Open Access Article
Open Access ArticleIdentification of γ-butyrolactone signalling molecules in diverse actinomycetes using resin-assisted isolation and chemoenzymatic synthesis†
Yuta
Kudo
 *ab,
Keiichi
Konoki
b and
Mari
Yotsu-Yamashita
*ab,
Keiichi
Konoki
b and
Mari
Yotsu-Yamashita
 b
b
aFrontier Research Institute for Interdisciplinary Sciences, Tohoku University, 6-3 Aramaki-Aza-Aoba, Aoba-ku, Sendai, Miyagi 980-8578, Japan. E-mail: yuta.kudo.d5@tohoku.ac.jp
bGraduate School of Agricultural Science, Tohoku University, 468-1 Aramaki-Aza-Aoba, Aoba-ku, Sendai, Miyagi 980-8572, Japan
First published on 25th February 2025
Abstract
Actinomycetes are prolific producers of secondary metabolites with diverse bioactivities. Secondary metabolism in actinomycetes is regulated by signalling molecules, often termed “bacterial hormones.” In Streptomyces griseus, the γ-butyrolactone (GBL) A-factor (1) plays a key role in regulating secondary metabolism, including streptomycin production. The widespread presence of afsA, the gene encoding A-factor synthase, suggests that GBLs are a major class of signalling molecules in actinomycetes. However, their identification is hindered by the requirement for large-scale cultures. This study presents two methodologies for identifying natural GBLs. First, a resin-assisted culture method combined with MS-guided screening enabled the isolation and structural determination of GBLs (2–5) from smaller-scale cultures. Second, a chemoenzymatic synthesis method involving one-pot three enzymatic reactions was developed, allowing the production of GBL standards (10a–10l). Using these standards, HR-LCMS analysis of 31 strains across 10 actinomycetes genera identified GBLs in nearly half of the tested strains, including genera where GBLs were detected for the first time. Chiral HPLC analysis further revealed the presence of the (3S)-isomer of GBL (11), an enantiomer of known GBLs. This study uncovers the widespread distribution and structural diversity of GBLs among actinomycetes, providing insights into their regulatory roles and potential for activating secondary metabolism, which may facilitate the discovery of new natural products.
Introduction
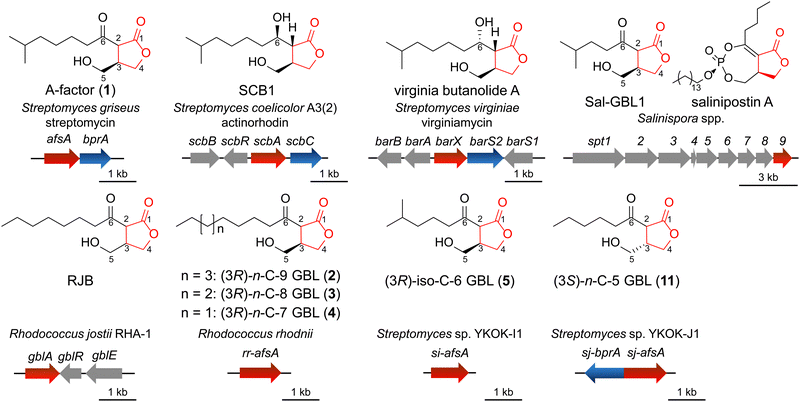
Actinobacteria are a vital source of secondary metabolites with unique chemical structures and diverse biological activities, including antibiotic, antifungal, antitumour, and immunosuppressant activities.1 These specialized metabolites have been developed into pharmaceuticals, beneficial chemicals, and biological probes. Owing to their remarkable metabolic versatility, actinomycetes are of great medical significance and remain an indispensable platform for drug discovery. Genome analyses have revealed that actinomycetes possess a vast yet largely untapped potential for secondary metabolite production. In recent years, the emergence and spread of antibiotic-resistant bacteria have become a global public health concern. Addressing this challenge requires a continuous supply of new antibiotics. Many antibiotics, including streptomycin, daptomycin, retapamulin, telavancin, ceftaroline fosamil, fidaxomicin and dalbavancin were produced industrially via fed-batch fermentation of actinomycetes, which highlights their industrial significance, and importance of regulation of secondary metabolism.2–4 Actinobacteria are also characterized by a complex life cycle that includes aerial mycelium and spore formation. Actinobacteria regulate these features, exhibiting exceptional secondary metabolism and morphological development, using low-molecular-weight signalling molecules. In Streptomyces griseus, the signalling molecule A-factor (1, Fig. 1) triggers secondary metabolism, including antibiotic streptomycin production and morphological differentiation, by activating a signalling pathway.5,6 The A-factor is recognized by the receptor protein ArpA, which dissociates from DNA upon binding to the A-factor, leading to the expression of the central transcriptional activator AdpA. AdpA activates its regulons, including pathway-specific transcription factors for streptomycin biosynthesis genes and morphological differentiation.Signalling molecules have been identified in Streptomyces species. The majority of known signalling molecules have 2,3-disubstituted γ-butyrolactone (GBL) structures (Fig. 1).6–11 Other chemotypes, such as butanolide-type avenolide and SRB-1, as well as furan-type methylenomycin furan signalling molecules have also been identified.11–14 Normal, iso- and anteiso-type fatty acyl side chains in signalling molecules have been reported to have various chain lengths. In the case of GBL signalling molecules, the 6-keto GBLs, which includes the A-factor (1), and the 6-hydroxy GBLs, represented by SCB1 in Streptomyces coelicolor A3(2), have been reported. Both (6R) and (6S) stereochemistry have been reported in 6-hydroxy GBLs (Fig. 1). All GBL signalling molecules with defined stereochemistry have been reported in the (3R)-form, although some early papers illustrated (3S)-form GBL structures without experimental verification. The stereochemistry at the C-2 position of 6-keto GBLs is uncertain due to the occurrence of epimerization via keto–enol tautomerization. Signalling molecules have been less commonly identified in genera other than Streptomyces. Kudo et al. identified GBLs in the marine actinomycete Salinispora spp.15 A GBL named RJB was identified in Rhodococcus jostii RHA-1 using LCMS, but the stereochemistry at C-3 in RJB was not determined.16 The signalling molecule B-factor has been reported in Nocardia,17 but no GBLs have been confirmed.
The A-factor is biosynthesized by the key biosynthetic gene afsA, which encodes A-factor synthase (AfsA). Homologues of afsA are widely distributed among actinomycetes.11 Although signalling pathways vary among species,5,18–21 they share a common feature: the initial recognition of signalling molecules by specific receptors. Early studies suggested that GBL induces secondary metabolite production in various Streptomyces species.22–25 Bioinformatic analysis further suggested that approximately 60% of Streptomyces strains are capable of producing GBL-type compounds.26 Despite their significance, only a limited number of signalling molecules have been structurally characterized. Actinomycetes represent a vast group of microorganisms encompassing numerous genera, with the genus Streptomyces alone comprising over 1000 species. However, fewer than 20 GBLs have been unambiguously identified using analytical chemistry methods such as NMR and high-resolution liquid chromatography-mass spectrometry (HR-LCMS).11 Signalling molecules are typically produced in low quantities, possibly because they act in vivo at nanomolar concentrations (e.g., 10−9 M for the A-factor). Conventional methods for isolation and structural elucidation require immense effort. The isolation of known signalling molecules often involves the processing of hundreds of litres of liquid cultures of actinomycetes. Notable examples include the isolation of SCB1 from 300 litres,8 virginiae butanolide A from 1150 litres,27 and IM-2 from 1150 litres of culture.28 The significant effort required for the identification of signalling molecules may explain why many signalling molecules remain unidentified. While these molecules are critical regulatory factors, no practical and universally applicable methods currently exist for their systematic identification. As a result, knowledge of their chemical structures and distribution among actinomycetes is still limited.
