Open Access Article
Open Access ArticleA versatile approach for geometry-based self-assembly of DNA–protein hybrid nanostructures using histone–DNA interactions†
Hajar
Al-Zarah
,
Maged F.
Serag
,
Faisal
Alkhaldi
and
Satoshi
Habuchi
 *
*
King Abdullah University of Science and Technology, Biological and Environmental Science and Engineering Division, Thuwal 23955-6900, Saudi Arabia. E-mail: satoshi.habuchi@kaust.edu.sa
First published on 3rd December 2024
Abstract
We report an enhanced versatility in constructing DNA–protein hybrid nanostructures using histone–DNA complexes (HDs). By leveraging HDs, we demonstrate precise and scalable assembly of DNA origami tiles and a 2D triangular nanostructure. Our results extend the potential applications of DNA nanotechnology from the nanoscale to the microscale without the need for complex pre-designs.
DNA nanotechnology has seen significant advancements, transitioning from nano- and micro-sized structures such as DNA origami,1 DNA crystals,2 and DNA bricks3 to developing hybrid structures incorporating DNA with other molecules, such as proteins.4,5 The integration of proteins into DNA nanotechnology has significantly enriched the field, driven by sequence-specific6–9 and non-sequence-based mechanisms.10,11 However, one of the critical challenges, particularly in DNA origami, is its inherent limitation in size scalability due to the reliance on scaffold length and sequence, restricting the complexity and size of the resulting structures.
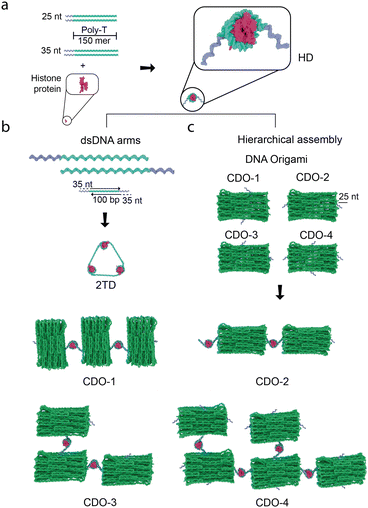
While various strategies have been explored, highly scalable building blocks for assembling diverse nanostructures remain to be developed. In this study, we report a versatile approach to assembling DNA–protein hybrid nanostructures using histone-based DNA–protein complexes (HDs) (Fig. 1a) as a flexible, scaffold-independent tool.12,13 HDs are unique in that they offer a geometry-based, non-sequence-specific interaction between DNA and proteins and, thus, would allow for the creation of complex DNA assemblies without the constraints of traditional DNA origami.
We designed two types of nanostructures to demonstrate the versatility of HDs in precise and size-scalable assembly. We first developed a triangle-shaped DNA nanostructure (2TD, Fig. 1b), which is structurally related to a tetrahedral DNA nanostructure (TDN). TDN is well known for its potential in biomedical applications, including drug delivery and antioxidant activity.14 The broad applicability of HDs for the precise assembly of different types of DNA nanostructures can be evaluated by constructing the HDs-mediated pre-designed 2TD without templates. Building on this, we investigated the applicability of HDs for the hierarchical scalable assembly of DNA nanostructures by characterizing the HDs-mediated assembly of DNA origami tiles (Fig. 1b).
In the formation of standard DNA-only TDNs, four ssDNA molecules pair specifically with one another, forming triangles. Each ssDNA molecule contributes a side that pairs with complementary sequences from three other ssDNA molecules, ensuring precise TDN assembly.15 In 2TD, we designed the triangular shape by incorporating HDs at the apex of the triangle (Fig. 1b). We previously reported that HDs were formed upon assembling histone proteins (i.e., H2, H3A, H3B, and H4) with ssDNA strand (e.g., poly-thymine (poly-T)) of a total length of 300-nucleotide (nt) at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio.12 This self-assembly resulted in the formation of nucleosome-like nanostructure (i.e., histone octamer wrapped by the ssDNA). To incorporate HDs into the apex of the triangular DNA nanostructure, we assembled HDs using 150-nt long poly-T ssDNA strands conjugated to 25 or 35-nt that serve as a sticky end (Fig. 1a). Two ssDNA strands with a total length of 300-nt are incorporated into HDs under this condition through a geometry-based, non-sequence-specific DNA–histone interaction,12,13 leaving two unbound sticky ends that bind to complementary handles on double-stranded DNA (dsDNA) arms and form 2TD (Fig. 1b). Orthogonal sequences were chosen for the sticky ends to prevent non-specific interactions. The GC content in the sticky ends was increased to approximately 60% to enhance the dsDNA stability by incorporating more hydrogen bonds (refer to the Materials and methods section (ESI†) for the exact sequences used).
