 162(9), 1662–1671 CrossRef CAS PubMed.
- (a) G. K. Schroeder, C. Lad, P. Wyman, N. H. Williams and R. Wolfenden, Proc. Natl. Acad. Sci. U. S. A., 2006, 103(11), 4052–4055 CrossRef CAS PubMed; (b) K. S. Gates, Chem. Res. Toxicol., 2009, 22(11), 1747–1760 Search PubMed; (c) K. Temburnikar and K. L. Seley-Radtke, Beilstein J. Org. Chem., 2018, 14, 772–785 Search PubMed; (d) S. S. Bag, A. Banerjee and S. Sinha, Synlett, 2024, 1195–1227 CAS; (e) K. W. Wellington and S. A. Benner, Nucleosides, Nucleotides Nucleic Acids, 2006, 25(12), 1309–1333 CrossRef CAS PubMed; (f) E. De Clercq, J. Med. Chem., 2016, 59(6), 2301–2311 CrossRef CAS PubMed; (g) J. Štambaský, M. Hocek and P. Kočovský, Chem. Rev., 2009, 109(12), 6729–6764 CrossRef PubMed.
- R. Dörrenhaus, P. K. Wagner and S. Kath-Schorr, Biol. Chem., 2023, 404(10), 883–896 CrossRef PubMed.
- (a) M. D. Lewis, J. K. Cha and Y. Kishi, J. Am. Chem. Soc., 1982, 104(18), 4976–4978 CrossRef CAS; (b) M. Terauchi, H. Abe, A. Matsuda and S. Shuto, Org. Lett., 2004, 6(21), 3751–3754 CrossRef CAS PubMed.
- (a) Y. Jiang, Y. Zhang, B. C. Lee and M. J. Koh, Angew. Chem., Int. Ed., 2023, 62(38), e202305138 CrossRef CAS PubMed; (b) C. Zhang, S.-Y. Xu, H. Zuo, X. Zhang, Q.-D. Dang and D. Niu, Nat. Synth., 2023, 2, 251–260 CrossRef CAS.
- E. S. Hoffmann, M. C. De Pascali, L. Neu, C. Domnick, A. Soldà and S. Kath-Schorr, RSC Chem. Biol., 2024, 5(6), 556–566 RSC.
- Y. Li, Z. Wang, L. Li, X. Tian, F. Shao and C. Li, Angew. Chem., Int. Ed., 2022, 61(1), e202110391 CrossRef CAS PubMed.
- A. S. Bhanu Prasad, T. M. Stevenson, J. R. Citineni, V. Nyzam and P. Knochel, Tetrahedron, 1997, 53(21), 7237–7254 CrossRef.
- Y. Wei, B. Benzvi and T. Diao, Angew. Chem., Int. Ed., 2021, 60(17), 9433–9438 CrossRef CAS PubMed.
- E. Mao, C. K. Chung, Y. Ji, Y.-H. Lam and P. E. Maligres, J. Org. Chem., 2021, 86(11), 7529–7536 CrossRef CAS PubMed.
- S. Häcker, M. Schrödter, A. Kuhlmann and H.-A. Wagenknecht, JACS Au, 2023, 3(7), 1843–1850 CrossRef PubMed.
- M. Merkel, L. Dehmel, N. P. Ernsting and H.-A. Wagenknecht, Angew. Chem., Int. Ed., 2017, 56(1), 384–388 CrossRef CAS PubMed.
- (a) Y. Gholamalipour, A. Karunanayake Mudiyanselage and C. T. Martin, Nucleic Acids Res., 2018, 46(18), 9253–9263 CrossRef CAS PubMed; (b) S. N. Sarcar and D. L. Miller, Sci. Rep., 2018, 8(1), 13885 CrossRef PubMed.
- L. Bornewasser and S. Kath-Schorr, Methods Mol. Biol., 2022, 2439, 223–240 CrossRef CAS PubMed.
- (a) A. V. Le and M. C. Hartman, RSC Chem. Biol., 2024, 5(11), 1111–1121 RSC; (b) A. Yadav, K. Khera, R. Mehrotra, K. Kaur, Y. Agrawal, P. Srivastava and S. Thakur, Arch. Clin. Biomed. Res., 2022, 6(6), 916–932 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available: Synthesis routes and analytical data to dCTPT3 TP and rCTPT3 TP, details of biochemical studies, supplementary LC-MS results. See DOI: https://doi.org/10.1039/d5cc02749g |
| This journal is © The Royal Society of Chemistry 2025 |