The biosynthetic pathway of the A-factor (1) has been elucidated primarily through in vitro reactions involving AfsA, butanolide phosphate reductase (BprA), and phosphatase.29 In this pathway, AfsA catalyses the condensation of dihydroxyacetone phosphate (DHAP) and β-ketoacyl-ACP (acyl carrier protein), followed by spontaneous aldol condensation to form butenolide phosphate. Subsequently, BprA reduces the double bond between C-2 and C-3, resulting in the formation of GBL phosphate. The final step involves dephosphorylation, producing the GBL A-factor (1). A homologue of AfsA, ScbA, performs a similar function in S. coelicolor A3(2), catalysing the biosynthesis of SCB1.30 Another homologue, MmfL, is employed in the biosynthesis of methylenomycin furan, even though the chemical structures of the final products differ.31 The biosynthesis of salinipostin, a highly unusual phosphotriester compound with antimalarial activity, also begins with the formation of a GBL structure catalysed by an AfsA homologue, Spt9.15 Additionally, GBLs such as Sal-GBL1 and Sal-GBL2 have been detected in salinipostin-producing Salinispore strains.15 These strains also produce volatile GBL compounds, known as salinilactones, which exhibit cytotoxicity.32–34 As shown in previous analyses, the afsA gene not only is essential for the biosynthesis of GBLs but also plays a critical role in the production of related compounds. The afsA gene in each strain may also accept different β-ketoacyl-ACPs. With respect to the synthesis of GBL, the first asymmetric total synthesis of the A-factor was achieved by Mori in 1981.35–37 Brown et al. later reported an efficient biomimetic racemic synthesis of the A-factor,38 and Parkinson et al. recently demonstrated the asymmetric synthesis of SCBs.39
The fatty acyl chain structure, 6-hydroxy group, and stereochemistry of signalling molecules are rigorously recognized by the ArpA receptor. Actinomycete signalling molecules, such as the A-factor and SCBs, are commonly considered species-specific due to their high receptor specificity and are often referred to as “bacterial hormones.” These molecules have been studied for their regulatory function in producer strains and closely related strains. However, some studies have shown that signalling molecules can influence secondary metabolism in nonproducer strains. For example, supplementation with SCBs induced paulomycin production in Streptomyces albus,40 and butenolides stimulated avermectin production in Streptomyces avermitilis.41 Recently, actinomycete signalling molecules have been reported to activate quorum sensing in Gram-negative bacteria and enhance secondary metabolite production.42 These findings suggest that actinomycetes utilize signalling molecules not only as self-inducing regulators but also as potential chemical tools for interspecies communication. On the other hand, the addition of plant hormones has been reported to induce antibiotic actinorhodin production in Streptomyces, implying that signalling pathways may be influenced by chemical signals from other organisms.43 In the field of synthetic biology, the artificial construction of signalling pathways has garnered attention as a promising tool for developing regulatory systems.44–46
Given the specific ability of signalling molecules to regulate secondary metabolism and morphological development, identifying such molecules in individual species is crucial for understanding and controlling the production of natural compounds in actinomycetes. While most research on signalling molecules has focused on the genus Streptomyces, studies on other genera remain limited, as described above. Nevertheless, actinomycetes outside the genus Streptomyces are also known to produce valuable secondary metabolites. For example, antitumour compounds tested in clinical trials, such as salinosporamide and rebeccamycin, were isolated from the genera Salinispora and Lentzea (formerly designated Nocardia), respectively.47–49 Anti-trypanosomal macrolides actinoallolides were found in the endophytic actinomycete Actinoallomurus.50,51 These examples demonstrate that actinomycetes other than Streptomyces are also promising sources of bioactive natural compounds. Approaches such as cocultivation52 and heterologous expression53–56 have been demonstrated to be effective in activating dormant secondary metabolism and discovery of the new natural compounds. However, actinomycetes still possess largely untapped potential for secondary metabolism, and exploring strategies to harness this potential remains a critical challenge. Understanding the signalling molecules in a broad range of actinomycetes is fundamental for unlocking their metabolic potential and optimizing the production of beneficial compounds. The use of signalling molecules to activate silent secondary metabolic pathways would represent a viable strategy for discovering new natural products, potentially paving the way for resupplying novel pharmaceuticals derived from actinomycetes.
In this study, we aimed to establish a methodology for the rapid identification of GBLs, which are major signalling molecules in actinomycetes (Fig. 2). Our first objective was to achieve efficient isolation and structural determination of GBLs without culturing hundreds of litres of actinomycetes. Over the past few decades, mass spectrometry (MS)-guided screening has become a practical and widely employed technique in natural product chemistry.57–60 We cocultured actinomycetes with absorbent resin to enhance GBL production and subjected the extracts to MS-guided screening focused on the GBL structure. This approach enabled the isolation and structural determination of GBLs (2–5) as new natural products from a culture volume of less than 7.5 litres. To further accelerate the identification process, we developed a concise chemoenzymatic synthesis method for synthesizing a series of GBLs with varying side chain structures. Using both the synthesized and isolated GBLs, we analysed extracts from 31 strains across 10 actinomycete genera with HR-LCMS, revealing the widespread occurrence of GBLs among diverse actinomycetes. Unexpectedly, a GBL isolated from Streptomyces sp. YKOK-J1 was identified as the (3S)-n-C-5 GBL (11), marking the first experimental confirmation of (3S)-isomer of GBL. This study expands our understanding of the chemical diversity and distribution of GBLs in actinomycetes, providing insights into their metabolic potential and role as regulatory molecules.
Results and discussion
Resin-assisted isolation and structure determination of γ-butyrolactone compounds
We cocultured actinomycetes with resin and subjected the crude extracts to MS-guided screening to rapidly identify GBL compounds. GBLs were isolated from the actinomycetes showing significant production, and their structures, including their absolute stereochemistry, were determined. A total of 31 strains were analysed in this study (Table S1, ESI†). We first selected the actinomycetes containing afsA homologues (pairwise AA similarity >30%) from various genera, including Streptomyces, Nocardia, Rhodococcous, Kutzneria, Gordonia, Kitasatospora, Dietzia and Salinispora, which were obtained from bacterial collections. As described above, the salinipostin biosynthetic gene cluster (spt) corresponds to the production of GBLs;11,15,61 therefore, 14 strains containing an spt-like biosynthetic gene cluster were included. Strains lacking high-similarity afsA homologues were also selected from the Mycobacterium and Mycolicibacterium genera. Five Streptomyces strains were isolated from soil collected in Okinawa, Japan, and were also subjected to analysis. The 16S rRNA sequences of these strains were amplified via PCR and analysed, and the whole genomes of the two strains were sequenced via next-generation sequencing (NGS). In a previous study, GBL production in marine actinomycete Salinispora strains was enhanced by cocultivation with the absorbance resin Amberlite XAD7HP (Sigma-Aldrich).15,62 Although definitive evidence is lacking, it is presumed that the resin enhances GBL production in actinomycetes by adsorbing GBL in the culture. Based on those findings, the 31 strains representing 10 genera from 6 families were fermented in liquid media with XAD7HP resin. The cells and resin were extracted with acetone and pretreated with a C-18 SPE column. Eluent fractions were analysed using HR-LCMS in positive and negative ion modes, and the m/z values corresponding to [CxHyO4]+ and [CxHyO4]− were detected. As a result, notable peaks (2–4) were detected in Rhodococcus rhodnii JCM 3203, and peak (5) was detected in the isolated Streptomyces sp. YKOK-I1 (5) (Fig. 3a and b). These potential GBLs were then subjected to isolation and spectroscopic analysis.Compound 2 was extracted with acetone from a 2.5 litres culture of R. rhodnii JCM 3203 fermented with resin and isolated using a Cosmosil 140C18-OPN column and an InertSustain C18 column (2: 0.30 mg). GBLs 3 and 4 were obtained from an additional long-term 5 litres culture of R. rhodnii JCM 3203 and then purified using a similar method. The isolation of 3 and 4 was achieved with a Kinetex XB-C18 column (3: 0.15 mg, 4: 0.11 mg). The molecular formulae of these compounds were determined to be C15H26O4 (2), C14H24O4 (3) and C13H22O4 (4) on the basis of their ESI-TOF-MS monoisotopic peaks, suggesting side chains C-9, C-8, and C-7, respectively (Fig. S1–S4, ESI†). The 1H NMR spectrum of 2 in CDCl3 showed diagnostic double triplet signals at δH 2.99 and 2.64 ppm (Fig. S67 and S68, ESI†). The methyl triplet signal at δH 0.88 indicated the presence of a n-alkyl side chain. Signal sets derived from keto–enol tautomerization were also observed. 2D NMR (COSY, TOCSY, HSQC, and HMBC) clarified the planar structure of 2 (Fig. 3 and Fig. S69–S72, ESI†). All the proton and carbon signals in the major keto form were assigned (Table S2, ESI†). Similarly, the structures of 3 and 4 were characterized using 1D- and 2D-NMR spectra (Fig. S73–S84, Tables S3 and S4, ESI†). Compounds 3 and 4 resemble GBL structures and contain n-octyl and n-heptyl chains, respectively (Fig. 3). The absolute configurations of these compounds were analysed using CD spectroscopy. Compound 2 displayed positive Cotton effects (284 nm, Δε + 0.446, 218 nm, Δε + 0.265, Fig. S6, ESI†), closely resembling the (3R)-A-factor (283.5 nm, Δε + 0.699, 221.0 nm, Δε + 0.420).35,37 Thus, 2 was assigned as (3R)-n-C-9 GBL (Fig. 3), the longest side-chain natural GBL known to date. Compounds 3 and 4 were assigned as (3R)-n-C-8 GBL and (3R)-n-C-7 GBL, respectively, on the basis of their CD spectra (Fig. S7 and S8, ESI†). Compound 5 was isolated from a 5 litres culture of Streptomyces sp. YKOK-I1 fermented with resin (5, 0.15 mg). The molecular formula of 5 was determined to be C12H20O4 (5) on the basis of the monoisotopic peaks in the ESI-TOF-MS data, suggesting a C-6 side chain (Fig. S1 and S5, ESI†). NMR analysis revealed a planar structure, and CD analysis confirmed a (3R)-configuration (Fig. 3, Fig. S9 and S85–S90, ESI†). Compounds 2 and 3 represent the first naturally occurring 6-keto GBLs with these side chain lengths. A compound with an n-C-7 side chain, which was identified via LCMS with a racemic synthetic standard, was previously reported as RJB in Rhodococcus jostii RHA-1.16 However, this RJB was not analysed using NMR, and the absolute configuration was not investigated. Compound 5 has been reported to be a biosynthetic intermediate of virginiae butanolide A, a signalling molecule in S. virginiae,63 but was first isolated as a natural product from Streptomyces sp. YKOK-I1. HR-LCMS analysis confirmed that Streptomyces sp. YKOK-I1 lacked any possible peaks for virginiae butanolide A.