1 ratio.12 This self-assembly resulted in the formation of nucleosome-like nanostructure (i.e., histone octamer wrapped by the ssDNA). To incorporate HDs into the apex of the triangular DNA nanostructure, we assembled HDs using 150-nt long poly-T ssDNA strands conjugated to 25 or 35-nt that serve as a sticky end (Fig. 1a). Two ssDNA strands with a total length of 300-nt are incorporated into HDs under this condition through a geometry-based, non-sequence-specific DNA–histone interaction,12,13 leaving two unbound sticky ends that bind to complementary handles on double-stranded DNA (dsDNA) arms and form 2TD (Fig. 1b). Orthogonal sequences were chosen for the sticky ends to prevent non-specific interactions. The GC content in the sticky ends was increased to approximately 60% to enhance the dsDNA stability by incorporating more hydrogen bonds (refer to the Materials and methods section (ESI†) for the exact sequences used).
100-base pairs (bp) dsDNA with 35-nt complementary handles at both ends were used as the arms of 2TD (Fig. 1b, refer to the Materials and methods section (ESI†) for the exact sequences used). Agarose gel electrophoresis of the dsDNA arms without the handles (i.e., annealed 100-bp-long dsDNA without 35-nt handles) revealed a distinct band corresponding to the 100-bp length (Fig. S1a, ESI†). Atomic force microscopy (AFM) analysis of these dsDNA arms in liquid showed homogeneous dsDNA arms without aggregation (Fig. S1b, ESI†). The dsDNA had a mean height and length of 2 nm and 34 nm (Fig. S1b and c, ESI†), consistent with the expected dimensions (i.e., 2 nm thickness and 34 nm length for the 100-bp dsDNA).16 In addition, the frequency histogram of approximately 70 dsDNA arm lengths showed a narrow distribution around the expected size of 34 nm (Fig. S1c, ESI†). These results confirm the integrity and homogeneity of the annealed dsDNA arms.
The AFM micrographs also show some curvature in the dsDNA arms. Since the persistence length of dsDNA is approximately 50 nm,17 the 34 nm-long dsDNA arms are expected to exhibit reduced flexibility. The observed curvature could thus be attributed to the high salt concentration (200 mM Mg(CH3COO)2 and 100 mM NiCl2) used, which is known to reduce the persistence length to approximately 35 nm.18 Additionally, the mechanical force applied during AFM imaging may have contributed to this observed curvature.19
The pre-assembled dsDNA arms with 35-nt handles and HDs formed under the optimum conditions (see Materials and methods section (ESI†) for the details) were mixed at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio to obtain 2TD. Agarose gel electrophoresis of the annealed dsDNA arms showed a band at 170 bp, corresponding to the 100-bp dsDNA with 70 nt ssDNA overhangs (Fig. S2, ESI†). A subsequent gel analysis of dsDNA arms and HDs revealed two bands near 300 bp and around 170 bp (Fig. 2, lane 3). The former corresponds to the expected 2TD structure, while the latter likely represents excess unannealed dsDNA arms. The bands observed for a mixture of 2TD and the dsDNA arms without the handles also support this assignment (i.e., a band at 100 bp for the dsDNA arm without the handles and two bands at 300 bp and around 170 bp corresponding to 2TD and excess dsDNA arms with the 35-nt handles) (Fig. 2, lane 1, 2 and 4). Excess unannealed dsDNA arms could be excluded through gel extraction.
1 ratio to obtain 2TD. Agarose gel electrophoresis of the annealed dsDNA arms showed a band at 170 bp, corresponding to the 100-bp dsDNA with 70 nt ssDNA overhangs (Fig. S2, ESI†). A subsequent gel analysis of dsDNA arms and HDs revealed two bands near 300 bp and around 170 bp (Fig. 2, lane 3). The former corresponds to the expected 2TD structure, while the latter likely represents excess unannealed dsDNA arms. The bands observed for a mixture of 2TD and the dsDNA arms without the handles also support this assignment (i.e., a band at 100 bp for the dsDNA arm without the handles and two bands at 300 bp and around 170 bp corresponding to 2TD and excess dsDNA arms with the 35-nt handles) (Fig. 2, lane 1, 2 and 4). Excess unannealed dsDNA arms could be excluded through gel extraction.
