 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
SAM-III aptamer enables detection of enzymatic SAM analogue generation†
Jonas
Schöning
 ab,
Aileen
Tekath
ab,
Aileen
Tekath
 b,
Nicolas V.
Cornelissen
b,
Nicolas V.
Cornelissen
 b,
Arne
Hoffmann
b,
Arne
Hoffmann
 ab and
Andrea
Rentmeister
ab and
Andrea
Rentmeister
 *ab
*ab
aDepartment of Chemistry, Ludwig-Maximilians-University Munich, Butenandtstr. 5-13, Haus F, D-81377 Munich, Germany. E-mail: a.rentmeister@lmu.de
bInstitute of Biochemistry, University of Münster, Corrensstr. 36, D-48149 Muenster, Germany
First published on 2nd December 2024
Abstract
S-Adenosyl-L-methionine (SAM) is crucial for methylation and tightly controlled in cells. We examined SAM-III riboswitch response to 17 SAM analogues and used a Spinach/SAM aptasensor to monitor their enzymatic formation in situ. Most SAM analogues were recognized, unless they featured an ortho-substituted benzyl ring, indicating potential regulatory effects via SAM riboswitches.
Riboswitches are regulatory elements found primarily in the 5′-untranslated regions of RNA from biosynthetic genes.1 Upon binding to their ligand, they undergo structural changes that affect gene expression levels. One of the most prominent natural riboswitches senses S-adenosyl-L-methionine (SAM)1,2 and regulates gene expression in the sulfur and SAM metabolism, including methionine adenosyltransferase (MAT).3 Currently eight distinct riboswitch classes have been reported for SAM, which can be divided into the SAM-I superfamily (SAM-I,4,5 SAM-IV,6 SAM-I/IV7), the SAM-II superfamily (SAM-II,8,9 SAM-V10,11), the SAM-III12 riboswitches, the SAM-VI13 riboswitches and the SAM/SAH14 riboswitches. All SAM riboswitches recognize their ligand by hydrogen bonding to the nucleobase moiety and strong electrostatic interactions with the sulfonium center (Table S1, ESI†). However, the shape of the binding cleft surrounding the methyl group differs considerably among the classes. Few SAM-like metabolites, like S-adenosyl-L-homocysteine (SAH), have been tested regarding binding to SAM riboswitches, however, no data on SAM analogues (also called AdoMet analogues) with larger alkyl or benzyl groups instead of methyl were published.15,16
Aptasensors consist of a metabolite-binding aptamer domain and a signaling domain. Fluorescent light-up aptamers (FLAPs) are particularly useful and popular signaling domains.17–20 SAM-dependent aptasensors with various FLAPs are available and have been used to monitor SAM dynamics and SAM levels in cells.15,21
SAM is the primary methylation agent and second most abundant cofactor after ATP. SAM analogues, in which the methyl group has been replaced by other residues, allow site-specific alkylation or benzylation by promiscuous methyltransferases. In particular, the enzymatic formation from methionine (analogues) and ATP catalyzed by methionine adenosyltransferase (MAT) has been demonstrated also in cells.22 Recent work on engineered MATs greatly expanded the scope of enzymatically available SAM analogues to now include a plethora of benzylic groups.23,24 However, monitoring MAT activity for SAM analogue synthesis requires HPLC analysis. Alternative approaches, like detection by phosphomolybdate, suffer from inaccuracy.25 Methods for fast and easily applicable analysis of SAM analogue detection, preferably for monitoring their in situ formation, are therefore highly required. Aptasensors could provide a convenient approach, but no data is available for the acceptance of SAM analogues.
In this study, we investigated the promiscuity of the SAM-III riboswitch for 17 SAM analogues and implemented the Spinach/SAM aptasensor for in situ detection of chemo-enzymatic synthesis of SAM analogues by an engineered MAT enzyme.