Using an MS-guided screening method combined with resin fermentation, we successfully achieved concise identification of new natural GBLs from small-scale cultures of actinomycetes. This method provides solutions to the challenges faced in previous studies, which required the processing of hundreds of litres of cultures to isolate GBLs. In addition to the identified GBLs, potential natural GBL compounds were detected in the actinomycetes extracts during screening. While most known GBL compounds are 6-hydroxy GBLs, our results suggest that 6-keto GBL compounds are more widely distributed than previously recognized. Despite enhanced production through resin cocultivation, the amount of GBLs in some extracts was estimated to be insufficient for NMR and CD analysis based on the peak intensities. In addition, in Streptomyces sp. YKOK-J1 (Fig. S10, ESI†), the GBL yield was notably lower in 500 mL cultures (N = 2) than in 50 mL small-scale cultures, which presented high concentrations of GBLs (N = 4).
Chemoenzymatic synthesis of GBL compounds
MS-guided screening suggested a wide distribution of 6-keto GBL compounds. To expand the rapid identification methodology, we synthesized a series of 6-keto GBL compounds via a chemoenzymatic approach (Scheme 1). Previous studies have reported the biomimetic racemic synthesis of the A-factor.38 Horinouchi et al. reported key reactions catalysed by AfsA and BprA in the A-factor biosynthetic pathway. In recent years, technological advancements in enzyme structure prediction and heterologous expression of enzymes have facilitated the development of enzyme-mediated synthesis of natural products and pseudo-natural products.64–66 In this study, we performed enzymatic reactions using the AfsA homologues, BprA and phosphatase with synthesized substrate mimics. Docking models were constructed using AutoDock Vina67,68 to evaluate the interaction between the AfsA homologue protein structures generated by AlphaFold269 and β-ketoacyl-N-acetyl cysteamine (SNAC) substrates, which serve as mimics of β-ketoacyl-ACP. On the basis of this in silico analysis, two AfsA homologues, rr-afsA from R. rhodnii 3203 and sj-afsA from Streptomyces sp. YKOK-J1, were selected as biocatalysts (Fig. S11, ESI†). rr-afsA was expected to recognize substrates with long n-acyl chains. On the basis of the LCMS analysis (Fig. S10, ESI†), Streptomyces sp. YKOK-J1 was estimated to produce n- and iso-C-5 GBL; therefore, SJ-AfsA might recognize substrates with branched and middle-length acyl chains. However, sj-afsA showed low sequence homology with AfsA in S. griseus (Table 1). Similarly, based on the docking model, it was predicted that the known BprA in Streptomyces griseus29 recognizes various butanolide phosphates as substrates (Fig. S11, ESI†). rr-afsA, sj-afsA, and bprA were cloned into pET28a and pET28-MBP-TEV70 vectors. With these constructs, three enzymes were heterologously expressed in E. coli and then purified using Ni-NTA affinity chromatography (Fig. S12, ESI†). Twelve β-ketoacyl-SNAC substrates (6a–6l), mimics of the substrate β-ketoacyl-ACP, were prepared via concise organic synthesis (see ESI†). Another substrate, DHAP (7), was purchased or synthesized. With the substrates and enzymes in hand, in vitro enzymatic reactions were conducted. Both MBP-Rr-AfsA and MBP-SJ-AfsA gave possible butanolide phosphate products (8, Fig. S13, ESI†) with exact masses matching the expected values (substrate: n-C-4 β-ketoacyl-SNACs (6c), m/z 277.0481, [M − H]− calcd. 277.0483). However, the products were unstable.15 Chemical reduction of the C–C double bond in butenolide phosphate with NaBH3CN yielded relatively stable products (m/z 279.0644, [M − H]− calcd. 279.0639), estimated to be GBL phosphate (9, Fig. S13, ESI†). Considering the instability of butenolide phosphate (8), we adopted successive enzymatic reactions to avoid handling unstable butenolide intermediates. A reaction with AfsA was performed, followed by a reaction with BprA and the cofactor NADPH. LCMS analysis using C-18- and C-8-reversed-phase columns confirmed that all the tested substrates were converted into corresponding GBL-phosphates (9) via successive MBP-Rr-AfsA and MBP-BprA enzymatic reactions (Fig. S14–S26, ESI†). Compared with Rr-AfsA, SJ-AfsA exhibited narrower substrate tolerance, but higher conversion rates were observed with n-C-4, n-C-5, and iso-C-5 side chain substrates (Table S6, ESI†). Since compounds 2–4 were identified in R. rhodnii, the genuine substrates of Rr-AfsA are likely to have these chain lengths. However, Rr-AfsA showed higher conversion efficiency for substrates with shorter chain lengths under the reaction condition (25 °C for 1 h). This discrepancy might be attributed to the low solubility of the β-ketoacyl-SNAC substrate (6) in the reaction buffer. In line with the natural compound profile of Streptomyces sp. YKOK-J1 (Fig. S10, ESI†), SJ-AfsA accepted substrates with n- and iso-C-5 side chains and similar substrates. However, substrates with longer side chains were poorly converted into the corresponding phosphates by SJ-AfsA (Table S6, ESI†), probably due to the spatial constraints in the binding pocket of SJ-AfsA. MBP-BprA reduced all the tested butenolide to butyrolactone, although substrate specificity was not evaluated in detail in this study because of the instability of the butanolide substrate under the reaction conditions. | ||
| Scheme 1 Chemoenzymatic synthesis of 6-keto γ-butyrolactones (10a–10l) using three types of enzymes. | ||
| a Partial 16S rRNA sequence shows 100% identity with Streptomyces wuyuanensis. b Partial 16S rRNA sequence shows 100% identity with Streptomyces sporoverrucosus and some Streptomyces spp. c Partial 16S rRNA sequence shows 99.8% identity with Streptomyces cinereoruber subsp. fructofermentans. d Whole 16S rRNA sequence shows 98.8% identity with Streptomyces acidiscabies. e Whole 16S rRNA sequence shows 98.6% identity with Streptomyces cyaneochromogenes. f Strains with AfsA homology higher than 30% but no detectable GBL are shown in blue. g iso-C-6 GBL was detected, although its absolute configuration was not determined. h Strains with AfsA homology higher than 30% are shown in yellow. i Genome sequence was not available. |
|---|

|
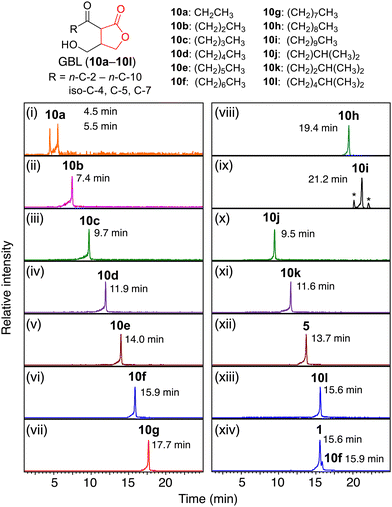
The AfsA and BprA reaction products, GBL phosphates (9), were subjected to dephosphorylation using a commercially available bacterial alkaline phosphatase (BAP). Dephosphorylation of all the tested GBL-phosphates was confirmed by detecting their exact masses via LCMS (Fig. S27–S38, ESI†). Exploiting the substrate capability of the two AfsA, BprA, and BAP enzymes, we synthesized twelve 6-keto GBL compounds with side chains, including n-C-2, n-C-3, n-C-4, n-C-5, n-C-6, n-C-7, n-C-8, n-C-9, n-C-10, iso-C-4, iso-C-5 and iso-C-7 side chains (Fig. 4, 10a–10l).