We further characterized the assembled 2TD using transmission electron microscopy (TEM). Negative-stained TEM images showed a triangular-shape nanostructure (Fig. 3a and b) with a perimeter of around 100 nm (Fig. 3c). Given the length of the arms, 2TD is expected to have its perimeter of about 100 nm (i.e., 34 nm + 34 nm + 34 nm). Thus, the TEM data demonstrate the HD-mediated successful assembly of 2TD.
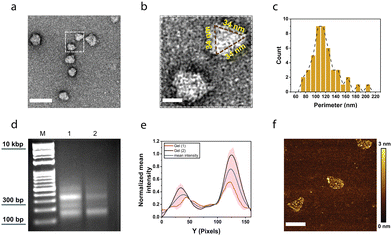 | ||
| Fig. 3 Self-assembly of the two-dimensional triangular DNA structure (2TD) using histone–DNA hybrid nanostructures (HDs). (a) Negative staining transmission electron microscopy image displaying 2TD shapes. The scale bar represents 100 nm. (b) The white box highlights an enlarged view of the region in (a). (c) Distribution of the perimeter of 2TD based on electron microscopy images. (d) Agarose gel electrophoresis (AGE) image comparing the control sample (i.e., dsDNA arms and ssDNA without histone) in lane 1 and 2TD in lane 2. The molecular weight ladder is indicated as (M). (e) Normalized intensities of the two bands present in lane 2 in (d) and Fig. 2 lane 3. The curve was smoothed using an FFT filter with a 5-point window. (f) AFM micrograph showing the successful formation of 2TD. The scale bar represents 200 nm. | ||
To further demonstrate the role of HDs in guiding 2TD assembly, we compared the 2TD with a control sample containing 100-bp dsDNA arms and 185-nucleotide ssDNA that was used to assemble HDs (i.e., 150-nt long poly-T with 35-nt handle) by agarose gel electrophoresis. The control sample showed a laddering effect with sharp bands between 100–400 bp, indicating various DNA fragments (Fig. 3d, lane 1). In contrast, 2TD displayed two sharp bands: one slightly above 100 bp that corresponds to the excess dsDNA arms with the 35-nt handles and another at around 300 bp, representing the 2TD structure (Fig. 3d, lane 2). These results suggest that HDs are critical for assembling the triangular-shaped 2TD nanostructures, further supported by imaging data. The assembly efficiency, calculated as the ratio of unannealed dsDNA arms to fully assembled 2TD structures, was approximately 36%. This value was extracted from the area under the curve of the mean normalized intensity profile of the gels (Fig. 3e) and is reasonably consistent with the 48% assembly efficiency estimated from TEM images (Fig. S3, ESI†). This calculation is based on the expected perimeter of the assembled 2TD. While minor shape variations may arise due to the structure's low rigidity, we consider perimeter measurements to be a reliable indicator, with an anticipated size variation of within ±5%.
Negative-staining TEM micrographs of the control sample revealed dsDNA arms of the expected 34 nm size, large random DNA nanostructures, and smaller DNA fragments associated with ssDNA (Fig. S4, ESI†). These observations confirm that the control sample did not form 2TD and highlight the necessity of HDs for guided 2TD assembly. Aggregation with positively charged uranyl acetate during the staining process may have contributed to the absence of large structures in the gel results (Fig. S4b, ESI†). The structure of 2TD measured by AFM showed a slight discrepancy. While the height of 2TD was consistent with the expected 2 nm, the lateral size was three times larger than expected (Fig. 3f). The discrepancy in the observed size of the 2TD structures in AFM images can be attributed to a combination of factors. The zeta sizer analysis indicates that the high salt medium used in liquid-cell imaging promotes aggregation of 2TD (Fig. S5, ESI†). The size might also be overestimated due to liquid drag forces, along with an electrical double layer surrounding the negatively charged DNA sample. These factors can lead to slight deformations and lateral expansion of the structure in the imaging medium. Additionally, the absence of expected voids within the 2TD structures supports the interpretation that these forces contribute to the observed dimensional variation.20
The successful formation and characterization of the 2TD DNA nanostructure using HDs demonstrate the potential construction of intricate structures with relative simplicity and efficiency, paving the way for more complex 3D nanostructures. The robustness and versatility of the HD-mediated assembly approach suggest its potential for precisely synthesizing a wide variety of DNA-based architectures, extending the utility of DNA nanostructures from the nano- to the micro-scale. However, the results indicate some degree of heterogeneity in the formed structures due to the non-sequence-specific interaction between histone and DNA. The 300-nt DNA segment wrapped around the histone could potentially include the sticky ends. Assembling the structure externally, followed by purification of the desired structure, will help mitigate this issue. Additionally, high rigidity is not anticipated, given the non-covalent nature of the bonds within the structure.
