Open Access Article
Open Access ArticleConstruction of a dual-color fluorescent light-up biosensor based on sequential coding for label-free and sensitive detection of multiple Piwi-interacting RNAs†
Jinping
Hu‡
,
Jie
Zhang‡
,
Juan
Hu
 * and
Chun-yang
Zhang
* and
Chun-yang
Zhang
 *
*
School of Chemistry and Chemical Engineering, State Key Laboratory of Digital Medical Engineering, Southeast University, Nanjing 211189, China. E-mail: hujuan@seu.edu.cn; zhangcy@seu.edu.cn
First published on 5th March 2025
Abstract
We construct a dual-color light-up biosensor for label-free and sensitive detection of multiple piRNAs. This biosensor exhibits high sensitivity with a limit of detection of 3.19 aM for piRNA-36026 and 4.71 aM for piRNA-823. Moreover, it can accurately measure cellular piRNA levels with single-cell sensitivity, and discriminate breast cancer tissues from healthy tissues.
Piwi-interacting RNAs (piRNAs) are an emerging category of 24–32 nt small non-coding RNAs that are associated with the Piwi subfamily of Argonaute proteins to form a piRNA-induced silencing complex.1 Accumulating evidence reveals that the dysregulated piRNAs are implicated in the occurrence and progression of various diseases (e.g., diabetes, cardiovascular diseases, neurological disorders, and cancers),2 making them valuable biomarkers for disease diagnosis and targeted therapeutics. Consequently, sensitive and accurate quantification of circRNAs is of great significance for biomedical research and clinical diagnosis.
Conventional methods for piRNA assay mainly include Northern blotting,1b RNA sequencing (RNA-seq),3 and stem-loop quantitative reverse transcription-polymerase chain reaction (qRT-PCR).4 However, they usually involve hazardous radiation, sophisticated instruments, large sample consumption, and tedious operating procedures. Recently, a series of nucleic acid amplification techniques have been developed for piRNA assay.5 Among them, loop-mediated isothermal amplification (LAMP) has attracted more and more attention due to its ultrahigh efficiency compared with PCR.6 However, LAMP has some inescapable limitations: (1) LAMP utilizes 4–6 primers to recognize 6–8 target sites, making primer design extremely difficult; (2) both inner and loop primers contain two target recognition sites with approximately 40 nt in length, making them susceptible to nonspecific interactions (e.g., formation of primer dimers).
To circumvent these limitations, we develop a dual-color light-up biosensor based on base-stacking hybridization-powered sequential coding for label-free detection of multiple piRNAs (Scheme 1). As a proof-of-concept, we selected piRNA-36026 and piRNA-823 as the model targets. The detailed molecular structure of constructed branch-shaped probe is shown in Fig. S1 (ESI†). Because the stability of short duplex can be significantly improved through the stacking bases when two or more short oligonucleotides hybridize with the same DNA template in close proximity,7 the hybridization of piRNA-36026 and piRNA-823 with the domains a and b of branch-shaped probe can stabilize the short 5-bp metastable primer duplexes (c/c* and d/d*). As a result, the primer can be polymerized along the template to form structure I, accompanied by the disintegration of branch-shaped probe. Moreover, the 3′-terminus of template-36026 and template-823 can be extended to form structure II with the assistance of Bst 3.0 DNA polymerase, accompanied by the release of target piRNAs that can initiate new hybridization-polymerization cycles. Due to the reduced thermostability of PS-DNA/DNA,8 the palindromic duplex of structure II is denatured under the reaction temperature of 55 °C, facilitating the self-folding of terminal palindrome to initiate new extension reaction, resulting in the production of structure III and the displacement of structure IV. Notably, the primer-templated 3′-terminal strand extension of template-36026 and template-823 leads to the generation of sequences complementary to the foldback-binding domains, facilitating the self-folding of 3′-terminal to form stable hairpin structures. The self-folded hairpin-primed polymerization can generate structure V with PS-DNA/DNA palindromic tails that can be repeatedly folded and elongated to form long concatemer amplicons (structure VI). Both structure III and structure VI contain intact T7 promoter sequence that can be recognized by T7 RNA polymerase to activate the transcriptional synthesis of Spinach aptamers and Mango aptamers. Subsequently, DFHBI and TO1-biotin fluorogens specifically incorporate into the Spinach and Mango aptamers, respectively, to produce enhanced dual-color fluorescence signals. On the contrary, in the absence of target piRNAs, the primer cannot form a stable duplex, and no polymerization reaction occurs. Consequently, neither the synthesis of dsDNA concatermers nor the transcription of fluorescent RNA aptamers occurs, and no fluorescence signal is observed.