Next, we developed a one-pot chemoenzymatic synthesis of GBLs using three successive enzymatic reactions involving AfsA, BprA, and BAP. The reaction began with n-C-5 β-ketoacyl-SNAC (6d) and DHAP (7) incubated with MBP-SJ-AfsA. To this mixture, MBP-BprA, NADPH, and MgCl2 were added, and the mixture was incubated at 25 °C for 2 h. To address the low solubility of the substrate, the mixture was further incubated at 37 °C for an additional hour. Afterwards, dephosphorylation using the BAP enzyme was performed at 37 °C for another hour. The reaction product was subjected to a C-18 SPE column, yielding a semipurified product sufficient for NMR analysis. A single HPLC purification using a C-18 column enabled the isolation of GBL (10). The isolated product (10d) was fully characterized via 1D- and 2D-NMR analyses (Fig. S91–S95, ESI†). The yield of the isolated compound was approximately 30% (2.6 mg per single batch) when the n-C-5-SNAC (6d) substrate was used, with a total reaction time of approximately 4 h. This method was also applied to n-C-6, n-C-7, iso-C-5 and iso-C-7 β-ketoacyl-SNACs (6e, 6f, 6k, and 6l), and their structures and purity were confirmed by NMR (Fig. S96–S108, ESI†). The NMR spectra of natural compound 4 and the enzymatically synthesized n-C-7 GBL were identical (10f, Fig. S102, ESI†). Although organic synthesis of these GBL compounds has been reported previously,27,35,39,71 this chemoenzymatic synthesis method may offer a simpler and faster approach for synthesizing a variety of GBLs. Modifying the structure of the SNAC substrate, which can be prepared in 2–3 steps within a few days, would facilitate the efficient synthesis of structurally diverse GBLs.
Configurational analysis of enzymatic products
Horinouchi et al. first reported an in vitro BprA reaction and suggested the stereoselective reduction of butenolide based on a biological activity assay.29 In this study, the low optical purity of the enzymatic product (10d) was indicated by the small Cotton effect in the CD spectrum. Hence, we investigated the chirality of the enzymatic products of GBLs using a chiral column chromatography. After the chiral column screening, four peaks estimated to be (3R)- and (3S)-n-C-5 GBL with keto–enol tautomers were detected in the enzymatic product (10d) via HPLC-DAD (Fig. 5(i), 254 nm, DAICEL CHIRALPAK AD-H). Fractionation and CD spectral analysis confirmed the separation of the (3R)- and (3S)-GBL enantiomers ((3R)-n-C-5 GBL; 285 nm, Δε + 0.643, 219 nm, Δε + 0.311, (3S)-n-C-5 GBL; 285 nm, Δε − 0.419, 219 nm, Δε − 0.209; Fig. S39 and S40, ESI†). The n-C-5 GBL (10d) synthesized using both MBP-BprA and tag-free BprA showed peaks with a ratio of (3R)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) (3S) = 62
(3S) = 62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 38. This result suggested that the MBP tag did not affect the stereoselectivity of the product. The n-C-7 GBL (10f) synthesized using MBP-BprA showed the same ratio of (3R)
38. This result suggested that the MBP tag did not affect the stereoselectivity of the product. The n-C-7 GBL (10f) synthesized using MBP-BprA showed the same ratio of (3R)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) (3S) = 62
(3S) = 62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 38, whereas the natural n-C-7 GBL (4) isolated from R. rhodnii JCM 3203 exhibited a significantly higher (3R)-ratio ((3R)
38, whereas the natural n-C-7 GBL (4) isolated from R. rhodnii JCM 3203 exhibited a significantly higher (3R)-ratio ((3R)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) (3S) = 95
(3S) = 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 (Fig. S41, ESI†)). Using the substrate iso-C-7 β-ketoacyl-SNAC, which has the genuine acyl chain structure of BprA, the selectivity was slightly improved, but the product still consisted of a mixture of enantiomers ((3R)
5 (Fig. S41, ESI†)). Using the substrate iso-C-7 β-ketoacyl-SNAC, which has the genuine acyl chain structure of BprA, the selectivity was slightly improved, but the product still consisted of a mixture of enantiomers ((3R)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) (3S) = 72
(3S) = 72![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 28, Fig. S42, ESI†).
28, Fig. S42, ESI†).
The stereochemistry at C-3 should be established during the reduction of butenolide by BprA. To improve stereoselectivity, we attempted to identify BprA homologues capable of achieving high stereoselectivity. bprA is occasionally found in close proximity to afsA, as observed in Streptomyces griseus,29S. coelicolor A3(2),45S. venezuelae (JadW2),72 and S. virginiae (BarS2)73 (Fig. 1). In Streptomyces sp. YKOK-J1, the sj-afsA and bprA homologues (sj-bprA) are located in opposite directions (Fig. 1). Additionally, an afsA homologue was identified in Streptomyces sp. YKOK-I1 (si-afsA) through BLAST searches. No corresponding bprA homologue was found in the flanking region of rr-afsA and si-afsA, despite the high (3R)-enantiomeric ratio observed in the GBL compounds isolated from R. rhodnii JCM 3203 and Streptomyces sp. YKOK-I1 (2–5). Possible butenolide reductases were found in these strains via BLAST searches (rr-bprA and si-bprA), although these candidates were located far from afsA homologues. To test these candidates, SJ-BprA, Rr-BprA, and SI-BprA were expressed in E. coli. An enzymatic assay with MBP-Rr-AfsA and Rr-BprA showed only a trace product peak of n-C-7 GBL phosphate (9f) in the LCMS data. Similarly, an enzymatic assay using MBP-SJ-AfsA and SI-BprA produced only a trace product peak of n-C-5 GBL phosphate (9d) in the LCMS data. MBP-SJ-BprA exhibited a low conversion rate (approximately 20%), and the TEV protease was unable to effectively cleave the MBP tag from the fusion protein. Therefore, SJ-BprA was expressed without an MBP tag, although the yield of soluble protein was low. Eventually, SJ-BprA fully catalysed the conversion of n-C-5 butenolide phosphate (8d) into GBL phosphate (9d). The enzymatically produced GBL (10d) using SJ-BprA was analysed using chiral HPLC-DAD. Unexpectedly, the product exhibited a higher (3S)-enantiomeric ratio ((3R)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) (3S) = 37
(3S) = 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 63, Fig. S43, ESI†). Thus, high stereoselectivity in the reduction of butenolide phosphate could not be achieved in vitro in this study. The isolated natural compounds did not exhibit 100% optical purity. It is possible that racemization through intramolecular transesterification, as observed in the GBL synthesis process,35 might have occurred to a very small extent within the actinomycete cells or during the isolation process.