We first asked which SAM riboswitch domains may exhibit promiscuity for SAM analogues by examining their crystal structures (Fig. S1, ESI†). Among all SAM riboswitches, the SAM-III riboswitch, also known as the SMK box, has the largest binding cleft surrounding the methyl group of SAM (Fig. 1A).26 The methyl group is situated adjacent to the P2 helix, while helices P1 and P4 form the binding pocket. These helices are positioned further from the methyl group, creating a binding cleft that is open toward the riboswitch surface. This unoccupied space suggests the potential to accommodate larger substituents at the sulfonium center.
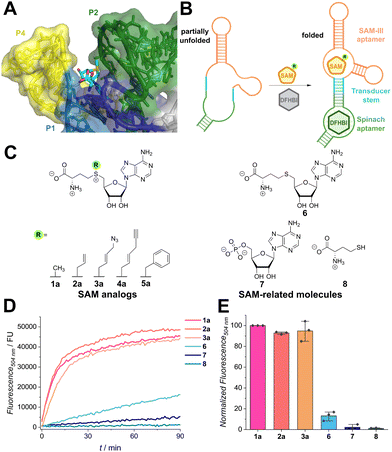 | ||
Fig. 1 Recognition of SAM analogues by the Spinach/SAM aptasensor. (A) Crystal structure of the SAM-III aptamer of Enterococcus faecalis with SAM bound (shown in sticks) (PDB: 3E5C). The methyl group at the sulfonium center (yellow) is in proximity to the P2 helix (green) and framed by the P1 (blue) and P4 helices (yellow). (B) Scheme illustrating the function of the aptasensor. Binding of SAM (analogue) induces hybridization of the transducer stem and folding of the Spinach aptamer domain. DFHBI ((Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one) can then bind and the fluorescence signal increases. (C) Chemical structures of SAM analogues (1–5) and structurally related molecules used as controls (6–8). (D) Time course showing activation of fluorescence of the Spinach/SAM aptasensor by indicated SAM analogues (500 μM) and controls (500 μM) during incubation at 25 °C. Fluorescence was measured in 1 min intervals. The net fluorescence without ligand was subtracted from the raw data to exclude analogue-independent signal increase by fluorophore binding. Assay conditions: 120 μM DFHBI, 3 μM Spinach sensor, 100 flashes, manual z-position 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 709 μm, manual gain 200 and with λEx = 460 nm/λEm = 504 nm. (E) Bar plot depicting intensity measurements recorded after 30 min. Fluorescence intensities were normalized to SAM and repeated in n = 3 independent experiments. 709 μm, manual gain 200 and with λEx = 460 nm/λEm = 504 nm. (E) Bar plot depicting intensity measurements recorded after 30 min. Fluorescence intensities were normalized to SAM and repeated in n = 3 independent experiments. | ||
To investigate the promiscuity, we tested the Spinach/SAM aptasensor designed by Jaffrey et al. that is based on the SMK box from Enterococcus faecalis.15 When SAM is bound, the transducer hybridizes, allowing the Spinach aptamer to adopt its active conformation and bind the fluorophore DFHBI (i.e., ((Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one)). Binding leads to suppression of intramolecular rotation and thus an increase in fluorescence (Fig. 1B). We first measured binding affinities for SAM and a panel of chemically synthesized SAM analogues (1a–5a, Fig. S3, ESI†) by incubating the analogues for 30 min with the aptasensor. The obtained EC50-value of 2.26 μM ± 0.10 μM is slightly higher than the literature value for the natural riboswitch (KD = 0.57 μM).16 This difference may be attributed to alterations of the transducer and Spinach domains. All tested SAM analogues were accepted by the aptasensor and with growing size of the moiety, the binding affinity decreased (EC50 = 1a: 2.26 μM < 2a: 4.1 μM < 4a: 11.0 μM < 3a: 11.6 μM < 5a: 39.5 μM). Interestingly, SAM analogue concentrations of approximately 1 mM led to a signal decrease. Similar observations were reported by Chen et al. for the Spinach aptasensor, but not for the Pepper-based construct, which might hint on SAM-interference with the Spinach aptamer.27
To prove that SAM-related metabolites do not interfere with aptasensor signaling, several metabolites were tested (Fig. 1D and E). To this end, the metabolites and the aptasensor were directly mixed and time-dependent measurements were performed. While SAM analogues show a fast increase and reach a plateau after 15 min, SAH (6) and AMP (7) only show a slow increase in fluorescence, indicating weak binding to the RNA (Fig. 1D). The amino acid L-methionine (8) does not induce fluorescence. Strong preference for SAM in contrast to SAM-related molecules after 30 min of incubation (Fig. 1E) is in line with experiments reported by Paige et al.15