 | ||
| Scheme 1 Schematic illustration of a dual-color light-up biosensor based on base-stacking hybridization-powered sequential coding for label-free and sensitive monitoring of multiple piRNAs. | ||
We performed 2% AGE to verify the products of piRNA-triggered self-priming LAMP reaction (Fig. 1A and Fig. S2, ESI†). Distinct bands of amplification products with different molecular weights are observed in the presence of branch-shaped probe + piRNA-36026 + piRNA-823 (Fig. 1A, lane 1), template-36026 + primer-36026 + piRNA-36026 (Fig. 1A, lane 3), and template-823 + primer-823 + piRNA-823 (Fig. 1A, lane 5), indicating the occurrence of target piRNA-triggered continuous polymerization for the generation of long concatemer amplicons. In contrast, no characteristic bands of amplification products are observed in the presence of branch-shaped probe (Fig. 1A, lane 2), template-36026 + primer-36026 (Fig. 1A, lane 4), template-823 + primer-823 (Fig. 1A, lane 6), template-36026 + primer-36026 + piRNA-823 (Fig. 1A, lane 5), and template-823 + primer-823 + piRNA-36026 (Fig. 1A, lane 5). Moreover, we employed transmission electron microscope (TEM) to characterize the amplification products of self-priming LAMP (Fig. S2, ESI†). These results are consistent with those obtained by AGE analysis. We further used fluorescence measurement to validate the target-initiated T7 polymerase-mediated transcriptional synthesis of Spinach aptamers and Mango aptamers. The piRNA-36026 induces enhanced DFHBI fluorescence (Fig. 1B, green curve), and piRNA-823 induces enhanced TO1-biotin fluorescence (Fig. 1C, red curve). On the contrary, the absence of piRNA-36026 generates negligible DFHBI fluorescence signal (Fig. 1B, gray curve), and piRNA-823 generates negligible TO1-biotin fluorescence signal (Fig. 1C, blue curve). These results reveal the feasibility of this biosensor for quantitative detection of multiple piRNAs.
Under the optimized conditions (Fig. S3–S7, ESI†), we evaluated the sensitivity of the proposed method (Fig. 2A and D). The piRNA-36026 induces enhanced DFHBI fluorescence signal in a dose-dependent manner (Fig. 2B). In logarithmic coordinates, the DFHBI fluorescence intensity (F) is linearly dependent on the piRNA-36026 concentration (C) in the range from 1 × 10−17 to 1 × 10−11 M (inset of Fig. 2C). The regression equation is F = 984.34 + 91.48 lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C (R2 = 0.9991), and the limit of detection (LOD) is calculated to be 3.19 × 10−18 M for piRNA-36026 assay. Similarly, the fluorescence signal of TO1-biotin improves when the concentration of piRNA-823 increases from 0 to 2 × 10−8 M (Fig. 2E). The TO1-biotin fluorescence intensity (F) exhibits a good linear correlation with the logarithm of piRNA-823 concentration (C) in the range from 1 × 10−17 to 1 × 10−11 M (inset of Fig. 2F), with an equation of F = 1031.17 + 111.06 lg
C (R2 = 0.9991), and the limit of detection (LOD) is calculated to be 3.19 × 10−18 M for piRNA-36026 assay. Similarly, the fluorescence signal of TO1-biotin improves when the concentration of piRNA-823 increases from 0 to 2 × 10−8 M (Fig. 2E). The TO1-biotin fluorescence intensity (F) exhibits a good linear correlation with the logarithm of piRNA-823 concentration (C) in the range from 1 × 10−17 to 1 × 10−11 M (inset of Fig. 2F), with an equation of F = 1031.17 + 111.06 lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C (R2 = 0.9995). The LOD is determined to be 4.71 × 10−18 M for piRNA-823 assay.