63, Fig. S43, ESI†). Thus, high stereoselectivity in the reduction of butenolide phosphate could not be achieved in vitro in this study. The isolated natural compounds did not exhibit 100% optical purity. It is possible that racemization through intramolecular transesterification, as observed in the GBL synthesis process,35 might have occurred to a very small extent within the actinomycete cells or during the isolation process.
Identification of natural (3S)-isomer of GBL
The stereochemistry at the C-3 position of GBL has been suggested to be crucial for its biological activity. To date, all unambiguously determined GBLs have been reported to have (3R) stereochemistry. Although (3S)-form GBL compounds were described in early studies, no experimental evidence, such as optical rotation data, CD spectra, or results of chiral column analysis using authentic standards, was provided. In this study, SJ-BprA was found to produce GBL with a relatively high proportion of the (3S)-isomer, leading us to investigate the absolute stereochemistry of GBL produced by Streptomyces sp. YKOK-J1. LCMS analysis indicated the presence of n-C-5 GBL in this strain (Fig. 6c and Fig. S48, ESI†). To confirm the stereochemistry, n-C-5 GBL (11) was extracted with acetone from a total of 255 mL of culture of Streptomyces sp. YKOK-J1 fermented with resin. The extract was purified using Strata C18-E, and the fractions containing n-C-5 GBL (11) were further purified using an InertSustain C-18 column. The purified n-C-5 GBL was then analysed using chiral HPLC-DAD, along with standards of (3S)-n-C-5 GBL and racemic n-C-5 GBL. The purified n-C-5 GBL from Streptomyces sp. YKOK-J1 presented retention times identical to those of (3S)-n-C-5 GBL, with an enantiomeric ratio of (3R)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) (3S) = 1
(3S) = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 (Fig. 5). This study represents the first determination of a natural (3S)-form GBL from an actinomycete strain, which may expand the known chemical diversity of signalling molecules. Both enantiomers are present in animal pheromones such as olean and frontalin, and their ratio can provide information about sex, age, and other characteristics.74,75 The discovery of enantiomers in bacterial signalling molecules such as GBL, often referred to as “bacterial hormones”, is intriguing and may suggest a similarly complex role in microbial communication.
9 (Fig. 5). This study represents the first determination of a natural (3S)-form GBL from an actinomycete strain, which may expand the known chemical diversity of signalling molecules. Both enantiomers are present in animal pheromones such as olean and frontalin, and their ratio can provide information about sex, age, and other characteristics.74,75 The discovery of enantiomers in bacterial signalling molecules such as GBL, often referred to as “bacterial hormones”, is intriguing and may suggest a similarly complex role in microbial communication.
GBL distribution in diverse actinomycete
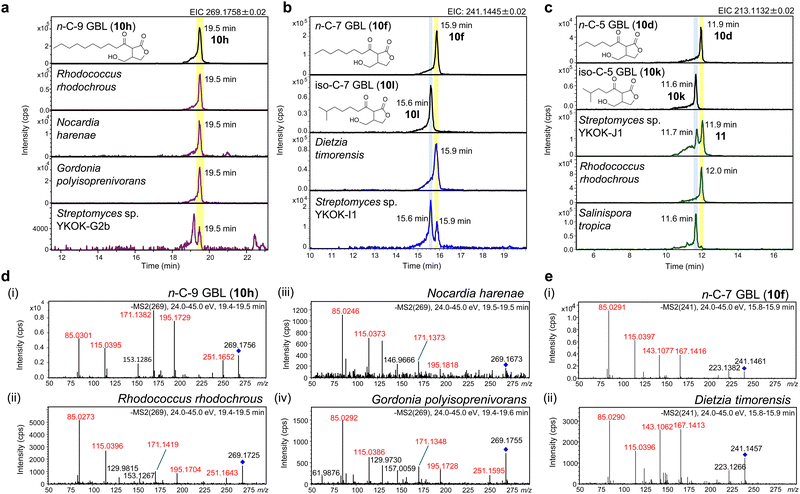
With the isolated and synthetic GBL standards in hand, we investigated the presence of GBL compounds in various actinomycetes. Extracts from 31 actinomycete strains (Table 1 and Table S1, ESI†) were reanalyzed via HR-LCMS with 13 GBL standards (5, 10a–10l). The results are summarized in Table 1. The A-factor, iso-C-7 GBL, was clearly detected in the extract of Streptomyces griseus at a retention time identical to that of the synthetic standard (10l), validating the accuracy of this analytical method (Fig. 4 (xiii, xiv)). Previously reported n-C-4 and iso-C-5 GBL (Sal-GBL1 and Sal-GBL2)15 were confirmed in Salinispora tropica, and iso-C-4 GBL (10b) was newly identified as a natural product in this strain. The n-C-5, n-C-6, and n-C-10 GBLs (10f, 10e, 10i) were also newly identified as natural products in the genera Rhodococcus and/or Streptomyces. The n-C-7 GBL (10f) and n-C-9 GBL (10h) were detected in the genera Dietzia, Nocardia, Gordonia, Rhodococcus, and Streptomyces (Fig. 6a and b). Notably, the detection of GBLs in Nocardia, Gordonia, and Dietzia using HR-LCMS is reported here for the first time. The MSMS spectra of the detected GBLs (10h, 10f) in these genera were identical to the spectra of their synthetic standards (Fig. 6d and e). This finding suggests that GBLs also function as signalling molecules in these genera of actinomycetes. These GBL-producing strains harbour afsA homologues with more than 30% AA similarity (Table 1). GBLs were not detected in the strains with low-similarity afsA homologues (<30% similarity), including Mycobacterium cookii, Mycolicibacterium austroafricanum, Mycolicibacterium chlorophenolicum, and Streptomyces griseoaurantiacus, although Streptomyces sp. YKOK-J1 exhibited the production GBLs (Table 1). Among the four Rhodococcus strains, only R. erythropolis did not produce detectable GBLs, probably due to the partial deletion of the hot-dog domain in its afsA homologue. While the presence of afsA homologues is a strong indicator of GBL production, GBLs were not detected in eight strains with plausible afsA homologues showing more than 30% AA similarity (Table 1). n-C-2 and n-C-3 GBL (10a and 10b) were not detected in any of the tested strains, but the high volatility of these compounds may have led to their loss during culture and extraction. Overall, GBL compounds were detected in 15 out of 31 tested strains, representing 6 of the 10 tested genera (Table 1). These results revealed that various actinomycetes produce GBL compounds. Among the GBL-producing strains, seven strains contained spt-like gene clusters, but phosphotriester compounds were not detected in these strains by MS-guided screening of phosphotriester compounds focusing on their characteristic fragment ions, a method we developed.626-Hydroxy GBLs have been reported to be the major signalling molecules in Streptomyces species.11,26 This study revealed that 6-keto GBLs are also distributed in Streptomyces species, although the known natural 6-keto GBLs are limited to examples such as the A-factor (1) in S. griseus. 6-Hydroxy GBLs are biosynthesized from 6-keto GBLs via the reduction of a ketone at C-6.39,76 The reduction of 6-keto GBLs (10e, 10l) with NaBH4 yielded a diastereomeric mixture of 6-hydroxy GBLs. Analysis of these 6-hydroxy GBLs revealed identical peaks in some actinomycete strains lacking 6-keto GBLs, such as the SCB-producing strains Streptomyces coelicolor A3(2) (S. violaceoruber) and Streptomyces sp. YKOK-D1 (Fig. S49–S53, ESI†). However, no possible 6-hydroxy GBLs were observed in the 15 actinomycete strains containing 6-keto GBLs. This observation suggested that the 6-keto GBLs detected in these strains were not biosynthetic precursors of 6-hydroxy GBL. Recent work by Parkinson et al. demonstrated the stereoselective synthesis of SCBs, which contain a (6R)-hydroxy group, from 6-keto GBLs via the ketoreductase ScbB.39 Similarly, the stereoselective reduction of a ketone at C-6 to a hydroxy group was reported for BarS1, which produces virginiae butanolide A with a (6S)-hydroxy group.76 These ketoreductases could be used to chemoenzymatically synthesize a series of 6-hydroxy GBLs.
