Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Nanophotonic platform for the highly sensitive detection of trypsin enzyme in human urine†
Shrishty
Bakshi
 *,
Kezheng
Li
*,
Kezheng
Li
 ,
Pankaj K.
Sahoo
and
Thomas F.
Krauss
,
Pankaj K.
Sahoo
and
Thomas F.
Krauss
School of Physics, Engineering and Technology, University of York, York, YO10 5DD, UK. E-mail: shrishty.bakshi@york.ac.uk
First published on 13th March 2025
Abstract
Trypsin is an important indicator of pancreatic disease. We introduce a nanophotonic test for the detection of trypsin. Our approach is based on the hydrolysis of β-casein by trypsin. We demonstrate 0.1 ng mL−1 of trypsin detection in human urine and across the clinically relevant range.
Pancreatic diseases are some of the most common causes of illness around the globe, affecting a significant number of people globally.1,2 The incidence of pancreatitis, one of the most common pancreas diseases, results in 12 000 hospital admissions per year in the UK alone3 and the problem is rapidly increasing worldwide.4 These observations call for a better, highly sensitive testing technology that is easily accessible and that can help detect pancreatic diseases at an early stage for better management and timely treatment. Trypsin is an important enzyme, used for studying pancreatic diseases.5,6 Trypsin is involved in the hydrolysis of protein molecules into shorter peptide fragments to enhance their absorption.7,8 Studies have found that there is no trypsin found in the urine of healthy people. However, the urine of patients who have undergone pancreas transplants contains high levels of trypsin, i.e. concentrations on the order of 10's of μg mL−1.9,10 We also note that trypsin plays an important role in the progression of pancreatic cancer and development of diseases such cystic fibrosis, viral diseases, etc.11–13
The techniques commonly used for trypsin detection are enzyme-linked immunosorbent assay (ELISA),14 mass spectrometry,15 fluorescent-based assays,9 radioimmunoassay16 and gel electrophoresis.17 The major drawback of these methods is the requirement for labelling and the need for bulky and sensitive instrumentation as well as skilled labour, which results in high costs and lengthy analysis times, thus reducing the uptake of such tests. Efforts to develop point-of-care biosensing devices for trypsin detection are underway,18 but most approaches are based on fluorescence,11,19–21 which requires labelling and more complicated read-out instruments. Alternatively, tests that are more suitable for near-patient use tend to have lower sensitivity and a limited dynamic range.22 The ESI,† Table S1 provides a more detailed comparison of different label-free sensing technologies developed for the detection of trypsin. Overall, we observe that a highly sensitive trypsin biosensor with a large dynamic range that is easy to use and can be realised cost-effectively has not yet been demonstrated.
Here, we introduce such a biosensor. Our modality is based on a wavelength-scale grating that supports guided mode resonances (GMRs), which can be readily excited with collimated light from an LED. This modality is much easier to implement than many other photonic sensors that require the difficult coupling of laser light into a waveguide of submicron cross-section. By continuously varying (“chirping”) the period of the grating, we translate spectral information into spatial information such that the readout can be performed with a simple CMOS camera, thereby removing the requirement for a high-resolution spectrometer.23 In addition, to address any drift caused by the light source or temperature-related fluctuations, a reference channel is placed in close proximity to the measurement channel. We have also shown that the sensor can be realised in a handheld format without loss of performance24 and that it is robust against thermal fluctuations and mechanical vibration;25 hence, it is ideally suited for near-patient deployment. Here, we present evidence that this modality is also effective for monitoring trypsin levels in clinical fluids, especially human urine. The sensing principle is based on the hydrolysis of a β-casein layer immobilised on the sensor surface by the trypsin enzyme.