C (R2 = 0.9995). The LOD is determined to be 4.71 × 10−18 M for piRNA-823 assay.
To investigate the selectivity of this biosensor toward piRNA-36026 and piRNA-823, we employed irrelevant piRNAs including piRNA-651, piRNA-36743, and piRNA-31143 as the interferences. As depicted in Fig. 2G, when piRNA-36026 and piRNA-823 coexist, significantly improved DFHBI and TO1-biotin fluorescence signals are observed. In the presence of piRNA-36026 alone, only a distinct DFHBI fluorescence signal can be detected. In the presence of piRNA-823 alone, only a distinct TO1-biotin fluorescence signal can be observed. We further evaluated the anti-interference capability of the proposed biosensor by measuring the fluorescence signals generated by different-concentration target piRNAs spiked in diluted sera and interfering RNA mixture. No significant change is observed in the DFHBI and TO1-biotin fluorescence signals (Fig. 2H and I).
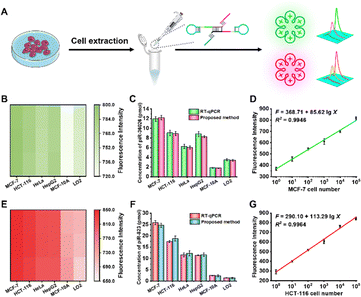
To demonstrate the feasibility of this biosensor for real sample analysis, we measured the piRNA-36026 and piRNA-823 expression levels in MCF-7 cells, HCT-116 cells, HeLa cells, HepG2 cells, MCF-10A cells, and LO2 cells (Fig. 3A). As depicted in Fig. 3B and E, high expression levels of piRNA-36026 and piRNA-823 are observed in cancer cells (i.e., MCF-7 cells, HCT-116 cells, HeLa cells, and HepG2 cells) compared with normal cells (i.e., MCF-10A cells and LO2 cells). These results are further confirmed by stem-loop qRT-PCR (Fig. 3C and F and Fig. S8 and S9, ESI†). Moreover, we measured the DFHBI and TO1-biotin fluorescence signals generated by various numbers of MCF-7 cells and HCT-116 cells, respectively. The DFHBI fluorescence signal (F) shows a linear dependent on the logarithmic numbers of MCF-7 cells (X) from 1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, with an equation of F = 368.71 + 85.62 lg
000, with an equation of F = 368.71 + 85.62 lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) X (R2 = 0.9946) (Fig. 3D). Similarly, in the logarithmic scale, the TO1-biotin fluorescence signal (F) is linearly correlated with the numbers of HCT-116 cells (X) from 1 to 1
X (R2 = 0.9946) (Fig. 3D). Similarly, in the logarithmic scale, the TO1-biotin fluorescence signal (F) is linearly correlated with the numbers of HCT-116 cells (X) from 1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, with an equation of F = 290.10 + 113.29 lg
000, with an equation of F = 290.10 + 113.29 lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) X (R2 = 0.9964) (Fig. 3G). Notably, the LOD is measured to be 1 cell for both piRNA-36026 and piRNA-823 assay.
X (R2 = 0.9964) (Fig. 3G). Notably, the LOD is measured to be 1 cell for both piRNA-36026 and piRNA-823 assay.