In terms of chemical structure, this study expands the range of known side chain lengths of natural GBLs. Furthermore, the identification of (3S)-isomer of GBL highlights additional stereochemical diversity among natural GBLs. Bioinformatic analyses have revealed the widespread presence of afsA homologues across actinomycetes, suggesting that GBL compounds are key signalling molecules.11 However, only analytical chemistry can be used to determine their exact structures. This study provides formal evidence for the widespread occurrence of GBL compounds in actinomycete strains spanning multiple genera. Resin-assisted isolation involves simply adding commercially available resin to the culture, making it easily applicable to cultivable actinomycetes. Similarly, chemoenzymatic GBL synthesis can be reconstituted using AfsA and BprA. By applying analyses using GBL standards to a library of actinomycete extracts, valuable insights into the distribution of GBL can be obtained. The application of synthesized GBLs has significant potential for activating secondary metabolic pathways across these diverse actinomycetes. This approach could lead to the discovery of novel bioactive compounds and further expand our understanding of the chemical and biological diversity within this microbial group.
Experimental
Compound screening using LCMS
Bacteria were cultured with 2% starter culture in a 300 mL Erlenmeyer flask containing 50 mL of medium at 28 °C or 30 °C with shaking at 180 rpm using an EYELA MMS-3020 device. After 36–48 hours, sterile Amberlite XAD7HP resin (1 g, Sigma-Aldrich) was added to the flask, and fermentation was allowed to proceed for an additional 2–4 days. The cells and resin were collected by centrifugation at 4000×g for 10 min and then soaked in 50 mL of acetone for 30 min. After filtration in vacuo, the acetone extract was concentrated using a rotary evaporator. The resulting residue was dissolved in methanol (MeOH), and the solution was applied to a Strata C18-E SPE (2 g, Phenomenex) or Cosmosil 140C18-OPN column (1 g, 10 mm internal diameter (i.d.)). The column was then washed with three volumes of H2O, and eluate fractions were obtained using three volumes of H2O/MeOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v), three volumes of H2O/MeOH (1
1, v/v), three volumes of H2O/MeOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, v/v), and four volumes of pure MeOH. Aliquots of the fractions were subjected to HR-LCMS analysis. HR-LCMS was performed using a Kinetex XB-C18 column (2.6 μm, 4.6 mm i.d. × 100 mm, 100 Å, Phenomenex) with the following conditions: positive and negative mode, mobile phase A: 0.1% formic acid in H2O (v/v), mobile phase B: 0.1% formic acid in acetonitrile (MeCN) (v/v), 0.3 mL min−1, 0–25 min (20–100% B), 25–35 min (100% B), and 30 °C. The MS1 extracted ion chromatograms (EICs) corresponding to the GBLs were generated, and the molecular formula of each compound was determined on the basis of the high-resolution mass of the monoisotopic peaks.
4, v/v), and four volumes of pure MeOH. Aliquots of the fractions were subjected to HR-LCMS analysis. HR-LCMS was performed using a Kinetex XB-C18 column (2.6 μm, 4.6 mm i.d. × 100 mm, 100 Å, Phenomenex) with the following conditions: positive and negative mode, mobile phase A: 0.1% formic acid in H2O (v/v), mobile phase B: 0.1% formic acid in acetonitrile (MeCN) (v/v), 0.3 mL min−1, 0–25 min (20–100% B), 25–35 min (100% B), and 30 °C. The MS1 extracted ion chromatograms (EICs) corresponding to the GBLs were generated, and the molecular formula of each compound was determined on the basis of the high-resolution mass of the monoisotopic peaks.
Identification of GBLs from actinomycetes
Extracts of the 31 strains of bacteria were analysed using HR-LCMS with the isolated and synthesized GBLs (5 and 10a–10l). Actinomycete extracts prepared for compound screening were analysed under the same LCMS conditions. The molecular formulae of the detected compounds were determined on the basis of the high-resolution mass of monoisotopic peaks. Selected extracts were further analysed using LC-MSMS. MSMS spectra of representative detected peaks in Rhodococcus rhodochrous, Nocardia harenae, Gordonia polyisoprenivorans and Dietzia timorensis were analysed in automsms mode in otofControl with precursor ions: m/z 269 and 241 and a collision energy of 24–45 eV.One-pot chemoenzymatic reaction using AfsA, BprA, and BAP
The reaction mixture containing 2.5 to 12.5 μM MBP-SJ-AfsA, 10 mM n-C-5 β-ketoacyl-SNAC (6d), and 11 mM DHAP (7) was incubated in 1 or 2 mL of 50 mM McIlvaine buffer (pH 7.0) at 25 °C for 15 min. To this AfsA reaction, a BprA reaction mixture containing 5 μM BprA, 11 mM MgCl2, and 11 mM NADPH in an equal volume of 50 mM McIlvaine buffer (pH 7.0) was added, and the mixture was subsequently incubated at 25 °C for 2 h and 37 °C for 1 h. After adding 10× BAP buffer and BAP enzyme (10 units per mL), the mixture was incubated at 37 °C for 1 h. One volume of EtOH was added to the reaction mixture and then centrifuged at 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 3 min to remove insoluble debris. The supernatants from several reaction batches were combined and evaporated in vacuo. The residue was applied to a Strata C18-E SPE column (2 g, Phenomenex), and then the column was washed with three volumes of H2O. The eluate fractions were obtained using three volumes of H2O/MeOH (4
000×g for 3 min to remove insoluble debris. The supernatants from several reaction batches were combined and evaporated in vacuo. The residue was applied to a Strata C18-E SPE column (2 g, Phenomenex), and then the column was washed with three volumes of H2O. The eluate fractions were obtained using three volumes of H2O/MeOH (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v), three volumes of H2O/MeOH (1
1, v/v), three volumes of H2O/MeOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v), three volumes of H2O/MeOH (1
1, v/v), three volumes of H2O/MeOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, v/v) and four volumes of pure MeOH. Fractions containing GBL were identified via ESI-MS and evaporated in vacuo. The resulting concentrate was applied to an InertSustain C18 column (10 mm i.d. × 250 mm, 5 μm, GL Sciences) with the following gradient conditions: mobile phase A, 0.1% formic acid in H2O; mobile phase B, 0.1% formic acid in MeCN; and gradient, 40% B, hold for 10 min, 40–70% B in 60 min, at a flow rate of 2.0 mL min−1. Compound elution was monitored by DAD and confirmed by ESI-MS. The structure and purity of 10d were confirmed using NMR (Fig. S91–S95, ESI†). The pure compound 10d in mg-scale was obtained within one day after initiating the one-pot reaction. The average isolated yield was 27.8% (23.3–30.8%, n = 6). Keto–enol tautomerization in GBLs caused a low yield during HPLC purification.
4, v/v) and four volumes of pure MeOH. Fractions containing GBL were identified via ESI-MS and evaporated in vacuo. The resulting concentrate was applied to an InertSustain C18 column (10 mm i.d. × 250 mm, 5 μm, GL Sciences) with the following gradient conditions: mobile phase A, 0.1% formic acid in H2O; mobile phase B, 0.1% formic acid in MeCN; and gradient, 40% B, hold for 10 min, 40–70% B in 60 min, at a flow rate of 2.0 mL min−1. Compound elution was monitored by DAD and confirmed by ESI-MS. The structure and purity of 10d were confirmed using NMR (Fig. S91–S95, ESI†). The pure compound 10d in mg-scale was obtained within one day after initiating the one-pot reaction. The average isolated yield was 27.8% (23.3–30.8%, n = 6). Keto–enol tautomerization in GBLs caused a low yield during HPLC purification.
