 Open Access Article
Open Access ArticleAn esterase-sensitive persulfide/hydrogen sulfide generating fluorogenic probe enhances antioxidant response†
Bharat S.
Choudhary
 a,
Akshi
Vashistha
a,
Akshi
Vashistha
 b,
Ankan
Ghosh
b,
Rachit
Agarwal
b,
Ankan
Ghosh
b,
Rachit
Agarwal
 b and
Harinath
Chakrapani
b and
Harinath
Chakrapani
 *a
*a
aDepartment of Chemistry, Indian Institute of Science Education and Research Pune, Pune 411 008, Maharashtra, India. E-mail: harinath@iiserpune.ac.in
bDepartment of Bioengineering, Indian Institute of Science, Bengaluru 560 012, Karnataka, India
First published on 27th June 2025
Abstract
Hydrogen sulfide (H2S) and sulfane sulfur species such as persulfides and polysulfides protect the cells from oxidative stress and are central to the endogenous antioxidant response. Here, we designed and developed a tool that is cleaved by esterase to generate phenacylthiol, which is an artificial substrate for 3-mercaptopyruvate sulfurtransferase (3-MST) and non-electrophilic lactone as a by-product. The esterase-sensitive persulfide generator enhances H2S and sulfane sulfur and protects chondrocytes from the lethality induced by elevated reactive oxygen species (ROS).
Hydrogen sulfide (H2S), a gaseous signaling molecule, is responsible for maintaining redox homeostasis in the cells, and is involved in primary metabolism, antibiotic response, and aging, and is a potential therapeutic agent involved in several diseases such as Parkinson's, Alzheimer's, and Osteoarthritis.1–4 Several physiological effects of H2S are mediated via protein persulfidation, a post-translational modification of cysteine residues (RSH) to persulfides (RS-SH), which has protective effects against oxidative stress induced by reactive oxygen species (ROS).4–6 Persulfides (RS-SH) and polysulfides (RS–(S)nH) collectively form the sulfane sulfur pool and have emerged as crucial mediators of the oxidative stress response.7 Persulfides exhibit superior reactivity in scavenging reactive oxygen species (ROS) and mitigating oxidative stress.8 Hence, to enhance the cellular persulfides, several persulfide-generating probes that are activated by enzymes,9–13 light,14–16 ROS,17–20 and peroxynitrite21 have been developed (Fig. 1a). Several of the aforementioned probes have exhibited potential in alleviating oxidative stress. However, many of them produce electrophilic byproducts, which may contribute to the electrophilic stress,22 and also, the disulfide bonds are susceptible to reaction with thiols.
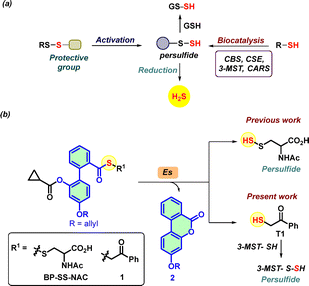 | ||
| Fig. 1 (a) Methods of persulfide generation via enzymatic or persulfide generating probes, sulfur transfer to GSH, and generation of H2S. (b) Reported biphenyl-based persulfide generator in the presence of esterase (Es) generates persulfide and non-electrophilic lactone 2 as a by-product. Based on a similar system, compound 1 generates thiol T1, which reacts with 3-MST to generate a persulfide. Previously, phenacyl thioacetate (AS) (see ESI,† Scheme S1) was shown to generate T1 in the presence of esterase.32 | ||
Persulfides are biosynthesized by the cystathionine-β-synthase (CBS),8 cystathionine γ-lyase (CSE),23 cysteinyl-tRNA synthetases (CARS),24 and 3-mercaptopyruvate sulfur transferase (3-MST)25,26 (Fig. 1a). We considered 3-MST, an endogenous enzyme that generates persulfides and H2S (Fig. 1b).25,27–29 3-MST utilizes 3-mercaptopyruvate (3-MP) as the natural substrate and accepts the sulfur from it to form a transient persulfide intermediate in its active site cysteine and an enolate of pyruvate as the byproduct.30 The sulfur from the persulfide intermediate can be transferred to thiols or proteins and produces H2S under reducing conditions.31 Recently, we developed phenacyl thiol as an artificial substrate for 3-MST and found that it efficiently produces persulfide and H2S in the presence of the enzyme (Fig. 1b).32–34
Our lab also developed a biphenyl-based esterase-sensitive persulfide-generating probe BP-SS-NAC that generates persulfide along with a non-electrophilic lactone 2 as a by-product (Fig. 1b).35 By leveraging the biphenyl scaffold, here, we developed an esterase-sensitive compound that generates phenacyl thiol (T1, Fig. 1b). Here, we report results of this compound to generate persulfide and H2S in the presence of 3-MST, and protect cells from oxidative stress in a chondrocyte model.