A robust and stable surface chemistry ensures the sensitive detection of analytes of interest from a complex matrix. Fig. 1(a–d) describes the preparation and operation of the biological interface used here. The interface employs polydopamine surface chemistry, which we have shown to be stable and effective.26,27 The polydopamine layer is applied to the grating to immobilise the β-casein substrate. The grating is fabricated using electron beam lithography, and it exhibits a resonance when illuminated by light at 650 nm wavelength (Fig. 2(a)). As we have already shown, the sensors can also be fabricated using nanoimprint lithography for larger-scale commercial purposes.24
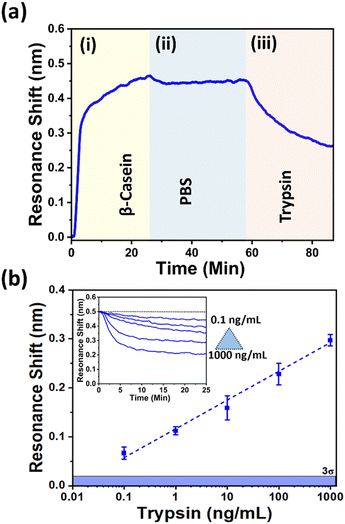
We first perform the measurements in phosphate buffer saline (PBS) solution. Typically, GMR sensors exhibit an increase in resonance wavelength (red shift) when proteins bind to the sensor surface. This occurs because the bound material increases the refractive index of the cladding, thereby causing the resonant mode to shift. Since we chirp the grating's period and illuminate with a fixed wavelength, this will translate the resonant position towards a lower period as the refractive index increases. The position of the resonance is then captured by a CMOS camera (Fig. 2a). In the case of detecting enzymatic activity, the resonance moves in the opposite direction (blue-shift), because trypsin digests the β-casein immobilised on the surface, resulting in a mass loss on the sensor surface. By tracking the resonance position over time, the kinetics of the digestion of the β-casein layer is monitored. Fig. 3a shows the sequence of the full assay, starting from the immobilisation of β-casein, resulting in mass gain, to washing, to the introduction of trypsin (10 ng mL−1), resulting in mass loss. Fig. 3b then shows the dynamic range for different trypsin concentrations, varying between 0.1 ng mL−1 and 1000 ng mL−1. We note that for a concentration of 0.1 ng mL−1, the observed shift is approx. 0.066 nm, which is well above the 3σ (mean 3σ = 0.019 nm ± 0.01 nm (n = 31 ± SD)) value of the system. For the higher concentrations of up to 1000 ng mL−1, we observe a linear trend with an R2 value of 0.99383; this high R2 value highlights that our sensor is suitable for quantifying different trypsin concentrations. The inset in Fig. 3b displays the individual unbinding curves for various concentrations. To demonstrate the specificity of the sensor against interfering molecules in human urine, we also studied the effect of urea and glucose. For reference, when using 100 ng mL−1 of trypsin in PBS, we observe a resonance shift of 0.228 ± 0.02 nm (n = 3). When adding 2% urea, the shift is 0.231 ± 0.01 nm (n = 3) for 100 ng mL−1 trypsin concentration and when adding 100 μg mL−1 of glucose, the shift is 0.232 ± 0.03 nm (n = 3) for 100 ng mL−1 trypsin concentration. Given that all values are comparable we can be confident that the main interferences in human urine only have a negligible effect on the sensor readout.
Finally, we assess the suitability of our sensor to perform trypsin detection in a real human matrix, here undiluted human urine. As before, we show the sequence of the assay for enzyme detection in human urine (Fig. 4a). The first step of β-casein immobilisation is the same, as shown in Fig. 3a(i). The second step, urine introduction, causes an upward shift because urine has a slightly higher refractive index than PBS Fig. 4a(ii). We note that the resonance curve is stable, indicating that there is neither fouling nor any other enzymatic reaction from interfering molecules (note that the urine was pooled from a sample of healthy volunteers, which may contain a variety of different molecules), which gives us confidence that our assay is specific to trypsin. When we introduce the same urine spiked with trypsin (Fig. 4a(iii)), we see a clear unbinding curve, indicating that the trypsin digests the substrate.
We used the same concentration range as before, 0.1 ng mL−1 to 1000 ng mL−1, and Fig. 4b shows the corresponding resonance shifts. The shift observed for 0.1 ng mL−1 is approximately 0.065 nm, which is above the 3 Sigma value. As before, we observe a linear trend with a high R2 value of 0.99469.
In conclusion, we have demonstrated a diagnostic test based on guided mode resonances that is able to detect the trypsin enzyme at concentrations as low as 0.1 ng mL−1. The response of the test is linear over a concentration range spanning 4 orders of magnitude, which includes the clinically relevant range for the diagnosis of pancreatic disease. The test performs equally well in phosphate buffered saline (PBS) as in undiluted human urine, which gives us confidence in its robustness and its ability to cope with the large variety of patient samples that are encountered in practice. The test can be used for the screening and monitoring at the point of care as we have previously demonstrated its realisation as a handheld instrument.24 In addition to the specific pancreatic disease application we describe here, the work opens up the possibility of detecting other types of enzymes with the choice of appropriate substrates. By combining the high performance of laboratory instruments with the convenience of a near-patient test, we aim to make this important diagnostic more widely available to improve the early detection and improved management of disease. In this scenario, we note that long-term storage of the functionalised sensors is required, which can be achieved by stabilising the sensor surface with sugars and polyhydric alcohols.28,29
We acknowledge funding by UK Research & Innovation under grants EP/V047434/1, EP/X037770/1 and EP/V047663/1.
Data availability
Data will be made available on reasonable request.Conflicts of interest
The authors declare there are no conflicts of interest.Notes and references
- J. Huang, V. Lok, C. H. Ngai, L. Zhang, J. Yuan, X. Q. Lao, K. Ng, C. Chong, Z. J. Zheng and M. C. S. Wong, Gastroenterology, 2021, 160, 744–754 CrossRef PubMed.
- A. Pourshams, et al. , Lancet Gastroenterol. Hepatol., 2019, 4, 934–947 CrossRef PubMed.