A similar approach was applied for assembling DNA origami tiles, a model system highlighting how HDs facilitate DNA nanostructure assembly. Here, HDs serve as versatile connectors for the hierarchical assembly. We used a multi-layer cuboid DNA origami (CDO) as a structural unit to assemble larger tiles and extended structures. To investigate how HDs promote the assembly of complex CDO tiles, we attached 25-nt long handles, which are complementary to the sticky ends on the HDs, at various locations (see Materials and methods section (ESI†) for the exact sequences used for the 25-nt long handles) (Fig. 1b); two handles positioned on each long side (CDO-1), two overhangs with one on each adjacent short and long side (CDO-3), two overhangs with one on each opposite short side (CDO-2), and four overhangs with one on each side (CDO-4).
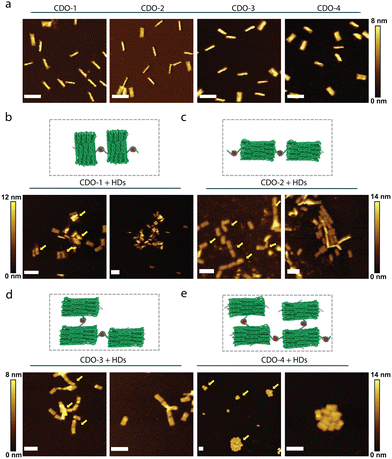
We confirmed the integrity of CDOs using AFM (Fig. 4a). Despite the variability in handle placement, the differences in the structures were not observable in the AFM micrographs due to the flexible nature of the 25-nt handles (i.e., the handles are not visible under the imaging conditions of the AFM experiment).
We then mixed CDOs with HDs with 25nt-long sticky ends complementary to the handles attached to CDOs at a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. Generally, the handles positioned on the short side of CDOs would tend to favor the formation of longitudinal tiles. In contrast, the handles on the long sides would increase the likelihood of side-by-side complexes.
1 ratio. Generally, the handles positioned on the short side of CDOs would tend to favor the formation of longitudinal tiles. In contrast, the handles on the long sides would increase the likelihood of side-by-side complexes.
After adding HDs and the self-assembly of tiles, we characterized the resulting CDO tiles using AFM. AFM micrographs of CDO-1, with two handles placed on the long sides, showed the formation of side-by-side tiles (Fig. 4b). In contrast, CDO-2, with the handles on the short sides, displayed clear longitudinal tiles (Fig. 4c). CDO-3, featuring the handles on adjacent short and long sides, exhibited bidirectional tiles and indications of crossed DNA origami (Fig. 4d). CDO-4, with the handles on all four sides, began to form a bulk structure (Fig. 4e). Mono-layered squared DNA origami (SDO) was used as control samples, including SDO alone, SDO with ssDNA (150-nt long poly-T ssDNA strands conjugated to 25-nt long sticky ends), and SDO with histones. None of these conditions showed signs of assembly into origami tiles (Fig. S6, ESI†). We selected SDO based on our previous work,13 which demonstrated the superiority of mono-layered DNA origami in visualizing binding interactions when compared to HDs. The results rule out the assembly of CDOs into tiles through non-specific interactions with histones or among CDOs (Fig. S6, ESI†) and confirm the HD-mediated assembly of the DNA origami tiles.
These results demonstrate that the strategic placement of the 25-nt handles on the cuboid DNA origami constructs directly influences the self-assembly process and the resulting structural morphology of the tiles. The ability to guide the formation of side-by-side or longitudinal assemblies by altering handle positions highlights the modularity and tunability of DNA origami-based nanostructures. This control over handle placement allows for the engineering of DNA origami tiles with specific orientations and hierarchical architectures, which is crucial for applications requiring precise nanoscale organization. Additionally, the observation that HDs can link multiple CDOs through multiple sticky ends suggests the potential for constructing more complex and interconnected nanostructures, which could overcome the traditional limitations of scaffold length and sequence.21 This insight may pave the way for more sophisticated synthetic biology and nanotechnology designs.