Compound 3 was synthesized following a reported protocol.35 Compound 3 was reacted with freshly prepared phenacyl thiol (T1) in the presence of DCC and DMAP to afford the biphenyl-based esterase-sensitive 3-MST artificial substrate 1 in 20% yield (Scheme 1).
First, to understand if 1 was stable in pH 7.4 buffer, HPLC analysis was carried out. The thioester was found to be stable over 120 min (see ESI,† Fig. S1). In the presence of esterase, as expected, compound 1 produced lactone 2 and T1 (Fig. 2A). Upon treatment of 1 with esterase, we observed the complete disappearance of 1 (RT 16.7 min) within 60 min, along with the formation of 2 (RT 14.9 min) and T1 (RT 10.0 min) (Fig. 2B and see ESI,† Fig. S2). HPLC analysis revealed the quantitative formation of 2 and thiol T1 (Fig. 2C and see ESI,† Fig. S2). In the case of the persulfide analogue BP-SS-NAC that we previously reported, the persulfide bond is susceptible to cleavage and thiol exchange, and the perthioester itself was cleaved by Es, leading to a diminished yield of the lactone.35 Hence, the thioester 1 has advantages in this regard since we find that the yield of the lactone is nearly quantitative.
Since lactone 2 is fluorescent, it can be monitored using fluorescence-based experiments (limit of detection (LOD) ≈ 0.3 μM, see ESI,† Fig. S3). The compound 1 was itself non-fluorescent, but the addition of Es (1 U mL−1) led to a significant increase in fluorescence intensity attributable to the formation of 2 (λex = 320 nm; λem = 432 nm) after incubation for 1 h in pH 7.4 buffer (see ESI,† Fig. S4A). Furthermore, a dose-dependent increase in fluorescence was seen during 1 h (see ESI,† Fig. S4B). Curve fitting for the formation of 2 to a first-order exponential equation yielded rate constants ranging from 0.10 to 0.15 min−1. The yield of 2 when 1 with Es for 1 h was nearly quantitative, as determined by a fluorescence experiment (see ESI,† Fig. S5). Pre-treatment with PMSF (phenylmethanesulfonyl fluoride), an inhibitor of esterase,36 resulted in a diminished yield of 2 (see ESI,† Fig. S5). We evaluated the cleavage of compound 1 under cell culture conditions using human chondrocyte C28/I2 cells and observed that 1 was cleaved in whole cells or cell lysates, resulting in a fluorescence signal corresponding to the formation of 2 (see ESI,† Fig. S6 and S7). Imaging using confocal microscopy in MEF cells treated with 1 further confirmed the formation of the fluorescent lactone 2 (see ESI,† Fig. S8). Next, the selectivity of 1 for esterase over other biologically relevant species was evaluated, and no significant increase in fluorescence intensity was seen except when 1 was treated with esterase (see ESI,† Fig. S9). Overall, these results supported that 1 underwent lactonization to produce 2 and thiol T1 following esterase-mediated cleavage.