To assess the applicability of this biosensor for clinical diagnosis, we measured the expression levels of piRNA-36026 and piRNA-823 in 5 pairs of breast cancer tissues and healthy para-carcinoma tissues (Fig. 4A). As depicted in Fig. 4B and D, the expressions of piRNA-36026 and piRNA-823 in breast cancer tissues are much higher than those in healthy para-carcinoma tissues, consistent with the upregulation of piRNA-36026 and piRNA-823 in breast cancer.9 The average expressions of piRNA-36026 and piRNA-823 in breast cancer tissues are measured to be 8.23 ± 3.29 pM and 28.56 ± 7.99 pM, respectively, which are 4.57-fold and 6.01-fold higher than those in healthy para-carcinoma tissues (1.88 ± 0.59 pM for piRNA-36026 and 4.75 ± 2.72 pM for piRNA-823) (t test, P < 0.001, Fig. 4C and E), consistent with the results of stem-loop qRT-PCR (Fig. 4F and G). We further employed receiver operator characteristic (ROC) analysis to evaluate the diagnosis accuracy of this biosensor. The area under the curve (AUC) value is calculated to be 1 (Fig. 4H), suggesting that this biosensor can distinguish breast cancer patients from healthy persons with good accuracy.
In summary, we developed a dual-color fluorescent light-up biosensor based on base-stacking hybridization-powered sequential coding for label-free and sensitive detection of multiple piRNAs. In comparison with the reported piRNA assays (Table S2, ESI†),5,10 this biosensor possesses distinct merits: (1) the incorporation of PS backbone into nucleic acid substrates facilitates the self-folding of palindromic duplex, without the involvement of multiple primers in canonical LAMP; and (2) T7 RNA polymerase is highly T7 promoter-specific to initiate the transcription of downstream DNA sequences, endowing this assay with good accuracy. This biosensor is capable of sensitively quantifying cellular piRNA expressions at the single-cell level, and accurately distinguishing breast cancer patients from healthy individuals, providing a new paradigm for piRNA-related oncology, clinical diagnosis, and biomedical research.
This work was supported by the Frontier Technologies R&D Program of Jiangsu (BF2024063).
Data availability
Data are available upon the request.Conflicts of interest
There are no conflicts to declare.Notes and references
- (a) A. Girard, R. Sachidanandam, G. J. Hannon and M. A. Carmell, Nature, 2006, 442, 199–202 CrossRef PubMed; (b) N. C. Lau, A. G. Seto, J. Kim, S. Kuramochi-Miyagawa, T. Nakano, D. P. Bartel and R. E. Kingston, Science, 2006, 313, 363–367 CrossRef CAS PubMed; (c) D. M. Ozata, I. Gainetdinov, A. Zoch, D. O’Carroll and P. D. Zamore, Nat. Rev. Genet., 2019, 20, 89–108 CrossRef CAS PubMed.
- (a) A. Muhammad, R. Waheed, N. A. Khan, H. Jiang and X. Song, Database, 2019, 2019, baz052 CrossRef PubMed; (b) R. Suzuki, S. Honda and Y. Kirino, Front. Genet., 2012, 3, 204 CAS; (c) C. B. Assumpçao, D. Q. Calcagno, T. M. T. Araújo, S. E. B. dos Santos, A. K. C. R. dos Santos, G. J. Riggins, R. R. Burbano and P. P. Assumpçao, Epigenomics, 2015, 7, 975–984 CrossRef; (d) K. W. Ng, C. Anderson, E. A. Marshall, B. C. Minatel, K. S. Enfield, H. L. Saprunoff, W. L. Lam and V. D. Martinez, Mol. Cancer, 2016, 15, 5 CrossRef PubMed.
- W. H. Weng, N. Liu, Y. Toiyama, M. Kusunoki, T. Nagasaka, T. Fujiwara, Q. Wei, H. L. Qin, H. F. Lin, Y. L. Ma and A. Goel, Mol. Cancer, 2018, 17, 16 CrossRef.
- F. Tang, K. Hayashi, M. Kaneda, K. Lao and M. A. Surani, Biochem. Biophys. Res. Commun., 2008, 369, 1190–1194 CrossRef CAS.