Others
General experimental procedures, bacterial strains, isolation of GBLs from actinomycetes (2–5), purification of (3S)-n-C-5 GBL (11) from Streptomyces sp. YKOK-J1, chiral column analysis, NMR measurements, CD spectral measurements, synthesis of β-ketoacyl-SNAC (6), DHAP synthesis (7), docking study, isolation of Streptomyces sp. strains and their 16S rRNA sequence analysis and whole genome analysis, accession number, DNA synthesis, expression and purification of proteins, enzymatic reactions, reduction of C-6 ketone in GBLs were described in ESI.†Conclusions
Signalling molecules such as GBLs are pivotal regulators of secondary metabolism in actinomycetes, which are prolific sources for drug discovery. This study established two methodologies for the rapid identification of GBLs in actinomycetes. The first approach utilizes coculture with an adsorbent resin, enabling the structural determination of GBLs (2–5) from small-scale cultures. The second approach involves the concise chemoenzymatic synthesis of 6-keto GBLs, which facilitates their identification across a broad range of actinomycetes via HR-LCMS. This study revealed the structural diversity of natural GBLs, including variations in side chain length and stereochemistry at the C-3 position. The methodologies developed here represent valuable tools for systematically identifying GBLs and will provide insights into their roles as signalling molecules. Furthermore, these findings contribute foundational knowledge for elucidating the regulatory mechanisms of secondary metabolism in actinomycetes, paving the way for harnessing these pathways to discover novel bioactive compounds.Author contributions
Conceptualization, methodology, investigation, visualization and writing – original draft by Y. K. resources and funding acquisition by Y. K. and M. Y.-Y. writing – review & editing by Y. K., M. Y.-Y. and K. K.Data availability
The data supporting this article have been included as part of the ESI.† NMR spectra, CD spectra, LCMS chromatograms, MS spectra, NMR tables, bacterial strain list, accession numbers, docking models, SDS-PAGE, Chrial HPLC chromatograms.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Japan Society for the Promotion of Science (JSPS) under KAKENHI Grant-in-Aid for Early-Career Scientists no. JP22K14833 and Grant-in-Aid for Scientific Research no. JP24K08723 to Y. K., and no. JP23H02146 and JP23K26839 to M. Y.-Y. This work was supported by Yazaki Memorial Foundation of Science and Technology and The Naito Foundation (to Y. K). The authors thank Prof. T. Fujii, Graduate School of Agricultural Science, Tohoku University, for supporting the CD spectrum measurement using a JASCO J-700WI. pET28-MBP-TEV was a gift from Zita Balklava & Thomas Wassmer (Addgene plasmid #69929; https://n2t.net/addgene:69929). Bacterial strains were provided by Japan Collection of Microorganisms, RIKEN BRC which is participating in the National BioResource Project of the MEXT, Japan, and by the National Institute of Technology and Evaluation (NITE), NBRC (NITE Biological Resource Center). A part of this study was supported by Support system for young researchers to use research equipment, instruments, and devices in Tohoku University. Support from the FRIS CoRE, which is a shared research environment, is acknowledged.References
- E. A. Barka, P. Vatsa, L. Sanchez, N. Gaveau-Vaillant, C. Jacquard, J. P. Meier-Kolthoff, H.-P. Klenk, C. Clément, Y. Ouhdouch and G. P. van Wezel, Microbiol. Mol. Biol. Rev., 2016, 80, 1–43 CrossRef PubMed.
- G. P. van Wezel and K. J. McDowall, Nat. Prod. Rep., 2011, 28, 1311–1333 RSC.
- V. Fedorenko, O. Genilloud, L. Horbal, G. L. Marcone, F. Marinelli, Y. Paitan and E. Z. Ron, BioMed Res. Int., 2015, 2015, 591349 Search PubMed.
- H. U. Van Der Heul, B. L. Bilyk, K. J. McDowall, R. F. Seipke and G. P. Van Wezel, Nat. Prod. Rep., 2018, 35, 575–604 RSC.
- S. Horinouchi, Biosci., Biotechnol., Biochem., 2007, 71, 283–299 CrossRef CAS PubMed.
- S. Horinouchi and T. Beppu, Proc. Jpn. Acad., Ser. B, 2007, 83, 277–295 CrossRef CAS PubMed.
- S. Horinouchi and T. Beppu, Mol. Microbiol., 1994, 12, 859–864 CrossRef CAS PubMed.
- E. Takano, T. Nihira, Y. Hara, J. J. Jones, C. J. Gershater, Y. Yamada and M. Bibb, J. Biol. Chem., 2000, 275, 11010–11016 CrossRef CAS PubMed.
- N. H. Hsiao, S. Nakayama, M. E. Merlo, M. de Vries, R. Bunet, S. Kitani, T. Nihira and E. Takano, Chem. Biol., 2009, 16, 951–960 CrossRef CAS PubMed.
- J. D. Sidda, V. Poon, L. Song, W. Wang, K. Yang and C. Corre, Org. Biomol. Chem., 2016, 14, 6390–6393 RSC.
- K. E. Creamer, Y. Kudo, B. S. Moore and P. R. Jensen, Microb. Genomics, 2021, 7, 1–14 Search PubMed.
- K. Arakawa, N. Tsuda, A. Taniguchi and H. Kinashi, ChemBioChem, 2012, 13, 1447–1457 CrossRef CAS PubMed.
- C. Corre, L. Song, S. O’Rourke, K. F. Chater and G. L. Challis, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 17510–17515 Search PubMed.
- S. Kitani, K. T. Miyamoto, S. Takamatsu, E. Herawati, H. Iguchi, K. Nishitomi, M. Uchida, T. Nagamitsu, S. Omura, H. Ikeda and T. Nihira, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 16410–16415 Search PubMed.
- Y. Kudo, T. Awakawa, Y. L. Du, P. A. Jordan, K. E. Creamer, P. R. Jensen, R. G. Linington, K. S. Ryan and B. S. Moore, ACS Chem. Biol., 2020, 15, 3253–3261 CrossRef CAS PubMed.
- A. Ceniceros, L. Dijkhuizen and M. Petrusma, Sci. Rep., 2017, 7, 17743 CrossRef PubMed.
- T. Kawaguchi, T. Asahi, T. Satoh, T. Uozumi and T. Beppu, J. Antibiot., 1984, 37, 1587–1595 CrossRef CAS PubMed.
- I. Kapoor, P. Olivares and S. K. Nair, eLife, 2020, 9, e57824 CrossRef CAS PubMed.
- S. Zhou, H. Bhukya, N. Malet, P. J. Harrison, D. Rea, M. J. Belousoff, H. Venugopal, P. K. Sydor, K. M. Styles, L. Song, M. J. Cryle, L. M. Alkhalaf, V. Fülöp, G. L. Challis and C. Corre, Nature, 2021, 590, 463–467 CrossRef CAS PubMed.
- E. Takano, Curr. Opin. Microbiol., 2006, 9, 287–294 CrossRef CAS PubMed.
- K. Arakawa, Biosci., Biotechnol., Biochem., 2014, 78, 183–189 CrossRef CAS PubMed.
- O. Hara and T. Beppu, J. Antibiot., 1982, 35, 349–358 CrossRef CAS PubMed.
- H. Ohashi, Y.-H. Zheng, T. Nihira and Y. Yamada, J. Antibiot., 1989, 42, 1191–1195 CrossRef CAS PubMed.
- K. Hashimoto, T. Nihira and Y. Yamada, J. Ferment. Bioeng., 1992, 73, 61–65 CrossRef CAS.
- Y. Yamada, Nippon Hosenkin Gakkaishi, 1995, 9, 57–65 Search PubMed.
- A. V. Polkade, S. S. Mantri, U. J. Patwekar and K. Jangid, Front. Microbiol., 2016, 7, 131 Search PubMed.
- Y. Yamada, K. Sugamura, M. Kondo, H. Yanagimoto and H. Okada, J. Antibiot., 1987, 40, 496–504 CrossRef CAS PubMed.
- K. Sato, T. Nihira, S. Sakuda, M. Yanagimoto and Y. Yamada, J. Ferment. Bioeng., 1989, 68, 170–173 CrossRef CAS.
- J.-Y. Kato, N. Funa, H. Watanabe, Y. Ohnishi and S. Horinouchi, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2378–2383 Search PubMed.
- N. H. Hsiao, J. Söding, D. Linke, C. Lange, C. Hertweck, W. Wohlleben and E. Takano, Microbiology, 2007, 153, 1394–1404 CrossRef CAS PubMed.
- S. Zhou, N. R. Malet, L. Song, C. Corre and G. L. Challis, Chem. Commun., 2020, 56, 14443–14446 RSC.
- C. Schlawis, S. Kern, Y. Kudo, J. Grunenberg, B. S. Moore and S. Schulz, Angew. Chem., Int. Ed., 2018, 57, 14921–14925 CrossRef CAS PubMed.