Next, compound 1 was tested for its ability to generate 3-MST persulfide in the presence of Es and 3-MST, and the sulfur transfer to a small molecule thiol using two independent assays was studied (Fig. 3A). Sulfane sulfur detection was carried out using a well-established fluorogenic probe, SSP-2 (see ESI,† Scheme S2).37 When the reaction mixture of 1 + Es+ 3-MST was incubated with the SSP-2, a significant increase in fluorescence intensity (λex = 482 nm; λem = 518 nm), corresponding to the generation of sulfane sulfur. As DTT is known to cleave persulfides, pre-treatment with DTT followed by SSP-2, a reduction in the fluorescence signal was observed (Fig. 3B). The protein persulfide 3-MST-S-S− is known to transfer the sulfhydryl group to the small molecule thiol acceptor such as GSH. A standard LC/MS assay was conducted using an established HPE-IAM electrophile as a persulfide trapping agent (see ESI,† Scheme S3).38 When the reaction mixture of 1 + Es + 3MST was treated with GSH in the presence of HPE-IAM, a peak for the formation of the GSS-HPE-AM adduct (expected m/z = 517.1427; observed m/z = 517.1429) was observed at 7.3 min, indicating the formation of GS-SH (Fig. 3C, D, and see ESI,† Fig. S10) Under these conditions, we also observe Bis-S-HPE-AM (expected m/z = 389.1530; observed m/z = 389.1537), presumably due to the reaction of HPE-IAM with H2S formed by the reaction of GS-SH persulfide with GSH (Fig. 3E, and see ESI,† Fig. S11). Under cellular conditions, 3-MST persulfide is reduced by thioredoxin reductase to produce H2S and 3-MST.31 The H2S formation from compound 1 in the presence of Es, 3-MST, and DTT (a mimic of thioredoxin reductase) was evaluated using two independent assays (Fig. 3A).32 Firstly, the formation of H2S from compound 1, lead acetate assay, was employed (see ESI,† Scheme S4). The addition of an aliquot of the reaction mixture containing 1, Es, 3-MST, and DTT to lead acetate paper resulted in a dark coloration, indicating the formation of lead sulfide (Fig. 3F and see ESI,† Fig. S12). As expected, positive control AS gave a good signal for H2S, whereas lactone 2 did not produce any H2S (Fig. 3F and see ESI,† Fig. S12).
Next, we measured H2S formation using a standard methylene blue (MB) colorimetric assay (see ESI,† Scheme S5); indeed, the results of this assay corroborated with the lead acetate assay (Fig. 3G).
Osteoarthritis (OA) is the most common joint disease and is characterized by a gradual loss of articular cartilage and joint hypertrophy.39 A study has shown that the H2S generated from 3-MST protects against joint calcification and severe osteoarthritis.3 So, enhancing the H2S and sulfane sulfur in chondrocytes may represent a potential way to treat OA. We first used human chondrocyte C28/I2 cells in culture and found that 1 was well tolerated up to 100 μM after 24 h of incubation (Fig. 4A). Given the central role of oxidative stress in the progression of OA,40,41 we determined the cytoprotective effects of 1 in C28/I2 cells against oxidative stress induced by a cell-permeable ROS generator (MGR1).42 Cells were pre-incubated with 1 for 3 h and then exposed to MGR-1 (15 μM), after which cell metabolic activity was assessed. As expected, exposure of cells to MGR-1 resulted in a drastic reduction in cell metabolic activity. However, pre-treatment of cells with 1 demonstrated significant protective effects, rescuing cells from ROS-induced oxidative stress in a concentration-dependent manner (Fig. 4B). This result was also seen with BP-SS-NAC, and supports the applicability of the phenacylthiol-based strategy for mitigating oxidative stress.35
Next, we utilized micromass cultures derived from the C28/I2 cell line, an in vitro model for cartilage,43 to evaluate the ability of 1 to sustain sGAG production under oxidative stress induced by MGR-1 and found that cells were capable of sustaining sGAG when treated with compound 1 (see ESI,† Fig. S13). Overall, these results suggest that 1 was capable of protecting cells from oxidative stress.
In summary, we report a new persulfide generator that is cleaved by esterase to generate phenacylthiol, an artificial substrate for 3-MST, along with a non-electrophilic and fluorescent by-product. The compound was able to generate lactone quantitatively, and the resulting fluorescence can be useful for use in cellular experiments. The antioxidant property of 1 in chondrocytes serves as a proof-of-principle for this strategy, while further work is needed to characterize precise mechanisms.
Financial assistance for this project was from IISER Pune and ANRF Science and Engineering Research Board (HC, CRG/2023/003892). The manuscript was written with inputs from all authors. BSC, AV, and AG carried out all experiments under the supervision of RA and HC.
Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the ESI.†Notes and references
- T. V. Mishanina, M. Libiad and R. Banerjee, Nat. Chem. Biol., 2015, 11, 457–464 CrossRef CAS.
- M. R. Filipovic, J. Zivanovic, B. Alvarez and R. Banerjee, Chem. Rev., 2018, 118, 1253–1337 CrossRef CAS.
- S. Nasi, D. Ehirchiou, A. Chatzianastasiou, N. Nagahara, A. Papapetropoulos, J. Bertrand, G. Cirino, A. So and N. Busso, Arthritis Res. Ther., 2020, 22, 49 CrossRef CAS.