- T. C. Hall, G. Garcea, M. A. Webb, D. Al-Leswas, M. S. Metcalfe and A. R. Dennison, J. Eval. Clin. Pract., 2014, 20, 203–207 CrossRef PubMed.
- G. Ouyang, G. Pan, Q. Liu, Y. Wu, Z. Liu, W. Lu, S. Li, Z. Zhou and Y. Wen, BMC Med., 2020, 18, 388 CrossRef PubMed.
- J. Yin, T. Cui, Y. Yang and T.-L. Ren, Chemosensors, 2023, 11, 469 CrossRef CAS.
- V. V. Gummadi and T. Gonska, Pancreas, 2024, 10–97 Search PubMed.
- J. Kaur and P. K. Singh, Crit. Rev. Anal. Chem., 2022, 52, 949–967 CrossRef CAS PubMed.
- V. G. Vertiprakhov and N. V. Ovchinnikova, Front. Physiol., 2022, 13, 874664 CrossRef PubMed.
- J. Xu, E. Prost, K. Haupt and B. Tse Sum Bui, Sens. Actuators, B, 2018, 258, 10–17 CrossRef CAS.
- W. A. See and J. L. Smith, Transplantation, 1991, 52, 630–633 CrossRef CAS PubMed.
- C. Munoz-San Martin, M. Pedrero, M. Gamella, A. Montero-Calle, R. Barderas, S. Campuzano and J. M. Pingarron, Anal. Bioanal. Chem., 2020, 412, 6177–6188 CrossRef CAS PubMed.
- P. Nyberg, M. Ylipalosaari, T. Sorsa and T. Salo, Exp. Cell Res., 2006, 312, 1219–1228 CrossRef CAS PubMed.
- O. G. Famutimi, V. G. Adebiyi, B. G. Akinmolu, O. V. Dada and I. O. Adewale, Futur. J. Pharm. Sci., 2024, 10, 126 CrossRef.
- M. A. Seia, P. W. Stege, S. V. Pereira, I. E. De Vito, J. Raba and G. A. Messina, Anal. Biochem., 2014, 463, 31–37 CrossRef CAS PubMed.
- P. A. Everley, C. A. Gartner, W. Haas, A. Saghatelian, J. E. Elias, B. F. Cravatt, B. R. Zetter and S. P. Gygi, Mol. Cell. Proteomics, 2007, 6, 1771–1777 CrossRef CAS PubMed.
- C. O'Connor, M. O'Donnell and K. McGeeney, Clin. Chim. Acta, 1981, 114, 29–35 CrossRef PubMed.
- R. B. Lefkowitz, G. W. Schmid-Schonbein and M. J. Heller, Electrophoresis, 2010, 31, 2442–2451 CrossRef CAS PubMed.
- G. Erturk, M. Hedstrom and B. Mattiasson, Biosens. Bioelectron., 2016, 86, 557–565 CrossRef CAS PubMed.
- S. Xu, F. Zhang, L. Xu, X. Liu, P. Ma, Y. Sun, X. Wang and D. Song, Sens. Actuators, B, 2018, 273, 1015–1021 CrossRef CAS.
- L. Hu, S. Han, S. Parveen, Y. Yuan, L. Zhang and G. Xu, Biosens. Bioelectron., 2012, 32, 297–299 CrossRef CAS PubMed.
- M. Wang, D. Su, G. Wang and X. Su, Anal. Bioanal. Chem., 2018, 410, 6891–6900 CrossRef CAS PubMed.
- M. Nemati, A. Santos, T. Kumeria and D. Losic, Anal. Chem., 2015, 87, 9016–9024 CrossRef CAS PubMed.
- G. J. Triggs, Y. Wang, C. P. Reardon, M. Fischer, G. J. O. Evans and T. F. Krauss, Optica, 2017, 4, 229–234 CrossRef CAS PubMed.
- A. Drayton, K. Li, M. Simmons, C. Reardon and T. F. Krauss, Opt. Express, 2020, 28, 32239–32248 CrossRef PubMed.
- K. Li, N. J. Suliali, P. K. Sahoo, C. D. Silver, M. Davrandi, K. Wright, C. Reardon, S. D. Johnson and T. F. Krauss, ACS Sens., 2024, 9, 1857–1865 CrossRef CAS PubMed.
- S. Bakshi, K. Li, P. Dong, I. Barth, C. Kunstmann-Olsen, S. Johnson and T. F. Krauss, Talanta, 2024, 268, 125300 CrossRef CAS PubMed.
- S. Bakshi, P. K. Sahoo, K. Li, S. Johnson, M. J. Raxworthy and T. F. Krauss, Biosens. Bioelectron., 2023, 242, 115743 CrossRef CAS PubMed.
- S. Gerlsma, J. Biol. Chem., 1968, 243, 957–961 CrossRef CAS PubMed.
- J. F. Back, D. Oakenfull and M. B. Smith, Biochemistry, 1979, 18, 5191–5196 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cc04840g |
| This journal is © The Royal Society of Chemistry 2025 |