We then asked if the aptasensor can be used to monitor direct in situ generation of SAM analogues. PC-MjMAT is a promiscuous MAT variant from Methanocaldococcus jannaschii that cannot only react methionine and ATP, but also various methionine analogues (Fig. 2A).23 PC-MjMAT has been shown to synthesize SAM analogues bearing small alkyl and a range of benzylic groups, including the photocleavable ortho-nitrobenzyl group.23,24 PC-MjMAT and the aptasensor were combined in one-pot reactions and a panel of 17 methionine analogues was tested (Fig. 2B). Methionine and two small analogues containing the allyl or propargyl group (2b, 9b) and the selenium (introduced for higher stability)28 were well tolerated. When we tested the benzylic methionine analogues (5b, 10b–22b), we observed that out of the 14 compounds tested, 10 were accepted by the aptasensor, leading to a fluorescent signal (Fig. 2B). We observed that the position of the substituent provides the major contribution to their acceptance by the riboswitch, following the order para > meta > ortho. One exception is the pyridinylmethyl 10b with a lower aptasensor activation that might form an additional hydrogen bond with the aptasensor. While all para- and meta-modified analogues bound to the aptamer, ortho-modified analogues did not show significant binding to the aptasensor. This included ortho-nitrobenzyl groups of the photo-cleavable compounds 19b–21b, which could be interesting for reversible blocking of methyltransferase target sites.29,30 To exclude chemically specific effects or quenching by the nitro group, the ortho-trifluoromethyl-modified analogues 11 (para), 16 (meta) and 22 (ortho) were tested. Again, modification with an ortho-modified substituent inhibited binding to the RNA whereas para- and meta-modified analogues were accepted. The ortho-substituent is closer to the nucleic acid than meta- and para-substituents, suggesting steric hindrance as main reason preventing binding.
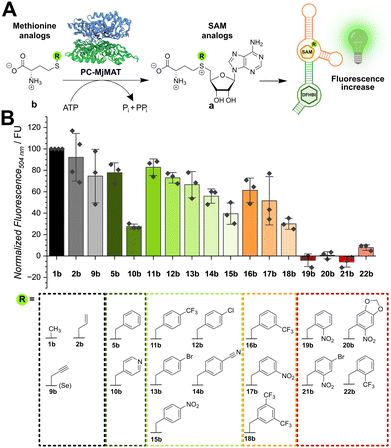 | ||
| Fig. 2 In situ detection of SAM analogue formation in enzymatic reactions. (A) Scheme for the chemo-enzymatic synthesis of SAM analogues by PC-MjMAT starting from methionine analogues. (B) Screening of SAM analogue acceptance by the SAM aptasensor by PC-MjMAT chemoenzymatic synthesis starting from methionine analogues. The bar plot shows normalized fluorescence intensities after 90 min of measurement for all SAM analogues tested. Graphs for the time-dependent fluorescence increase for all analogues are shown in Fig. S4 (ESI†). Conditions: 1 mM L-methionine analogue, 1 mM ATP, 100 μM PC-MjMAT, 120 μM DFHBI, 3 μM aptasensor, MAT/aptasensor buffer (50 mM HEPES, 100 mM KCl, 10 mM MgCl2, pH = 7.4), 25 °C. The control signal without analogue was subtracted to exclude SAM analogue independent signal increase by fluorophore binding. As 15b is a fluorophore, the control signal here included the methionine analogue but no PC-MjMAT to correct for background fluorescence. The assay was performed n = 3 times for each analog. | ||
This study investigated for the first time how SAM riboswitches respond to analogues of the cofactor SAM. The results show that the SMK box can accommodate a diverse panel of SAM analogues harboring larger alkyl or various benzyl groups. This is in line with the crystal structure showing a large solvent-exposed binding-cleft, which had led us to hypothesize that diverse SAM analogues might be accepted. Out of the 19 tested compounds, 15 were binding and led to a good signal in the SAM/Spinach aptasensor. We conclude that steric rather than electronic effects provide the key contribution to recognition of SAM analogues with benzylic groups by the SAM riboswitch. This is supported by the observation that ortho-modified benzylic analogues are not accepted.