- A. K. Mustafa, M. M. Gadalla, N. Sen, S. Kim, W. Mu, S. K. Gazi, R. K. Barrow, G. Yang, R. Wang and S. H. Snyder, Sci. Signaling, 2009, 2, ra72 Search PubMed.
- C. Yang, N. O. Devarie-Baez, A. Hamsath, X. Fu and M. Xian, Antioxid. Redox Signaling, 2020, 33, 1092–1114 CrossRef CAS.
- H. Kimura, Br. J. Pharmacol., 2020, 177, 720–733 CrossRef CAS.
- T. Zhang, H. Tsutsuki, K. Ono, T. Akaike and T. Sawa, J. Clin. Biochem. Nutr., 2021, 68, 5–8 CrossRef CAS.
- T. Ida, T. Sawa, H. Ihara, Y. Tsuchiya, Y. Watanabe, Y. Kumagai, M. Suematsu, H. Motohashi, S. Fujii, T. Matsunaga, M. Yamamoto, K. Ono, N. O. Devarie-Baez, M. Xian, J. M. Fukuto and T. Akaike, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 7606–7611 CrossRef CAS.
- Y. Zheng, B. Yu, Z. Li, Z. Yuan, C. L. Organ, R. K. Trivedi, S. Wang, D. J. Lefer and B. Wang, Angew. Chem., Int. Ed., 2017, 56, 11749–11753 CrossRef CAS.
- Z. Yuan, Y. Zheng, B. Yu, S. Wang, X. Yang and B. Wang, Org. Lett., 2018, 20, 6364–6367 CrossRef CAS.
- K. M. Dillon, R. J. Carrazzone, Y. Wang, C. R. Powell and J. B. Matson, ACS Macro Lett., 2020, 9, 606–612 CrossRef CAS.
- K. M. Dillon, H. A. Morrison, C. R. Powell, R. J. Carrazzone, V. M. Ringel-Scaia, E. W. Winckler, R. M. Council-Troche, I. C. Allen and J. B. Matson, Angew. Chem., Int. Ed., 2021, 60, 6061–6067 CrossRef CAS.
- P. Bora, M. B. Sathian and H. Chakrapani, Chem. Commun., 2022, 58, 2987–2990 RSC.
- A. Chaudhuri, Y. Venkatesh, J. Das, M. Gangopadhyay, T. K. Maiti and N. D. P. Singh, J. Org. Chem., 2019, 84, 11441–11449 CrossRef CAS.
- A. Chaudhuri, Y. Venkatesh, B. C. Jena, K. K. Behara, M. Mandal and N. D. P. Singh, Org. Biomol. Chem., 2019, 17, 8800–8805 RSC.
- B. Roy, M. Shieh, T. Takata, M. Jung, E. Das, S. Xu, T. Akaike and M. Xian, J. Am. Chem. Soc., 2024, 146, 30502–30509 CrossRef CAS.
- C. R. Powell, K. M. Dillon, Y. Wang, R. J. Carrazzone and J. B. Matson, Angew. Chem., Int. Ed., 2018, 57, 6324–6328 CrossRef CAS.
- R. A. Hankins, S. I. Suarez, M. A. Kalk, N. M. Green, M. N. Harty and J. C. Lukesh, Angew. Chem., Int. Ed., 2020, 59, 22238–22245 CrossRef CAS.
- P. Bora, P. Chauhan, S. Manna and H. Chakrapani, Org. Lett., 2018, 20, 7916–7920 CrossRef CAS.
- Y. Wang, K. M. Dillon, Z. Li, E. W. Winckler and J. B. Matson, Angew. Chem., Int. Ed., 2020, 59, 16698–16704 CrossRef CAS.
- Y. Xu, B. Xu, J. Wang, H. Jin, S. Xu, G. Wang and L. Zhen, Chem. – Eur. J., 2022, 28, e202200540 CrossRef CAS.
- D. C. Thompson, J. A. Thompson, M. Sugumaran and P. Moldéus, Chem.-Biol. Interact., 1993, 86, 129–162 CrossRef CAS.
- T. Yamanishi and S. Tuboi, J. Biochem., 1981, 89, 1913–1921 CrossRef CAS.
