Open Access Article
Open Access ArticleLate-stage peptide modification with salicylaldehyde tag enhances affinity for nuclear factor-kappa B essential modulator
Mattia Mason a,
Kaliroi Peqini
a,
Kaliroi Peqini b,
Federico Uggeri
b,
Federico Uggeri a,
Diego Rondellibc,
Sara Sattin
a,
Diego Rondellibc,
Sara Sattin a,
Luca Pignataro
a,
Luca Pignataro a,
Sara Pellegrino
a,
Sara Pellegrino b,
Laura Belvisi
b,
Laura Belvisi a,
Edoardo Scarpabc,
Raffaella Bucci
a,
Edoardo Scarpabc,
Raffaella Bucci *b and
Alberto Dal Corso
*b and
Alberto Dal Corso *a
*a
aUniversità degli Studi di Milano, Dipartimento di Chimica, 20133 Milano, Italy. E-mail: alberto.dalcorso@unimi.it
bUniversità degli Studi di Milano, Dipartimento di Scienze Farmaceutiche, 20133 Milano, Italy. E-mail: raffaella.bucci@unimi.it
cIstituto Nazionale di Genetica Molecolare (INGM), 20122 Milano, Italy
First published on 1st December 2025
Abstract
The late-stage peptide functionalization with salicylaldehyde (SA) tags is described here as a versatile design of potential Lys-engaging, reversible-covalent ligands. This approach was applied to a known binder for NEMO, a protein involved in the activation of the pro-inflammatory transcription factor NFκB. Fluorescence anisotropy screening led to the identification of SA-tagged peptides with higher affinity than the wild-type sequence.
Introduction
In recent years, growing attention has been devoted to the identification of peptide and peptidomimetic ligands capable of selectively targeting proteins of interest,1 particularly those forming covalent bonds with their targets.2 This emerging class of covalent peptide binders offers unique opportunities for designing potent and durable modulators of protein functions. Salicylaldehyde (SA) derivatives have emerged as powerful amine-engaging units and, stimulated by the high frequency of Lys residues in the proteome,3 they have been incorporated in various small molecule drugs,4 glycomimetic5 and peptide6 ligands.In the latter context, considering that protein–peptide binding interactions typically span a wide interface area, the precise insertion of the SA tag at suitable peptide positions could be crucial to covalently engage a specific Lys(ε-NH2) group of the target protein. Ideally, this additional covalent interaction could synergize with the original non-covalent forces, resulting in a highly stable ligand–protein complex.
With this aim, we recently reported on the synthesis of non-coded amino acids, substituting the homoserine side chain with a SA tag. These new scaffolds, endowed with suitable protecting groups, could be inserted in primary sequences during peptide synthesis.7 Unfortunately, these modified amino acids could be obtained only with long and inefficient multistep synthetic sequences, which strongly limit their use as building blocks for peptide production. For this reason, we looked at more practical and versatile design of SA-tagged peptides.
“Click” chemistry approaches, such as the copper-catalysed azide–alkyne cycloaddition (CuAAC), offer unique opportunities to connect two highly functionalized molecules in an efficient and chemoselective way.8 In particular, CuAAC proved a robust approach to connect the azido group in biomolecules (i.e. nucleic acids9 and peptides N and C-termini6) to alkyne-bearing SA derivatives, devoid of protecting groups on the aldehyde and phenol units.
Propargylglycine (Pra) is a non-coded amino acid featuring a terminal alkyne group on its side chain: its N-Fmoc protected version is commercially available in both enantiomeric forms, enabling the insertion of alkyne units in peptide sequences through solid phase peptide synthesis (SPPS).
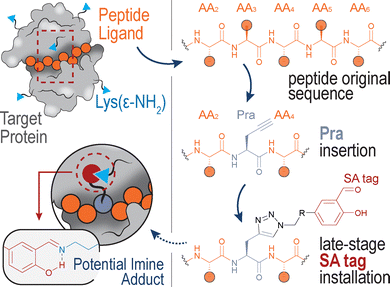
Inspired by the high versatility of the Pra amino acid, we designed a practical strategy for the preparation of potential Lys-engaging peptides (Fig. 1), consisting in the Pra insertion within the peptide sequence and subsequent installation of the SA tag through CuAAC.
A known peptide-protein interaction was selected as a case study to verify the feasibility of our approach, demonstrating that the generation of a panel of SA-tagged peptides and their screening against the protein of interest can lead to hit compounds with enhanced binding affinity compared to the wild-type peptide sequence.