We implemented the aptasensor for in situ detection of SAM analogues generated in enzymatic reactions. Formation of SAM analogues led directly to a fluorescent signal that was detected in 384 well format. This assay can be a valuable tool to assess the promiscuity of SAM-synthesizing enzymes and facilitate their engineering, which currently requires HPLC analyses. As the fluorescence signal by the riboswitch can be reverted by depletion of SAM (analogue), the assay might also be used to monitor the activity of SAM (analogue) consuming enzymes. In future work, the assay may also be implemented in flow cytometry to allow for high-throughput screening. As SAM-aptasensors have been utilized to measure SAM and in cells, both in bacteria and eukaryotes,15,27 our results suggest that imaging of SAM analogues may also be possible, although careful controls accounting for natural SAM would have to be performed.
A. R. thanks the DFG (CRC1459 – Project-ID 433682494 and RE2796/10-1) for financial support. The mass spectrometry and NMR facilities of the Organic Chemistry Institute at the University of Münster and of the Department of Chemistry at the LMU Munich are gratefully acknowledged for analytical services.
Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Notes and references
- R. R. Breaker, Biochemistry, 2022, 61, 137–149 CrossRef CAS.
- P. J. McCown, K. A. Corbino, S. Stav, M. E. Sherlock and R. R. Breaker, RNA, 2017, 23, 995–1011 CrossRef CAS PubMed.
- F. J. Grundy and T. M. Henkin, Mol. Microbiol., 1998, 30, 737–749 CrossRef CAS PubMed.
- W. C. Winkler, A. Nahvi, N. Sudarsan, J. E. Barrick and R. R. Breaker, Nat. Struct. Biol., 2003, 10, 701–707 CrossRef CAS.
- W. C. Winkler, A. Nahvi, A. Roth, J. A. Collins and R. R. Breaker, Nature, 2004, 428, 281–286 CrossRef CAS PubMed.
- Z. Weinberg, E. E. Regulski, M. C. Hammond, J. E. Barrick, Z. Yao, W. L. Ruzzo and R. R. Breaker, RNA, 2008, 14, 822–828 CrossRef CAS PubMed.
- J. J. Trausch, Z. Xu, A. L. Edwards, F. E. Reyes, P. E. Ross, R. Knight and R. T. Batey, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 6624–6629 CrossRef CAS PubMed.
- K. A. Corbino, J. E. Barrick, J. Lim, R. Welz, B. J. Tucker, I. Puskarz, M. Mandal, N. D. Rudnick and R. R. Breaker, Genome Biol., 2005, 6, R70 CrossRef PubMed.
- S. D. Gilbert, R. P. Rambo, D. van Tyne and R. T. Batey, Nat. Struct. Mol. Biol., 2008, 15, 177–182 CrossRef CAS PubMed.
- E. Poiata, M. M. Meyer, T. D. Ames and R. R. Breaker, RNA, 2009, 15, 2046–2056 CrossRef CAS PubMed.
- L. Huang and D. Lilley, Nucleic Acids Res., 2018, 46, 6869–6879 CrossRef CAS PubMed.