- T. Akaike, T. Ida, F. Y. Wei, M. Nishida, Y. Kumagai, M. M. Alam, H. Ihara, T. Sawa, T. Matsunaga, S. Kasamatsu, A. Nishimura, M. Morita, K. Tomizawa, A. Nishimura, S. Watanabe, K. Inaba, H. Shima, N. Tanuma, M. Jung, S. Fujii, Y. Watanabe, M. Ohmuraya, P. Nagy, M. Feelisch, J. M. Fukuto and H. Motohashi, Nat. Commun., 2017, 8, 1177 CrossRef.
- N. Nagahara, T. Yoshii, Y. Abe and T. Matsumura, J. Biol. Chem., 2007, 282, 1561–1569 CrossRef CAS.
- N. Nagahara, M. Tanaka, Y. Tanaka and T. Ito, Antioxidants, 2019, 8, 116 CrossRef CAS.
- Y. Kimura, S. Koike, N. Shibuya, D. Lefer, Y. Ogasawara and H. Kimura, Sci. Rep., 2017, 7, 10459 CrossRef.
- B. Pedre and T. P. Dick, Biol. Chem., 2021, 402, 223–237 CrossRef CAS.
- B. Pedre, D. Talwar, U. Barayeu, D. Schilling, M. Luzarowski, M. Sokolowski, S. Glatt and T. P. Dick, Nat. Chem. Biol., 2023, 19, 507–517 CrossRef CAS.
- N. Nagahara and T. Nishino, J. Biol. Chem., 1996, 271, 27395–27401 CrossRef CAS.
- P. K. Yadav, K. Yamada, T. Chiku, M. Koutmos and R. Banerjee, J. Biol. Chem., 2013, 288, 20002–20013 CrossRef CAS.
- P. Bora, S. Manna, M. A. Nair, R. R. M. Sathe, S. Singh, V. S. Sreyas Adury, K. Gupta, A. Mukherjee, D. K. Saini, S. S. Kamat, A. B. Hazra and H. Chakrapani, Chem. Sci., 2021, 12, 12939–12949 RSC.
- S. Manna, R. Agrawal, T. Yadav, T. A. Kumar, P. Kumari, A. Dalai, S. Kanade, N. Balasubramanian, A. Singh and H. Chakrapani, Angew. Chem., Int. Ed., 2024, 63, e202411133 CrossRef CAS.
- S. M. Gupta, P. S. Mohite and H. Chakrapani, Chem. Sci., 2025, 16, 4695–4702 RSC.
- B. S. Choudhary, T. A. Kumar, A. Vashishtha, S. Tejasri, A. S. Kumar, R. Agarwal and H. Chakrapani, Chem. Commun., 2024, 60, 1727–1730 RSC.
- P. C. Smith, A. F. McDonagh and L. Z. Benet, J. Pharmacol. Exp. Ther., 1990, 252, 218–224 CrossRef CAS.
- W. Chen, C. Liu, B. Peng, Y. Zhao, A. Pacheco and M. Xian, Chem. Sci., 2013, 4, 2892–2896 RSC.
- H. A. Hamid, A. Tanaka, T. Ida, A. Nishimura, T. Matsunaga, S. Fujii, M. Morita, T. Sawa, J. M. Fukuto, P. Nagy, R. Tsutsumi, H. Motohashi, H. Ihara and T. Akaike, Redox Biol., 2019, 21, 101096 CrossRef.
- J. Martel-Pelletier, Osteoarthr. Cartil., 1998, 6, 374–376 CrossRef CAS.
- M. Y. Ansari, N. Ahmad and T. M. Haqqi, Biomed. Pharmacother., 2020, 129, 110452 CrossRef CAS.
- L. Liu, P. Luo, M. Yang, J. Wang, W. Hou and P. Xu, Front. Mol. Biosci., 2022, 9 DOI:10.3389/fmolb.2022.1001212.
- D. S. Kelkar, G. Ravikumar, N. Mehendale, S. Singh, A. Joshi, A. K. Sharma, A. Mhetre, A. Rajendran, H. Chakrapani and S. S. Kamat, Nat. Chem. Biol., 2019, 15, 169–178 CrossRef CAS.
- K. V. Greco, A. J. Iqbal, L. Rattazzi, G. Nalesso, N. Moradi-Bidhendi, A. R. Moore, M. B. Goldring, F. Dell’Accio and M. Perretti, Biochem. Pharmacol., 2011, 82, 1919–1929 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5cc01523e |
| This journal is © The Royal Society of Chemistry 2025 |