Results and discussion
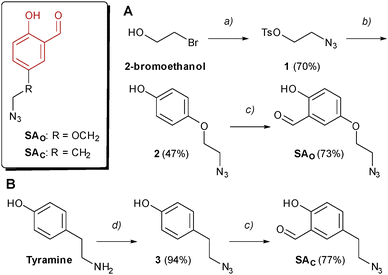
Two SA derivatives bearing an azido group were designed and prepared as highlighted in Scheme 1. In particular, the azido group was connected to the aromatic ring through either a phenol ether bond (SAO) or a short alkyl chain (SAC). SAO was prepared starting from 2-bromoethanol, which was transformed into the corresponding azide through an SN2 reaction and then treated with tosyl chloride, to give compound 1 in good yields. Following the same strategy reported by our group in 2021,10 the tosyl ester was reacted with hydroquinone, to give ether 2, and then subjected to a regioselective phenol ortho-formylation under the Skattebøl conditions. The second SA-azide unit (SAC) was prepared in only two synthetic steps, starting from commercially available tyramine. Following a published protocol,11 the diazo-transfer reagent imidazole-1-sulfonylazide was prepared as a hydrogen sulphate salt, which can be handled safely as a solid. This reagent converted tyramine into the corresponding azide 3 in excellent yield (94%). Finally, a phenol ortho-selective formylation led to the desired SA-azide derivative SAC. The detailed reaction procedures and analytical data are included in the SI.To compare the reactivity of aldehydes SAO and SAC towards imine bond formation in aqueous solution, a nuclear magnetic resonance (NMR) experiment was performed, following published protocols.9,12 Stock solutions of the SA-azide derivatives in DMSO-d6 were diluted in phosphate buffer prepared in deuterium oxide (pH 7.4), and 1H-NMR spectra were recorded both in the presence or absence of Nα-acetyl-lysine in 10-fold molar excess. As shown in Fig. S1 (SI), after 1 h incubation at room temperature, the presence of Lys in the mixture led to approx. 33% conversion of both aldehydes into the corresponding imines, indicating that the SAO and SAC are similarly reactive toward the Lys(ε-NH2) group.
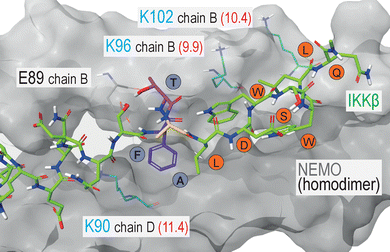
The synthesized SA-azide derivatives were then installed in a model peptide binder. As a case study, we selected the protein–protein interaction (PPI) between the Ser/Thr-specific IκB kinase (IKK) and the nuclear factor-kappa B essential modulator (NEMO), which represents an important step in the activation of the pro-inflammatory transcription factor NFκB. Given this relevant role in the inflammatory signalling cascade, the IKK:NEMO PPI has been identified as an attractive clinical target and as subject of various peptide discovery campaigns.13 The TALDWSWLQTE sequence of IKKβ proved a fundamental player of this PPI, leading to the identification of this 11-mer fragment as NEMO-binding peptide (NBP).14 Interestingly, while NBP does not include Lys residues, the crystal structure of the NEMO-IKKβ complex revealed the presence of three Lys(ε-NH2) groups in NEMO, proximal to the interface with the 11-mer IKKβ fragment (Fig. 2).15
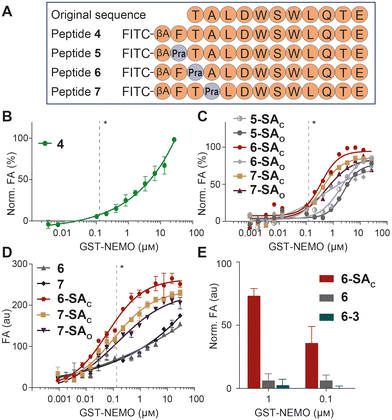
The analysis of Lys(ε-NH3+) pKa, performed with the Rosetta software, led to pKa values of 11.4, 9.9 and 10.4 for NEMO K90, K96 and K102, respectively (shown in Fig. 2), indicating that K96 may be the most nucleophilic site of the series. This residue, together with K90, is particularly close to IKKβ NBD (the measured distances vary from approximately 7 and 11 Å, all details are shown in Fig. S2), lying in the surrounding of the peptide N-terminus. In this region, the short distance between the amide proton in the backbone of IKKβ F734 and the side chain of NEMO E89 indicates the formation of an intermolecular H-bond (Fig. S2). In addition, F734 shows hydrophobic interactions with NEMO L93. As these non-covalent forces conceivably play a significant role in the complex stabilization, we included the F734 residues as the peptides’ N-termini, resulting in the final FTALDWSWLQTE sequence. Finally, moving towards the peptide C-terminus, the hydrophobic features and the peculiar conformation of the WSWL fragment appear crucial for the PPI stabilization. In light of these observations, the three N-terminal residues of the FTALDWSWLQTE sequence (i.e., IKKβ residues F734, T735 and A736) were identified as potential anchoring points for the SA tag, ideally aimed at the engagement of either K90 or K96, without disrupting the relevant non-covalent interactions of the wild-type sequence (Fig. 3A). As a final feature of the target peptides, a β-alanine unit was located at the N-termini as a spacer for the peptides labelling with fluorescein isothiocyanate (FITC), an essential dye for fluorescent anisotropy (FA) studies.