- R. T. Fuchs, F. J. Grundy and T. M. Henkin, Nat. Struct. Mol. Biol., 2006, 13, 226–233 CrossRef CAS PubMed.
- G. Mirihana Arachchilage, M. E. Sherlock, Z. Weinberg and R. R. Breaker, RNA Biol., 2018, 15, 371–378 CrossRef PubMed.
- J. X. Wang, E. R. Lee, D. R. Morales, J. Lim and R. R. Breaker, Mol. Cell, 2008, 29, 691–702 CrossRef CAS.
- J. S. Paige, T. Nguyen-Duc, W. Song and S. R. Jaffrey, Science, 2012, 335, 1194 CrossRef CAS PubMed.
- C. Lu, A. M. Smith, R. T. Fuchs, F. Ding, K. Rajashankar, T. M. Henkin and A. Ke, Nat. Struct. Mol. Biol., 2008, 15, 1076–1083 CrossRef CAS PubMed.
- X. Lu, K. Y. S. Kong and P. J. Unrau, Chem. Soc. Rev., 2023, 52, 4071–4098 RSC.
- F. Zuo, L. Jiang, N. Su, Y. Zhang, B. Bao, L. Wang, Y. Shi, H. Yang, X. Huang, R. Li, Q. Zeng, Z. Chen, Q. Lin, Y. Zhuang, Y. Zhao, X. Chen, L. Zhu and Y. Yang, Nat. Chem. Biol., 2024, 20, 1272–1281 CrossRef CAS PubMed.
- A. Autour, S. Jeng, A. D. Cawte, A. Abdolahzadeh, A. Galli, S. Panchapakesan, D. Rueda, M. Ryckelynck and P. J. Unrau, Nat. Commun., 2018, 9, 656 CrossRef PubMed.
- M. Mieczkowski, C. Steinmetzger, I. Bessi, A.-K. Lenz, A. Schmiedel, M. Holzapfel, C. Lambert, V. Pena and C. Höbartner, Nat. Commun., 2021, 12, 3549 CrossRef CAS PubMed.
- M. You, J. L. Litke and S. R. Jaffrey, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E2756–E2765 CAS.
- K. Islam, I. Bothwell, Y. Chen, C. Sengelaub, R. Wang, H. Deng and M. Luo, J. Am. Chem. Soc., 2012, 134, 5909–5915 CrossRef CAS PubMed.
- A. Peters, E. Herrmann, N. V. Cornelissen, N. Klöcker, D. Kümmel and A. Rentmeister, ChemBioChem, 2022, 23, e202100437 CrossRef CAS PubMed.
- N. V. Cornelissen, R. Mineikaitė, M. Erguven, N. Muthmann, A. Peters, A. Bartels and A. Rentmeister, Chem. Sci., 2023, 14, 10962–10970 RSC.
- I. S. Yunus, A. Palma, D. L. Trudeau, D. S. Tawfik and P. R. Jones, Metab. Eng., 2020, 57, 217–227 CrossRef CAS.
- S. Gong, Y. Wang, Z. Wang and W. Zhang, J. Phys. Chem. B, 2016, 120, 12305–12311 CrossRef CAS PubMed.
- Z. Chen, W. Chen, Z. Reheman, H. Jiang, J. Wu and X. Li, Nucleic Acids Res., 2023, 51, 8322–8336 CrossRef CAS PubMed.
- S. Willnow, M. Martin, B. Lüscher and E. Weinhold, ChemBioChem, 2012, 13, 1167–1173 CrossRef CAS PubMed.
- J. Hemphill, J. Govan, R. Uprety, M. Tsang and A. Deiters, J. Am. Chem. Soc., 2014, 136, 7152–7158 CrossRef CAS PubMed.
- N. Klöcker, F. P. Weissenboeck, M. van Dülmen, P. Špaček, S. Hüwel and A. Rentmeister, Nat. Chem., 2022, 14, 905–913 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cc04860a |
| This journal is © The Royal Society of Chemistry 2025 |