In summary, we successfully used histone-ssDNA building blocks (HDs) to create versatile, self-assembled DNA nanostructures without relying on sequence specificity or chemical modifications. This approach enhances our geometry-based assembly of DNA–protein hybrid nanostructures, offering simplicity, flexibility, and size scalability, significantly advancing DNA nanotechnology. Future work will focus on optimizing conditions for dynamic higher-order assembly and complex structures (e.g., 3D TDN) for practical applications. Our findings highlight the broad utility of HDs in constructing hierarchical DNA origami and pre-designed structures, paving the way for more complex and functional nanostructures while addressing the limitations of traditional scaffold-based methods.
This study was supported by King Abdullah University of Science and Technology (KAUST).
Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There is no conflict to declare.References
- P. W. K. Rothemund, Nature, 2006, 440, 297–302 CrossRef CAS.
- E. Winfree, F. Liu, L. A. Wenzler and N. C. Seeman, Nature, 1998, 394, 539–544 CrossRef CAS PubMed.
- Y. Ke, L. L. Ong, W. M. Shih and P. Yin, Science, 2012, 338, 1177–1183 CrossRef CAS PubMed.
- M. B. Danielsen, H. Mao and C. Lou, Cell Rep. Phys. Sci., 2023, 4, 101620 CrossRef CAS.
- N. Stephanopoulos, Chemistry, 2020, 6, 364–405 CrossRef CAS.
- K. Zhou, Y. Ke and Q. Wang, J. Am. Chem. Soc., 2018, 140, 8074–8077 CrossRef CAS PubMed.
- R. Sharma, A. G. Davies and C. Wälti, ACS Nano, 2014, 8, 3322–3330 CrossRef CAS PubMed.
- F. Praetorius and H. Dietz, Science, 2017, 355, eaam5488 CrossRef PubMed.
- K.-W. Wong, Z. Wang, D. He and H.-W. Li, Chem. Eng. J., 2024, 483, 149228 CrossRef CAS.
- Y. Tian, Y. He, A. E. Ribbe and C. Mao, Org. Biomol. Chem., 2006, 4, 3404 RSC.
- Y.-Y. Kim, Y. Bang, A.-H. Lee and Y.-K. Song, ACS Nano, 2019, 13, 1183–1194 CrossRef CAS PubMed.
- M. F. Serag, A. Aikeremu, R. Tsukamoto, H. Piwoński, M. Abadi, N. Kaji, J. R. Dwyer, Y. Baba and S. Habuchi, ACS Nano, 2019, 13, 8155–8168 CrossRef CAS.
- H. Al-Zarah, M. F. Serag, M. Abadi and S. Habuchi, ACS Appl. Nano Mater., 2023, 6, 9515–9522 CrossRef CAS.
- Q. Zhang, S. Lin, S. Shi, T. Zhang, Q. Ma, T. Tian, T. Zhou, X. Cai and Y. Lin, ACS Appl. Mater. Interfaces, 2018, 10, 3421–3430 CrossRef CAS.
- X. Shao, S. Lin, Q. Peng, S. Shi, X. Wei, T. Zhang and Y. Lin, Small, 2017, 13, 1602770 CrossRef.
- I. Krstić, B. Endeward, D. Margraf, A. Marko and T. F. Prisner, in EPR Spectroscopy, ed. M. Drescher and G. Jeschke, Springer Berlin Heidelberg, Berlin, Heidelberg, 2011, vol. 321, pp. 159–198 Search PubMed.
- P. J. Hagerman, Annu. Rev. Biophys. Biophys. Chem., 1988, 17, 265–286 CrossRef CAS.
- S. Brinkers, H. R. C. Dietrich, F. H. De Groote, I. T. Young and B. Rieger, J. Chem. Phys., 2009, 130, 215105 CrossRef.
- A. Stylianou, S.-V. Kontomaris, C. Grant and E. Alexandratou, Scanning, 2019, 2019, 1–25 CrossRef.
- O. Teschke, R. A. Douglas and T. A. Prolla, Appl. Phys. Lett., 1997, 70, 1977–1979 CrossRef CAS.
- A. E. Marras, MRS Commun., 2022, 12, 543–551 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Methods and materials and supplementary figures. See DOI: https://doi.org/10.1039/d4cc05253f |
| This journal is © The Royal Society of Chemistry 2025 |