The designed peptide sequences are represented in Fig. 3A, and include the wild-type peptide 4 and its three analogues with single-point mutations at N-terminal F, T and A residues with Pra (peptides 5, 6 and 7, respectively). These peptides were prepared by automated, microwave-assisted SPPS, an efficient technique for peptide production within a short period of time.16 Following the HPLC purification of Pra-bearing peptides 5-7, the alkyne group of each compound was easily connected to either SAO or SAC azides through CuAAC, to give the corresponding SA-tagged peptides. All compounds were purified by HPLC and the chromatograms of crude reaction mixtures (see Fig. S3 in the SI) indicated quantitative conversions of all alkynes 5-7 into the corresponding triazoles. Isolated products were characterized by LC-MS, as detailed in the SI.
Following a published protocol,13b FA assays were performed using recombinant NEMO, tagged with glutathione-S-transferase (GST) and produced by a commercial supplier. The resulting peptide (compound 4 in Fig. 3A) showed low affinity for GST-NEMO (Kd > 25 µM, as no FA saturation was observed at high protein concentrations, see Fig. 3B), in line with data reported elsewhere for NBP.17 At this stage, the six SA derivatives were screened in parallel against GST-NEMO in FA binding studies, and the results are shown in Fig. 3C. Among these compounds, 6-SAC, 7-SAC and 7-SAO showed the best binding performances, with saturation of FA values over 1 µM concentration. Of note, we observed no significant differences among FA data recorded at various incubation times. For homogeneity, all data described here were obtained upon 30 minutes of incubation.
This screening prompted us to investigate the affinity of these hit compounds in comparison with their cognate alkyne precursors 6 and 7. The results of this experiment, performed in duplicate, are shown in Fig. 3D, unveiling a remarkably higher binding affinity of SA derivatives compared to the original Pra-modified peptides. Compound 6-SAC proved particularly promising, with an apparent Kd value of 60 ± 9 nM. Finally, to verify the impact of the peculiar SA reactivity on the observed affinity, the alkyne group of peptide 6 was reacted with the azide unit of phenol 3, devoid of the formyl group. This led to an analogue of lead compound 6-SAC (named 6-3), with suppressed aminophilic behaviour. In a dedicated FA study, including peptide 6 and its derivatives 6-SAC and 6-3, the absence of the aldehyde proved detrimental for the adhesion properties of the peptide (Fig. 3E), thus confirming the importance of the SA reactivity for the peptide binding potency.
These FA data confirmed our hypothesis that the tailored insertion of the SA tag into a peptide binder can significantly increase its affinity for the protein target. With the present data, we can only presume that the enhanced affinity is due to the formation of an intermolecular imine adduct, as only in-depth investigations through mass spectrometry or X-ray crystallography can ultimately prove this interaction.
At this stage, we focused on the characterization of our peptide's solubility properties, as the renowned solubility issues of peptides can hinder the follow-up investigations, both in cellular and biochemical assays in vitro. For this purpose, the aldehyde-bearing peptide 6-SAC and its alkyne precursor (6) were dissolved in DMSO at various concentrations and then diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in PBS, to give samples at 5 µM, 125 nM, and 10 nM in PBS (+1% DMSO). These samples were then subjected to dynamic light scattering (DLS) studies and the results are shown in Fig. S4 (SI). In particular, while peptides formed large and stable aggregates at 5 µM concentrations, the 125 and 10 nM solutions resulted in a rapid decay of correlograms and a shift of size distributions toward small aggregates. These data indicate the 125 nM concentration (i.e., the value adopted in FA studies) as a solubility limit for these species in solution. As a consequence, future studies involving higher sample concentrations will require structural optimization or a fine tuning of sample preparation protocols (e.g., screening of buffers, surfactants, etc.).
