Open Access Article
Open Access ArticleBioinformatic, structural, and biochemical analysis leads to the discovery of novel isonitrilases and decodes their substrate selectivity†
Tyler
Hostetler
,
Tzu-Yu
Chen
 and
Wei-chen
Chang
and
Wei-chen
Chang
 *
*
Department of Chemistry, North Carolina State University, Raleigh, NC 27695, USA. E-mail: wchang6@ncsu.edu
First published on 29th January 2025
Abstract
Bacterial species, such as Mycobacterium tuberculosis, utilize isonitrile-containing peptides (INPs) for trace metal trafficking, e.g., copper or zinc. Despite their importance, very few INP structures have been characterized to date. Reported INPs consist of a peptide backbone and β-isonitrile amide moieties. While the peptide backbone can be annotated using an adenylation domain predictor of non-ribosomal peptide synthetase (NRPS), determining the alkyl chain of β-isonitrile amide moieties remains challenging via conventional analytical techniques. In this study, we focus on non-heme iron and 2-oxoglutarate (Fe/2OG) dependent isonitrilases that exhibit inherent selectivity toward the alkyl chain length of the substrate, thus enabling the structural elucidation of INPs. Based on two known isonitrilase structures, we identified eight residue positions that control substrate selectivity. Using a custom Python program that we developed, BioSynthNexus, over 350 Fe/2OG isonitrilase genes were identified. One of these enzymes was engineered through mutations at eight selected positions, effectively modifying its substrate preference to favor either a shorter or a longer alkyl chain. Furthermore, by examining several annotated isonitrilases at eight selected positions, substrate preferences of several isonitrilases were predicted and validated through biochemical assays. Together, these findings allow for effective identification of isonitrilases and INPs, and establish a predictive framework for determining the preferred alkyl chain of β-isonitrile amide moieties.
Introduction
Isonitrile (R–N![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C)-containing peptides (INPs) are known to be produced by several bacteria, including Streptomyces and Mycobacterium.1–3 Due to the coordination property of the isonitrile group, INPs function as metallophores in metal trafficking, such as copper and zinc.1–7 INP from M. tuberculosis (Mtb) is responsible for metal uptake, which is critical for cell survival and virulence.2,6–9 Despite their importance, only a limited number of INPs (1–5) have been isolated and characterized to date (Fig. 1A). Due to similar biosynthetic pathways used, INPs are constituted of a peptide backbone and β-isonitrile amide moieties (Fig. 1B and C).1–3 Briefly, INP biosynthesis starts with the ligation of an α,β-unsaturated fatty acid to an acyl carrier protein (ACP) by an ACP-ligase, where the ACP-bound substrate undergoes Michael addition with glycine. Subsequent hydrolysis by a dual-functional thioesterase yields 6.10,11 The isonitrile group is installed by a non-heme iron and 2-oxoglutarate (Fe/2OG) dependent isonitrilase via decarboxylation-assisted desaturation (6 → 7).2–4,11–16 Compound 7 is then incorporated onto the peptide backbone via a non-ribosomal peptide synthetase (NRPS) followed by reductive cleavage to afford INPs.
C)-containing peptides (INPs) are known to be produced by several bacteria, including Streptomyces and Mycobacterium.1–3 Due to the coordination property of the isonitrile group, INPs function as metallophores in metal trafficking, such as copper and zinc.1–7 INP from M. tuberculosis (Mtb) is responsible for metal uptake, which is critical for cell survival and virulence.2,6–9 Despite their importance, only a limited number of INPs (1–5) have been isolated and characterized to date (Fig. 1A). Due to similar biosynthetic pathways used, INPs are constituted of a peptide backbone and β-isonitrile amide moieties (Fig. 1B and C).1–3 Briefly, INP biosynthesis starts with the ligation of an α,β-unsaturated fatty acid to an acyl carrier protein (ACP) by an ACP-ligase, where the ACP-bound substrate undergoes Michael addition with glycine. Subsequent hydrolysis by a dual-functional thioesterase yields 6.10,11 The isonitrile group is installed by a non-heme iron and 2-oxoglutarate (Fe/2OG) dependent isonitrilase via decarboxylation-assisted desaturation (6 → 7).2–4,11–16 Compound 7 is then incorporated onto the peptide backbone via a non-ribosomal peptide synthetase (NRPS) followed by reductive cleavage to afford INPs.
Despite employing a similar biosynthetic strategy, INPs produced from Streptomyces and Mycobacterium exhibit distinct structural features in both the peptide backbone and β-isonitrile amide moieties. For example, INPs (1, 2, and 5) from Streptomyces consist of a single peptide derived from either L-lysine or L-ornithine, with β-isonitrile amide moieties that contain short alkyl side chains (n = 1).1,3 In contrast, while the dipeptide scaffold of INP from Mtb was annotated based on the NRPS adenylation (A) domain and MS analysis, the alkyl chain length of the β-isonitrile amide moieties remains unclear.2 To bridge this gap, our previous research utilized in vitro assays with Rv0097, the corresponding Fe/2OG isonitrilase from Mtb, and synthetic substrates with varying alkyl chain lengths (6, n = 1–15) to determine the alkyl side chain length in 3.4 These findings suggest that Fe/2OG isonitrilases may play a key role in regulating the alkyl chain length of the β-isonitrile amide moieties in INPs.
Using substrate-bound protein structures of Fe/2OG isonitrilases, e.g., ScoE, Rv0097, and MmaE,4,12–14 mutagenesis studies to identify residues responsible for the alkyl chain specificity were reported.4,15 A single mutation, such as F203A in ScoE and G204F in Rv0097, led to a shift in the preferred alkyl side chain length.4,15 However, the selectivity profile distribution of these variants remains slightly different from their counterparts. For example, even with three residues mutated in Rv0097 (A202E/T203G/G204F), the preferred alkyl chain length (n = 3) still differs from that of ScoE (n = 1). These pioneering studies suggested that there might be additional residues governing alkyl chain length selectivity. Considering the function of INPs as metallophores in Streptomyces and Mycobacterium, we speculated if they are also used by other bacteria. Herein, we developed a custom Python program, BioSynthNexus,17 to search for uncharacterized isonitrilases that are related to uncharacterized isonitrile-containing natural products. Through structural and in silico analysis, we identified several residues that play an important role in the selectivity profiles of isonitrilases. The function of these residues was validated through systematic mutagenesis. To further test if the predicted substrate preference can be applied, several potential isonitrilases found using our program were overexpressed and their substrate scopes were confirmed biochemically. Together, this work establishes an easy-to-use tool to search for Fe/2OG isonitrilases and isonitrile-containing natural product biosynthetic gene clusters, and provides a roadmap for substrate scope prediction of Fe/2OG isonitrilases.
Results and discussion
Bioinformatic analysis reveals putative Fe/2OG isonitrilases
To date, all reported biosynthetic gene clusters (BGCs) of INPs found in Streptomyces and Mycobacterium encode a Fe/2OG enzyme responsible for isonitrile installation.1–3,11 Considering the critical function of INPs used for metal trafficking, it is likely that other bacteria might also deploy similar approaches for isonitrile installation en route to INP biosynthesis. To search for novel Fe/2OG isonitrilases, a previously characterized isonitrilase ScoE was used to generate a sequence similarity network (SSN), followed by a genome neighborhood network (GNN) analysis.18,19 With this approach, over 9000 BGCs were identified as encoding an annotated Fe/2OG gene (∼20–96% sequence identity to ScoE). However, the functions of the majority of genes are not assigned. Several literature studies reported that Fe/2OG enzyme homologs can produce diverse reaction outcomes,20–24 suggesting the challenges in using sequence identity to predict enzyme function within the Fe/2OG enzyme family. To search for targeted function, i.e., isonitrilase, we filtered 9000 Fe/2OG genes based on their co-occurrence with an upstream gene, the dual-functional thioesterase, because it provides the glycinyl substrate 6 for isonitrilase (Fig. 1B).11,13 To improve in silico analysis of the 9000 genes, we developed a custom Python program, BioSynthNexus (https://github.com/Tyler-Hostetler/BioSynthNexus).17 With BioSynthNexus, we can search through BGCs encoding Fe/2OG genes and filter them based on the presence of specific protein family domain(s) (see examples and step-by-step instructions in Fig. S1–S6, ESI†). After applying the thioesterase (PF10862) as a filter, only 365 BGCs remain (Fig. 2). To help functional annotation and further engineering, our program can export amino acid sequences of these genes in FASTA format for other applications. For example, we used this function to build a multiple sequence alignment and showed that key residues including the 2-histidine/1-carboxylate triad for iron chelation, residues responsible for substrate positioning, and a conserved tyrosine essential for the production of an aldimine intermediate reported in the previous studies are conserved across 365 genes (Fig. S7 and S8, ESI†).12–14 Notably, quite a few (∼143) of these Fe/2OG isonitrilases are found in pathogenic bacteria including Mtb, Nocardia brasiliensis and Mycobacterium lepromatosis (Fig. S9, ESI†). Interestingly, beyond INPs, other isonitrilases involved in the polyketide antibiotic aerocyanidin and amycomicin biosynthetic pathways, i.e., AecA and AmcA, are also identified.13,25–27 | ||
| Fig. 2 SSN of 365 putative Fe/2OG isonitrilases. Nodes of the major genera are colored accordingly. The SSN is visualized with Cytoscape28 where edges indicate at least 60% identity. | ||
Identification of key residues governing alkyl chain length selectivity in isonitrilases
To identify key residues governing alkyl chain length selectivity in isonitrilases, we examined the substrate bound structures of two isonitrilases that utilize different substrates. ScoE converts methyl-6 to the corresponding methyl-7 (6 → 7, n = 1), while 6 with a longer alkyl chain (n = 7) is bound in the Rv0097 structure.4,12 In these structures, the 2-His/1-Asp coordination triad is overlapped. Hydrogen bonding interactions of Tyr, Lys and Arg with the carboxylate groups of 6 and the Tyr with the amine group are observed in both ScoE and Rv0097, thus revealing a very similar substrate positioning in the active site of ScoE and Rv0097 (Fig. S8, ESI†). However, the size of the hydrophobic pocket for alkyl chain accommodation differs (Fig. 3A and B). A detailed comparison of the hydrophobic pocket led to the identification of several residues that could potentially affect substrate selection. Overall, eight positions were found (P1–P8, Fig. 3D, E and Fig. S10, ESI†). Among them, P1–P3 were previously identified by our group and others,4,15 where the P1–P3 result in significant steric differences between the two enzymes, i.e., E237/G238/F239 in ScoE and A202/T203/G204 in Rv0097 (Fig. S10A, ESI†). P4–P7, located at part of the hydrophobic pocket, were identified. Specifically, compared to ScoE, an α-helix observed in Rv0097 shifts the entire loop away from the active site, enlarging the pocket to accommodate substrates with a longer alkyl chain (Fig. S10B, ESI†). This structural difference may arise from the disruption caused by a proline residue (P5) in Rv0097. Consequently, D152 (P4) in Rv0097 shows hydrogen bonding with a predominately conserved arginine, which is not observed for S187 in ScoE (Fig. S10C, ESI†). In contrast, a π–π interaction between Y191 (P6) and a conserved phenylalanine in ScoE reduces the pocket size (Fig. S10D, ESI†), enabling the binding of substrates with a short alkyl chain. Furthermore, P8, a cysteine in Rv0097 and a methionine in ScoE on the other side of the pocket, shows slightly different interactions with surrounding residues (Fig. S10E, ESI†). | ||
| Fig. 3 (A)–(C) Comparison of the pocket sizes for the alkyl chain in Fe/2OG isonitrilases. (A) Crystal structure of Rv0097 bound with heptyl-6 (n = 7, PDB ID: 8KHT). (B) Crystal structure of ScoE bound with methyl-6 (n = 1, PDB ID: 6L6X). (C) Predicted structure of Srug docked with heptyl-6 (n = 7). (D) and (E) Eight positions were selected based on structure-guided analysis. These selected residues are positioned around the alkyl chain pocket, illustrated using Rv0097 as an example (D). (F) General scheme for analyzing the isonitrile product. Substrate 6 with various alkyl chains (n = 1–11) was incubated with isonitrilases followed by derivatization using tetrazine to generate a common pyrazole product 8. (G) Substrate selectivity profiles of isonitrilases, where ScoE, Srug, and Rv0097 prefer short, medium, and long alkyl chain substrates, respectively. Protein structures were visualized in ChimeraX.29 | ||
To validate if these eight positions are responsible for substrate recognition and can be further used to guide the rational design of isonitrilase selectivity, we first identified an isonitrilase that has partial identity with both ScoE and Rv0097 among these eight positions. To accomplish this, the constructed sequence alignment was simplified to include only the eight positions we selected (Fig. S11, ESI†). With the simplified alignment, a potential isonitrilase from the pathogenic species Segniliparus rugosus30 was selected. This potential isonitrilase, Srug, has identical resides at P3, P5 and P7 with Rv0097, while its P1, P4, P6 and P8 are conserved with ScoE (Fig. 3E). At the P2 site, Srug has an alanine residue which is similar to the glycine in ScoE. Based on partially overlapped residues with both Rv0097 and ScoE, Srug should have a distinctive substrate scope. In the predicted structure of Srug docked with heptyl-6 (n = 7), the hydrophobic pocket for alkyl chain accommodation is different from that of ScoE and Rv0097 (Fig. 3C). To test this hypothesis, Srug was codon-optimized and heterologously expressed in E. coli. To develop a method for product detection and quantification, the samples were quenched with an equal volume of methanol and analyzed using liquid chromatography mass spectrometry (LC-MS) (Fig. S12, ESI†). However, variations in the alkyl chain lengths of the substrates and products affect retention times and cause potential differences in MS response, which could introduce errors in quantification. To address this issue, 3,6-di-2-pyridyl-1,2,4,5-tetrazine was used to convert the isonitrile group into a common pyrazole product for analysis (Fig. 3F and Fig. S13, ESI†).4,31 With this strategy, the same pyrazole product 8 is generated and monitored for comparison, regardless of the substrate used. Following the incubation of Srug, ScoE and Rv0097 with substrates that have different alkyl chain lengths (6, n = 1–11), the derivatized products were subjected to LC-MS analysis (Fig. S14, see ESI,† for Experimental details). As shown in Fig. 3G, ScoE can only accommodate substrates that have a short alkyl chain, e.g., a methyl and propyl group, while Rv0097 prefers a longer alkyl chain (n = 11). Notably, Srug exhibits substrate preference for medium-length alkyl chains (n = 5–9). This result supports our hypothesis that these eight selected positions delineate isonitrilase substrate preference regarding alkyl chain length. Despite Srug sharing ∼76% and ∼44% sequence identity with Rv0097 and ScoE, their substrate profiles are different.
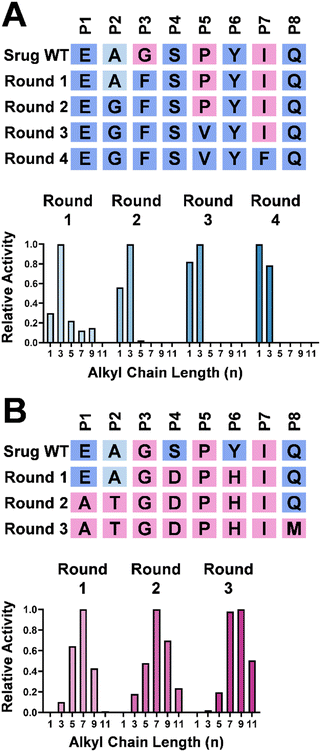
Systematic mutagenesis of Srug
To validate the effect of these eight positions, systematic mutagenesis experiments were carried out (Fig. 4). To shift the substrate preference of Srug toward a shorter alkyl chain, P3 was mutated to incorporate a sterically hindered amino acid, e.g., Gly to Phe. This alteration reshaped the substrate profile, making 6 with a propyl group (n = 3) the optimal substrate for this single variant (Fig. 4A). This finding aligns with the literature that P1–P3 positions play a critical role in differentiating substrate profiles.4,15 After subsequent multiple-round mutagenesis, the corresponding residues were introduced to mimic ScoE. In this variant, a substrate preference toward the methyl group (n = 1) over the propyl group (n = 3) was achieved (Fig. 4A).In parallel, Srug was mutated to accommodate a substrate with a longer alkyl chain length (Fig. 4B). First, mutations were conducted at the P4–P7 positions to enlarge the pocket size by shifting the entire loop away from the active site. Although the heptyl substrate (n = 7) remains the optimal substrate, its preference for shorter alkyl chain lengths (n = 1–5) decreases. In the second round of mutagenesis at P1–P3 sites, the undecyl and propyl substrates (n = 11 and 3) have similar activity, while the heptyl (n = 7) substrate is still preferred. After incorporating Met at P8 position, the relative activity toward the undecyl substrate (n = 11) increased ca. 36-fold compared to the round-one mutant (Fig. S15, ESI†). However, compared to Rv0097 which prefers the undecyl group (n = 11), this Srug variant shows a preference for the nonyl group (n = 9). This result suggests that other residues around the hydrophobic pocket or those with distal interactions may also contribute to substrate preference. Nevertheless, these observations highlight the importance of the eight selected positions in governing alkyl chain length selectivity in Fe/2OG isonitrilases.
Forecast of the alkyl chain length selectivity using eight selected positions
Fe/2OG isonitrilases have been reported to install isonitrile moieties en route to INPs and polyketide biosynthesis.1,3 However, compared to 365 BGCs found through in silico analysis (Fig. 2), only a limited number of isonitrile-containing natural products have been characterized to date. This is partially due to the lability of the isonitrile group that degrades to a N-formyl group during the isolation processes.16,27,32,33 To help access the possible structure of potential INPs from 365 BGCs, we utilized the eight selected positions that we identified to forecast the composition of β-isonitrile amide moieties. They are biosynthesized by isonitrilases and then appended onto the peptide backbone by NRPS (Fig. 1C). To test if our model can be used to predict the alkyl chain length of uncharacterized isonitrilases, we randomly picked a few Fe/2OG enzymes from the SSN analysis with their names simplified to the corresponding species. One candidate that captured our attention is from Mycobacterium lepromatosis, the causative agent of Hansen's disease, also known as leprosy.34–36 Similar to Rv and Sco BGCs, an NRPS is encoded in the gene cluster of M. lepromatosis, indicating this BGC is associated with an INP. The isonitrilase from M. lepromatosis (Mlep) has a nearly identical amino acid composition at the selected positions with Srug (Fig. 5A), suggesting similar substrate profiles. We overexpressed Mlep in E. coli, and it showed a substrate preference toward a medium-length alkyl chain substrate (n = 7) (Fig. 5B). Furthermore, two homologs, Nfla and Rfas, from pathogenic Nocardia flavorosea37,38 and Rhodococcus fascians39 were selected and tested. Although the corresponding biosynthetic pathways for these two species have not been elucidated, co-occurrences of NRPS with isonitrilase in their BGCs imply that they are again associated with INP biosynthesis. After examining the eight selected positions of Nfla and Rfas, the components closely resemble those in the Srug round 3 variant (Fig. 4A and 5A). This finding suggests that short-length alkyl chain substrates are used with the propyl group (n = 3) likely being preferred. We heterologously expressed Nfla and Rfas, and in vitro assays revealed that both enzymes indeed show a preference toward the propyl alkyl chain (Fig. 5B). | ||
| Fig. 5 (A) Eight selected positions of uncharacterized Fe/2OG isonitrilases found through in silico analysis. (B) Substrate selectivity profiles of selected isonitrilases. | ||
Inspired by isonitrilase genes, e.g., AecA and AmcA, that co-occurred with polyketide genes in aerocyanidin and amycomicin biosynthesis,13,25–27 we were curious whether these eight positions governing substrate selectivity can be applied to isonitrile-containing natural products beyond INPs. The eight selected positions in AecA and AmcA are highly similar to ScoE, suggesting that the short-length alkyl chains of 6 are utilized (Fig. S16A, ESI†). Furthermore, the deviation at P5 in AmcA toward Rv0097 supports the presence of a propyl moiety in the amycomicin,11 contrasting with the methyl moiety in aerocyanidin (Fig. S16B, ESI†).26,27 Next, we selected two uncharacterized isonitrilases from Halothiobacillus neapolitanus (Hneap) and Corynebacterium provencense (Cpro). In the BGC of H. neapolitanus, polyketide synthase (PKS) is encoded while the corresponding product has not been reported. Interestingly, neither NRPS nor PKS can be identified in the C. provencense cluster, suggesting the unique structural scaffold for its potential isonitrile-containing product. The selected eight positions of Hneap and Cpro are highly similar with those of ScoE (Fig. 5A), thus suggesting that they prefer substrates with a shorter alkyl chain. Indeed, both enzymes showed preference toward methyl and propyl groups under in vitro conditions (Fig. 5B). Together, we demonstrated that these eight positions can be used to annotate the alkyl chain length of the isonitrile moiety, i.e. short (n = 1–3), medium (n = 5–9), or long (n ≥ 11), thus aiding the elucidation of plausible chemical structures of corresponding natural products.
Conclusion
Bacteria such as Mycobacterium and Streptomyces produce INPs to scavenge rare metals for their survival in metal-restrictive environments.1,2,6,7 However, only a limited number of INPs have been isolated and characterized to date. Due to the lability of the isonitrile moiety, isolating these natural products without isonitrile degradation remains a challenging task. To help identify novel INPs and to elucidate possible chemical structures, we developed a custom Python program, BioSynthNexus, to facilitate in silico analysis for isonitrilase and BGC identification.17 The co-occurrence of a dual-functional thioesterase is utilized to confidently identify Fe/2OG enzymes with isonitrilase activity (Fig. 2) for which several were verified in terms of function in vitro. Next, we identified key residues that are responsible for the alkyl chain recognition of isonitrilases. Using structural analysis of the substrate-bound isonitrilase structures, eight positions (P1–P8) that influence substrate preferences were identified (Fig. 3). To test and forecast substrate scope using these eight positions, we reconstituted the activity of a previously unknown enzyme Srug and conducted systematic mutagenesis to achieve substrate scope shifting (Fig. 4). Furthermore, we demonstrated that these eight positions can be used to predict the preferred length of alkyl chain, i.e. short (n = 1–3), medium (n = 5–9), or long (n ≥ 11), of several previously unknown Fe/2OG isonitrilases from pathogenic bacteria (Fig. 5).Furthermore, while numerous tools exist for genome visualization and comparison, such as EFI-GND,18,19 AnnoView,40 Artemis comparison tool,41 and JBrowse 2,42 challenges persist when exploring protein homologs that catalyze similar reaction types. BioSynthNexus is designed to search for homologs within a given GNN by efficiently filtering the protein family (Pfam) IDs in the neighborhoods. While a similar strategy was applied in the literature,43,44 manual comparison is required to analyze the results from a given GNN. With BioSynthNexus, the process can be completed in a time-efficient manner, and it can remotely access the UniProt database to retrieve sequence information for neighborhoods of interest in FASTA format, enabling seamless integration for further multiple sequence alignment.
Author contributions
Tyler Hostetler: conceptualisation, investigation, writing. Tzu-Yu Chen: conceptualisation, investigation, writing. Wei-chen Chang: writing, review & editing, project administration, supervision, funding acquisition.Data availability
Instructions towards BioSynthNexus installation, usage, and examples are all provided on the GitHub repository (https://github.com/Tyler-Hostetler/BioSynthNexus). Experimental protocols are reported in the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was also supported by the National Institutes of Health (NIH) GM127588 and the Lord Scholar and Goodnight Early Career Innovator.References
- L. Wang, M. Zhu, Q. Zhang, X. Zhang, P. Yang, Z. Liu, Y. Deng, Y. Zhu, X. Huang, L. Han, S. Li and J. He, ACS Chem. Biol., 2017, 12, 3067–3075 CrossRef PubMed.
- K. Mehdiratta, S. Singh, S. Sharma, R. S. Bhosale, R. Choudhury, D. P. Masal, A. Manocha, B. D. Dhamale, N. Khan, V. Asokachandran, P. Sharma, M. Ikeh, A. C. Brown, T. Parish, A. K. Ojha, J. S. Michael, M. Faruq, G. R. Medigeshi, D. Mohanty, D. S. Reddy, V. T. Natarajan, S. S. Kamat and R. S. Gokhale, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2110293119 CrossRef.
- N. C. Harris, M. Sato, N. A. Herman, F. Twigg, W. Cai, J. Liu, X. Zhu, J. Downey, R. Khalaf, J. Martin, H. Koshino and W. Zhang, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 7025–7030 CrossRef PubMed.
- T.-Y. Chen, J. Chen, M. W. Ruszczycky, D. Hilovsky, T. Hostetler, X. Liu, J. Zhou and W.-C. Chang, ACS Catal., 2024, 14, 4975–4983 CrossRef PubMed.
- Y. Xu and D. S. Tan, Org. Lett., 2019, 21, 8731–8735 CrossRef PubMed.
- K. Bhatt, H. Machado, N. S. Osório, J. Sousa, F. Cardoso, C. Magalhães, B. Chen, M. Chen, J. Kim, A. Singh, C. M. Ferreira, A. G. Castro, E. Torrado, W. R. Jacobs, A. Bhatt and M. Saraiva, mSphere, 2018, 3, e00352–18 CrossRef PubMed.
- J. A. Buglino, Y. Ozakman, Y. Xu, F. Chowdhury, D. S. Tan and M. S. Glickman, mBio, 2022, 13, e02513–22 CrossRef PubMed.
- F. Wolschendorf, D. Ackart, T. B. Shrestha, L. Hascall-Dove, S. Nolan, G. Lamichhane, Y. Wang, S. H. Bossmann, R. J. Basaraba and M. Niederweis, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 1621–1626 CrossRef PubMed.
- N. Dhar and J. D. McKinney, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 12275–12280 CrossRef PubMed.
- T. Chisuga, A. Miyanaga, F. Kudo and T. Eguchi, J. Biol. Chem., 2017, 292, 10926–10937 CrossRef PubMed.
- N. C. Harris, D. A. Born, W. Cai, Y. Huang, J. Martin, R. Khalaf, C. L. Drennan and W. Zhang, Angew. Chem., Int. Ed., 2018, 57, 9707–9710 CrossRef PubMed.
- T.-Y. Chen, J. Chen, Y. Tang, J. Zhou, Y. Guo and W.-C. Chang, Angew. Chem., Int. Ed., 2020, 59, 7367–7371 CrossRef CAS PubMed.
- T.-Y. Chen, Z. Zheng, X. Zhang, J. Chen, L. Cha, Y. Tang, Y. Guo, J. Zhou, B. Wang, H.-W. Liu and W.-C. Chang, ACS Catal., 2022, 12, 2270–2279 CrossRef CAS PubMed.
- R. Jonnalagadda, A. Del Rio Flores, W. Cai, R. Mehmood, M. Narayanamoorthy, C. Ren, J. P. T. Zaragoza, H. J. Kulik, W. Zhang and C. L. Drennan, J. Biol. Chem., 2021, 296, 100231 CrossRef CAS PubMed.
- A. Del Rio Flores, M. Narayanamoorthy, W. Cai, R. Zhai, S. Yang, Y. Shen, K. Seshadri, K. De Matias, Z. Xue and W. Zhang, Biochemistry, 2023, 62, 824–834 CrossRef CAS PubMed.
- A. Del Rio Flores, D. W. Kastner, Y. Du, M. Narayanamoorthy, Y. Shen, W. Cai, V. Vennelakanti, N. A. Zill, L. B. Dell, R. Zhai, H. J. Kulik and W. Zhang, J. Am. Chem. Soc., 2022, 144, 5893–5901 CrossRef CAS PubMed.
- T. Hostetler, Zenodo, 2025 DOI:10.5281/zenodo.14675094.
- R. Zallot, N. Oberg and J. A. Gerlt, Biochemistry, 2019, 58, 4169–4182 CrossRef CAS PubMed.
- N. Oberg, R. Zallot and J. A. Gerlt, J. Mol. Biol., 2023, 435, 168018 CrossRef CAS PubMed.
- W. Kim, T.-Y. Chen, L. Cha, G. Zhou, K. Xing, N. K. Canty, Y. Zhang and W.-C. Chang, Nat. Commun., 2022, 13, 5343 CrossRef CAS PubMed.
- H. Tao, T. Mori, H. Chen, S. Lyu, A. Nonoyama, S. Lee and I. Abe, Nat. Commun., 2022, 13, 95 CrossRef CAS PubMed.
- V. Siitonen, B. Selvaraj, L. Niiranen, Y. Lindqvist, G. Schneider and M. Metsä-Ketelä, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 5251–5256 CrossRef CAS.
- Y. Matsuda, T. Iwabuchi, T. Fujimoto, T. Awakawa, Y. Nakashima, T. Mori, H. Zhang, F. Hayashi and I. Abe, J. Am. Chem. Soc., 2016, 138, 12671–12677 CrossRef CAS.
- M. Homma, K. Uchida, T. Wakabayashi, M. Mizutani, H. Takikawa and Y. Sugimoto, Front. Plant Sci., 2024, 15, 1392212 CrossRef.
- Z. Zheng, J. Clardy and H.-W. Liu, J. Am. Chem. Soc., 2024, 146, 21061–21068 CrossRef CAS PubMed.
- W. L. Parker, M. L. Rathnum, J. H. Johnson, J. S. Wells, P. A. Principe and R. B. Sykes, J. Antibiot., 1988, 41, 454–460 CrossRef CAS PubMed.
- G. Pishchany, E. Mevers, S. Ndousse-Fetter, D. J. Horvath, C. R. Paludo, E. A. Silva-Junior, S. Koren, E. P. Skaar, J. Clardy and R. Kolter, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 10124–10129 CrossRef CAS PubMed.
- P. Shannon, A. Markiel, O. Ozier, N. S. Baliga, J. T. Wang, D. Ramage, N. Amin, B. Schwikowski and T. Ideker, Genome Res., 2003, 13, 2498–2504 CrossRef CAS PubMed.
- E. C. Meng, T. D. Goddard, E. F. Pettersen, G. S. Couch, Z. J. Pearson, J. H. Morris and T. E. Ferrin, Protein Sci., 2023, 32, e4792 CrossRef CAS PubMed.
- W. R. Butler, C. A. Sheils, B. A. Brown-Elliott, N. Charles, A. A. Colin, M. J. Gant, J. Goodill, D. Hindman, S. R. Toney, R. J. Wallace and M. A. Yakrus, J. Clin. Microbiol., 2007, 45, 3449–3452 CrossRef CAS PubMed.
- Y.-B. Huang, W. Cai, A. Del Rio Flores, F. F. Twigg and W. Zhang, Anal. Chem., 2020, 92, 599–602 CrossRef CAS.
- G. Ernouf, I. K. Wilt, S. Zahim and W. M. Wuest, ChemBioChem, 2018, 19, 2448–2452 CrossRef CAS.
- S. F. Brady, J. D. Bauer, M. F. Clarke-Pearson and R. Daniels, J. Am. Chem. Soc., 2007, 129, 12102–12103 CrossRef CAS PubMed.
- A. Virk, B. Pritt, R. Patel, J. R. Uhl, S. A. Bezalel, L. E. Gibson, B. M. Stryjewska and M. S. Peters, Emerg. Infect. Dis., 2017, 23, 1864–1866 CrossRef PubMed.
- X. Y. Han, K. C. Sizer, J. S. Velarde-Félix, L. O. Frias-Castro and F. Vargas-Ocampo, Int. J. Dermatol., 2012, 51, 952–959 CrossRef PubMed.
- P. Deps and S. M. Collin, Front. Microbiol., 2021, 12, 698588 CrossRef PubMed.
- S. Valdezate, N. Garrido, G. Carrasco, M. J. Medina-Pascual, P. Villalón, A. M. Navarro and J. A. Saéz-Nieto, J. Antimicrob. Chemother., 2017, 72, 754–761 CAS.
- C.-K. Tan, C.-C. Lai, S.-H. Lin, C.-H. Liao, C.-H. Chou, H.-L. Hsu, Y.-T. Huang and P.-R. Hsueh, Clin. Microbiol. Infect., 2010, 16, 966–972 CrossRef PubMed.
- S. A. H. M. Dhaouadi and A. Rhouma, Ann. Appl. Biol., 2020, 177, 4–15 CrossRef.
- X. Wei, H. Tan, B. Lobb, W. Zhen, Z. Wu, D. H. Parks, J. D. Neufeld, G. Moreno-Hagelsieb and A. C. Doxey, Brief. Bioinform., 2024, 25, bbae229 CrossRef PubMed.
- T. J. Carver, K. M. Rutherford, M. Berriman, M.-A. Rajandream, B. G. Barrell and J. Parkhill, Bioinform, 2005, 21, 3422–3423 CrossRef CAS PubMed.
- C. Diesh, G. J. Stevens, P. Xie, T. D. J. Martinez, E. A. Hershberg, A. Leung, E. Guo, S. Dider, J. Zhang, C. Bridge, G. Hogue, A. Duncan, M. Morgan, T. Flores, B. N. Bimber, R. Haw, S. Cain, R. M. Buels., L. D. Stein and I. H. Holmes, Genome Biol., 2023, 24, 74 CrossRef PubMed.
- S. Zhao, A. Sakai, X. Zhang, M. W. Vetting, R. Kumar, B. Hillerich, B. S. Francisco, J. Solbiati, A. Steves, S. Brown, E. Akiva, A. Barber, R. D. Seidel, P. C. Babbitt, S. C. Almo, J. A. Gerlt and M. P. Jacobson, eLife, 2014, 3, e03275 CrossRef PubMed.
- J. D. Rudolf, X. Yan and B. Shen, J. Ind. Microbiol. Biotechnol., 2016, 43, 261–276 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cb00304g |
| This journal is © The Royal Society of Chemistry 2025 |