- C. Schlawis, T. Harig, S. Ehlers, D. G. Guillen-Matus, K. E. Creamer, P. Jensen and S. Schulz, ChemBioChem, 2020, 21, 1629–1632 CrossRef CAS PubMed.
- K. Jerye, H. Lüken, A. Steffen, C. Schlawis, L. Jänsch, S. Schulz and M. Brönstrup, Adv. Sci., 2024, 11, e2309515 CrossRef PubMed.
- K. Mori, Tetrahedron Lett., 1981, 22, 3431–3432 CrossRef CAS.
- K. Mori and K. Yamane, Tetrahedron, 1982, 38, 2919–2921 CrossRef CAS.
- K. Mori, Tetrahedron, 1983, 39, 3107–3109 CrossRef CAS.
- J. B. Morin, K. L. Adams and J. K. Sello, Org. Biomol. Chem., 2012, 10, 1517–1520 RSC.
- L. E. Wilbanks, H. E. Hennigan, C. D. Martinez-Brokaw, H. Lakkis, S. Thormann, A. S. Eggly, G. Buechel and E. I. Parkinson, ACS Chem. Biol., 2023, 18, 1624–1631 CrossRef CAS PubMed.
- Y. Zhang, M. Wang, J. Tian, J. Liu, Z. Guo, W. Tang and Y. Chen, Appl. Microbiol. Biotechnol., 2020, 104, 1695–1705 CrossRef CAS PubMed.
- T. B. Nguyen, S. Kitani, S. Shimma and T. Nihira, Appl. Environ. Microbiol., 2018, 84, e02791-17 CrossRef PubMed.
- X. Liu, W. Wang, J. Li, Y. Li, J. Zhang and H. Tan, Sci. China: Life Sci., 2021, 64, 1575–1589 CrossRef CAS PubMed.
- A. van der Meij, S. S. Elsayed, C. Du, J. Willemse, T. M. Wood, N. I. Martin, J. M. Raaijmakers and G. P. van Wezel, Appl. Environ. Microbiol., 2023, 89, e0123923 CrossRef PubMed.
- M. Biarnes-Carrera, R. Breitling and E. Takano, Curr. Opin. Chem. Biol., 2015, 28, 91–98 CrossRef CAS PubMed.
- M. Biarnes-Carrera, C.-K. Lee, T. Nihira, R. Breitling and E. Takano, ACS Synth. Biol., 2018, 7, 1043–1055 CrossRef CAS PubMed.
- J. E. Bowyer, E. Lc de Los Santos, K. M. Styles, A. Fullwood, C. Corre and D. G. Bates, J. Biol. Eng., 2017, 11, 30 CrossRef PubMed.
- R. H. Feling, G. O. Buchanan, T. J. Mincer, C. A. Kauffman, P. R. Jensen and W. Fenical, Angew. Chem., Int. Ed., 2003, 42, 355–357 CrossRef CAS PubMed.
- K. D. Bauman, V. V. Shende, P. Y. T. Chen, D. B. B. Trivella, T. A. M. Gulder, S. Vellalath, D. Romo and B. S. Moore, Nat. Chem. Biol., 2022, 18, 538–546 CrossRef CAS PubMed.
- D. E. Nettleton, T. W. Doyle, B. Krishnan, G. K. Matsumoto and J. Clardy, Tetrahedron Lett., 1985, 26, 4011–4014 CrossRef CAS.
- Y. Inahashi, M. Iwatsuki, A. Ishiyama, A. Matsumoto, T. Hirose, J. Oshita, T. Sunazuka, W. Panbangred, Y. Takahashi, M. Kaiser, K. Otoguro and S. Ōmura, Org. Lett., 2015, 17, 864–867 CrossRef CAS PubMed.
- Y. Inahashi, T. Shiraishi, A. Také, A. Matsumoto, Y. Takahashi, S. Ōmura, T. Kuzuyama and T. Nakashima, J. Antibiot., 2018, 71, 749–752 CrossRef CAS PubMed.
- S. Hoshino, H. Onaka and I. Abe, J. Ind. Microbiol. Biotechnol., 2019, 46, 363–374 CrossRef CAS PubMed.
- J. J. Zhang, X. Tang and B. S. Moore, Nat. Prod. Rep., 2019, 36, 1313–1332 RSC.
- D. Xue, Z. Shang, E. A. Older, Z. Zhong, C. Pulliam, K. Peter, M. Nagarkatti, P. Nagarkatti, Y.-X. Li and J. Li, ACS Chem. Biol., 2023, 18, 508–517 CrossRef CAS PubMed.
- C. Pulliam, D. Xue, A. Campbell, E. Older and J. Li, ChemBioChem, 2024, 25, e2024005 CrossRef PubMed.
- S. Arsın, M. Pollari, E. Delbaje, J. Jokela, M. Wahlsten, P. Permi and D. Fewer, RSC Chem. Biol., 2024, 5, 1035–1044 RSC.
- S. Komatsu, C. Tsumori, K. Ohnishi and K. Kai, ACS Chem. Biol., 2020, 15, 2860–2865 CrossRef CAS PubMed.
- Y. Kudo, C. T. Hanifin, Y. Kotaki and M. Yotsu-Yamashita, J. Nat. Prod., 2020, 83, 2706–2717 CrossRef CAS PubMed.
- Y. Kudo, C. T. Hanifin and M. Yotsu-Yamashita, Org. Lett., 2021, 23, 3513–3517 CrossRef CAS PubMed.
- F. Schalk, J. Fricke, S. Um, B. H. Conlon, H. Maus, N. Jäger, T. Heinzel, T. Schirmeister, M. Poulsen and C. Beemelmanns, RSC Adv., 2021, 11, 18748–18756 RSC.
- G. C. A. Amos, T. Awakawa, R. N. Tuttle, A. C. Letzel, M. C. Kim, Y. Kudo, W. Fenical, B. S. Moore and P. R. Jensen, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E11121–E11130 Search PubMed.
- Y. Kudo, K. Konoki and M. Yotsu-Yamashita, Biosci., Biotechnol., Biochem., 2022, 86, 1333–1342 CrossRef PubMed.
- N. Shikura, T. Nihira and Y. Yamada, FEMS Microbiol. Lett., 1999, 171, 183–189 CrossRef CAS PubMed.
- S. Galanie, D. Entwistle and J. Lalonde, Nat. Prod. Rep., 2020, 37, 1122–1143 RSC.
- M. Hall, RSC Chem. Biol., 2021, 2, 958–989 RSC.
- N. Cardullo, V. Muccilli and C. Tringali, RSC Chem. Biol., 2022, 3, 614–647 RSC.
- O. Trott and A. J. Olson, J. Comput. Chem., 2010, 31, 455–461 CrossRef CAS PubMed.
- J. Eberhardt, D. Santos-Martins, A. F. Tillack and S. Forli, J. Chem. Inf. Model., 2021, 61, 3891–3898 CrossRef CAS PubMed.
- M. Mirdita, K. Schütze, Y. Moriwaki, L. Heo, S. Ovchinnikov and M. Steinegger, Nat. Methods, 2022, 19, 679–682 CrossRef CAS PubMed.
- H. Currinn, B. Guscott, Z. Balklava, A. Rothnie and T. Wassmer, Cell. Mol. Life Sci., 2016, 73, 393–408 CrossRef CAS PubMed.
- J. B. Davis, J. D. Bailey and J. K. Sello, Org. Lett., 2009, 11, 2984–2987 CrossRef CAS PubMed.
- L. Wang and L. C. Vining, Microbiology, 2003, 149, 1991–2004 CrossRef CAS PubMed.
- Y. J. Lee, S. Kitani, H. Kinoshita and T. Nihira, Arch. Microbiol., 2008, 189, 429 CrossRef CAS.
- G. Haniotakis, W. Francke, K. Mori, H. Redlich and V. Schurig, J. Chem. Ecol., 1986, 12, 1559–1568 CrossRef CAS PubMed.
- L. Rasmussen and D. Greenwood, Chem. Senses, 2003, 28, 433–446 CrossRef CAS PubMed.
- N. Shikura, J. Yamamura and T. Nihira, J. Bacteriol., 2002, 184, 5151–5157 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5cb00007f |
| This journal is © The Royal Society of Chemistry 2025 |