100 in PBS, to give samples at 5 µM, 125 nM, and 10 nM in PBS (+1% DMSO). These samples were then subjected to dynamic light scattering (DLS) studies and the results are shown in Fig. S4 (SI). In particular, while peptides formed large and stable aggregates at 5 µM concentrations, the 125 and 10 nM solutions resulted in a rapid decay of correlograms and a shift of size distributions toward small aggregates. These data indicate the 125 nM concentration (i.e., the value adopted in FA studies) as a solubility limit for these species in solution. As a consequence, future studies involving higher sample concentrations will require structural optimization or a fine tuning of sample preparation protocols (e.g., screening of buffers, surfactants, etc.).
Conclusions
We have described herein a protocol for a general functionalization of peptide ligands with the SA tag, i.e., a well-known amine-engaging electrophile. Ideally, our design aims at the reversible-covalent engagement of amino groups in proteins and the generation of lead peptides with ultra-high affinity. The preliminary insertion of the Pra residue in a restricted area of the peptide's primary sequence was guided by visual inspection of crystal structures available in the PDB, but more systematic approaches are also feasible. The late-stage peptide modification with SA-azide fragments (SAO and SAC) through CuAAC avoids the use of protecting groups and generates the final products with high efficiency (see chromatograms in Fig. S3), ideally allowing the application of this strategy in highly diluted peptide solutions (e.g., in combinatorial libraries).In our NEMO-binding peptides, the increased binding affinity observed for our SA-tagged ligands compared to the control compounds supports the feasibility of SA-tagged “anti-inflammatory peptides”. However, the self-assembly tendency of these peptides, revealed by DLS, may limit their efficient cellular uptake and the interaction with the biological target. Therefore, future biological evaluation will consider peptide incorporation in copolymers or the installation of cell-penetrating sequences.
Considering the high abundance of Lys residues in the proteome and their preferential expression on protein surfaces, we believe that the SA insertion into peptide ligands holds promises for the development of synthetic ligands with unprecedented affinities and prolonged residence time on the target.
Author contributions
Conceptualization, supervision and writing: RB, ADC; methodology, investigation and data curation: MM, KP, FU, LB, DR, ES; resources: SS, LP, SP; funding acquisition: RB, ADC, ES, SS, LB.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5cb00282f.Acknowledgements
We thank M. Rossoni for its assistance with the computational analysis. The Università degli Studi di Milano is gratefully acknowledged for the PhD fellowship (to MM) and for financial contributions (to RB and ADC, My First SEED Grant 2023 “RECIPe”). E.S. acknowledges Fondazione Cariplo (grant number 2022-0438) for financial support. SS was funded by the European Union – Next Generation EU, Mission 4, Component 2, CUP B93D21010860004.References
- (a) L. Lombardi, V. Del Genio, F. Albericio and D. R. Williams, Chem. Rev., 2025, 125, 7099 CrossRef CAS PubMed; (b) D. Petrov, L. Plais, K. Schira, J. Cai, M. Keller, A. Lessing, G. Bassi, S. Cazzamalli, D. Neri, A. Gloger and J. Scheuermann, Nat. Commun., 2025, 16, 3273 CrossRef CAS PubMed; (c) K. Colas, D. Bindl and H. Suga, Chem. Rev., 2024, 124, 12213 CrossRef CAS PubMed; (d) M. L. Merz, S. Habeshian, B. Li, J. G. L. David, A. L. Nielsen, X. Ji, K. Il Khwildy, M. M. Duany Benitez, P. Phothirath and C. Heinis, Nat. Chem. Biol., 2024, 20, 624 CrossRef CAS PubMed; (e) B. D. Ellenbroek, J. P. Kahler, S. R. Evers and S. J. Pomplun, Angew. Chem., Int. Ed., 2024, 63, e202401704 CrossRef CAS PubMed; (f) R. Bucci, L. De Rosa, G. Bertoni, R. Di Stasi, M. della Valle, D. Diana, S. Peppicelli, K. Peqini, M. L. Gelmi, F. Bianchini and L. D. D'Andrea, Bioorg. Chem., 2025, 165, 109039 CrossRef CAS PubMed; (g) D. Di Lorenzo, N. Bisi, R. Bucci, I. Ennen, L. Lo Presti, V. Dodero, R. Brandt, S. Ongeri, M. L. Gelmi and N. Tonali, iScience, 2025, 28, 112272 CrossRef CAS PubMed.
- (a) F. M. Paulussen and T. N. Grossmann, J. Pept. Sci., 2023, 29, e3457 CrossRef CAS PubMed; (b) N. M. Grob, C. Remarcik, S. L. Rössler, J. Y. K. Wong, J. C. K. Wang, J. Tao, C. L. Smith, A. Loas, S. L. Buchwald, D. L. Eaton, M. Preciado López and B. L. Pentelute, ACS Chem. Biol., 2024, 19, 101 CrossRef CAS PubMed; (c) E. Oduro-Kwateng, M. Ali, I. O. Kehinde, L. Rao and M. E. S. Soliman, BMC Methods, 2025, 2, 6 CrossRef.
- G. Kim, J. Grams and K. L. Hsu, Chem. Rev., 2025, 125, 6653 CrossRef CAS PubMed.
- (a) C. Gampe and V. A. Verma, J. Med. Chem., 2020, 63, 14357 CrossRef CAS PubMed; (b) M. Mason, L. Belvisi, L. Pignataro and A. Dal Corso, ChemBioChem, 2024, 25, e202300743 CrossRef CAS PubMed.
- G. Antonini, A. Bernardi, E. Gillon, A. Dal Corso, M. Civera, L. Belvisi, A. Varrot and S. Mazzotta, J. Med. Chem., 2024, 67, 19546 CrossRef CAS PubMed.
- G. Sacco, D. Arosio, M. Paolillo, A. Gloger, J. Scheuermann, L. Pignataro, L. Belvisi, A. Dal Corso and C. Gennari, Chem. – Eur. J., 2023, 29, e202203768 CrossRef CAS PubMed.
- M. Mason, B. Nava, L. Belvisi, L. Pignataro and A. Dal Corso, Eur. J. Org. Chem., 2024, e202400229 CrossRef CAS.
- M. Meldal and C. Wenzel Tornøe, Chem. Rev., 2008, 108, 2952 CrossRef CAS PubMed.
- A. Dal Corso, M. Catalano, A. Schmid, J. Scheuermann and D. Neri, Angew. Chem., Int. Ed., 2018, 57, 17178 CrossRef CAS PubMed.
- G. Sacco, S. Stammwitz, L. Belvisi, L. Pignataro, A. Dal Corso and C. Gennari, Eur. J. Org. Chem., 2021, 1763–1767 CrossRef CAS.
- (a) G. T. Potter, G. C. Jayson, G. J. Miller and J. M. Gardiner, J. Org. Chem., 2016, 81, 3443 CrossRef CAS PubMed; (b) Y. Zhang, Q. Zhang, C. T. T. Wong and X. Li, J. Am. Chem. Soc., 2019, 141, 12274–12279 CrossRef CAS PubMed.
- C. Delmas, E. Sager, C. Henry, U. Hassiepen, P. R. Skaanderup and I. Kerschgens, Chimia, 2025, 79, 152 CrossRef CAS PubMed.
- (a) H. Yu, L. Lin, Z. Zhang, H. Zhang and H. Hu, Signal Transduction Targeted Ther., 2020, 5, 209 CrossRef CAS PubMed; (b) C. A. Rhodes, P. G. Dougherty, J. K. Cooper, Z. Qian, S. Lindert, Q. E. Wang and D. Pei, J. Am. Chem. Soc., 2018, 140, 12102 CrossRef CAS PubMed.
- (a) M. J. May, F. D’Acquisto, L. A. Madge, J. Glöckner, J. S. Pober and S. Ghosh, Science, 2000, 289, 1550 CrossRef CAS PubMed; (b) J. S. Orange and M. J. May, Cell. Mol. Life Sci., 2008, 65, 3564 CrossRef CAS PubMed; (c) L. Opazo-Ríos, A. Plaza, Y. er Sánchez Matus, S. Bernal, L. Lopez-Sanz, L. Jimenez-Castilla, D. Carpio, A. Droguett, S. Mezzano, J. Egido and C. Gomez-Guerrero, Int. J. Mol. Sci., 2020, 21, 4225 CrossRef PubMed.
- M. Rushe, L. Silvian, S. Bixler, L. L. Chen, A. Cheung, S. Bowes, H. Cuervo, S. Berkowitz, T. Zheng, K. Guckian, M. Pellegrini and A. Lugovskoy, Structure, 2008, 16, 798 CrossRef CAS PubMed.
- (a) K. Peqini, S. Attanasio, L. Feni, G. Cappelletti and S. Pellegrino, J. Pept. Sci., 2024, 30, e3556 CrossRef CAS PubMed; (b) F. Guzmán, M. Aróstica, T. Román, D. Beltrán, A. Gauna, F. Albericio and C. Cárdenas, Electron. J. Biotechnol., 2023, 64, 27 CrossRef.
- J. Becerra, Modulation of NF-κB Activity with Synthetic Inhibitors of NEMO, PhD thesis, University of Michigan, Ann Arbor, Michigan, USA, 2023 Search PubMed.
| This journal is © The Royal Society of Chemistry 2025 |