- (a) Y. Dou, Y. He, H. Zhang, M. Yang, Q. Liu, W. Ma, X. Fu and Y. Chen, Anal. Methods, 2024, 16, 6810–6818 RSC; (b) M. El Aamri, R. Zayani, S. Baachaoui, H. Mohammadi, A. Amine and N. Raouafi, Sens. Actuators, B, 2024, 399, 134749 CrossRef CAS; (c) Q. Q. Ge, Q. Han, Y. Han, F. Ma, C. Z. Li and C. Y. Zhang, Chem. Commun., 2024, 60, 408–411 Search PubMed; (d) R. Jia, X. He, W. Ma, Y. Lei, H. Cheng, H. Sun, J. Huang and K. Wang, Anal. Chem., 2019, 91, 15107–15113 CrossRef CAS PubMed.
- T. Notomi, H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino and T. Hase, Nucleic Acids Res., 2000, 28, 12 CrossRef PubMed.
- (a) Z. X. Lu, D. M. Duan, R. Cao, L. M. Zhang, K. X. Zheng and J. Li, Chem. Commun., 2011, 47, 7452–7454 RSC; (b) Y. X. Zhao, L. Qi, F. Chen, Y. Zhao and C. H. Fan, Biosens. Bioelectron., 2013, 41, 764–770 CrossRef CAS PubMed.
- (a) M. Boczkowska, P. Guga and W. J. Stec, Biochemistry, 2002, 41, 12483–12487 CrossRef CAS PubMed; (b) L. A. LaPlanche, T. L. James, C. Powell, W. D. Wilson, B. Uznanski, W. J. Stec, M. F. Summers and G. Zon, Nucleic Acids Res., 1986, 14, 9081–9093 CrossRef CAS PubMed.
- (a) Y. J. Lee, S. U. Moon, M. G. Park, W. Y. Jung, Y. K. Park, S. K. Song, J. G. Ryu, Y. S. Lee, H. J. Heo, H. N. Gu, S. J. Cho, B. A. Ali, A. A. Al-Khedhairy, I. Lee and S. Kim, Biomaterials, 2016, 101, 143–155 CrossRef CAS PubMed; (b) X. Ding, Y. Li, J. H. Lü, Q. Zhao, Y. F. Guo, Z. Y. Lu, W. J. Ma, P. F. Liu, R. G. Pestell, C. L. Liang and Z. R. Yu, Front. Cell Dev. Biol., 2021, 9, 641052 CrossRef.
- (a) C. Y. Hu, L. P. Cao, X. J. Wu, G. X. Chen, Y. F. Li, J. Wang, C. Z. Huang and L. Zhan, Biosens. Bioelectron., 2024, 255, 112623 CrossRef PubMed; (b) Y. Tong, B. X. Guan, Z. W. Sun, X. J. Dong, Y. Q. Chen, Y. R. Li, Y. Y. Jiang and J. Li, Talanta, 2023, 257, 124307 Search PubMed; (c) M. El Aamri, S. Baachaoui, H. Mohammadi, N. Raouafi and A. Amine, Anal. Chim. Acta, 2024, 1305, 342583 CrossRef CAS; (d) H. Chen, Z. Zhuang, Y. Chen, C. Qiu, Y. Qin, C. Tan, Y. Tan and Y. Jiang, Anal. Chim. Acta, 2023, 1246, 340896 CrossRef CAS PubMed; (e) J. Li, Y. Tong, Z. W. Sun, Y. Q. Chen, Y. S. Wang, L. Q. Zhou, Y. Y. Jiang, P. L. Li, C. X. Wang and L. T. Du, Acta Biomater., 2022, 149, 287–296 CrossRef CAS PubMed; (f) Z. W. Sun, Y. Tong, J. Li, Y. D. Wang, F. C. Gao, H. Li, C. X. Wang, L. T. Du and Y. Y. Jiang, Sens. Actuators, B, 2022, 368, 132244 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cc06421f |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |